Abstract
Objective
To assess experimental virulence among sequence type 131 (ST131) Escherichia coli bloodstream isolates in relation to virulence genotype and subclone.
Methods
We analysed 48 Spanish ST131 bloodstream isolates (2010) by PCR for ST131 subclone status (H30Rx, H30 non-Rx, or non-H30), virulence genes (VGs), and O-type. Then we compared these traits with virulence in a murine sepsis model, as measured by illness severity score (ISS) and rapid lethality (mean ISS ≥ 4).
Results
Of the 48 study isolates, 65% were H30Rx, 21% H30 non-Rx, and 15% non-H30; 44% produced ESBLs, 98% were O25b, and 83% qualified as extraintestinal pathogenic E. coli (ExPEC). Of 49 VGs, ibeA and iss were associated significantly with non-H30 isolates, and sat, iha and malX with H30 isolates. Median VG scores differed by subclone, i.e., 12 (H30Rx), 10 (H30 non-Rx), and 11 (non-H30) (p < 0.01). Nearly 80% of isolates represented a described virotype. In mice, H30Rx and non-H30 isolates were more virulent than H30 non-Rx isolates (according to ISS [p = 0.03] and rapid lethality [p = 0.03]), as were ExPEC isolates compared with non-ExPEC isolates (median ISS, 4.3 vs. 2.7: p = 0.03). In contrast, most individual VGs, VG scores, VG profiles, and virotypes were not associated with mouse virulence.
Conclusions
ST131 subclone and ExPEC status, but not individual VGs, VG scores or profiles, or virotypes, predicted mouse virulence. Given the lower virulence of non-Rx H30 isolates, hypervirulence probably cannot explain the ST131-H30 clade's epidemic emergence.
Introduction
The spread of a specific Escherichia coli clone called sequence type 131 (ST131) is one of the key drivers of the rising prevalence of resistance to quinolones and cephalosporins among E. coli that cause urinary tract infections, with cephalosporin resistance being mainly due to expression of extended-spectrum beta-lactamases (ESBLs), particularly CTX-M-15 [1–3]. The as-yet-unknown reasons for this globally distributed clone's epidemic success are under active investigation [4].
The ST131 pandemic is driven mainly by two nested subclones within ST131, i.e., H30 (or clade C), which contains allele 30 of fimH (type 1 fimbriae), and H30Rx (or clade C2), a sublineage within H30 that often expresses CTX-M-15 [4]. Strains that encode allele H30 but do not produce CTX-M-15 are usually grouped in the H30 non-Rx subclone (or Clade C1). Several studies have suggested that, apart from being extensively resistant, ST131 is also highly virulent, with a higher capability of causing infection than other strains [5,6]. ST131 virulence has been assessed in different animal models [7–9] with different conclusions. Moreover, these studies have not addressed ST131 subclone status.
ST131 usually contains a high number of putative virulence genes (VGs) which probably contributes to its virulence [7]. Indeed, in multiple studies, ST131 has been associated with specific VGs, including usp (uropathogenic specific protein), iutA (aerobactin receptor), fyuA (yersiniobactin receptor), ompT (outer membrane protein T), and iroN (salmochelin receptor) [10–14], which variously encode for toxins, adhesins, siderophores, or outer membrane proteins. Moreover, within ST131, based on specific combinations of VGs some authors have described virotypes A, B, C, D (with subsets 1–5), and E [10,15,16]. Of these virotypes, A, B and C have been associated with higher experimental virulence [15].
Hence, we sought to clarify these relationships among ST131 isolates from Spain by studying 48 ST131 isolates, including representatives of the different ST131 subclones. Specifically, we characterized our ST131 isolates extensively in vitro, assessed their virulence in a murine sepsis model, and sought correlations between experimental virulence and VG content, virotype, ExPEC status, and ST131 subclone (H30Rx, H30 non-Rx, and non-H30).
Material and methods
Isolates and detection of ST131 subclones and O types
The present ST131 study isolates represented a subset of the ITU-BRAS collection, the product of a Spanish multicentre study of bloodstream isolates from patients with bacteraemia of urinary origin who were admitted in 8 hospitals of different geographic areas from October 2010 to June 2011 [17]. This study was approved by the Ethics Commitee for Clinical Research of Hospital Ramón y Cajal. The collection's 425 E. coli isolates were characterized previously for ESBL production by PCR and sequencing [18]. Here, they were screened for ST131 status using PCR-based detection of ST131-specific single-nucleotide polymorphisms (SNPs) within mdh and gyrB [19]. ST131 isolates were further classified into the H30 and H30Rx subclones by PCR-based detection of subclone-specific SNPs [20] and were screened by PCR for the two most common O types found in ST131 strains, O25b and O16 [21]. For extended virulence genotyping and testing in the murine sepsis model we selected all ESBL-producing ST131 isolates (n = 21), plus 27 arbitrarily chosen non-ESBL-producing ST131 isolates, giving 48 isolates total. Moreover, to stablish the clonal relationship the 48 ST131 isolates were typed by PFGE-XbaI and a dendrogram was created using the PFGE-patterns (Bionumerics version 7.5, Applied Maths NV) according to the unweighted pair group mean arithmetic method based on pairwise Dice similarity coefficients.
Virulence gene scores and virotypes
Forty-nine putative ExPEC-associated VGs and variants thereof were determined by an established multiplex PCR assay (Table 1) [10,22–24]. The VG score was calculated as the total number of VGs detected, adjusted for multiple detection of the pap (pilus associated with pyelonephritis, P fimbriae), sfa/foc (S and F1C fimbriae), and kpsII (capsular polysaccharide synthesis group 2) operons [25]. Isolates were classified as ExPEC if they carried ≥ 2 of papAH and/or papC (counted as one), sfa/foc, afa/dra (Dr antigen specific adhesin), iutA, and kpsMII [25].
