Abstract
Probody™ therapeutics are recombinant, proteolytically-activated antibody prodrugs, engineered to remain inert until activated locally by tumor-associated proteases. Probody therapeutics exploit the fundamental dysregulation of extracellular protease activity that exists in tumors relative to healthy tissue. Leveraging the ability of a Probody therapeutic to bind its target at the site of disease after proteolytic cleavage, we developed a novel method for profiling protease activity in living animals. Using NIR optical imaging, we demonstrated that a non-labeled anti-EGFR Probody therapeutic can become activated and compete for binding to tumor cells in vivo with a labeled anti-EGFR monoclonal antibody. Furthermore, by inhibiting matriptase activity in vivo with a blocking-matriptase antibody, we show that the ability of the Probody therapeutic to bind EGFR in vivo was dependent on protease activity. These results demonstrate that in vivo imaging of Probody therapeutic activation can be used for screening and characterization of protease activity in living animals, and provide a method that avoids some of the limitations of prior methods. This approach can improve our understanding of the activity of proteases in disease models and help to develop efficient strategies for cancer diagnosis and treatment.
Keywords: Protease activity, Monoclonal antibody, In vivo imaging, Tumor targeting
1. Introduction
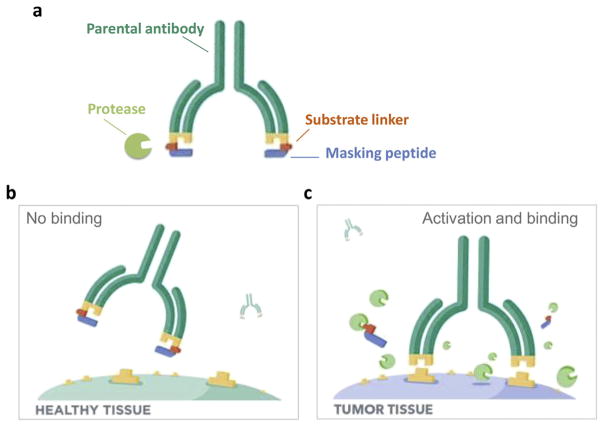
Proteases have long been associated with cancer invasion and metastasis due to their ability to degrade extracellular matrix components and their regulation of cleavage, processing, or shedding of cell signaling molecules [1]. The proteolytic tumor micro-environment is complex, characterized by structurally and functionally diverse proteases that include the matrix metalloproteinases (MMPs), serine proteases, and others [2,3]. The Pro-body technology leverages the upregulation of the activity of these proteases in the tumor microenvironment to achieve disease tissue-specific therapeutic activity. Probody therapeutics contain a masking peptide fused to the N-terminus of the light chain of the antibody through a protease-cleavable linker peptide (Fig. 1). In the intact form, the mask physically prevents the Probody therapeutic from binding to the target antigen in healthy tissues; however, in the diseased environment, the linker is cleaved and the masking peptide is released, resulting in a fully active antibody capable of binding to its target antigen. As such, the proteolytically cleavable linker, which contains a substrate sequence recognized by one or more proteases, can serve to profile the proteolytic environment of the tumor microenvironment.
Fig. 1.
Structure and design of Probody therapeutics. (A) A Probody therapeutic is a monoclonal antibody that contains a light chain extension consisting of a masking peptide (cyan) that blocks the antigen-binding site (yellow), and a protease-specific substrate-containing linker (orange). (B) In the absence of active protease, the Probody therapeutic is functionally masked and cannot effectively interact with target. (C) In the presence of the targeted active protease (green), the linker is cleaved, the masking peptide disassociates, and the Probody therapeutic becomes competent to bind to its target.
In order to develop substrates that are efficiently cleaved at sites of disease, a better understanding of the regulation of protease activity in tumors is needed. However, dissecting how proteases carry out their biological functions in vivo has been challenging, because their activities are regulated by redundant mechanisms, including regulation of biosynthesis at the transcription and translation levels, localization, activation of zymogens and binding of endogenous inhibitors and cofactors. Several methods have been developed to identify the presence of proteases and their activity, including activity-based probes [4,5], active site antibodies [6–8] and proteomics-based approaches [9]. Here we present a new approach for detection of in vivo protease activity, through in vivo optical imaging using Probody technology. Optical imaging has become a useful approach in biomedical sciences because it is a fast, sensitive, and cost-effective method to track and characterize expression of a target, detect enzyme activity and monitor cancer progression or regression and response to therapies in living animals. Leveraging the ability of a Probody therapeutic to bind to a target at the site of disease in a protease-dependent manner, we developed and applied a new technique for non-invasive imaging of protease activity in vivo.
