Supplemental Digital Content is available in the text
Keywords: blood, diagnosis, endocarditis, serology, specific PCR, valve
Abstract
Blood culture-negative endocarditis (BCNE) may represent up to 70% of all endocarditis cases, depending on series. From 2001 to 2009, we implemented in our laboratory a multimodal diagnostic strategy for BCNE that included systematized testing of blood, and when available, valvular biopsy specimens using serological, broad range molecular, and histopathological assays. A causative microorganism was identified in 62.7% of patients.
In this study from January 2010 to December 2015, in an effort to increase the number of identified causative microorganisms, we prospectively added to our diagnostic protocol specific real-time (RT) polymerase chain reaction (PCR) assays targeting various endocarditis agents, and applied them to all patients with BCNE admitted to the 4 public hospitals in Marseille, France.
A total of 283 patients with BCNE were included in the study. Of these, 177 were classified as having definite endocarditis. Using our new multimodal diagnostic strategy, we identified an etiology in 138 patients (78.0% of cases). Of these, 3 were not infective (2.2%) and 1 was diagnosed as having Mycobacterium bovis BCG endocarditis. By adding specific PCR assays from blood and valvular biopsies, which exhibited a significantly greater sensitivity (P < 10−2) than other methods, causative agents, mostly enterococci, streptococci, and zoonotic microorganisms, were identified in an additional 27 patients (14 from valves only, 11 from blood only, and 2 from both). Finally, in another 107 patients, a pathogen was detected using serology in 37, valve culture in 8, broad spectrum PCR from valvular biopsies and blood in 19 and 2, respectively, immunohistochemistry from valves in 3, and a combination of several assays in 38.
By adding specific RT-PCR assays to our systematic PCR testing of patients with BCNE, we increased the diagnostic efficiency by 24.3%, mostly by detecting enterococci and streptococci that had not been detected by other diagnostic methods, but also agents requiring specific management such as Mycoplasma hominis and Tropheryma whipplei.
1. Introduction
Blood culture-negative endocarditis (BCNE), that is, endocarditis in which blood cultures using usual laboratory methods remain sterile, may account for 2.5% to 70% of all cases of endocarditis, depending on countries.[1] This geographical variation in incidence may be explained by several factors including: differences in the diagnostic criteria used; specific epidemiological factors, as is the case for fastidious zoonotic agents; variations in early use of antibiotics prior to blood sampling; differences in sampling and testing strategies[2]; and involvement of unknown pathogens or noninfective etiologies.
Our center serves as a reference center for the diagnosis of BCNE. From 2010 to 2015, we received specimens from more than 1500 patients worldwide for the diagnosis of BCNE. In an effort to reduce the proportion of endocarditis with no identified etiology, we have continuously diversified the diagnostic tests used. In addition to systematic serological testing for the detection of fastidious agents, especially Coxiella burnetii and Bartonella spp,[2–4] we have demonstrated that, when available, valvular biopsies are the most useful specimens with regard to diagnostic performance, notably thanks to histological examination and broad range polymerase chain reaction (PCR).[3,5,6] In addition, we have demonstrated that syndrome-based sampling and testing was particularly suited to the diagnosis of BCNE.[3,7]
In a previous study of 759 BCNE cases, we identified an infectious and a noninfective etiology in 62.7% and 2.5% of cases, respectively, using systematic serological testing as well as broad range PCR from blood and/or cardiac valves.[3] However, in that study, patient recruitment was biased as patients’ specimens were referred to our laboratory from around the world, many for confirmation of a specific diagnosis.
Since 2010, in an effort to reduce the proportion of BCNE with no identified etiology, we added to our diagnostic scheme specific real-time (RT)-PCR assays for common agents of this disease such as Bartonella species, C burnetii, Enterococcus faecalis, E faecium, Escherichia coli, Mycoplasma hominis, Staphylococus aureus, streptococci from the gallolyticus and oralis groups, and Tropheryma whipplei. In this study, we report the results of prospective testing of all patients admitted to Marseille University Hospitals with a suspected diagnosis of BCNE from 2010 to 2015 using systematic specific RT-PCR assays from valves or blood.
2. Methods
2.1. Patients
From January 1st, 2010 to December 31st, 2015, we prospectively included all patients admitted with suspected BCNE to the 4 university hospitals in Marseille, France. Blood cultures were considered negative when no microorganism grew after 5 days of incubation. For each studied patient, a questionnaire was completed by the physician in charge. Requested information included age, sex, the involved cardiac valve and its type (native or prosthesis, and the type of preexisting valvular defect, if any), contact with cats or body lice, drug abuse, immunodeficiency (and its type), antibiotic uptake prior to blood cultures, clinical and echocardiographic data used in the Duke score,[8] clinical and biological data used in the Marseille score,[9] antibiotic treatment, and outcome (valvular surgery, recovery, and death). Signed informed consent was obtained from all patients. Systematic testing was performed for all BCNE patients using a diagnostic kit and, when available, the patients’ valvular biopsies. The study was approved by the ethics committee of the Institut Federatif de Recherche 48 under reference 07-015.
