Abstract
An alcohol dehydrogenase from the halophilic archaeon Haloferax volcanii (HvADH2) has been engineered by rational design to broaden its substrate scope towards the conversion of a range of aromatic substrates, including flurbiprofenol, that is an intermediate of the non-steroidal anti-inflammatory drug, flurbiprofen. Wild-type HvADH2 showed minimal activity with flurbiprofenol (11.1 mU/mg). A homology model of HvADH2 was built and docking experiments with this substrate revealed that the biphenyl rings of flurbiprofenol formed strong interactions with residues F85 and F108, preventing its optimal binding in the active site. Mutations at position 85 however did not increase activity. Site directed mutagenesis at position F108 allowed the identification of three variants showing a significant (up to 2.3-fold) enhancement of activity towards flurbiprofenol, when compared to wild-type HvADH2. Interestingly, F108G variant did not show the classic inhibition in the presence of (R)-enantiomer when tested with rac-1-phenylethanol, underling its potential in racemic resolution of secondary alcohols.
Introduction
Enzymes are appealing as a ‘green’ adjunct to chemical synthesis of pharmaceutical building blocks because of their broad specificity, enantioselectivity and ability to work under process conditions [1]. Found in all three domains of life, alcohol dehydrogenases (ADHs) are members of the oxidoreductase family, which catalyze the interconversion of primary and/or secondary alcohols into aldehydes and ketones, respectively [2]. A recent review details the staggering applications of ADHs in the production of pharmaceutical building blocks [3]. Codexis described the evolution of Lactobacillus kefir ADH towards the enantiopure intermediate of the anti-depressant, (S)-duloxetine, with yields as high as 150 g/L [4].
Many additional examples of enzyme engineering applied to ADHs, which have led to variants suitable for industrial applications, have been reported in the literature [5–7]. Sequence alignments of protein families identify potential ‘hot-spots’ for mutagenesis as non-conserved positions; they are then further probed by homology modelling and in silico docking [8]. A site-directed mutagenesis strategy has applied for redesigning substrate specificity in glutamate dehydrogenase from Halobacterium salinarum [9]. The substitutions K89L, A163L and S367A converted this enzyme into a dehydrogenase accepting L-methionine, L-norleucine and L-norvaline as substrates. Biocatalytic strategies employing ADHs have already been reported to produce 2-arylpropionic acids and the corresponding derivatives [10–13]. Hyperthermophilic SsADH-10 from Sulfolobus solfataricus was applied to the enzymatic reduction and racemization of 2-arylpropionaldehydes [14].
In a previous study, HvADH2 from Haloferax volcanii (wild-type, WT) showed an unusually broad substrate specificity, with good activity with medium-chain alcohols, modest activity with secondary alcohols and also significant activity with benzyl alcohol [15–17]. Later studies into the solvent tolerance and immobilization of HvADH2 prompted a deeper investigation of the substrate scope of HvADH2 [18–19]. HvADH2 showed some activity with flurbiprofenol in a low concentration salt buffer to facilitate solubility.
Results and discussion
HvADH2 homology model and docking analysis
Previous characterization of HvADH2 showed that the enzyme has a broad substrate scope because it can accept medium-chain alcohols, has modest activity with secondary alcohols and retained 50% activity with the aromatic substrate, benzyl alcohol [17]. The potential of HvADH2 to reduce prochiral aromatic ketones was also investigated and it was found that 2-phenylpropionaldehyde was readily accepted (S7 Fig). The model of the 3D structure of HvADH2 was obtained by the SWISS-MODEL web-based server [20, 21] using the formaldehyde dismutase from Pseudomonas putida (PDB: 2dph) as the template (27% sequence identity with HvADH2) [22]. The quality of the HvADH2 homology model was assessed using the software ERRAT [23]: 30.1% of the protein structure model could be rejected at a 95% confidence level (as compared to a threshold of 5% rejection for a high quality model), but these (less accurate) regions are located on the protein surface and do not affect the overall conformation and substrate binding at the active site (S1 Fig). The quality of the model was assessed by several bioinformatic tools that confirmed it as a reliable model (see S2 Fig).
The NAD+ cofactor and the conserved catalytic Zn2+ ion were modelled into the HvADH2 model based on their position and conformation observed in the structure of the formaldehyde dehydrogenase from Pseudomonas putida (PDB: 1kol, 26% sequence identity with HvADH2) [24]. HvADH2 possesses the typical Rossmann binding motif found in NAD-binding enzymes.