Table 1. Distribution of virulence genes by ST131 subclones.
| Virulence genesa, b | N° of isolates with genea,b (column %) | p valuec | |||||
|---|---|---|---|---|---|---|---|
| Functional category | Specific gene | Total (n = 48) | Non-H30 (n = 7) | H30 non-Rx (n = 10) | H30Rx (n = 31) | Non-H30 vs. H30d | H30-non-Rx vs. Rx |
| Adhesins | papAH | 16 (33) | 3 (43) | 0 (0) | 13 (42) | 0.67 | 0.02 |
| papC | 17 (35) | 5 (71) | 0 (0) | 12 (39) | 0.08 | 0.02 | |
| papEF | 17 (35) | 5 (71) | 0 (0) | 12 (39) | 0.08 | 0.02 | |
| papGe | 15 (31) | 3 (43) | 0 (0) | 12 (39) | 0.66 | 0.02 | |
| papGII | 13 (27) | 1 (14) | 0 (0) | 12 (39) | 0.66 | 0.02 | |
| papGIII | 1 (2) | 1 (14) | 0 (0) | 0 (0) | 0.15 | NA | |
| sfa/foc | 1 (2) | 1 (14) | 0 (0) | 0 (0) | 0.15 | NA | |
| iha | 41 (85) | 3 (43) | 8 (80) | 30 (97) | < 0.01 | 0.14 | |
| afa/dra | 13 (27) | 1 (14) | 0 (0) | 12 (39) | 0.66 | 0.02 | |
| hra | 17 (35) | 4 (57) | 0 (0) | 13 (42) | 0.23 | 0.02 | |
| Toxins | hlyD | 11 (23) | 4 (57) | 0 (0) | 7 (23) | 0.04 | 0.16 |
| hlyF | 7 (15) | 3 (43) | 1 (1) | 3 (10) | 0.05 | 1.0 | |
| cnf1 | 7 (15) | 0 (0) | 0 (0) | 7 (23) | 0.57 | 0.16 | |
| cdtB | 1 (2) | 1 (14) | 0 (0) | 0 (0) | 0.15 | NA | |
| sat | 41 (85) | 2 (29) | 9 (90) | 30 (97) | < 0.01 | 0.43 | |
| Siderophores | iroN | 6 (13) | 3 (43) | 0 (0) | 3 (10) | 0.03 | 0.56 |
| fyuA | 46 (96) | 5 (71) | 10 (100) | 31 (100) | 0.02 | NA | |
| iutA | 45 (94) | 5 (71) | 9 (90) | 31 (100) | 0.05 | 0.24 | |
| Protectins and invasins | kpsMII f | 40 (83) | 5 (71) | 5 (50) | 30 (97) | 0.33 | < 0.01 |
| K1 | 1 (2) | 1 (14) | 0 (0) | 0 (0) | 0.15 | NA | |
| K2 | 25 (52) | 2 (29) | 0 (0) | 23 (74) | 0.24 | < 0.01 | |
| K5 | 8 (17) | 0 (0) | 3 (30) | 5 (16) | 0.58 | 0.38 | |
| K15 | 1 (2) | 1 (14) | 0 (0) | 0 (0) | 0.15 | NA | |
| iss | 8 (17) | 4 (57) | 1 (10) | 3 (9.7) | 0.01 | 1.0 | |
| traT | 38 (79) | 4 (57) | 8 (80) | 26 (84) | 0.15 | 1.0 | |
| cvaC | 4 (8) | 1 (14) | 1 (10) | 2 (6) | 0.48 | 1.0 | |
| ibeA | 6 (12.5) | 6 (83) | 0 (0) | 0 (0) | < 0.01 | NA | |
| Other | ompT | 46 (96) | 5 (71) | 10 (100) | 31 (100) | 0.02 | NA |
| malX | 44 (92) | 3 (43) | 10 (100) | 31 (100) | < 0.01 | NA | |
aGenes detected in all isolates: fimH, yfcV, and usp.
bGenes not detected in any isolate: papGI/I’, sfaS, focG, afaE8, bmaE, gafD, F17, and clpG (adhesins); pic, vat, tsh, astA (toxins); ireA (siderophore); kpsMTIII and rfc (protectins); and H7 fliC, clbN, and clbB (other).
cP values obtained by chi2 test or Fisher’s test as appropriate. Statistically significant values are in bold.
dH30 included the non-Rx and Rx subsets.
eIsolates with at least one papG allelic variant or that are PCR-positive with generic papG primers.
fIsolates with at least one kpsII variant. NA: Not applicable.
Aggregate VG profiles were defined based on presence/absence of the studied VGs. The kpsMII and pap operons were considered to be present if any of the corresponding genes or their allelic variants were detected. Isolates were assigned to virotypes (A, B, C, D1-5, and E) as described by Mora et al. based on presence/absence of specific VGs: ibeA (invasion of brain endothelium), iroN, sat (secreted autotransporter toxin), afa/dra, papGII, papGIII, cnf1 (cytotoxic necrotizing factor 1), hlyD (ɑ-haemolysin), cdtB (cytolethal distending toxin), and K1 (group II capsule variant) [15]. Isolates not corresponding to a defined virotype were classified as “other”.