2.2. Diagnostic procedures
2.2.1. Serology
Indirect immunofluorescence assays to detect significant levels of antibodies to C burnetii (phase I IgG titer >1:800), Bartonella quintana, B henselae (IgG ≥ 1:800), and Legionella pneumophila (total antibody titer ≥1:256) were performed as previously described.[4] Specific antibodies to Brucella melitensis and Mycoplasma pneumoniae were detected with an immunoenzymatic antibody test (titer ≥1:200) and the Platellia M pneumoniae IgM kit (BioRad, Marnes-la-Coquette, France), respectively. When 1st rank tests were negative, we systematically performed a Western blot using Bartonella spp antigens as previously described.[10]
2.2.2. Detection of auto-antibodies
The presence of rheumatoid factor, antinuclear antibodies, and anti-DNA antibodies was determined using the Rheumatoid Factor IgM kit (Orgentec, Trappes, France), ANA Hep2 kit (BMD, Marne-la-Vallée, France), and MuST Connective kit (Inodiag, Signes, France), respectively.
Patients with porcine bioprostheses were systematically tested for total (TIgE) and specific immunoglobulin E to pork (SIgEp) using the FEIA ImmunoCAP kit (Phadia, Sweden).
2.2.3. Molecular detection methods
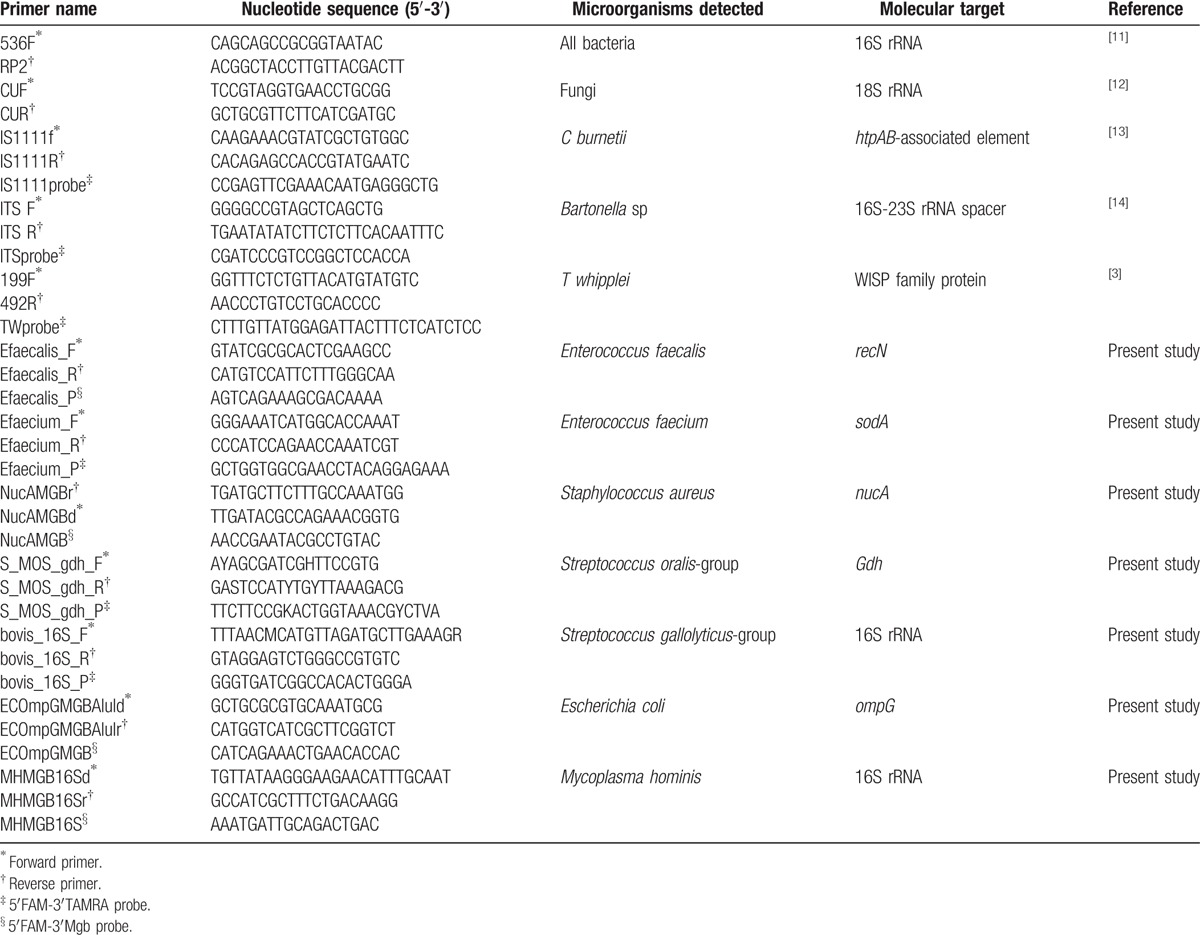
Bacterial DNA was extracted from surgically excised valves or EDTA blood, when no valve was available, using the QIAmp Tissue kit (QIAGEN, Hilden, Germany) as described by the manufacturer. Prosthetic valvular material for which no tissue was available for direct DNA extraction or histological examination (eg, some of the cardiovascular implantable electronic device [CIED] leads) were vortexed in 10 mL of sterile trypticase soy broth, and the DNA was extracted from this suspension. PCR and RT-PCR primers and targets are detailed in Table 1.