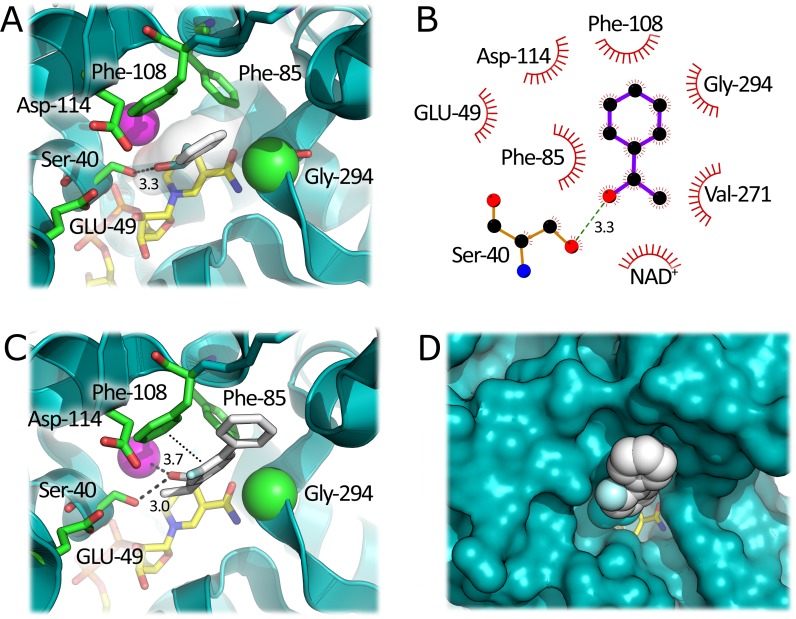
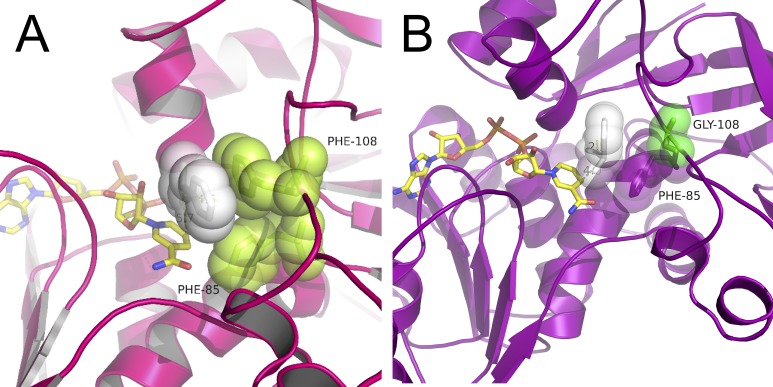
Docking studies with the secondary aromatic alcohol, (S)-1-phenylethanol, ((S)-1-PheOH) allowed the identification of the residues involved in the substrate binding at the active site of HvADH2. Two phenylalanine residues in position 85 and 108 (F85 and F108), at the top of the active site, were identified as being important for substrate binding. These residues, together with G294, form a hydrophobic pocket for the bulky aromatic ring of the substrate: in particular, F108 participates in π-π stacking interactions with the aromatic ring of (S)-1-PheOH. The polar hydroxyl group of the substrate is located in a polar region of the active site lined by E49, D144 and S40 which forms a H-bond with oxygen of (S)-1-PheOH (Fig 1). HvADH2 showed a specific activity of 1200 mU/mg with (S)-1-PheOH under standard assay conditions (while with 10 mM benzyl alcohol, 1 mM NADP+, 50 mM glycine-KOH, pH 10.0, the activity was 2300 mU/mg). The aspecific van der Waals interactions provided by the two Phe residues could explain the promiscuous activity of HvADH2 on aromatic substrates.
Fig 1. Three-dimensional model of HvADH2 active site in complex with different docked ligands.
A) Complex with (S)-1-PheOH. B) LigPlot analysis of the interactions between docked (S)-1-PheOH and HvADH2 model. The substrate is in purple. Hydrophobic contacts are shown as dark red arches. C) Complex with (S)-flurbiprofenol. D) Complex with (S)-flurbiprofenol, surface representation. The ligands are represented as white sticks or spheres. The NAD+ cofactor is in yellow and Zn2+ is represented as a pink sphere. Important residues are shown in green; the αC of G294 is shown as a sphere. The π-π staking interactions are shown by dotted lines and H-bonds by dashed lines.