2. Methods
2.1. Probody therapeutic expression, purification and labeling
Probody therapeutics were generated as previously described [10]. In brief, Probody therapeutics were expressed in a modified pcDNA3.1 mammalian expression vector (Life Technologies) and produced in CHO-S cells (Life Technologies). Probody therapeutics were affinity-purified with MabSelect SuRe protein A columns (GE Healthcare) coupled to an AKTA FPLC (GE Healthcare). The purity of purified Probody therapeutics was analyzed by SDS-PAGE, and their homogeneity was analyzed by size exclusion chromatography with a Superdex 200, 10/300 GL column (GE Healthcare).
Antibodies and Probody therapeutics were labeled with a near-infrared fluorescent Alexa Fluor® 750 dye (ThermoFisher Scientific, A20111) by incubation for 1 h at room temperature. The reaction was stopped with 1 M Tris–HCl buffer, pH 8.5 and labeled antibody and Probody therapeutics were separated from free dye using Zeba desalting columns (Life Technologies, 87768). Degree of labeling (DOL) was determined with NanoDrop spectrophotometer. Antibodies and Probody therapeutics with DOL of 2–3 were used in the imaging studies.
2.2. H292 xenograft model
All animal experiments were approved by our Institutional Animal Care Committee (IACUC). Mice were maintained with free access to standard chow and water. NCI-H292 human lung cancer cells were obtained from the American Type Culture Collection (ATCC). They were cultured in RPMI medium supplemented with 10% (v/v) fetal bovine serum (FBS). The cells were maintained in a humidified atmosphere of 5% CO2 at 37 °C. For H292 xenograft studies, 7- to 9-week-old female athymic nu/nu (Charles River Laboratories) mice were inoculated subcutaneously in the right hind flank with 5 × 106 NCI-H292 cells (ATCC) suspended 1:1 (v/v) with Matrigel in serum-free medium. Clinical observations, body weights, and digital caliper tumor volume measurements were made two times weekly once tumors became measureable. Tumor volumes were calculated with the formula (ab2)/2, where a is the longer and b is the smaller of two perpendicular diameters.
2.3. In vivo imaging of protease activity using Probody therapeutics and competition binding to EGFR
H292 xenograft tumor-bearing mice with tumor volumes of 250–500 mm3 were evenly distributed by tumor size into 3 groups with n = 3 per group. The animals were then pretreated by an intraperitoneal injection of 10 mg/kg of the anti-EGFR Probody therapeutic. Control group mice were intraperitoneally injected with PBS or a 10 mg/kg of cetuximab, an anti-EGFR monoclonal antibody. Forty-eight hours after pretreatment all mice were injected intraperitoneally with 10 mg/kg of AlexaFluor 750 (AF750)-conjugated cetuximab (Cetuximab-AF750).
Spectral fluorescence images of the mice were obtained with an IVIS Spectrum/CT imaging system (Caliper Life Sciences, PE) at 48 h and 72 h after Cetuximab-AF750 injection using excitation and emission wavelengths of 745 nm and 800 nm, respectively. During the procedure, the mice were kept under gaseous anesthesia (5% isofluorane) at 37 °C.
2.4. Protease inhibitor A11 blocking studies with Pb-Tx in vivo imaging
H292 xenograft tumor-bearing mice with tumor volumes of 350–750 mm3 were evenly distributed by tumor size into two groups with n = 3 per group. The animals were then pre-treated by an intraperitoneal injection of 15 mg/kg of A11, a human recombinant antibody that recognizes the active form of matriptase over the zymogen form and is a specific inhibitor of its activity. The control group mice were intraperitoneally injected with 15 mg/kg of monoclonal anti-CD20 antibody rituximab. Twenty-four hours after pretreatment all mice were injected intraperitoneally with 10 mg/kg of AlexaFluor 750 (AF750)-conjugated Probody therapeutic, (Pb-Tx-AF750). Spectral fluorescence images of the mice were obtained at 24 h and 72 h after Pb-Tx-AF750 injection.