Murine sepsis model
Isolates were tested for in vivo virulence using a modified protocol of an established murine subcutaneous sepsis model [26]. Experimentation guidelines of the Veterans Affairs Medical Centre of Minneapolis were followed in the conduct of the mouse model experiments under animal use protocol 120603, as approved by the local Institutional Animal Care and Use Committee (IACUC). Mice were kept in ventilated cages (maximum 10 mice per cage) with food and water ad libitum and following natural cycles of light and darkness. Briefly, female pathogen-free outbred Swiss-Webster mice (mean age 60 days, mean weight 23g) were inoculated subcutaneously in the nape of the neck with approximately 109 CFU/mL log-phase bacteria in 0.2 mL saline. Mouse health was assessed twice daily for 3 days post-challenge. Mice were classified daily as to maximal illness severity, which ranged from 1 (healthy) to 5 (dead), with scores 2 (barely ill), 3 (moderately ill) and 4 (severely ill) in between. Scores were based on external exam of the mice and their behaviour (i.e., redness around the eyes or nose, cleaning habits, huddling, and movement). The exams were performed concurrently by two experienced investigators who were unaware of the characteristics of the challenge strains, and then reached a consensus regarding the score. Each particular characteristic was not scored, rather, all were considered jointly to reach the final illness score. All mice that reached score 4 were euthanized to minimize suffering. All surviving mice were euthanized at the end of the study according to the local IACUC guidelines, using carbon dioxide inhalation. All efforts were taken to minimize suffering of the mice at all times, and no analgesic or anaesthetic was needed. No unexpected deaths were observed. Each experimental series included a positive control strain (urosepsis isolate CFT073; 80%-100% of mice dead by 3 days post-challenge) and a negative control strain (laboratory strain MG1655; no perceptible mouse illness or lethality) [26]. Each test strain was assessed initially in 5 mice, followed by another 5 mice if the initial testing did not yield a consistent result (i.e., lethality or survival for 4 or 5 of the initially challenged mice).
Daily illness severity scores for the mice challenged with a given isolate were averaged to give the isolate's mean illness severity score (ISS). The variables used for comparisons of virulence potential included overall mean ISS (as a continuous variable), rapid lethality (i.e., mean illness severity score ≥ 4, indicating that all mice were severely ill or dead by day 1 post-inoculation).
Statistical analysis
Continuous variables were described as medians and interquartile ranges (IQR), and were compared using the Mann-Whitney or Kruskal-Wallis test, as appropriate. Dichotomous variables were described using frequencies and percentages, and were compared using a chi-square test or Fisher’s exact test, as appropriate. The criterion for statistical significance was p < 0.05. The Bonferroni correction method was applied when multiple comparisons were performed. Data were analysed with software STATA version 11 (Stata Statistical Software: Release 11. College Station, TX: StataCorp LP). A heatmap representing presence/absence of the studied VGs in relation to strain characteristics (subclone, ExPEC status, ISS, virotype and virulence profile) was performed using RStudio software version 1.0.44 (RStudio Team 2016, RStudio: Integrated Development for R. RStudio, Inc., Boston, MA) with the pheatmap package.
Results
Study isolates
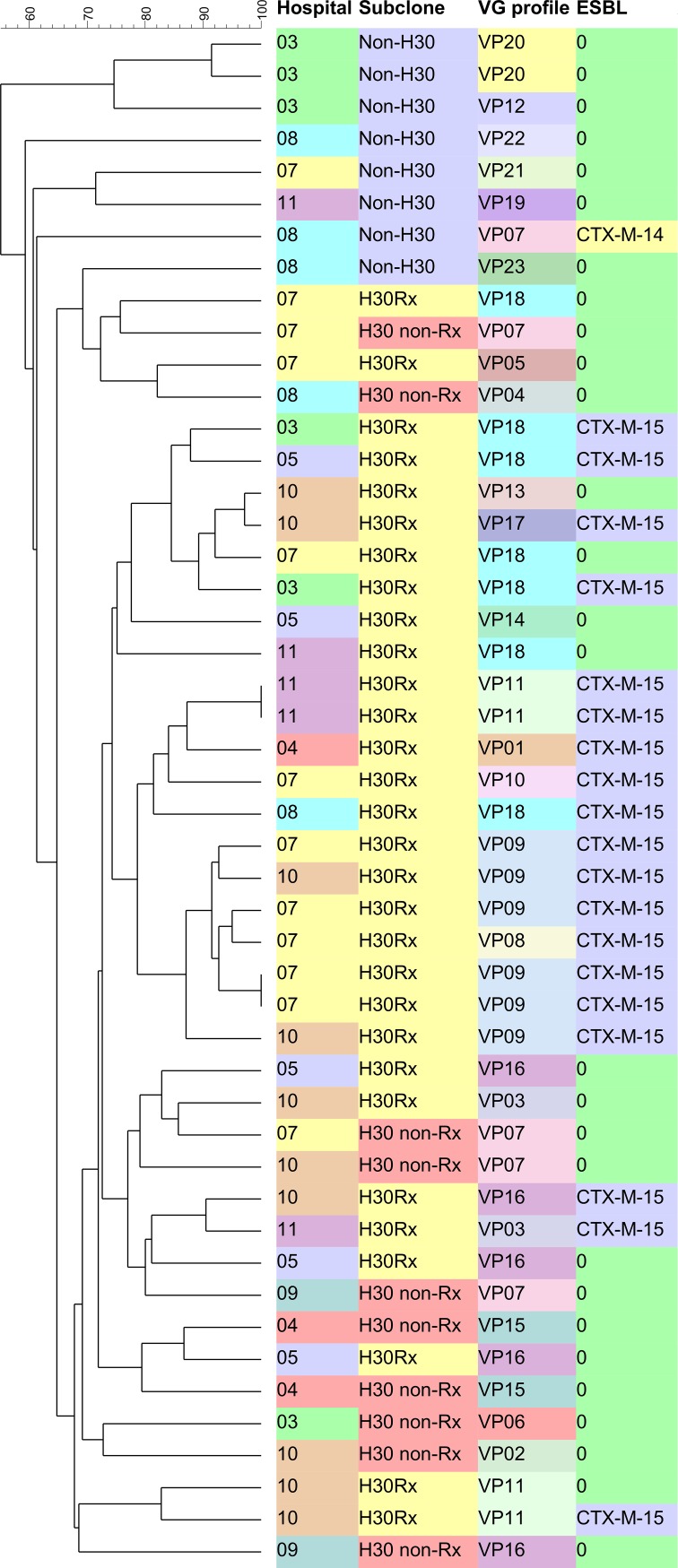
Of the 48 ST131 bloodstream isolates, 31 (65%) were H30Rx, 10 (21%) were H30 non-Rx and 7 (15%) isolates belonged to the non-H30 subclone. Twenty-one isolates (44%) were ESBL producers, of which all but one belonged to the H30Rx subclone (Fig 1). By O-type PCR, 47 isolates (98%) were O25b and one (2%) was O16. The dendrogram showed that ST131 subclones were evenly distributed across participant hospitals without a clonal overrepresentation (Fig 1).
