Abstract
The aim of this study was to investigate if 2 common single nucleotide polymorphisms (SNPs) in the interleukin-13 (IL-13) gene, rs1800925 and rs20541 are associated with chronic obstructive pulmonary disease (COPD) risk.
Case–control association studies were retrieved systematically from PubMed, Scopus, ISI Web of Science, China National Knowledge Infrastructure, and Wanfang databases using standardized subject terms.
Eleven studies including 3077 participants (1896 cases and 1181 controls) were analyzed. Evidence for a positive association between the T allele of the IL-13 SNP rs1800925 and COPD risk was found in the overall population (odds ratio [OR] = 1.57, 95% confidence interval [95% CI]: 1.21–2.04, Pz = .001). In subgroup analysis according to ethnicity, the T allele of rs1800925 was associated with an increased risk of COPD in Asians (OR = 1.88, 95% CI: 1.23–2.87, Pz = .004) and Caucasians (OR = 1.30, 95% CI: 1.01–1.67, Pz = .041), respectively. For rs20541, the results suggested an association between rs20541 and COPD risk in Caucasians under the recessive model (OR = 2.79, 95% CI: 1.13–6.92, Pz = .026), whereas this SNP was not associated with COPD in Asians.
This meta-analysis suggests that the T allele of rs1800925 is associated with the increased risk of COPD in both Asians and Caucasians, whereas rs20541 is associated with the risk of COPD in Caucasians but not in Asians.
Keywords: chronic obstructive pulmonary disease, interleukin-13, meta-analysis, polymorphism
1. Introduction
Chronic obstructive pulmonary disease (COPD) is a common, chronic inflammatory airway disease characterized by irreversible airflow obstruction. It is a leading cause of death worldwide and currently available treatments are largely ineffective.[1] Although exposure to cigarette smoke is the greatest risk factor for COPD, other environmental and genetic factors are likely involved. Familial aggregation studies and genome-wide association studies have suggested a strong genetic component to the risk of developing COPD.[2]
Interleukin-13 (IL-13) is a pleiotropic cytokine whose primary source are activated type 2 T helper (Th2) cells and to a lesser extent mast cells, macrophages, dendritic cells, and natural killer cells. IL-13 has a variety of functions, including regulation of proinflammatory cytokine expression from macrophages, upregulation of adhesion molecules, induction of B-cell production of immunoglobulin E (IgE), and effects on mucus synthesis.[3] Animal and clinical studies have indicated that IL-13 may be involved in the pathogenesis of COPD. Zheng and coworkers[4] reported that IL-13 overexpression in the adult murine lung resulted in emphysema with enhanced lung volumes and compliance, mucus metaplasia, and inflammation. In patients with emphysema, lung IL-13 expression was positively correlated with pulmonary function including lung distension, airway obstruction, and impaired gas exchange.[5] Moreover, there was an increasing number of IL-13-expressing cells in the bronchial submucosa of smokers with symptoms of chronic bronchitis, suggesting that IL-13 contributed to the hypersecretion of mucus.[6] Given the pivotal role of IL-13 in airways disease, the IL-13 gene was considered as a candidate gene for COPD susceptibility. Located on chromosome 5q31–33, the IL-13 gene has 2 common single-nucleotide polymorphisms (SNPs): the polymorphism rs1800925 is a C to T substitution in the 5′ promoter region (–1055C/T), whereas rs20541 is a G to A exchange in exon 4.[7] Multiple association studies have evaluated the relationship of these genetic variations with COPD.[8–17] However, previous research findings have been inconsistent and the contribution of these SNPs to COPD risk remains unclear. This meta-analysis aimed to evaluate the evidence for associations of rs1800925 and rs20541 with COPD risk.
2. Methods
2.1. Literature search strategy and selection of studies
A systematic search was conducted in accordance with recognized methods. We identified eligible studies by searching Scopus, Pubmed, ISI Web of Science, China National Knowledge Infrastructure (CNKI), and Wanfang databases for studies published before 8 May 2017. The search words were broad and included “chronic obstructive pulmonary disease,” “COPD,” “polymorphism,” “variant,” “genetics,” “risk,” “association,” “rs1800925,” “–1111C/T,” “–1112C/T,” “–1055,” “rs20541,” “R130Q,” and “G2044A.” There was no language restriction for the search. Two reviewers screened titles and abstracts to select potentially relevant studies. If the suitability of a paper was uncertain, the full text was reviewed. Reference lists of the included studies and previously published related meta-analyses were hand-searched to ensure that no relevant studies were missed. This meta-analysis conformed to the Meta-analysis of Observational Studies in Epidemiology (MOOSE) statement standards. Since we only used published data, we did not apply for ethics approval.