Table 1.
Primers, probes, and PCR conditions used in this study.
2.2.4. Histopathology
Paraffin-embedded heart valves were examined with hematoxylin-eosin for histopathologic features.[5] To detect microorganisms within tissues, the Giemsa, Gram (Brown-Brenn and Brown-Hopps), periodic-acid Schiff, Grocott-Gomori, Warthin-Starry, Gimenez, and Ziehl-Nielsen stains were systematically performed as described elsewhere.[15]Bartonella henselae and quintana, and C burnetii and T whipplei were tested for in valvular specimens with immunohistochemistry using specific polyclonal antibodies as previously described.[16–18] For patients for whom all other techniques remained negative, we performed auto-immunohistochemistry as previously described.[19]
2.2.5. Statistical methods
All comparisons were performed using the Mantel–Haenszel chi-square test and the EPI info software (version 3.3.2) (http://www.cdc.gov/epiinfo/index.htm). Observed differences were considered significant when P was < .05 for 2-tailed tests.
3. Results
3.1. Patients
Over the study period, 918 patients admitted to Marseille public hospitals were diagnosed as having definite or possible endocarditis, according to the modified Duke criteria.[8] Of these, blood cultures were positive in 635 patients (69.2%) (Table 2). Of the remaining 283 patients, 177 (62.5%) were classified as having definite endocarditis, including 138 in whom our diagnostic strategy (Fig. 1) allowed the identification of an etiology (Table 2, Figs. 1 and 2, Supplemental digital content). This classification was based on pathological criteria in 114 patients, an association of 2 major Duke criteria in 30 patients, 1 major and 3 minor criteria in 26 patients, and 5 minor criteria in 7 patients. The remaining 106 patients (37.5%) were classified as having possible endocarditis. The mean age (±standard deviation) of these 177 patients was 65.3 ± 15.3 years old (range 1–100 years old). Their sex ratio (M/F) was 131/46. Seven patients were admitted to the Nord hospital and the remaining 170 to the Timone hospital (Supplemental digital content). A total of 113 patients had received antibiotics prior to blood culture collection (Supplemental digital content). Valvular specimens were available for 119/177 patients (67.2%), including native valves in 93 patients, bioprosthetic valves in 8, mechanic prostheses in 7, and CIED leads in 11. Both EDTA blood and serum samples were available for all 177 patients. The involved valves were the aortic, mitral, tricuspid, both the aortic and mitral valves, and a CIED in 83, 70, 4, 4, and 16 cases, respectively.
Table 2.
Comparison of microorganisms identified in patients with positive (n = 635) or negative (n = 283) blood cultures.
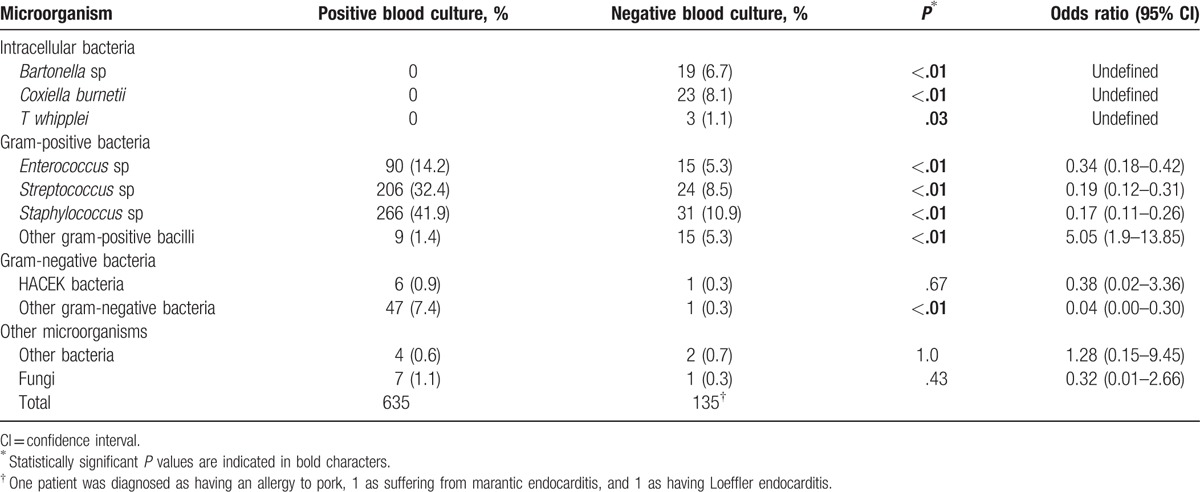
Figure 1.