Fig 1. Dendrogram of the 48 ST131-E. coli.
Hospitals: 03-Hospital Gregorio Marañón (Madrid); 04-Hospital del Mar (Barcelona) 05-Hospital Marqués de Valdecilla (Santander); 07-Hospital Ramón y Cajal (Madrid); 08-Hospital de la Santa Creu i San Pau (Barcelona); 09-Hospital Virgen Macarena (Sevilla); 10-Complejo Sanitario de Pontevedra (Pontevedra); 11-Hospital Mutua Tarrassa (Barcelona). VG profile: Virulence genes profile. VP: Virulence profile.
Virulence genes, scores, and ExPEC status
Of the 49 VGs tested, 32 (65%) were detected in at least 1 isolate, at frequencies ranging from 2% to 100% (Table 1) and all isolates encoded at least 8 VGs. All isolates contained fimH, usp, and yfcV (putative fimbrial subunit). Apart from these, the most prevalent VGs of each functional category were iha (adhesin, 85%), sat (toxin, 85%), fyuA (siderophore, 96%), kpsMII (protectin, 83%), and ompT (other, 96%).
The three ST131 clonal subsets (H30Rx, H30 non-Rx, and non-H30) differed significantly for the prevalence of certain VGs (Table 1). Whereas ibeA (83%) and iss (57%) were found mainly in non-H30 isolates (versus 0% and 10% in H30-positive isolates respectively, p ≤ 0.01), malX (100%), sat (95%) and iha (93%), were detected more commonly among the H30 isolates (versus 43%, 29% and 43% respectively among the non-H30 isolates, p < 0.01). Moreover, the VGs kpsII and K2 differed significantly in prevalence between the H30Rx and H30 non-Rx isolates (97% versus 50% and 74% versus 0% respectively, p < 0.01). Also of note, the H30 non-Rx isolates did not encode any VG of the pap operon and in fact the only adhesins found in this subclone were fimH, yfcV and iha.
Overall, VG scores ranged from 8 to 17 (median 12, IQR 10.5–14). Scores differed by subclone, with medians (IQR) of 12 (11–15) for H30Rx, 11 (10–14) for non-H30 isolates, and 10 (10–10) for H30 non-Rx (p < 0.01 for the three-group comparison). ESBL production was associated with higher VG score with median (IQR) VG score of 13 (12–15) in ESBL producers versus 10 (10–13) in non-ESBL producers (p <0.01).
Forty (83%) study isolates fulfilled molecular criteria for ExPEC, including all 31 H30Rx isolates (100%) and 71% (5/7) of non-H30 isolates but only 40% (4/10) of H30 non-Rx isolates (versus H30Rx: p < 0.01) (Table 2). ExPEC isolates showed higher VG scores than non-ExPEC isolates (median 12 versus 10: p < 0.01).
Table 2. Virulence genes scores, ExPEC status, and experimental virulence according to subclone and virotype.
| Virulence-related genotypes | Mouse sepsis model outcomes | ||||
|---|---|---|---|---|---|
| ST131 Subclone | Virotypes (no. isolates) | Median VGa score (IQRd) | ExPEC (row %) | Median ISSb (IQRd) | Rapid lethality (row %) |
| Non-H30 | C (n = 1) | 10 (0) | 0 (0) | 4.4 (0) | 1 (100) |
| D3 (n = 1) | 11 (0) | 1 (100) | 4.5 (0) | 1 (100) | |
| D4 (n = 1) | 10 (0) | 0 (0) | 1.7 (0) | 0(0) | |
| Otherc (n = 4) | 13.5 (3.5) | 4 (100) | 4.5 (1.4) | 3 (75) | |
| Total (n = 7) | 11 (3) | 5 (71) | 4.4 (2.4) | 5 (71) | |
| H30 non-Rx | C (n = 9) | 10 (0) | 4 (44) | 2.9 (1.6) | 3 (33) |
| Other (n = 1) | 8 (0) | 0 (0) | 1.5 (0) | 0 (0) | |
| Total (n = 10) | 10 (0) | 4 (40) | 2.75 (1.9) | 3 (30) | |
| H30Rx | A (n = 12) | 12 (1) | 12 (100) | 4.5 (0.75) | 9 (75) |
| B (n = 2) | 15 (0) | 2 (100) | 3.4 (1.8) | 1 (50) | |
| C (n = 5) | 11 (0) | 5 (100) | 4.1 (2) | 3 (60) | |
| E (n = 7) | 15 (0) | 7 (100) | 3.9 (1.7) | 3 (43) | |
| Other (n = 5) | 13 (0) | 5 (100) | 4.7 (0.3) | 5 (100) | |
| Total (n = 31) | 12 (4) | 31 (100) | 4.3 (1.5) | 21 (68) | |
aVG: virulence gene.