Any study was considered to be eligible for inclusion in our meta-analysis if it met the following criteria: published in a peer-reviewed journal, assessing the association of rs1800925 and/or rs20541 with COPD risk, case–control design, published in English or Chinese, and reported data for genotypic or allelic frequency in both cases and controls.
2.2. Data extraction
Data from included studies were extracted by 2 reviewers using a data extraction form. Inconsistencies were resolved in a consensus meeting. The following variables were obtained: first author, country, year of publication, ethnicity of participants, number of COPD patients and control subjects, and Hardy–Weinberg equilibrium (HWE; calculated, when not reported). We did not contact authors to obtain additional data.
2.3. Quality assessment
Methodological quality was assessed by 2 reviewers using the Newcastle–Ottawa Scale (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). Disagreements were resolved by consensus.
2.4. Statistical analyses
We calculated unadjusted odds ratios (ORs) with the corresponding 95% confidence interval (95% CI) from raw genotype frequency data and combined these for meta-analysis. According to the Single Nucleotide Polymorphism database (http://www.ncbi.nlm.nih.gov/snp), the minor allele is the T in rs1800925, while it is the A allele in rs20541. The Z-test was used to calculate the P-value of the overall effect for the meta-analysis and P <.05 was considered as statistically significant. Forest plots were used to display OR and 95% CI for each study and the pooled summary effect. Between-study heterogeneity was assessed by the Q-statistic and was considered statistically significant at P <.10. In case of high between-study heterogeneity, the DerSimonian–Laird random effects model is the model of choice to calculate the pooled effect size,[18] rather than the fixed-effects model. HWE was assessed for each dataset using the χ2 goodness-of-fit test. Sensitivity analysis involving omission of 1 study at a time and recalculating the pooled OR was conducted to test for robustness of the pooled effects. Publication bias was evaluated with the Begg rank correlation test (Begg test). Since there were fewer than 10 studies qualified for each SNP, We did not perform funnel plots to evaluate publication bias.[19] Stata software version 11.0 was used for all statistical analyses. P values <.05 were considered statistically significant for all tests, except for Q test for heterogeneity.
3. Results
3.1. Literature search and studies’ characteristics
The literature search in Scopus, PubMed, ISI Web of Science, CNKI, and Wanfang databases yielded 632 records, and 1 study was identified through lists of references (Fig. 1). After screening of the abstracts of these studies, 619 duplicate and/or irrelevant studies were removed. The remaining studies were retrieved for full-text evaluations. Among these, 11 studies from 10 papers met the inclusion criteria and were included in the meta-analysis. These studies were published in a relatively limited time span (2002–2013), witnessing the recent interest in this field. The polymorphisms rs1800925 and rs20541 were in HWE for all studies. The characteristics of the individual studies included in the meta-analysis are provided in Tables 1 and 2.
Figure 1.

Flow chart of study identification, screening, and selection.
Table 1.
Characteristics of the studies of rs1800925 and COPD risk.

Table 2.
Characteristics of the studies of rs20541 and COPD risk.