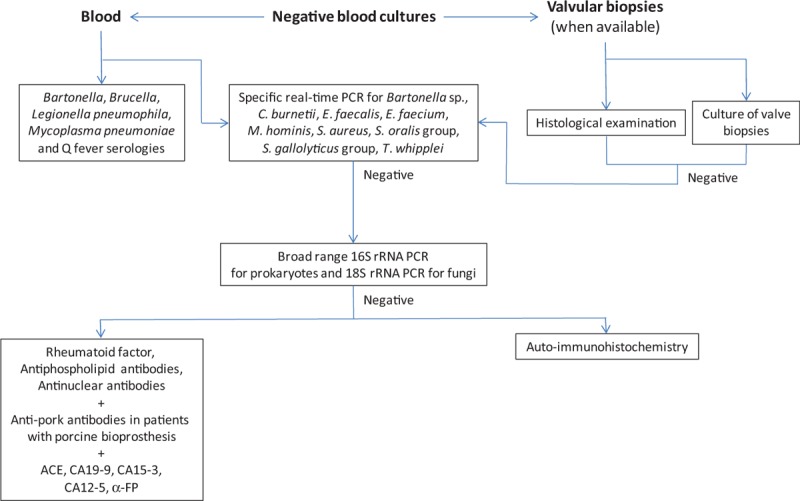
Proposed diagnostic strategy for patients with blood culture-negative endocarditis (BCNE).
Figure 2.
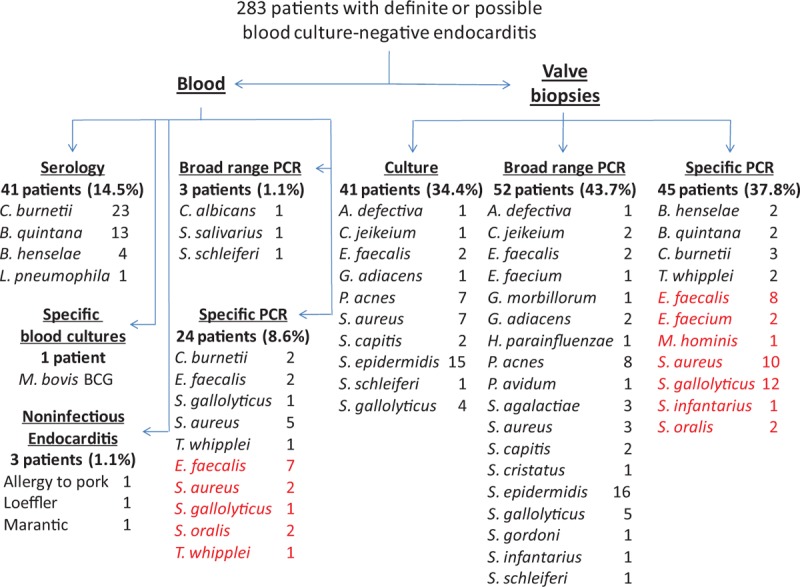
Distribution of identified etiological agents according to the diagnostic method used. The percentages of positive specimens per diagnostic method are indicated in parentheses. Etiological agents identified using newly added specific polymerase chain reaction (PCR) assays are indicated in red.
3.2. Diagnostic procedures
The overall sensitivity of our diagnostic strategy (138 etiologies identified/177 patients with definite BCNE, Fig. 2) was 78.0%. Serology using immunofluorescence assay (Fig. 1) provided a diagnosis in 41/177 patients (23.2%) (Fig. 2). Chronic Q fever (IgG titer to phase I C burnetii >1:800) was diagnosed in 23 patients. Seventeen patients had an IgG titer to B quintana and/or B henselae ≥1:800. Of these, cross-adsorption was able to identify 13 B quintana and 4 B henselae infections. Lastly, 1 patient had positive L pneumophila serology, with an Ig titer of 1:512.