2.5. In vivo imaging data analysis
Bright-field photographs were obtained with each image. The merged bright-field photographs and fluorescence images were generated using Living Image software 4.1.3 (Caliper Life Sciences). Tumor-associated fluorescence intensities were quantified in the region of interest (ROI). Identical illumination settings (lamp voltage, filters, f/stop, field of views, binning) were used for acquiring all images, and the fluorescence emission within each ROI was determined as the average photons per second per centimeter square per steradian (p/s/cm2/sr) in the quantitative analysis. Fluorescent intensities detected in tumors were further normalized with their corresponding background signals (tumor-to-background ratio, TBR). Data are presented as the mean ± SEM of three individual mice.
2.6. Statistical analysis
All mice used for the in vivo imaging studies were included in the analysis. Mean NIR fluorescence signals as represented by tumor to background ratios (TBR) of average radiant efficiency with SEM were plotted. 48 h and 72 h TBR values were calculated for one mouse in the A11/Pb-Tx-AF750 group by interpolation of linear regression analysis based on 0 h, 24 h and 96 h TBR data. A two-tailed Student’s t test was performed with Microsoft Excel to assess the statistical significance of TBR differences between treated and control groups. P values of ≤0.05 were considered statistically significant.
3. Results
3.1. In vivo imaging of Probody therapeutic by use of competitive target binding
A Probody therapeutic is a fully recombinant biotherapeutic comprised of a monoclonal antibody whose binding to target antigen is blocked by an extension of the NH2-terminus of the light chain, called a masking peptide (Fig. 1a and b). The masking peptide is connected to the light chain by a linker containing a substrate for one or more proteases. Upon cleavage of the substrate-linker by tumor-associated proteases, the mask is removed, and the activated Probody therapeutic binds its target, resulting in tumor-localized activity (Fig. 1c). We previously described a novel anti-epidermal growth factor receptor (EGFR)–directed Probody therapeutic (Pb-Tx) that is efficacious in mouse xenograft models and contains a substrate linker LSGRSDNH cleavable by the tumor-associated serine proteases matriptase (MT-SP1) and urokinase plasminogen activator (uPA) and by the cysteine protease legumain [10]. To evaluate the kinetics of Pb-Tx activation in the tumor microenvi-ronment of living animals, we previously used optical in vivo imaging with Pb-Tx constructs directly labeled with Alexa750, and demonstrated that the Probody therapeutic could be detected bound to tumors in a protease-dependent manner. However, this direct imaging technique has several limitations, including passive accumulation of the labeled Probody therapeutic in the highly vascularized tumor compartment due to the enhanced permeability and retention (EPR) effect, which can lead to high, non-specific background in the images. To address this limitation, we have developed an indirect approach that involves imaging using a labeled EGFR antibody, cetuximab, and competing for binding of this agent in vivo using a non-labeled Pb-Tx. In this way, specific, receptor-mediated binding of the activated Pb-Tx can be distinguished from non-specific EPR. Moreover, in contrast to conventional in vivo imaging, which requires direct labeling of several compounds, our competitive target binding approach uses only a single labeled agent, the EGFR antibody, which minimizes variability that is often associated with labeling of multiple testing agents, and eliminates potential effects of labeling on compound distribution, circulation half-life or binding.