3.2. Association of rs1800925 with COPD
Nine studies from 8 papers containing 1477 cases and 891 controls investigated the association between rs1800925 and COPD risk. In terms of ethnicity, 4 studies with 922 COPD patients and 347 controls assessed the SNP in Caucasians,[8,10,12,15] whereas 5 studies including 555 cases and 544 controls investigated rs1800925 in Asians.[9–11,13,17] In each study, 4 ORs and their corresponding 95% CIs were calculated: OR T allele versus C allele; dominant OR (TT + CT versus CC); recessive OR (TT versus CT + CC); and additive OR (TT versus CT versus CC). The pooled analysis showed that the T allele of rs1800925 was associated with increased risk of COPD in the overall population (Caucasians and Asians) (OR = 1.57, 95% CI: 1.21–2.04, Pz = .001) (Fig. 2 and Table 3), and this association was statistically significant under dominant (OR = 1.58, 95% CI: 1.19–2.10, Pz = .002), recessive (OR = 2.49, 95% CI: 1.59–3.88, Pz <.001) and additive (OR = 2.39, 95% CI: 1.50–3.81, Pz <.001) models of the genotype (Table 3). We further performed subgroup analysis based on ethnicity, finding that the T allele of rs1800925 was associated with increased risk of COPD in Asians (OR = 1.88, 95% CI: 1.23–2.87, Pz = .004) and Caucasians (OR = 1.30, 95% CI: 1.01–1.67, Pz = .041), respectively (Fig. 2 and Table 3). Compared to all studies together, between-study heterogeneity was reduced in Caucasians (Ph = .185 for dominant model and Ph = .314 for allelic contrast) but not in Asians (Ph = .059 for dominant model and Ph = .016 for allelic contrast) (Table 3). The results of Begg test were not significant, suggesting no evidence of publication bias (Table 3). Sensitivity analysis did not appreciably change the results of our meta-analyses (Fig. 3).
Figure 2.

Meta-analysis of the association between rs1800925 and COPD risk in allele contrast (T allele vs C allele).
Table 3.
Meta-analysis of the association between rs1800925 and COPD risk.

Figure 3.
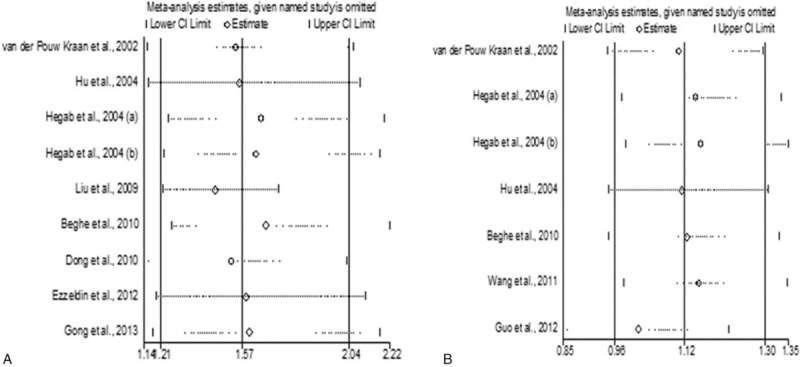
(A) Sensitivity analysis of the association between rs1800925 and COPD risk in allele contrast (T allele vs C allele). (B) Sensitivity analysis of the association between rs20541 and COPD risk in allele contrast (A allele vs G allele). COPD = chronic obstructive pulmonary disease.
3.3. Association between rs20541 and COPD risk
Seven studies from 6 papers with a total of 1418 COPD patients and 763 control subjects were included in the meta-analysis for rs20541.[8–10,12,14,16] In each study, 4 ORs and their corresponding 95% CIs were calculated: OR A allele versus G allele; dominant OR (AA + AG versus GG); recessive OR (AA versus AG + GG); and additive OR (AA versus AG versus GG). In the primary pooled analysis, we found no association between this polymorphism and COPD risk with all eligible studies (Caucasians and Asians) under dominant (OR = 1.12, 95% CI: 0.93–1.34, Pz = .251), recessive (OR = 1.26, 95% CI: 0.87–1.83, Pz = .221), and additive (OR = 1.30, 95% CI: 0.86–1.96, Pz = .208) models (Table 4). In addition, we did not find associations in allele contrast (OR = 1.12, 95% CI: 0.96–1.30, Pz = .152) (Table 4). However, in subgroup analysis according to ethnicity, we found that rs20541 was associated with COPD risk in Caucasians under the recessive model (OR = 2.79, 95% CI: 1.13–6.92, Pz = .026), whereas this SNP was not associated with COPD in Asians (Table 4). Sensitivity analysis demonstrated that omission of 1 study did not significantly influence the summary ORs (Fig. 3). We did not find between-study heterogeneity in any of the genetic risk models (Table 4). We did not identify publication bias for this SNP (Table 4).
Table 4.
Meta-analysis of the association between rs20541 and COPD risk.