Although flurbiprofenol has significantly increased bulk in the side chain, the docking analysis shows that the mode of binding is very similar to (S)-1-PheOH. This is because the additional aryl ring of the ligand is placed at the active site entrance, in contact with bulk solvent. The hydroxyl group of the substrate is still within H-bonding distance to S40 and is 3.7 Å from the active site Zn2+ ion. The activity of purified wild-type HvADH2 toward 1 mM rac-flurbiprofenol (2 M KCl, 30% MeOH to facilitate solubility) is 11.1 mU/mg. The racemic mixture of this alcohol was used since it was produced by the synthetic procedure employed (S1 File). Based on the HvADH2-substrate complex model, this >100-fold drop in specific activity relative to (S)-1-PheOH could be explained by non-optimal positions of the reactive carbon atom of the substrate and the C4 of the cofactor NAD+ (Fig 1C). Mutagenesis of F85 and F108 (i.e., substitution with a smaller residue) could result in a shorter distance between these two reactive atoms, thus affecting the catalytic activity of the enzyme by facilitating hydride transfer.
Sequence alignment analysis
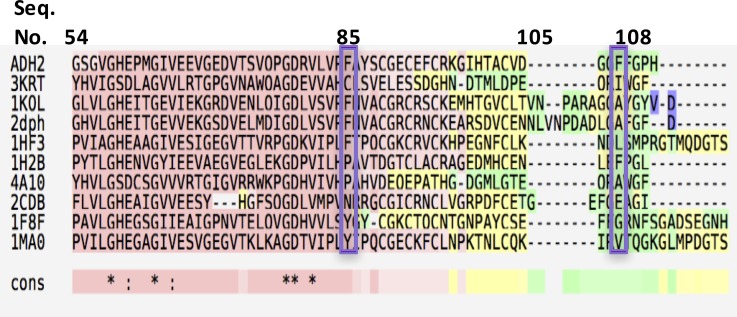
Bio-prodict 3DM database was used for the alignment of over 14000 sequences (HvADH2 no. D4GP73) to determine the conservation degree of the two identified phenylalanine residues (top 9 homologues shown in Fig 2) [25]. Across the alignment, the amino acid distribution at position 88 (numbering is per the 3DM database corresponding to F85 in HvADH2) was 17% phenylalanine, 16% proline and 28% was a gap. Position i1e (corresponding to F108) was less conserved in the alignment since it was found in only a limited pool of 400 sequences: the percentage of phenylalanine residues at this position was 9% (39% lysine, 21% arginine, and 9% phenylalanine with no gaps). Both positions seem to show a low degree of conservation pointing to a role in definition of the enzyme’s substrate preference: both positions were subjected to mutagenesis to investigate their role in substrate binding.
Fig 2. Sequence alignment of HvADH2 with top 9 homologues.
3KRT: putative crotonyl CoA reductase from Streptomyces coelicolor. 1KOL: Formaldehyde dehydrogenase from Pseudomonas putida. 2DPH: Formaldehyde dismutase from Pseudomonas putida. 1HF3: liver alcohol dehydrogenase from Equus caballus. 1H2B alcohol dehydrogenase from Aeropyrum pernix. 4A10: 2-octenoyl-CoA carboxylase reductase from Streptomyces sp. JS360. 2CDB: glucose dehydrogenase from Sulfolobus solfataricus. 1F8F: benzyl alcohol dehydrogenase from Acinetobacter calcoaceticus. 1MAO: glutathione-dependent formaldehyde dehydrogenase from Homo sapiens.
Site-directed mutagenesis of F85 and simultaneous site saturation of F85 and F108
To investigate the significance of F85, four mutations were investigated: a conservative F85Y variant as well as more challenging F85A, F85V and F85R substitutions. F85Y was the only variant which retained activity and was tested with a range of substrates, including flurbiprofenol. Activities were determined for HvADH2 wild-type and F85Y variant crude lysates; substrate concentration was fixed at 10 mM for benzyl alcohol and enantiopure molecules or 20 mM for racemic ones. No activity was detected with flurbiprofenol and the variant enzyme. The reference 100% activity with BzOH and wild-type HvADH2 refers to 247 mU/mg whereas 280 mU/mg with F85Y. In all cases, activity for F85Y variant was lower compared to wild-type: F85Y retained 50% activity with (S)-1-PheOH, whereas wild-type retained 73%. F-MBA was poorly accepted by both wild-type (17%) and F85Y (9%). While 2-Phe-1-Prop was an excellent substrate for wild-type (98%), activity dropped with F85Y (16%). Wild-type activity with 4-Phe-2-But was 53% compared to just 8% with F85Y.