Broad spectrum and specific PCR assays from EDTA blood detected pathogens in 3/177 (1.7%) and 24/177 (13.5%) patients, respectively, for a total of 27 diagnoses (Fig. 1, Supplemental digital content). When applied to valvular specimens, broad spectrum and specific PCR assays were positive in 52/119 (43.7%) and 45/119 (37.8%) patients, respectively, for a total of 85 diagnoses (14% overlap). Negative controls were negative in all assays. Overall, broad range PCR identified a causative agent in 51 patients and specific PCR in 56 (12.1% overlap, for a total number of PCR-positive patients of 99). Culture from valvular biopsies or CIED leads was positive in 41 patients (34.4%), but did not detect any microorganism that had not been identified by PCR (Supplemental digital content). Immunohistochemistry was positive in valvular biopsies from 2 patients infected with B henselae, 2 with B quintana, 3 with C burnetii, and 2 with T whipplei. Although useful in previous studies,[3,19] auto-immunohistochemistry did not provide any additional diagnosis in this series.
The microorganisms detected using culture and/or PCR included Bartonella species in 4 patients, C burnetii in 4, T whipplei in 3 (Fig. 1, Supplemental digital content), Enterococcus spp in 15, S aureus in 12, other staphylococci in 19, streptococci from the gallolyticus and oralis groups in 12 and 4, respectively, other streptococci in 7, other gram-positive germs in 15 (Abiotrophia defectiva, Corynebacterium jeikeium, Gemella morbillorum, Granulicatella adiacens, and Propionibacterium spp), gram-negative bacteria in 1 (Haemophilus parainfluenzae), other bacteria in 1 (M hominis), and fungi in 1 (Candida albicans). In addition, in an 86-year-old male who developed aortic BCNE following intrabladder BCG instillations for bladder cancer, specific blood cultures for mycobacteria in Middlebrook liquid medium grew M bovis BCG. With the exception of this latter case, cultures did not provide any diagnosis that was not made by another method. A 53-year-old male with recurrent mitral BCNE episodes and who had undergone 4 valve replacements with porcine bioprostheses was diagnosed as having allergy to pork[20]; an 83-year-old male who had a bladder cancer with elevated CA19-9 serum levels was diagnosed with mitral marantic endocarditis by histopathological examination of valvular biopsies; and a 34-year-old male who was admitted for BCNE with mitral valve vegetation was diagnosed as suffering from eosinophilic leukemia complicated with Loeffler endocarditis as confirmed by histopathological examination of the removed valve.
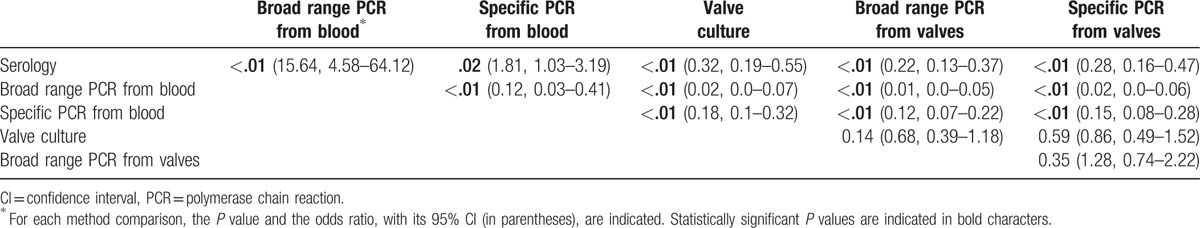
Although broad spectrum and specific PCR assays from blood and valves were complementary, each being the only one to identify an etiological agent in 2, 12, 11, and 14 patients, respectively (Supplemental digital content), broad range PCR from blood was statistically less sensitive than all other diagnostic methods used (Table 3). Similarly, specific PCR from blood provided significantly fewer diagnoses than serology and culture and PCR from valves (Table 3). In contrast, culture, broad range, and specific PCR from valve samples did not significantly differ (Table 3). Bartonella spp and C burnetii were only diagnosed by serology and/or specific PCR and/or immunohistochemistry, and T whipplei by specific PCR and/or immunohistochemistry. Among the 177 patients with definite BCNE (Supplemental digital content), enterococci were more likely to be detected by specific PCR than valve culture (15 vs 2, P < 10−2, OR 8.44, range 1.88–37.77) or broad range PCR (15 vs 3, P < 10−2, OR 5.57, range 1.57–19.81). Streptococci were detected significantly more in valves using specific PCR assays than culture (18 vs 4, P < 10−2, OR 5.12, range 1.67–15.64), but not more than broad spectrum PCR (18 vs 12, P = .32, OR 1.58, range 0.73–3.46), although the detected species differed. Streptococci from the gallolyticus and oralis groups were detected more by specific than broad range PCR (18 vs 6, P = .01, OR 3.35, range 1.28–8.78). The detection of staphylococci using broad spectrum PCR or culture from valvular specimens was not statistically different (23 vs 24, P = .88, OR 0.95, range 0.5–1.79). However, S aureus was statistically more identified by specific than broad spectrum PCR (12 vs 3, P = .02, OR 4.33, range 1.19–15.78). Thus, overall, specific RT-PCR was significantly more sensitive than broad range PCR (58 vs 12, P < 10−2, OR 8.47, range 4.22–17.0) and valve culture (58 vs 13, P < 10−2, OR 7.75, range 3.93–15.28).