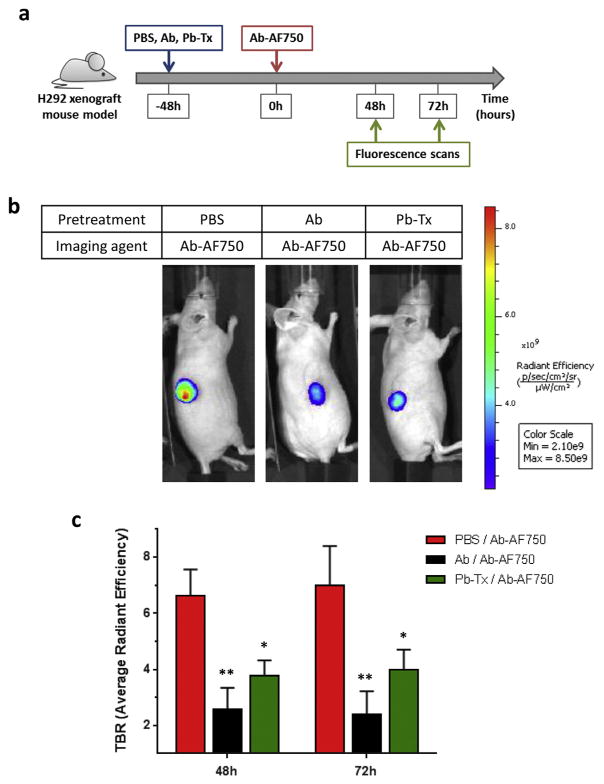
To evaluate this new approach, we used the H292 EGFR-dependent lung cancer xenograft model, due to its high expression of EGFR [11] and our prior experience with in vivo imaging using directly labeled Pb-Tx in this model [10]. In that previous study, significant Probody therapeutic activation and binding in the tumor microenvironment was detected 48 h after administration of labeled Pb-Tx. Therefore, in the current study, 48 h pre-treatment of tumor-bearing mice with non-labeled Pb-Tx was used, followed by in vivo imaging with the parental anti-EGFR antibody (Ab) conjugated with the near-infrared (NIR) fluorescent dye Alexa Fluor® 750 (Ab-AF750) (Fig. 2a). The H292 xenograft mouse model was developed by subcutaneous inoculation of 5 × 106 NCI-H292 cells suspended 1:1 (v/v) with Matrigel. After tumors reached a volume of 250–500 mm3, animals were pre-treated with 10 mg/kg of unlabeled Pb-Tx. In addition, control groups of animals were pre-treated either with PBS or 10 mg/kg of unlabeled parental anti-EGFR antibody (Ab) to demonstrate the maximal detectable dynamic range of antigen-dependent accumulation of the labeled antibody. To evaluate the antigen-dependent accumulation of Probody therapeutics, all mice were administered Ab-AF750 48 h after pretreatment with cold Probody therapeutics, thus enabling the detection of non-occupied EGFR. Spectral fluorescence images of the mice obtained with an IVIS Spectrum/CT imaging system at 48 h and 72 h after administration of Ab-AF750 demonstrated a lower level of tumor-associated NIR fluorescence in the groups pretreated with anti-EGFR Ab or Pb-Tx than in mice pre-treated with PBS (Fig. 2b). Tumor-to-background ratios (TBR) were calculated based on the average radiant efficiency detected at the tumor site and a specified non-tumor area. A significant decrease of TBR was demonstrated for mice pre-treated with Probody therapeutics when compared to PBS pre-treated animals at both the 48 h and 72 h time points (Fig. 2c). These results demonstrate that both anti-EGFR antibody and Probody therapeutics can effectively compete for binding to tumor EGFR with the labeled anti-EGFR antibody in vivo, as detected by optical imaging. Previously, we demonstrated that Pb-Tx does not detectably bind EGFR in the absence of protease activity in vitro, and that the binding of directly labeled Pb-Tx to EGFR in vivo in this tumor model is dependent of the presence of the protease substrate [10]. Taken together, these data suggest that our indirect, in vivo competition binding method can image pro-tease activity associated with H292 tumors in vivo.
Fig. 2.
In vivo imaging of Pb-Tx EFGR binding in an H292 xenograft model. (A) Schematic of the in vivo optical imaging method. Imaging was performed at 48 and 72 h following injection of cetuximab-AF750 into H292 tumor-bearing mice pretreated with PBS or a 10 mg/kg blocking dose of cetuximab or Pb-Tx. (B) Representative images of mice (n = 3) from each treatment group at 48 h. (C) Comparison of mean tumor-to-background ratios (TBR) based on average radiant efficiency values from cetuximab-AF750 in each group. Each bar represents mean TBR ± SEM; n = 3 for each group. *P < 0.05, **P < 0.01.