4. Discussion
In the literature, there were apparently conflicting associations reported between polymorphisms of the IL-13 gene and COPD risk. This study combined data from studies from Caucasian and Asian populations to investigate the relationship of 2 common IL-13 SNPs, rs1800925 and rs20541, with COPD. The main findings of our meta-analysis are the following: (1) the T allele of rs1800925 is associated increased risk of COPD in both Caucasians and Asians. In Asians, this was confirmed for dominant (TT + CT vs CC), recessive (TT vs CT + CC), and additive (TT vs CT vs CC) models, (2) and rs20541 is associated with increased risk of COPD in Caucasians under the recessive model (AA vs GA + GG), whereas there is no association of rs20541 with COPD risk in Asians.
IL-13 is a multifunctional cytokine which exerts important effects on both inflammatory and structural cells within the airway. It was observed that cells expressing IL-13 dramatically increased in the bronchial submucosa of smokers with symptoms of chronic bronchitis.[6] IL-13 induced mucus production in human airway epithelial cells through chloride channel calcium-activated 1-mediated activation of mitogen-activated protein kinase 13.[20] It also stimulated human bronchial epithelial cells to release proinflammatory cytokine IL-8, which played an important role in attracting neutrophils and monocytes in airway inflammation.[21] In addition, IL-13 enhanced monocyte-derived macrophages’ expression of osteopontin, a phosphorylated acidic glycoprotein that correlated with the degree of airflow limitation in smokers.[22] In vivo studies showed that the overexpression of IL-13 in the adult murine lung caused a phenotype that mirrored human COPD with prominent emphysema.[4] These lines of evidence suggested that IL-13 was implicated in the pathogenesis of COPD and genetic variations in the IL-13 gene may modify the risk for developing COPD. In this meta-analysis, we found an association of rs1800925 and rs20541 with COPD. The polymorphism rs1800925 is located in the promoter region of the IL-13 gene, which results in enhanced promoter activity and increased IL-13 transcription.[23] Previous studies demonstrated that rs1800925 enhanced the adverse effect of cigarette smoking on lung function, leading to the development of COPD.[24] The SNP rs20541 in exon 4 leads to the nonconservative replacement of a arginine with a glutamine in α-helix D, the region critical for interactions between IL-13 and its receptors.[25] Compared to wild-type (WT) IL-13, rs20541 was shown to be associated with higher IL-13 activity. Structure/function analyses found that rs20541 was significantly more active than WT IL-13 in inducing signal transducer and activator of transcription 6 phosphorylation, an essential step in the IL-13-dependent regulation of gene expression in inflammatory cells.[26] Carriers of the A allele of rs20541 were associated with lower lung function measured by forced expiratory volume in 1 second (FEV1).[12] Therefore, the adverse effects mediated by rs1800925 and rs20541 may increase the risk of developing COPD in individuals under certain environmental conditions. For rs20541, we noticed that the association with COPD was different between Caucasians and Asians. Several reasons may account for this difference. First, Caucasians and Asians have different genetic background. The minor allele frequencies of rs20541 in control subjects from Caucasian studies and Asian studies were 0.171 (95% CI: 0.144–0.202) and 0.295 (95% CI: 0.266–0.326), respectively. Second, COPD is a complex disease affected by interactions between genetic, epigenetic, and environmental factors. These factors may modify COPD risk in a distinct way in different populations.
Our results contrasted with those of a prior meta-analysis on the role of rs1800925 in COPD, finding that the T allele was not associated with increased risk of COPD in Asians.[27] We noticed that the prior meta-analysis included an Asian study showing deviation from HWE in the pooled analysis,[28] but it missed a qualified Asian study published in 2004 [9] which we included in our meta-analysis. In addition, the genotype distribution of rs1800925 was not correctly extracted from a case–control association study [12] in the prior meta-analysis. Besides these discrepancies, we included a recently published Asian study which provided new dataset for rs1800925.[17] Thus, the results of our analyses were more reliable to some extent. The IL-13 gene is located on chromosome 5q31–33 in the cluster of genes encoding Th2 cytokines. It has been reported that there is strong linkage disequilibrium (LD) between IL-13 SNPs,[29] which makes it difficult to identify the true genetic variation for COPD. Since most included studies did not perform haplotype analysis for the IL-13 polymorphisms, we were unable to address this issue in this meta-analysis. It will be extremely helpful to evaluate the relationship of COPD with haplotypes constructed by rs1800925 and rs20541 in large populations.