Table 3.
Statistical comparison of the sensitivities of the various diagnostic methods used.
All patients in this series received an empirical antibiotic treatment as per the guidelines of the European Society for Cardiology.[21] In 27 patients, the antibiotic regimen was adapted to the identified etiological agents. These included all patients infected with C albicans, C burnetii, L pneumophila, M bovis BCG, and M hominis. Five patients died within 6 to 30 days following admission. Of these, 4 were male. An etiological agent was identified by PCR in 2 patients, including E faecalis from a valvular biopsy in 1 and P acnes from a CIED lead in the other. Four died from heart failure and 1 from a cerebral embolism despite antibiotic treatment.
4. Discussion
We report here the results of the diagnostic strategy that we systematically applied to patients with BCNE from 2010 to 2015. To evaluate the effectiveness of our syndrome-based diagnostic strategy for BCNE without being biased by heterogeneous patient populations from areas with distinct epidemiologies, we only included patients admitted to Marseille public hospitals. Specific PCR assays were designed to target BCNE agents that had either been found to be common in Marseille[3] and/or that were fastidious, such as Bartonella spp, C burnetii, M hominis, and T whipplei.
Of 283 patients with BCNE, 177 patients were diagnosed as having definite endocarditis. An etiological diagnosis was made in 138 patients (78.0%), including 135 in whom a microorganism was identified. Of these, C burnetii, Bartonella spp, and T whipplei accounted for 32.8% of diagnoses and 15.9% of all BCNE cases (Table 2, Fig. 1). Such a high prevalence of these agents when compared to other series (4.9% of all IE cases from 2010 to 2015) may be explained by the endemicity of Q fever in the Marseille area and by the Marseille university hospitals’ role as reference center for these diseases. This may also, at least partially, explain the high rate of BCNE (30.8%) in our IE series during this period. Thus, we acknowledge the fact that the epidemiology of BCNE agents identified in the Marseille area may differ from those observed in other regions. Of the other identified BCNE agents, enterococci, streptococci, and staphylococci represented 10.9%, 17.5%, and 22.6% of diagnoses (8.5, 13.5, and 17.5% of BCNE cases, respectively). The major role of gram-positive cocci as BCNE agents has previously been observed in other series, most often resulting from the early administration of antibiotics.[3,22–24] In the present study, 56/70 patients infected with gram-positive cocci had received antibiotics prior to blood culture collection (80%, Supplemental digital content). Other gram-positive (A defectiva, C jeikeium, G morbillorum, G adiacens, and Propionibacterium spp) accounted for 10.9% of diagnoses (8.5% of BCNE cases). The high prevalence of S epidermidis and P acnes motivated us to add to our diagnostic strategy specific RT-PCR assays for these agents. In contrast, the low rate of gram-negative bacteria in BCNE patients (2.2% and 1.1% of diagnoses and BCNE cases, respectively) may be due to the fact that their prevalence is inferior to that of other bacteria and/or that they are cleared faster from the bloodstream by antibiotics. This also prompted the removal of the E coli-specific RT-PCR assay from our diagnostic protocol. In addition, unusual etiologies were identified, including 1 case each of L pneumophila, M bovis BCG, and M hominis infections, 1 of marantic endocarditis, 1 of Loeffler endocarditis, and 1 of allergy to pork.[20] The overall sensitivity of our diagnostic strategy (138 etiologies identified/177 patients with definite BCNE, 78.0%) was higher than that obtained in one of our previous series (65.2%[3]), in which we had included all patients with suspected BCNE,[3] but similar (77.9%[4]) to another study in which we also had considered only patients classified as definite endocarditis but that was biased by a high rate of patients referred to us from hospitals around the world with high suspicion of Q fever or Bartonella endocarditis.[4] When comparing the diagnostic yield of all laboratory techniques used for BCNE, we observed that the combination of specific PCR assays was significantly more sensitive than all other methods (Table 3). However, the specimen nature greatly influenced the diagnostic output, with specific PCR from valvular biopsies being significantly more sensitive than from blood (Table 3). In addition, broad range PCR from blood was significantly less sensitive than all other methods but identified microorganisms that were not in the detection spectrum of specific PCR assays in 2 patients for whom no valvular biopsy was available (Fig. 1). We observed that the detection overlap between diagnostic methods was globally low, except culture and immunohistochemistry from valvular specimens that did not detect any microorganism not identified by PCR. However, culture enables the antibiotic susceptibility testing of bacterial isolates and, thus, an optimization of patient management. Although auto-immunohistochemistry did not provide any additional diagnoses in this series, we have demonstrated its usefulness in previous studies.[3,19] As a consequence, we believe that the permanent addition or removal of diagnostic methods, which enabled the identification of an etiological agent in almost half of BCNE patients in this study, is needed to improve diagnostic output. On the basis of our results, we propose a refined diagnostic scheme in which specific PCR assays are used first, followed by other assays when the test is negative (Fig. 2).