bISS: illness severity score.
c”Other”: isolates that did not correspond with a defined virotype.
d IQR: Interquartile range
Virotypes and virulence profiles
Overall, 79% (38/48) of isolates corresponded with a previously described virotype, most commonly virotype C (31%) or A (25%) (Table 3). Virotype C occurred within all three subclones (16% of H30Rx, 90% of H30 non-Rx, and 14% of non-H30 isolates), while the other virotypes were found exclusively in certain subclones (Table 2). Moreover, virotypes differed significantly for VG scores (p < 0.01 overall) with virotype B and E showing the highest VG score with median of 15 (IQR 15–15) (Table 3).
Table 3. Prevalence of characteristics (column %) and median scores according to virotypes.
| Virotype | |||||||
|---|---|---|---|---|---|---|---|
| Charac.b | A (n = 12) | B (n = 2) | C (n = 15) | D3 (n = 1) | D4 (n = 1) | E (n = 7) | Othera (n = 10) |
| ExPEC | 12 (100) | 2 (100) | 9 (60) | 1 (100) | 0 (0) | 7 (100) | 9 (90) |
| VG score (IQR)c | 12 (1) | 15 (0) | 10 (1) | 10.5 (0) | 10 (0) | 15 (0) | 13 (1) |
| ISS (IQR)d | 4.5 (0.75) | 3.4 (1.8) | 3.8 (2) | 4.5 (0) | 1.7 (0) | 3.9 (1.7) | 4.6 (0.4) |
| RLe (col. %) | 9 (75) | 1 (50) | 7 (47) | 1 (100) | 0 (0) | 3 (43) | 8 (80) |
aOther: isolates did not correspond with a defined virotype.
bCharacteristics: For comparisons across all virotypes, the only statistically significant difference involved virulence gene scores (p < 0.01).
c VG score: Median virulence gene score (interquartile range).
d ISS: Median illness severity score (interquartile range).
e Rapid lethality.
Twenty-three different VG profiles (arbitrarily numbered P1 to P23) were observed (Fig 2). Diversity of profiles varied by clonal subset, that is, whereas collectively the 31 H30Rx isolates showed only 12 total profiles, and the 10 H30 non-Rx isolates only 6 total profiles, each of the 7 non-H30 isolates had a unique profile.
Fig 2. Heatmap with the virulence genes tested of the 48 Escherichia coli ST131 study isolates.
Only variably present virulence genes are shown, with all pap and kps variants counted as one; pap: isolates positive for any of the pap operon genes tested, i.e., papAH, papC, papEF, papG (with alleles I, I’, II, III), kps II: isolates positive for any of the kps II variants tested, i.e., kpsMII, K1, K2, K5 and K15. Virulence genes present or absent in all isolates are not shown. Red indicates presence and green indicates absence of gene. “Other” category (virotype column): a virotype could not be assigned. ISS: illness severity score. RL: rapid lethality. Profile (P): Aggregated virulence genes profile.
Murine sepsis model
Overall, the 48 study isolates showed a high level of morbidity and lethality in the murine sepsis model: the median ISS was 4.3 (IQR 2.8–4.7), and 29 (60%) were rapidly lethal. ESBL producers showed greater illness severity (median ISS, 4.4 for ESBL producers, versus 4.1 for non-ESBL producers: p = 0.04) and were more often rapidly lethal (71% versus 52%, p = 0.17).
By clonal subset, the H30Rx and non-H30 isolates exhibited the greatest illness severity (median ISS, 4.3 and 4.4, respectively), and H30 non-Rx isolates the lowest (median ISS, 2.75; versus other isolates: p = 0.04). Likewise, most H30Rx and non-H30 isolates exhibited rapid lethality (71% and 68%, respectively), as compared with only 30% of H30 non-Rx isolates (H30Rx versus H30 non-Rx: p = 0.06) (Table 2).
The illness severity score was also greater in ExPEC isolates (median ISS, 4.3 [ExPEC] versus 2.7 [non-ExPEC]: p = 0.03). In contrast, VG scores and aggregate VG profiles were not significantly associated with any sepsis model outcome, and among individual VG, only K2 was associated with higher ISS (median 4.45 in K2-positive isolates versus median 3.35 in K2-negative isolates, p < 0.01). Moreover, virotype A and D3 isolates showed the highest median ISS (4.5) although neither of them had the highest VG score and there was no significant overall association between virotype and these virulence outcomes (Table 2).
Discussion
In this study, we assessed 48 Spanish E. coli ST131 bloodstream isolates for VG content (expressed as individual VGs, ExPEC status, VG scores, VG profiles, and virotypes) and ST131 subclone status, and compared these bacterial characteristics with virulence in a murine sepsis model (expressed as ISS, and rapid lethality).
We found no significant association of in vivo virulence with VG scores, extended VG profiles, or virotypes. Moreover, among the 49 VG studied, the only one that was associated with a higher ISS was K2, which encodes certain capsular antigen variants K2. Although ESBL production seemed possibly to be associated with higher VG score and ISS, almost all (20/21) ESBL-producing isolates belonged to the H30Rx subclone, so this association could result from confounding factors. Also, the K2 gene was detected more often among H30Rx isolates (74%) than in the other isolates (17%), which also points to subclone classification as the reason for this association. In contrast, the ST131 subclones and ExPEC versus non-ExPEC isolates differed significantly for in vivo virulence. These differences suggest that the bacterial traits determining experimental virulence in this murine model are distributed differentially between ST131 clonal subsets, and between ExPEC versus non-ExPEC ST131 isolates, but do not necessarily include the VGs studied here or segregate reliably by virotype. Moreover, these data could indicate that the recent epidemic expansion of the H30 ST131 subclone–at least its large non-Rx component–must have explanations other than hyper-virulence, at least as measured in this murine sepsis model. Indeed, in this study the H30 non-Rx isolates showed a noticeable lack of VGs (mainly adhesins), specially compared with the phylogenetically related H30Rx subclone.
