Abstract
Therapy-related myelodysplastic syndrome is a long-term complication of cancer treatment in patients receiving cytotoxic therapy, characterized by high-risk genetics and poor outcomes. Allogeneic hematopoietic cell transplantation is the only potential cure for this disease, but the prognostic impact of pre-transplant genetics and clinical features has not yet been fully characterized. We report here the genetic and clinical characteristics and outcomes of a relatively large cohort of patients with therapy-related myelodysplastic syndrome (n=67) who underwent allogeneic transplantation, comparing these patients to similarly treated patients with de novo disease (n=199). The 5-year overall survival was not different between patients with therapy-related and de novo disease (49.9% versus 53.9%; P=0.61) despite a higher proportion of individuals with an Intermediate-2/High International Prognostic Scoring System classification (59.7% versus 43.7%; P=0.003) and high-risk karyotypes (61.2% versus 30.7%; P<0.01) among the patients with therapy-related disease. In mutational analysis, TP53 alteration was the most common abnormality in patients with therapy-related disease (n=18: 30%). Interestingly, the presence of mutations in TP53 or in any other of the high-risk genes (EZH2, ETV6, RUNX1, ASXL1: n=29: 48%) did not significantly affect either overall survival or relapse-free survival. Allogeneic stem-cell transplantation is, therefore, a curative treatment for patients with therapy-related myelodysplastic syndrome, conferring a similar long-term survival to that of patients with de novo disease despite higher-risk features. While TP53 alteration was the most common mutation in therapy-related myelodysplastic syndrome, the finding was not detrimental in our case-series.
Introduction
Therapy-related myelodysplastic syndrome (t-MDS) is a well-recognized clonal hematopoietic disorder occurring as a late complication following exposure to genotoxic chemotherapy and/or radiation therapy.1 Based on the 2016 World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia, t-MDS is recognized as part of therapy-related myeloid neoplasms.2,3 With recent advances in the management of early malignancies and prevalent use of adjuvant chemotherapy, the incidence of t-MDS seems to be increasing and the condition is becoming an increasing concern for cancer survivors.
t-MDS has an aggressive clinical course with generally dismal outcomes often accompanied by high-risk genetic features.4–7 Patients with t-MDS are usually elderly, have a poor performance status, and frequently manifest residual toxicity from prior therapies. Limiting factors, including adverse disease biology and poor clinical phenotype, commonly result in suboptimal responses to conventional chemotherapy, which consequently lead to low median survival rates for these patients.4,6–8 Allogeneic hematopoietic cell transplantation (HCT) is the only curative option for t-MDS, but to date, the long-term survival rate following this strategy has been relatively low with an excess of treatment-related mortality.9–12
With regards to pathogenesis, t-MDS is thought to be either secondary to genomic alterations induced by cytotoxic therapy, or to arise via outgrowth of pre-existing pre-leukemic myeloid clones after exposure to cytotoxic therapy.13,14 Patients with t-MDS usually present with a TP53 mutation,5,15–17 which is known to be associated with the aggressive disease course. While this mutation is less common in patients with de novo MDS and acute myeloid leukemia,5,16,18–22 the post-transplant relapse rate has been noted to be higher among patients with acute myeloid leukemia/MDS carrying the mutation.5,20,21 Bejar et al. reported no survivors beyond 5 years after allogeneic HCT among 18 patients with a TP53 mutation.20
In cohorts of mixed therapy-related and de novo MDS, correlations between several other mutations (ASXL1, RUNX1, EZH2, ETV6, TET2, and DNMT3A) and inferior transplant outcomes have been demonstrated.19–21 In one study, mutations in any of the five high-risk genes (TP53, EZH2, ETV6, RUNX1 or ASXL1) predicted lower overall survival for MDS patients, independent of other clinical factors.19 However, it is largely unknown whether the presence or absence of these mutations has a similar adverse predictive or prognostic impact when only patients with t-MDS who undergo allogeneic HCT are considered. Here, we report the outcome of one of the largest molecularly characterized cohorts of t-MDS patients who underwent allogeneic HCT.