There are several limitations to consider when interpreting the results of this meta-analysis. First, although we identified 9 studies for rs1800925 and 7 studies for rs20541 through extensive literature search from 5 major databases, the total number of subjects was still small. We expect that as more studies become available, a more comprehensive evaluation of the association of rs1800925 and rs20541 with COPD risk will be performed in the future. Second, the relationship of other IL-13 polymorphisms with COPD was assessed in prior studies, such as rs1295685 and rs1881457.[17] However, because the number of eligible studies was <3, we could not perform pooled analyses for these SNPs. Third, Asian studies evaluating rs1800925 showed between-study heterogeneity, which may be attributable to discrepancies in diagnostic criteria for COPD, selection of control subjects, sample size, and genotyping method. Fourth, we did not account for other factors including age, sex, and exposure to cigarette smoke in the pooled analyses, because we did not obtain original data from all included studies.
In conclusion, our meta-analysis indicates that the T allele of the IL-13 polymorphism rs1800925 is associated with increased risk of COPD in both Caucasians and Asians, whereas rs20541 is associated with COPD risk in Caucasians but not in Asians.
Footnotes
Abbreviations: CI = confidence interval, CNKI = China National Knowledge Infrastructure, COPD = chronic obstructive pulmonary disease, FEV1 = forced expiratory volume in 1 second, HWE = Hardy–Weinberg equilibrium, IL-13 = interleukin-13, LD = linkage disequilibrium, MOOSE = meta-analysis of observational studies in epidemiology, OR = odds ratio, SNP = single-nucleotide polymorphism, WT = wild type.
The authors have no funding and conflicts of interest to disclose.
References
- [1].Miniati M, Monti S, Pavlickova I, et al. Survival in COPD: impact of lung dysfunction and comorbidities. Medicine (Baltimore) 2014;93:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Barnes PJ, Burney PG, Silverman EK, et al. Chronic obstructive pulmonary disease. Nat Rev Dis Primers 2015;1:15076. [DOI] [PubMed] [Google Scholar]
- [3].Rayees S, Malik F, Bukhari SI, et al. Linking GATA-3 and interleukin-13: implications in asthma. Inflamm Res 2014;63:255–65. [DOI] [PubMed] [Google Scholar]
- [4].Zheng T, Zhu Z, Wang Z, et al. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J Clin Invest 2000;106:1081–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boutten A, Bonay M, Laribe S, et al. Decreased expression of interleukin 13 in human lung emphysema. Thorax 2004;59:850–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Miotto D, Ruggieri MP, Boschetto P, et al. Interleukin-13 and -4 expression in the central airways of smokers with chronic bronchitis. Eur Respir J 2003;22:602–8. [DOI] [PubMed] [Google Scholar]
- [7].Duffin KC, Freeny IC, Schrodi SJ, et al. Association between IL13 polymorphisms and psoriatic arthritis is modified by smoking. J Invest Dermatol 2009;129:2777–83. [DOI] [PubMed] [Google Scholar]
- [8].van der Pouw Kraan TC, Küçükaycan M, Bakker AM, et al. Chronic obstructive pulmonary disease is associated with the -1055 IL-13 promoter polymorphism. Genes Immun 2002;3:436–9. [DOI] [PubMed] [Google Scholar]
- [9].Hu RC, Xu YJ, Zhang ZX. Study on the correlation of interleukin-13 polymorphism and susceptibility to chronic obstructive pulmonary disease in Chinese Han population. Zhonghua Liu Xing Bing Xue Za Zhi 2004;25:607–11. (Chinese). [PubMed] [Google Scholar]
- [10].Hegab AE, Sakamoto T, Saitoh W, et al. Polymorphisms of IL4, IL13, and ADRB2 genes in COPD. Chest 2004;126:1832–9. [DOI] [PubMed] [Google Scholar]
- [11].Liu SF, Chen YC, Wang CC, et al. Il13 promoter (-1055) polymorphisms associated with chronic obstructive pulmonary disease in Taiwanese. Exp Lung Res 2009;35:807–16. [DOI] [PubMed] [Google Scholar]