Site-saturation mutagenesis was also performed at both sites, F85 and F108, simultaneously and screened for activity with flurbiprofenol (S1 File). From multiple rounds of screening, a double variant identified as F85AF108G was isolated as the best hit, and the substrate scope was further investigated. Docking (S)-flurbiprofenol with F85AF108G showed the correct conformation of the distal phenyl ring. Activity of HvADH2 F85AF108G in the crude extract was 30% higher with flurbiprofenol when compared to wild-type but when purified, this activity diminished and was therefore deemed a false positive. Since site directed and site saturation of F85 did not yield an improved variant, we focused on F108.
Site directed mutagenesis of F108 and screening with flurbiprofenol
Mutations at position F108 were evaluated initially by in silico modelling and docking with (S)-flurbiprofenol. HvADH2 variants F108A, F108G, F108L, F108P, F108V, F108W, and F108Y were modelled and tested in silico for affinity with (S)-flurbiprofenol, the main differences in theoretical binding energy are reported in Table ES.2. F108V was predicted as the best variant because of its ability to accommodate the biphenyl moiety of (S)-flurbiprofenol (S9A and S9B Fig). In silico results obtained with F108W, F108Y, F108P, and F108L variants were also indicative of improved binding. F108A substitution (S9C and S9D Fig) appeared to open the binding pocket and allow the distal aryl ring to point towards bulk solvent, which was not optimal in the wild-type enzyme.
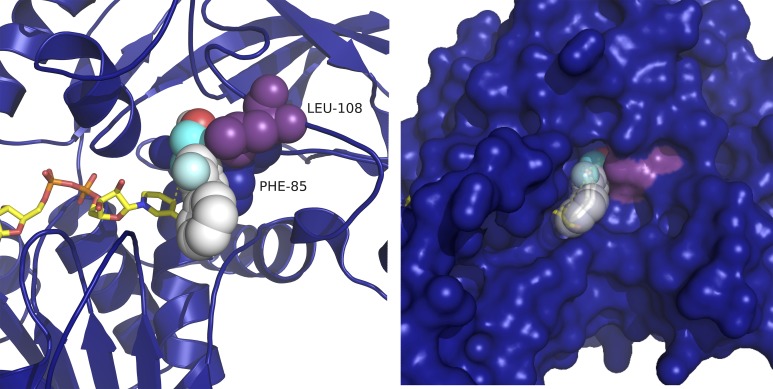
All these variants were engineered by site directed mutagenesis, expressed and purified (S1 File, small scale expression and purification) and activities are reported in Table 1. Contrary to the indications gathered from the docking, F108A and F108G showed no activity with rac-flurbiprofenol, whereas F108W, which introduces more steric bulk into the active site, was slightly more active than WT (13.8 vs. 11.1 mU/mg). The specific activity of F108Y variant (the most conservative substitution) on the latter compound was virtually identical to WT HvADH2. The F108L variant (docking shown in Fig 3) showed the highest specific activity of 25.4 mU/mg, a 2.3-fold increase in activity compared to WT enzyme. To investigate if a non-conserved mutation would be beneficial, a methionine variant was prepared (F108M). While F108M HvAFH2 was active with EtOH (1103 mU/mg), it did not show activity towards flurbiprofenol.
Table 1. Activity of purified wild-type and F108 variants of HvADH2 with rac-flurbiprofenol.
| Variant | Specific activity (mU/mg protein) |
|---|---|
| WT | 11.1 ± 0.2 |
| F108G | 0 |
| F108A | 0 |
| F108V | 16.2 ± 0.1 |
| F108W | 13.8 ± 0.3 |
| F108L | 25.4 ± 0.2 |
| F108Y | 11.7 ± 0.3 |
| F108P | 0 |
| F108M | b.d. |
b.d. = below detection
Fig 3. F108L docked (F85 blue spheres and L108 purple spheres; NAD+, yellow sticks) with (S)-flurbiprofenol (white spheres), surface view.
The distance from the hydroxyl oxygen to the catalytic zinc (O-Zn) is 4.4 Å, and the distance from the substrate α-carbon to the C4 of the nicotinamide ring (αC-C4), is 5.9.