3.2. Matriptase activity is required for activation of Pb-Tx in H292 xenograft model
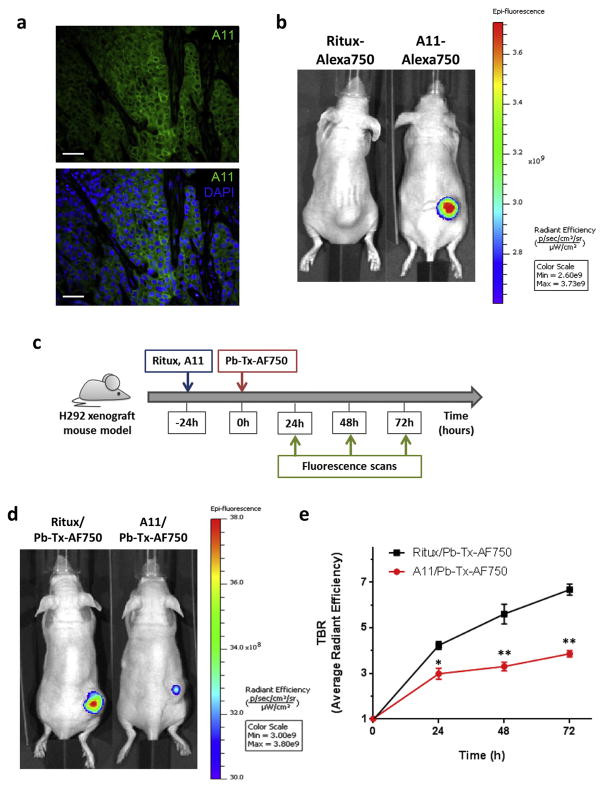
To further confirm that binding of Pb-Tx to EGFR in H292 tumors is dependent on protease activity, we characterized the protease profile of H292 xenograft tumors. Because Pb-Tx was designed to be activated by matriptase, we used an active-site–specific, recombinant human antibody for matriptase, called A11, which binds only to the active form of matriptase and inhibits its activity [7,12]. A11 was labeled with AlexaFluor 488 (AF7488) and immunofluorescence staining was performed on formalin-fixed paraffin-embedded (FFPE) H292 xenograft tumor sections. As shown in Fig. 3a, active matriptase is localized to H292 cancer cells, with a characteristic membranous pattern of staining (Fig. 3a). The presence of active matriptase was also qualitatively assessed in vivo using NIR optical imaging. A11 IgG labeled with the NIR fluorophore AlexaFluor 750 (A11-AF750) was administered to mice bearing H292 xenografts tumors and imaged at excitation and emission wavelengths of 745 nm and 800 nm, respectively. Significant accumulation of labeled A11 probe as compared to non-binding control anti-human CD20 antibody (Rituximab, Ritux-AF-750) was detected at the tumor site (Fig. 3b), indicating the presence of active matriptase in H292 xenografts in vivo. To test whether matriptase activity is necessary for Pb-Tx binding, we pre-treated H292 xenograft-bearing mice with 15 mg/kg of either unlabeled A11 to inhibit matriptase activity or a control antibody (Rituximab) and then performed NIR in vivo imaging with 10 mg/kg of Pb-Tx directly labeled with AF-750 (Pb-Tx-AF750) after 24 h, 48 h and 72 h (Fig. 3c). Pb-Tx accumulation in tumors and TBR of average radiant efficiency were calculated for each time point. Notably, a significant reduction of tumor-associated NIR fluorescence was detected in tumor-bearing mice pretreated with matriptase inhibitory antibody as compared to mice pre-treated with non-binding antibody control (Fig. 3d and e). This data provides further evidence that matriptase-dependent activation of Pb-Tx results in accumulation of Pb-Tx in H292 xenograft tumors.
Fig. 3.
Imaging of protease activity in an H292 xenograft model. (A) Immunofluorescence staining of matriptase activity using A11 active site-specific antibody in H292 xenograft tumor sections. A11 antibody staining is in green and nuclei are stained with DAPI (blue). Scale bar, 100 μm. (B) NIR optical imaging of A11-AF750 antibody accumulation in an H292 xenograft tumor model (right mouse). Rituximab-AF750 antibody was used as a negative, non-binding antibody control (left mouse). The images shown are representative of n = 3 mice/xenograft. (C) Schematic of the in vivo optical imaging study of Pb-Tx binding to tumor EGFR in the presence of matriptase inhibitor. Imaging was performed 24 h, 48 h and 72 h after injection of 10 mg/kg Pb-Tx-AF750 into H292 tumor-bearing mice pretreated with 15 mg/kg of either unlabeled A11 or a control antibody (Rituximab). (D) Representative images of mice (n = 3) from each treatment group at 48 h. (E) NIR optical imaging of Pb-Tx in H292 xenograft tumor bearing mice pre-treated with rituximab or the inhibitory matriptase matriptase antibody A11, followed by administration of labeled Pb-Tx-AF750. The TBR determined for both groups of mice (n = 3) demonstrate decrease of NIR fluorescence in tumors of mice pretreated with matriptase inhibitor A11. *P < 0.05, **P < 0.01.