- [12].Beghé B, Hall IP, Parker SG, et al. Polymorphisms in IL13 pathway genes in asthma and chronic obstructive pulmonary disease. Allergy 2010;65:474–81. [DOI] [PubMed] [Google Scholar]
- [13].Dong S, Tuohayi J, Hu X, et al. Correlation between polymorphism of interleukin-13 gene and susceptibility to chronic obstructive pulmonary disease in Uygur nationality in Xinjiang. J Chin Pract Diagn Ther 2010;24:984–6. Chinese. [Google Scholar]
- [14].Wang W, Yu Y, Qian R, et al. Association of polymorphisms of IL-13, IL-4, and β-adrenoceptor genes with susceptibility to chronic obstructive pulmonary disease. J Clin Intern Med 2011;28:332–4. (Chinese). [Google Scholar]
- [15].Ezzeldin N, Shalaby A, Saad-Hussein A, et al. Association of TNF-α-308G/A, SP-B 1580 C/T, IL-13-1055 C/T gene polymorphisms and latent adenoviral infection with chronic obstructive pulmonary disease in an Egyptian population. Arch Med Sci 2012;8:286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Guo Y, Gong Y, Pan C, et al. Association of genetic polymorphisms with chronic obstructive pulmonary disease in the Chinese Han population: a case-control study. BMC Med Genomics 2012;5:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gong Y, Shi GC, Wan HY, et al. Association between the interleukin-13 gene and development of chronic obstructive pulmonary disease in southern Chinese Han population: a case-control study. Chin Med J (Engl) 2013;126:4403–8. [PubMed] [Google Scholar]
- [18].DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986;7:177–88. [DOI] [PubMed] [Google Scholar]
- [19].Higgins JP, Altman DG, Gøtzsche PC, et al. The Cochrane Collaboration's tool for assessing risk of bias in randomised trials. BMJ 2011;343:d5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Alevy YG, Patel AC, Romero AG, et al. IL-13-induced airway mucus production is attenuated by MAPK13 inhibition. J Clin Invest 2012;122:4555–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Stríz I, Mio T, Adachi Y, et al. IL-4 and IL-13 stimulate human bronchial epithelial cells to release IL-8. Inflammation 1999;23:545–55. [DOI] [PubMed] [Google Scholar]
- [22].Maneechotesuwan K, Kasetsinsombat K, Wongkajornsilp A, et al. Simvastatin up-regulates adenosine deaminase and suppresses osteopontin expression in COPD patients through an IL-13-dependent mechanism. Respir Res 2016;17:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cameron L, Webster RB, Strempel JM, et al. Th2 cell-selective enhancement of human IL13 transcription by IL13-1112C>T, a polymorphism associated with allergic inflammation. J Immunol 2006;177:8633–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sadeghnejad A, Meyers DA, Bottai M, et al. IL13 promoter polymorphism 1112C/T modulates the adverse effect of tobacco smoking on lung function. Am J Respir Crit Care Med 2007;176:748–52. [DOI] [PubMed] [Google Scholar]
- [25].Madhankumar AB, Mintz A, Debinski W. Alanine-scanning mutagenesis of alpha-helix D segment of interleukin-13 reveals new functionally important residues of the cytokine. J Biol Chem 2002;277:43194–205. [DOI] [PubMed] [Google Scholar]
- [26].Vladich FD, Brazille SM, Stern D, et al. IL-13 R130Q, a common variant associated with allergy and asthma, enhances effector mechanisms essential for human allergic inflammation. J Clin Invest 2005;115:747–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen L, Shen Y, Liu L, et al. Interleukin-13-1112 C/T promoter polymorphism confers risk for COPD: a meta-analysis. PLoS One 2013;8:e68222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jiang L, He B, Zhao MW, et al. Association of gene polymorphisms of tumour necrosis factor-alpha and interleukin-13 with chronic obstructive pulmonary disease in Han nationality in Beijing. Chin Med J (English) 2005;118:541–7. [PubMed] [Google Scholar]
- [29].Alasandagutti ML, Ansari MS, Sagurthi SR, et al. Role of IL-13 genetic variants in signalling of asthma. Inflammation 2017;40:566–77. [DOI] [PubMed] [Google Scholar]