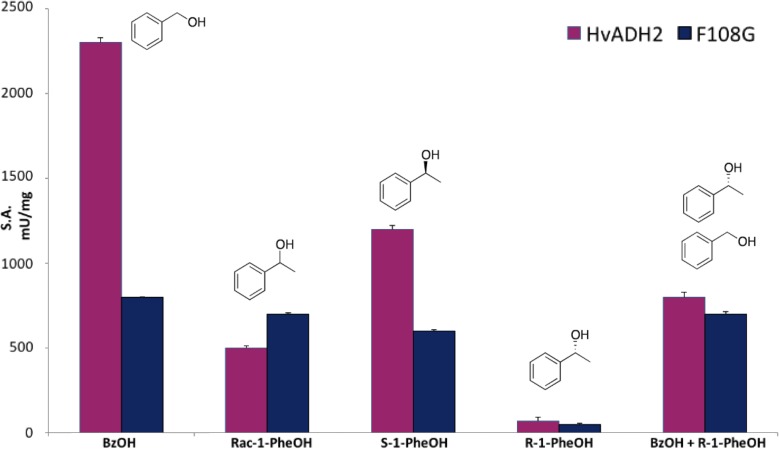
In order to clarify the structure-function relationships modulating HvADH2, purified F108G variant and wild-type HvADH2 were assayed with BzOH, rac-1-PheOH, (S)-1-PheOH and (R)-1-PheOH (Fig 4). Even if F108G HvADH2 was not active with flurbiprofenol, the dramatic change induced in the active site yielded a fully folded protein with 40% activity with respect to the WT, i.e. 800 mU/mg with 10 mM benzyl alcohol (see S1 File for details about the expression and purification).
Fig 4. Substrate specificity of purified F108G variant compared to WT HvADH2.
Buffer conditions: 4 M KCl, 50 mM Gly-KOH, pH 10.0.
Remarkably, F108G showed an increase in activity with rac-1-PheOH with respect to the WT (680 and 460 mU/mg respectively). However, with optically pure (S)-1-PheOH, WT was clearly more active (1200 mU/mg), whereas F108G maintained almost unaltered activity (620 mU/mg). To investigate if the opposite enantiomer was accepted as substrate, (R)-1-PheOH was tested with both WT and F108G HvADH2 but showed negligible activity (70 and 50 mU/mg, respectively). Addition of 10 mM of (R)-1-PheOH to the standard reaction mixture (10 mM benzyl alcohol) reduced the WT HvADH2 specific activity down to 840 mU/mg (in comparison to the 2,300 mU/mg of the original one), while, in the case of the F108G variant, the activity was virtually unaffected. Docking of (R)-1-PheOH to wild-type HvADH2 shows a clear interaction between the aromatic side chain of the substrate and F108 (Fig 5A) which is not present in the variant harbouring F108G (Fig 5B). On closer inspection of the docking, in the wild-type model, the distance between the reactive carbon atom of the substrate and the C4 of the cofactor NAD+ is 5.7 Å. This distance is shortened to 4.2 Å in the F108G model. The experimental evidence together with the in silico predictions strongly suggest that removal of the bulky side-chain from F108 in the glycine variant creates a cavity in the active site. This space could allow the binding of the preferred enantiomer while still housing the (R)-1-PheOH without hampering catalytic efficiency of the enzyme in the presence of a racemic mixture.
Fig 5. Docking of (R)-1-PheOH (white spheres).
A) Catalytic site of the WT enzyme (F85 and F108 green spheres, NAD+, yellow sticks) shows stabilization of the substrate aromatic ring by F108. The distance from the hydroxyl oxygen to the catalytic zinc (O-Zn) is 4.6 Å, and the distance from the substrate α-carbon to the C4 of the nicotinamide ring (αC-C4), is 5.7 Å. B) Catalytic site of the F108G variant. The distance from the hydroxyl oxygen to the catalytic zinc (O-Zn) is 2.5 Å, and the distance from the substrate α-carbon to the C4 of the nicotinamide ring (αC-C4), is 4.0 Å.
Experimental
Expression and purification of wild-type and variants of HvADH2 in Haloferax volcanii
The transformation, production, purification and identification of wild-type and variants of HvADH2 were performed as described previously [16, 17]. Small scale expression and purification are detailed in S1 File.
Enzyme assays
Enzyme activity was assayed as production of the NADPH cofactor detected at 340 nm, measured in intervals of 1 min for 20 min at 50°C (Epoch 2 microplate reader, BioTek, Bad Friedrichshall, Germany; 96 Well Clear Flat Bottom UV-Transparent Microplate Corning®, 3635). All kinetic assays were performed in triplicate. The blank was treated by adding the storage buffer (3 M KCl, 100 mM Tris-HCl, pH 8.0) instead of enzyme.
HvADH2 homology modelling
The web-based server SWISS-MODEL was used to build the model of HvADH2 with a GMQE score of 0.68 [20, 21]. The details for the docking and inspection are mentioned in S1 File.
HvADH2 mutant generation
The adh2 gene harboured in the pTA963 plasmid was mutated using the QuikChange Lightning Multi Site-Directed Mutagenesis Kit provided by Agilent Technologies®. Details of the PCR reaction and the oligonucleotide primers are detailed in S1 File and S1 Table.