4. Conclusions
In summary, these results support that in vivo imaging with Probody therapeutics can be a powerful tool for detection of pro-tease activity in vivo. Furthermore, the novel in vivo competition binding method presented in this work demonstrates the potential for detection of protease activity in vivo without the limitations of methods that use directly labeled Probody therapeutics. The dependence of Pb-Tx tumor accumulation on protease activity was confirmed by reduced tumor target binding of Pb-Tx in the presence of a specific protease inhibitor. This conclusion is further supported by previously published data that demonstrated that Pb-Tx binding depends on the presence of the protease substrate in the Probody therapeutic construct.
It should be noted that the ability to obtain data on protease activity non-invasively and longitudinally from the same live animal is an important advantage of the in vivo imaging approach, enabling generation of more data points from fewer animals. In addition, the non-invasive assessment of protease activity in vivo minimizes the possibility of data being confounded by changes in protease activity from tissue/cellular damage during tissue processing. Taken together, in vivo imaging with Probody therapeutics represents a valuable approach for monitoring protease activities in living animals, and enables evaluation of their role in development and progression of diseases, such as cancer. This information would be very useful for the validation of proteases as therapeutic targets and development of potent protease activatable drugs, such as Probody therapeutics.
Acknowledgments
We thank our CytomX colleagues Clayton White, Daniel R. Hostetter, Linnea Diep, Shouchun Liu and Jennifer H. Richardson for insightful discussions and help in preparation of material used in this work. CSC was supported by NIH grant 1P41CA196276.
Footnotes
Conflict of interest
K.R.W., E.M., W.M.K. and O.V. were employees of, and C.S.C. was a compensated advisor to, CytomX Therapeutics, Inc. at the time this work was performed.
References
- 1.Sevenich L, Joyce JA. Pericellular proteolysis in cancer. Genes Dev. 2014;28:2331–2347. doi: 10.1101/gad.250647.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overall CM, Kleifeld O. Tumour microenvironment - opinion: validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat Rev Cancer. 2006;6:227–239. doi: 10.1038/nrc1821. [DOI] [PubMed] [Google Scholar]
- 3.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deu E, Verdoes M, Bogyo M. New approaches for dissecting protease functions to improve probe development and drug discovery. Nat Struct Mol Biol. 2012;19:9–16. doi: 10.1038/nsmb.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godinat A, Budin G, Morales AR, et al. A biocompatible “split luciferin” reaction and its application for non-invasive bioluminescent imaging of protease activity in living animals. Curr Protoc Chem Biol. 2014;6:169–189. doi: 10.1002/9780470559277.ch140047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sela-Passwell N, Kikkeri R, Dym O, Rozenberg H, et al. Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential. Nat Med. 2012;18:143–147. doi: 10.1038/nm.2582. [DOI] [PubMed] [Google Scholar]
- 7.Darragh MR, Schneider EL, Lou J, Phojanakong PJ, Farady CJ, Marks JD, Hann BC, Craik CS. Specific targeting of proteolytic activity for tumor detection in vivo. Cancer Res. 2010;70:1505–1512. doi: 10.1158/0008-5472.CAN-09-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.LeBeau AM, Sevillano N, Markham K, et al. Imaging active urokinase plasminogen activator in prostate cancer. Cancer Res. 2015;75:1225–1235. doi: 10.1158/0008-5472.CAN-14-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleifeld O, Doucet A, auf dem Keller U, et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat Biotechnol. 2010;28:281–288. doi: 10.1038/nbt.1611. [DOI] [PubMed] [Google Scholar]
- 10.Desnoyers LR, Vasiljeva O, Richardson JH, et al. Tumor-specific activation of an EGFR-targeting Probody enhances therapeutic index. Sci Transl Med. 2013;5:207ra144. doi: 10.1126/scitranslmed.3006682. [DOI] [PubMed] [Google Scholar]
- 11.Raben D, Helfrich B, Chan DC, et al. The effects of cetuximab alone and in combination with radiation and/or chemotherapy in lung cancer. Clin Cancer Res. 2005;11:795–805. [PubMed] [Google Scholar]
- 12.LeBeau AM, Lee M, Murphy ST, et al. Imaging a functional tumorigenic biomarker in the transformed epithelium. Proc Natl Acad Sci U S A. 2013;110:93–98. doi: 10.1073/pnas.1218694110. [DOI] [PMC free article] [PubMed] [Google Scholar]