In our study, specific PCR increased the diagnostic yield by 24.3%. RT-PCR targeting specific microorganisms has been demonstrated in many previous studies to be more sensitive than conventional PCR for the diagnosis of IE.[7,25] In addition, to date, several authors have reported using the commercially available LightCycler SeptiFast (LCSF) system for diagnosing IE (Roche, Rotkreuz, Switzerland).[26–29] This multiplex PCR kit combines specific PCR assays that detect 19 bacteria and 6 fungi. When applied to blood from patients with IE, LCSF was demonstrated to exhibit a higher sensitivity than blood culture in case of previous antibiotic uptake.[28,29] Similarly, LCSF may detect significantly more pathogens from resected cardiac valves than culture.[26,27] However, LCSF suffers from a major limitation, that is, it cannot detect pathogens that are not in its target panel and thus is complementary to, but cannot replace, broad range PCR. In addition, unlike our specific PCR panel, LCSF does not take into account the specific epidemiology of IE, particularly in our region. When considering the 135 causative microorganisms identified in our series, 91 were in the spectrum of our specific PCR panel versus only 71 for LCSF (P = .01, OR 0.53, range 0.33–0.88).
Regarding the impact of our diagnostic approach to patient management, the results prompted a specific adaptation of the antibiotic treatment in 27 patients, all of whom recovered from their infections, and a continuation of the empirical treatment in the other 256. Among the latter group, 5 patients died within 30 days of admission, including 2 in whom a microorganism was identified. However, in these 2 patients, the administered empirical antibiotic treatment was active on the detected agents.
5. Conclusion
In summary, we demonstrated that using specific PCR assays targeting the most common pathogens in a given area has the potential to significantly increase the diagnostic yield of BCNE, but that these assays should be included in a global diagnostic strategy involving other methods such as serology, broad range PCR, and valve culture.
Supplementary Material
Acknowledgments
The authors thank the Méditerranée-Infection foundation and “Investissements d’avenir” program from the French “Agence Nationale de Recherche” under reference Méditerranée Infection 10-IAHU-03 for the support.
Footnotes
Abbreviations: BCNE = blood culture-negative endocarditis, CIED = cardiovascular implantable electronic device, LCSF = LightCycler SeptiFast, PCR = polymerase chain reaction, RT-PCR = real time polymerase chain reaction.
Funding/support: The study was funded by the Méditerranée-Infection foundation and benefited from the support of the “Investissements d’avenir” program from the French “Agence Nationale de Recherche” under reference Méditerranée Infection 10-IAHU-03.
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
References
- [1].Brouqui P, Raoult D. Endocarditis due to rare and fastidious bacteria. Clin Microbiol Rev 2001;14:177–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Raoult D, Casalta JP, Richet H, et al. Contribution of systematic serological testing in diagnosis of infective endocarditis. J Clin Microbiol 2005;43:5238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Fournier PE, Thuny F, Richet H, et al. Comprehensive diagnostic strategy for blood culture-negative endocarditis: a prospective study of 819 new cases. Clin Infect Dis 2010;51:131–40. [DOI] [PubMed] [Google Scholar]
- [4].Houpikian P, Raoult D. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore) 2005;84:162–73. [DOI] [PubMed] [Google Scholar]
- [5].Lepidi H, Durack DT, Raoult D. Diagnostic methods current best practices and guidelines for histologic evaluation in infective endocarditis. Infect Clin North Am 2002;16:339–61. ix. [DOI] [PubMed] [Google Scholar]
- [6].Greub G, Lepidi H, Rovery C, et al. Diagnosis of infectious endocarditis in patients undergoing valve surgery. Am J Med 2005;118:230–8. [DOI] [PubMed] [Google Scholar]
- [7].Morel AS, Dubourg G, Prudent E, et al. Complementarity between targeted real-time specific PCR and conventional broad-range 16S rDNA PCR in the syndrome-driven diagnosis of infectious diseases. Eur J Clin Microbiol Infect Dis 2015;34:561–70. [DOI] [PubMed] [Google Scholar]
- [8].Li JS, Sexton DJ, Mick N, et al. Proposed modifications to the duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis 2000;30:633–8. [DOI] [PubMed] [Google Scholar]