Overall, the present ST131 isolates exhibited high in vivo virulence, as described previously for other ST131 isolates [5,15]. However, we found in vivo virulence to vary considerably both between and within the different ST131 subclones, consistent with the findings of Johnson et al [9]. This variability in experimental virulence between and within ST131 subsets may explain why no consistent lethality differences have been found between ST131 isolates generally and non-ST131 isolates [9,15].
The H30 subset within ST131, with its Rx and non-Rx components, is the leading cause of ST131 expansion [20]. Interestingly, in the present comparison of experimental virulence and VG content, the H30Rx isolates more closely resembled the non-H30 isolates than they did the H30 non-Rx isolates, which conflicts with the underlying phylogenetic relationships. Subclone differentiation was not considered in previous reports from several authors that described variability among experimental virulence in ST131 strains and the lack of correlation between virulence gene content and experimental virulence [15,16,21]. Our results indicate that subclones should be studied individually to assess correlation between VG and virulence.
Olesen et al. suggested that the expansion of ST131, and of its H30Rx subclone in particular, could be due more to a specific subset of genes (not necessarily encoding exclusively antibiotic resistance or virulence) than to total number of VGs [20]. Recent whole genome analysis has pointed to the acquisition of certain accessory genes as the starting point for the clade C (or H30 subset) expansion [4]. Our results further refine these hypotheses, since although the H30Rx isolates exhibited some of the highest VG and lethality scores, the H30 non-Rx (i.e., clade C1) isolates had lower VG scores and were significantly less virulent than even the non-H30 isolates. Our results suggest that these subclone-specific differences in experimental virulence may result from genetic differences between subclones that involve sequences other than classic VGs. The acquisition of virulence traits, therefore, may not be the evolutionary event that drove the clade C expansion. The distinctive accessory traits of non-Rx H30 isolates, although seemingly unlikely to confer enhanced virulence, may enhance the organism's ability to colonize the host or to perform other non-directly pathogenic functions that favour clonal expansion, and so could be called more appropriately fitness traits. The limited number of strains in our study, however, does not allow for robust conclusions, and further studies clearly are needed.
A previous study that compared the virulence of ST131 and non-ST131 clinical isolates by using the same murine sepsis model as used here showed that the presence of certain genes (i.e., papAH, kpsMII, papGIII, vat, K1, and clbB/N), rather than ST131 status, was associated with higher lethality [9]. Conversely, a recent study that compared urinary and bloodstream isolates virulence in an insect larva model found an association of ST131 status with higher experimental virulence [7]. Notably, however, that model has been shown to yield results that do not correspond with those obtained in the murine sepsis model [27]. Here, in comparing different ST131 isolates in the murine sepsis model, despite considerable overall variability in virulence genotypes and virulence outcomes we found no significant associations between specific virulence genes and experimental virulence with the exception of the K2 gene.
Our inability to identify associations of virotype with virulence outcomes in the murine sepsis model also differs from previous findings [10,28]. Here, although virotype C was most common, it did not exhibit exceptionally high illness severity, lethality, or VG scores, whereas in two previous studies by different groups of investigators it was associated with increased mouse lethality [15,28]. We found that ST131 subclone and ExPEC status were better predictors of experimental virulence than was virotype, with the association of ExPEC status and higher lethality possibly being due to all H30Rx isolates qualifying as ExPEC. The inconsistent in vivo virulence differences observed here among virotypes suggest that the VG combinations that define the established virotypes do not reliably classify ST131 isolates according to their experimental virulence in the murine sepsis model.
This study's main limitations included the relatively small number of isolates (especially representing the H30 non-Rx subclone), which restricted statistical power. Moreover, almost half of the isolates were provided by two of the eight participating hospitals, although minimal evidence of intra-hospital clonality was found. However, few studies have addressed experimental virulence in relation to ST131 subclone classification, as our study does. Moreover, our main objective was to describe the findings and generate new hypothesis for subsequent more in-depth testing. Another possible limitation was the focus on bloodstream isolates from Spain, which restricted generalizability. This murine model does not address completely the pathogenesis of bloodstream infections such as previous colonization or persistence. Moreover, although this murine sepsis model is well-established, the use of presence/absence of virulence genes as a marker, without considering levels of expression, could affect the in vivo virulence results.
Our study’s main strengths were the representative study population (which was drawn broadly from different geographic areas of Spain), the inclusion of both ESBL-producing and non-producing isolates, the wide range of tested VGs, the attention to key ST131 subclones, and the use of a well-established murine sepsis model.
Conclusion
We found that experimental virulence in the murine sepsis model varies considerably among ST131 isolates, even within previously defined virotypes, whereas subclone and, to a lesser extent, ExPEC status identifies isolates with similar levels of experimental virulence. The comparatively high lethality of H30Rx isolates suggests that as-yet undefined sequences associated with this subclone might be responsible for its ability to cause severe infections. In contrast, the comparatively low virulence of non-Rx H30 isolates point away from virulence as an explanation for this subgroup's impressive epidemic emergence. Further study is needed to understand the basis for ST131's successful expansion and to find markers of experimental lethality that also can predict clinical outcome in infected patients.