Conclusions
Rational design coupled with molecular modelling were applied here for the generation of several site directed variants of HvADH2. Building a homology model of HvADH2 allowed the identification of two phenylalanine residues, at position 85 and 108, which were proposed to be critical residues for the binding of 1-PheOH due to π-π stacking interactions. Docking (S)-flurbiprofenol (an intermediate for the non-steroidal anti-inflammatory drug, flurbiprofen) into the wild-type HvADH2 model showed the unfavourable conformation of the distal aryl ring due to the interactions with the two Phe residues.
F85 appears critical for the stabilization in the binding pocket of small aromatic substrates, whereas the absence of the side chain of F108 facilitates the binding of secondary aromatic alcohols (i.e., 1-PheOH). Site saturation mutagenesis was performed at both sites to make a small, diverse library. However, several rounds of screening failed to identify an improved variant. The best hit, F85AF108G lost all activity after purification. It was then decided to perform site directed mutagenesis at each site, independently of each other. F85 tolerated only conservative mutations such as F85Y which was active with all tested substrates albeit less than the WT HvADH2. Sequence alignments confirmed that F108 was indeed a less conserved position and site directed mutagenesis was performed following in silico modelling and docking to predict improved variants. Among the generated single point variants, F108W, F108Y and F108L accepted flurbiprofenol with enhanced activity compared to wild-type HvADH2; specifically, F108L had a 2.3-fold improvement in activity. The F108G variant showed surprisingly no activity with flurbiprofenol while retaining 40% of the WT activity with benzyl alcohol. Further testing of this variant with rac-1-PheOH indicated that the enzyme performed significantly better than the WT possibly due to the larger binding site created. (R)-1-PheOH is not a substrate for the glycine variant nor for the WT HvADH2. This compound is a strong competitive inhibitor of the latter; whereas it is not able to bind to the F108G variant possibly due to the key role of F108 in stabilising the aromatic moiety of the substrate. In silico docking and site directed mutagenesis were successfully applied to improve enzymatic activity with a bulky, aromatic substrate which was poorly accepted by the wild-type enzyme. This model-guided mutagenesis was performed on a protein for which the closest structural analogue had less than 30% similarity in the sequence, underlying the power of this technique.
Supporting information
(PDF)
(TIFF)
(TIFF)
(TIFF)
Regions of the structure that can be rejected at the 95% and 99% confidence level are represented in yellow and red respectively.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Substrate concentration was fixed at 10 mM in 4 M KCl, 50 mM glycine buffer, pH 10.0.
(TIFF)
Lane 1: broad range protein marker Precision Plus Kaleidoscope, (10–250 kDa); Lane 2: WT; Lane 3: F108Y; Lane 4; F108L Lane 5; F108W.
(TIFF)
A-D. Docking analysis of (S)-flurbiprofenol to F108x HvADH2 variants. Panel A: Docking of (S)-flurbiprofenol to F108V HvADH2; panel B: surface view of panel A. The distance from the hydroxyl oxygen to the catalytic zinc (O-Zn) is 4.3 Å, and the distance from the substrate α-carbon to the C4 of the nicotinamide ring (αC-C4), is 6.7; panel C: docking of (S)-flurbiprofenol to F108A HvADH2; panel D: surface view of panel C. F85 is represented in purple spheres and F108 by lilac spheres, NAD+ by yellow sticks and (S)-flurbiprofenol by white spheres. The distance from the hydroxyl oxygen to the catalytic zinc (O-Zn) is 4.8 Å, and the distance from the substrate α-carbon to the C4 of the nicotinamide ring (αC-C4), is 7.1.
(TIFF)
Lane 1: broad range protein marker P7702S, (2–212 kDa); Lane 2: crude lysate; Lane 3–10: eluted fractions 1–8 respectively. The band corresponding to F108G is indicated by the arrow.
(TIFF)
Acknowledgments
The authors wish to acknowledge the support from Science Foundation Ireland through the Synthesis and Solid State Pharmaceutical Centre (12/RC/2275). We also acknowledge the support from the COST Action CM1303 “SysBiocat”.