- [9].Richet H, Casalta JP, Thuny F, et al. Development and assessment of a new early scoring system using non-specific clinical signs and biological results to identify children and adult patients with a high probability of infective endocarditis on admission. J Antimicrob Chemother 2008;62:1434–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Houpikian P, Raoult D. Western immunoblotting for Bartonella endocarditis. Clin Diagn Lab Immunol 2003;10:95–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Goldenberger D, Kunzli A, Vogt P, et al. Molecular diagnosis of bacterial endocarditis by broad-range PCR amplification and direct sequencing. J Clin Microbiol 1997;35:2733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].White TJ, Bruns T, Lee S. Innis MA, Gelfand DH, Sninsky JJ, White TJ, et al. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications. New York: Academic Press, Inc; 1990. 315–22. [Google Scholar]
- [13].Willems H, Thiele D, Frolich-Ritter R, et al. Detection of Coxiella burnetii in cow's milk using the polymerase chain reaction. J Vet Med B 1994;41:580–7. [DOI] [PubMed] [Google Scholar]
- [14].Roux V, Raoult D. Inter-and intraspecies identification of Bartonella (Rochalimea) species. J Clin Microbiol 1995;33:1573–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bruneval P, Choucair J, Paraf F, et al. Detection of fastidious bacteria in cardiac valves in cases of blood culture negative endocarditis. J Clin Pathol 2001;54:238–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lepidi H, Fournier PE, Raoult D. Quantitative analysis of valvular lesions during Bartonella endocarditis. A case control study. Am J Clin Pathol 2000;114:880–9. [DOI] [PubMed] [Google Scholar]
- [17].Lepidi H, Houpikian P, Liang Z, et al. Cardiac valves in patients with Q fever endocarditis: microbiological, molecular, and histologic studies. J Infect Dis 2003;187:1097–106. [DOI] [PubMed] [Google Scholar]
- [18].Lepidi H, Fenollar F, Dumler JS, et al. Cardiac valves in patients with Whipple endocarditis: microbiological, molecular, quantitative histologic, and immunohistochemical studies of five patients. J Infect Dis 2004;190:935–45. [DOI] [PubMed] [Google Scholar]
- [19].Lepidi H, Coulibaly B, Casalta JP, et al. Autoimmunohistochemistry: a new method for the histologic diagnosis of infective endocarditis. J Infect Dis 2006;193:1711–7. [DOI] [PubMed] [Google Scholar]
- [20].Loyens M, Thuny F, Grisoli D, et al. Link between endocarditis on porcine bioprosthetic valves and allergy to pork. Int J Cardiol 2013;167:600–2. [DOI] [PubMed] [Google Scholar]
- [21].Habib G, Lancellotti P, Antunes MJ, et al. 2015 ESC Guidelines for the management of infective endocarditis: The Task Force for the Management of Infective Endocarditis of the European Society of Cardiology (ESC). Endorsed by: European Association for Cardio-Thoracic Surgery (EACTS), the European Association of Nuclear Medicine (EANM). Eur Heart J 2015;36:3075–128. [DOI] [PubMed] [Google Scholar]
- [22].Tattevin P, Watt G, Revest M, et al. Update on blood culture-negative endocarditis. Med Mal Infect 2015;45:1–8. [DOI] [PubMed] [Google Scholar]
- [23].Ferrera C, Vilacosta I, Fernandez C, et al. Reassessment of blood culture-negative endocarditis: its profile is similar to that of blood culture-positive endocarditis. Rev Esp Cardiol (Engl Ed) 2012;65:891–900. [DOI] [PubMed] [Google Scholar]
- [24].Hoen B, Selton-Suty C, Lacassin F, et al. Infective endocarditis in patients with negative blood cultures: analysis of 88 cases from a one-year nationwide survey in France. Clin Infect Dis 1995;20:501–6. [DOI] [PubMed] [Google Scholar]
- [25].Lang S, Watkin RW, Lambert PA, et al. Evaluation of PCR in the molecular diagnosis of endocarditis. J Infect 2004;48:269–75. [DOI] [PubMed] [Google Scholar]
- [26].Fernandez AL, Varela E, Martinez L, et al. Evaluation of a multiplex real-time PCR assay for detecting pathogens in cardiac valve tissue in patients with endocarditis. Rev Esp Cardiol 2010;63:1205–8. [DOI] [PubMed] [Google Scholar]
- [27].Leli C, Moretti A, Pasticci MB, et al. A commercially available multiplex real-time PCR for detection of pathogens in cardiac valves from patients with infective endocarditis. Diagn Microbiol Infect Dis 2014;79:98–101. [DOI] [PubMed] [Google Scholar]
- [28].Casalta JP, Gouriet F, Roux V, et al. Evaluation of the LightCycler SeptiFast test in the rapid etiologic diagnosis of infectious endocarditis. Eur J Clin Microbiol Infect Dis 2009;28:569–73. [DOI] [PubMed] [Google Scholar]
- [29].Mencacci A, Leli C, Montagna P, et al. Diagnosis of infective endocarditis: comparison of the LightCycler SeptiFast real-time PCR with blood culture. J Med Microbiol 2012;61:881–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