Acknowledgments
The ITUBRAS-GEIH group are: JP. Horcajada (Hospital del Mar, Barcelona, Spain); E. Shaw (Hospital Universitario de Bellvitge-IDIBELL, Barcelona, Spain); E. Bunshow, E. Cercenado, B. Padilla and C. Sánchez-Carrillo (Hospital Gregorio Marañón, Madrid, Spain); MC. Fariñas, M. Gonzalo and L. Martínez-Martínez (Hospital Marqués de Valdecilla, Santander, Spain); E. Calbo, M. Riera, and M. Xercavins (Hospital Universitario Mútua de Terrassa, Barcelona, Spain); R. Gamallo and MA. Pallarés (Complexo Hospitalario de Pontevedra, Pontevedra, Spain); R Cantón, P. Ruiz-Garbajosa, and V. Pintado (Hospital Universitario Ramón y Cajal, Madrid, Spain); J. Gomez (Laboratorio de Referencia de Cataluña, Barcelona, Spain); N. Benito and B. Mirelis (Hospital Santa Creu i Sant Pau, Barcelona, Spain); and M. de Cueto, A. Pascual and J. Rodríguez-Baño (Hospital Virgen Macarena, Sevilla, Spain). Lead autor of the ITU-BRAS group: Juan Pablo Horcajada, Hospital del Mar-Medical Research Institute of Hospital del Mar (IMIM)-CEXS, Universitat Pompeu Fabra, Barcelona, Spain. Contact email address: jhorcajada@parcdesalutmar.cat.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by the Instituto de Salud Carlos III of Spain (www.isciii.es) with the grant PI13/02092; and Spanish Network for Research in Infectious Diseases or REIPI (www.reipi.org): RD12/0015/0004 and RD16/0016/0011; and co-financed by the European Development Regional Fund (ERDF), “A Way to Achieve Europe”. IMV is supported by a research contract from the REIPI (RD12/0015/0004). Part of this work was supported by a grant from the Spanish Society of Infectious Diseases and Clinical Microbiology (SEIMC) (Ayuda a la formación de la SEIMC 2014, IMV). This material also is based partly upon work supported by Office of Research and Development, Medical Research Service, Department of Veterans Affairs, grant # 1 I01 CX000192 01 (JRJ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Peirano G, Pitout JDD. Molecular epidemiology of Escherichia coli producing CTX-M beta-lactamases: the worldwide emergence of clone ST131 O25:H4. Int J Antimicrob Agents. 2010;35: 316–21. doi: 10.1016/j.ijantimicag.2009.11.003 [DOI] [PubMed] [Google Scholar]
- 2.Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, et al. Global Incidence of Carbapenemase-Producing Escherichia coli ST131. Emerg Infect Dis. 2014;20: 1928–1931. doi: 10.3201/eid2011.141388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lahlaoui H, Ben Haj Khalifa A, Ben Moussa M. Epidemiology of Enterobacteriaceae producing CTX-M type extended spectrum β-lactamase (ESBL). Med Mal Infect. 2014;44: 400–404. doi: 10.1016/j.medmal.2014.03.010 [DOI] [PubMed] [Google Scholar]
- 4.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Khanh Nhu NT, Roberts LW, Stanton-Cook M, et al. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. MBio. 2016;7: 1–12. doi: 10.1101/039123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clermont O, Lavollay M, Vimont S, Deschamps C, Forestier C, Branger C, et al. The CTX-M-15-producing Escherichia coli diffusing clone belongs to a highly virulent B2 phylogenetic subgroup. J Antimicrob Chemother. 2008;61: 1024–8. doi: 10.1093/jac/dkn084 [DOI] [PubMed] [Google Scholar]
- 6.Vimont S, Boyd A, Bleibtreu A, Bens M, Goujon J-M, Garry L, et al. The CTX-M-15-producing Escherichia coli clone O25b: H4-ST131 has high intestine colonization and urinary tract infection abilities. PLoS One. 2012;7: e46547 doi: 10.1371/journal.pone.0046547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciesielczuk H, Betts J, Phee L, Doumith M, Hope R, Woodford N, et al. Comparative virulence of urinary and bloodstream isolates of extra-intestinal pathogenic Escherichia coli in a Galleria mellonella model. Virulence. 2015;6: 145–51. doi: 10.4161/21505594.2014.988095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alghoribi MF, Gibreel TM, Dodgson AR, Beatson S a, Upton M. Galleria mellonella Infection Model Demonstrates High Lethality of ST69 and ST127 Uropathogenic E. coli. PLoS One. 2014;9: e101547 doi: 10.1371/journal.pone.0101547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson JR, Porter SB, Zhanel G, Kuskowski M a, Denamur E. Virulence of Escherichia coli clinical isolates in a murine sepsis model in relation to sequence type ST131 status, fluoroquinolone resistance, and virulence genotype. Infect Immun. 2012;80: 1554–62. doi: 10.1128/IAI.06388-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blanco J, Mora A, Mamani R, López C, Blanco M, Dahbi G, et al. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol. 2013;51: 3358–67. doi: 10.1128/JCM.01555-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicolas-Chanoine M-H, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, et al. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother. 2008;61: 273–81. doi: 10.1093/jac/dkm464 [DOI] [PubMed] [Google Scholar]
- 12.Johnson JR, Johnston B, Clabots C, Kuskowski MA, Castanheira M. Escherichia coli sequence type ST131 as the major cause of serious multidrug-resistant E. coli infections in the United States. Clin Infect Dis. 2010;51: 286–94. doi: 10.1086/653932 [DOI] [PubMed] [Google Scholar]