Abbreviations
- ADH
alcohol dehydrogenase
- F-MBA
fluoro-methyl benzyl alcohol
- 2-Phe-1-prop
2-phenyl-1-propanol
- 4-Phe-2-But
4-phenyl-2-butanol
- WT
wild-type
- HvADH2
ADH2 from Haloferax volcanii
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funded by Science Foundation Ireland through Synthesis and Solid State Pharmaceutical Centre (grant number 12/RC/2275) to FP and the European Cooperation in Science and Technology (COST Action CM1303 “SysBiocat” grant number ECOST-STSM-CM1303-010614-045245) to JC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pedragosa-Moreau S, Le Flohic A, Thienpondt V, Lefoulon F, Petit AM, Ríos-Lombardía N, et al. Exploiting the biocatalytic toolbox for the asymmetric synthesis of the heart-rate reducing agent Ivabradine. Adv Synth Catal. 2017;359: 485–493. [Google Scholar]
- 2.Wu X, Zhang C, Orita I, Imanaka T, Fukui T, Xing XH. Thermostable alcohol dehydrogenase from Thermococcus kodakarensis KOD1 for enantioselective bioconversion of aromatic secondary alcohols. Appl Environ Microbiol. 2013;79: 2209–2217. doi: 10.1128/AEM.03873-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng YG, Yin HH, Yu DF, Chen X, Tang XL, Zhang XJ, et al. Recent advances in biotechnological applications of alcohol dehydrogenases. Appl Microbiol Biotechnol. 2017;101: 987–1001. doi: 10.1007/s00253-016-8083-6 [DOI] [PubMed] [Google Scholar]
- 4.Savile C, Gruber JM, Mundorff E, Huisman G, Collier SJ. Ketoreductase polypeptides for the production of 3-aryl-3-hydroxypropanamine from a 3-aryl-3-ketopropanamine. US8288141: 2012.
- 5.Ziegelmann-Fjeld KI, Musa MM, Phillips RS, Zeikus JG, Vieille C. A Thermoanaerobacter ethanolicus secondary alcohol dehydrogenase mutant derivative highly active and stereoselective on phenylacetone and benzylacetone. Protein Eng Des Sel. 2007;20: 47–55. doi: 10.1093/protein/gzl052 [DOI] [PubMed] [Google Scholar]
- 6.Nealon CM, Welsh TP, Kim CS, Phillips RS. I86A/C295A mutant secondary alcohol dehydrogenase from Thermoanaerobacter ethanolicus has broadened substrate specificity for aryl ketones. Arch Biochem Biophys. 2016;606: 151–156. doi: 10.1016/j.abb.2016.08.002 [DOI] [PubMed] [Google Scholar]
- 7.Karume I, Takahashi M, Hamdan SM, Musa MM. Deracemization of secondary alcohols by using a singe alcohol dehydrogenase. ChemCatChem. 2016;8: 1459–1463. [Google Scholar]
- 8.Swint-Kruse L. Using evolution to guide protein engineering: the devil is in the details. Biophys J. 2016;111: 10–18. doi: 10.1016/j.bpj.2016.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Munawar N, Engel PC. Prospects for robust biocatalysis: engineering of novel specificity in a halophilic amino acid dehydrogenase. Extremophiles. 2013;17: 43–51. doi: 10.1007/s00792-012-0491-7 [DOI] [PubMed] [Google Scholar]
- 10.Quaglia D, Pori M, Galletti P, Emer P, Paradisi F, Giacomini D. His-tagged horse liver alcohol dehydrogenase: immobilization and application in the bio-based enantioselective synthesis of (S)-arylpropanols. Process Biochem. 2013;48: 810–818. [Google Scholar]
- 11.Konst P, Merkens H, Kara S, Kochius S, Vogel A, Zuhse R, et al. Enantioselective oxidation of aldehydes catalyzed by alcohol dehydrogenase. Angew Chem Int Ed Engl. 2012;51: 9914–9917. doi: 10.1002/anie.201203219 [DOI] [PubMed] [Google Scholar]
- 12.Giacomini D, Galletti P, Quintavalla A, Gucciardo G, Paradisi F. Highly efficient asymmetric reduction of arylpropionic aldehydes by horse liver alcohol dehydrogenase through dynamic kinetic resolution. Chem Commun. 2007;39: 4038–4040. [DOI] [PubMed] [Google Scholar]
- 13.Galletti P, Emer E, Gucciardo G, Quintavalla A, Pori M, Giacomini D. Chemoenzymatic synthesis of (2S)-2-arylpropanols through a dynamic kinetic resolution of 2-arylpropanals with alcohol dehydrogenases. Org Biomol Chem. 2010;8: 4117–4123. doi: 10.1039/c005098a [DOI] [PubMed] [Google Scholar]
- 14.Friest JA, Maezato Y, Broussy S, Blum P, Berkowitz DB. Use of a robust dehydrogenase from an archael hyperthermophile in asymmetric catalysis−dynamic reductive kinetic resolution entry into (S)-profens. J Am Chem Soc. 2010;132: 5930–5931. doi: 10.1021/ja910778p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liliensiek AK, Cassidy J, Gucciardo G, Whitely C, Paradisi F. Heterologous overexpression, purification and characterisation of an alcohol dehydrogenase (ADH2) from Halobacterium sp. NRC-1. Mol Biotechnol. 2013;55: 143–149. doi: 10.1007/s12033-013-9666-4 [DOI] [PubMed] [Google Scholar]
- 16.Timpson LM, Alsafadi D, Mac Donnchadha C, Liddell S, Sharkey MA, Paradisi F. Characterization of alcohol dehydrogenase (ADH12) from Haloarcula marismortui, an extreme halophile from the Dead Sea Extremophiles. 2012;16: 57–66. [DOI] [PubMed] [Google Scholar]
- 17.Timpson LM, Liliensiek AK, Alsafadi D, Cassidy J, Sharkey MA, Liddell S, et al. A comparison of two novel alcohol dehydrogenase enzymes (ADH1 and ADH2) from the extreme halophile Haloferax volcanii. Appl Microbiol Biotechnol. 2013;97: 195–203. doi: 10.1007/s00253-012-4074-4 [DOI] [PubMed] [Google Scholar]
- 18.Alsafadi D, Paradisi F. Effect of organic solvents on the activity and stability of halophilic alcohol dehydrogenase (ADH2) from Haloferax volcanii. Extremophiles. 2013;17: 115–122. doi: 10.1007/s00792-012-0498-0 [DOI] [PubMed] [Google Scholar]
- 19.Alsafadi D, Paradisi F. Covalent immobilization of alcohol dehydrogenase (ADH2) from Haloferax volcanii: how to maximize activity and optimize performance of halophilic enzymes. Mol Biotechnol. 2014;56: 240–247. doi: 10.1007/s12033-013-9701-5 [DOI] [PubMed] [Google Scholar]
- 20.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31: 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4: 1–13. doi: 10.1038/nprot.2008.197 [DOI] [PubMed] [Google Scholar]
- 22.Cheng JH. The x-ray crystal structure of formaldehyde dismutase at 2.3 Å resolution. Acta Crystallogr A Found Adv. 2002;58: 102. [Google Scholar]
- 23.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9): 1511–9. doi: 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tanaka N, Kusakabe Y, Ito K, Yoshimoto T, Nakamura KT. Crystal structure of formaldehyde dehydrogenase from Pseudomonas putida: the structural origin of the tightly bound cofactor in nicotinoprotein dehydrogenases. J Mol Biol. 2002;324: 519–533. [DOI] [PubMed] [Google Scholar]
- 25.Kuipers RK, Joosten HJ, Verwiel E, Paans S, Akerboom J, van der Oost J, et al. Correlated mutation analyses on super-family alignments reveal functionally important residues. Proteins. 2009;76: 608–616. doi: 10.1002/prot.22374 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(TIFF)
(TIFF)
(TIFF)
Regions of the structure that can be rejected at the 95% and 99% confidence level are represented in yellow and red respectively.
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Substrate concentration was fixed at 10 mM in 4 M KCl, 50 mM glycine buffer, pH 10.0.
(TIFF)
Lane 1: broad range protein marker Precision Plus Kaleidoscope, (10–250 kDa); Lane 2: WT; Lane 3: F108Y; Lane 4; F108L Lane 5; F108W.
(TIFF)
A-D. Docking analysis of (S)-flurbiprofenol to F108x HvADH2 variants. Panel A: Docking of (S)-flurbiprofenol to F108V HvADH2; panel B: surface view of panel A. The distance from the hydroxyl oxygen to the catalytic zinc (O-Zn) is 4.3 Å, and the distance from the substrate α-carbon to the C4 of the nicotinamide ring (αC-C4), is 6.7; panel C: docking of (S)-flurbiprofenol to F108A HvADH2; panel D: surface view of panel C. F85 is represented in purple spheres and F108 by lilac spheres, NAD+ by yellow sticks and (S)-flurbiprofenol by white spheres. The distance from the hydroxyl oxygen to the catalytic zinc (O-Zn) is 4.8 Å, and the distance from the substrate α-carbon to the C4 of the nicotinamide ring (αC-C4), is 7.1.
(TIFF)
Lane 1: broad range protein marker P7702S, (2–212 kDa); Lane 2: crude lysate; Lane 3–10: eluted fractions 1–8 respectively. The band corresponding to F108G is indicated by the arrow.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.