- 13.Johnson JR, Urban C, Weissman SJ, Jorgensen JH, Lewis JS, Hansen G, et al. Molecular epidemiological analysis of Escherichia coli sequence type ST131 (O25:H4) and blaCTX-M-15 among extended-spectrum-β-lactamase-producing E. coli from the United States, 2000 to 2009. Antimicrob Agents Chemother. 2012;56: 2364–2370. doi: 10.1128/AAC.05824-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lavigne J-P, Vergunst AC, Goret L, Sotto A, Combescure C, Blanco J, et al. Virulence potential and genomic mapping of the worldwide clone Escherichia coli ST131. PLoS One. 2012;7: e34294 doi: 10.1371/journal.pone.0034294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mora A, Dahbi G, López C, Mamani R, Marzoa J, Dion S, et al. Virulence patterns in a murine sepsis model of ST131 Escherichia coli clinical isolates belonging to serotypes O25b:H4 and O16:H5 are associated to specific virotypes. PLoS One. 2014;9: e87025 doi: 10.1371/journal.pone.0087025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahbi G, Mora A, Mamani R, López C, Alonso MP, Marzoa J, et al. Molecular epidemiology and virulence of Escherichia coli O16:H5-ST131: comparison with H30 and H30-Rx subclones of O25b:H4-ST131. Int J Med Microbiol. Elsevier GmbH.; 2014;304: 1247–57. doi: 10.1016/j.ijmm.2014.10.002 [DOI] [PubMed] [Google Scholar]
- 17.Horcajada JP, Shaw E, Padilla B, Pintado V, Calbo E, Benito N, et al. Healthcare-associated, community-acquired and hospital-acquired bacteraemic urinary tract infections in hospitalized patients: a prospective multicentre cohort study in the era of antimicrobial resistance. Clin Microbiol Infect. 2013;19: 962–8. doi: 10.1111/1469-0691.12089 [DOI] [PubMed] [Google Scholar]
- 18.Merino I, Shaw E, Horcajada JP, Cercenado E, Mirelis B, Pallarés MA, et al. CTX-M-15-H30Rx-ST131 subclone is one of the main causes of healthcare associated ESBL-producing Escherichia coli bacteraemia of urinary origin in Spain. J Antimicrob Chemother. 2016;71: 2125–2130. doi: 10.1093/jac/dkw133 [DOI] [PubMed] [Google Scholar]
- 19.Johnson JR, Menard M, Johnston B, Kuskowski M a, Nichol K, Zhanel GG. Epidemic clonal groups of Escherichia coli as a cause of antimicrobial-resistant urinary tract infections in Canada, 2002 to 2004. Antimicrob Agents Chemother. 2009;53: 2733–9. doi: 10.1128/AAC.00297-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olesen B, Frimodt-Møller J, Leihof RF, Struve C, Johnston B, Hansen DS, et al. Temporal Trends in Antimicrobial Resistance and Virulence-Associated Traits within the Escherichia coli Sequence Type 131 Clonal Group and Its H30 and H30-Rx Subclones, 1968 to 2012. Antimicrob Agents Chemother. 2014;58: 6886–95. doi: 10.1128/AAC.03679-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson JR, Clermont O, Johnston B, Clabots C, Tchesnokova V, Sokurenko E, et al. Rapid and specific detection, molecular epidemiology, and experimental virulence of the O16 subgroup within Escherichia coli sequence type 131. J Clin Microbiol. 2014;52: 1358–65. doi: 10.1128/JCM.03502-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JR, Stell AL. Extended Virulence Genotypes of Escherichia coli Strains from Patients with Urosepsis in Relation to Phylogeny and Host Compromise. J Infect Dis. 2000;181: 261–72. doi: 10.1086/315217 [DOI] [PubMed] [Google Scholar]
- 23.Moreno E, Andreu A, Pigrau C, Kuskowski MA, Johnson JR, Prats G. Relationship between Escherichia coli Strains Causing Acute Cystitis in Women and the Fecal E. coli Population of the Host. J Clin Microbiol. 2008;46: 2529–2534. doi: 10.1128/JCM.00813-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park HH-K, Woo S, Lee E, Cha J, Lee K-E, Lee SJ. Escherichia coli pap Genes as well as Adenovirus Type 11 and Type 21, and BK Virus were Involved with Severe Urinary Tract Infection in Infants. J Bacteriol Virol. 2011;41: 245 doi: 10.4167/jbv.2011.41.4.245 [Google Scholar]
- 25.Johnson JR, Kuskowski M a, Gajewski A, Sahm DF, Karlowsky J a. Virulence characteristics and phylogenetic background of multidrug-resistant and antimicrobial-susceptible clinical isolates of Escherichia coli from across the United States, 2000–2001. J Infect Dis. 2004;190: 1739–44. doi: 10.1086/425018 [DOI] [PubMed] [Google Scholar]
- 26.Johnson JR, Clermont O, Menard M, Kuskowski MA, Picard B, Denamur E. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J Infect Dis. 2006;194: 1141–50. doi: 10.1086/507305 [DOI] [PubMed] [Google Scholar]
- 27.Johnson JR, Porter S, Johnston B, Kuskowski MA, Spurbeck RR, Mobley HLT, et al. Host Characteristics and Bacterial Traits Predict Experimental Virulence for Escherichia coli Bloodstream Isolates From Patients With Urosepsis. Open forum Infect Dis. 2015;2: ofv083 doi: 10.1093/ofid/ofv083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peirano G, Van Der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, et al. Characteristics of Escherichia coli Sequence Type 131 Isolates That Produce Extended-Spectrum β-Lactamases: Global Distribution of the H30-Rx Sublineage. Antimicrob Agents Chemother. 2014;58: 3762–3767. doi: 10.1128/AAC.02428-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.