Methods
Study population
A total of 266 consecutive patients with a diagnosis of MDS who received an allogeneic transplant from a matched sibling or unrelated donor at the City of Hope between January 2000 and October 2014 were analyzed. Patients with chronic myelomonocytic leukemia, pancytopenia and/or dysplasia associated with paroxysmal nocturnal hemoglobinuria or aplastic anemia, and those who underwent cord blood or haploidentical transplantation were excluded. The diagnosis of t-MDS was based solely on a patient’s medical history of prior exposure to any cytotoxic chemotherapy and/or radiotherapy administered for prior malignant or non-malignant conditions. The study was approved by the City of Hope Institutional Review Board.
Data collection
Prior medical history, the patient’s demographic information, cytogenetic and molecular data, prior treatments including hypomethylating agents and transplant outcomes were collected through the institution’s electronic medical records, chart reviews and the blood and marrow transplant program database. An International Prognostic Scoring System (IPSS) score was generated for each patient, based on the percentage of myeloblasts in the marrow at the time of diagnosis, number of cytopenias, and risk classification of cytogenetics.
DNA samples for next-generation sequencing and microarrays
To include granulocytes, DNA was extracted from whole blood samples, following the manufacturer’s recommendations (Qiagen, Germantown, MD, USA). DNA samples were submitted for initial HLA-typing after the diagnosis of MDS and before HCT, based on the assumption that circulating myeloid cells in MDS patients are clonal and carry the same mutational profile of MDS. All patients had ≥20% circulating myeloid lineage cells at the time of the sample collection.
Next-generation sequencing library preparation and bioinformatics analysis
Next-generation sequencing (NGS) libraries were prepared from genomic DNA (40 ng) using the SureSelect target enrichment system (Agilent Technologies Inc.) after transposase-based fragmentation and adapter ligation. The adapter-ligated library was amplified by polymerase chain reaction and quality control was performed for sizing and concentration. Target regions were captured using a customized SureSelect library (Agilent Technologies) for all coding exons plus ten flanking bases of 72 genes (Online Supplementary Table S1). After hybridization of 750 ng of adapter-ligated library with biotin-labeled probes that are specific to target regions, the dual-index tag was added during post-capture polymerase chain reaction amplification. The amplified captured libraries were quality-controlled using a high sensitivity DNA Bioanalyzer kit (Agilent Technologies Inc.) then pooled and sequenced using Miseq V2 Reagent Kit/300 cycles with 150 bp paired-end sequencing. Alignment of sequence reads to the human genome (GRCh37/hg19), variant calling and annotation were performed independently using two software applications – CLCBiomedical Workbench (CLC Bio, Aarhus, Denmark) and NextGENE (Softgenetics, State Collage, PA, USA). Annotated variants were processed using previously published criteria.23,24 Synonymous variants, variants located >2 bp outside protein-coding regions, polymorphisms present in >1% in population databases including ExaC, Exome Variant Server and the 1000 Genomes Project, and variants with <30X coverage were filtered. The remaining variants were evaluated using tumor-specific databases (COSMIC, cBioportal), information retrieved from literature, sequence conservation, and in silico prediction algorithms, including SIFT, Polyphen-2, and FATHMM, for clinical significance.
Microarray methods
Cytogenomic microarray analysis was performed using the Affymetrix CytoScan HD platform, which consists of more than 2.6 million oligonucleotide probes across the genome including ~1.9 million unique non-polymorphic probes and 750,000 single nucleotide polymorphisms. Genomic linear positions in this microarray are given relative to GRCh37/hg19. The data were Initially analyzed with the Affymetrix Chromosome Analysis Suite. All pathogenic, likely pathogenic, large regions of copy neutral loss/absence of heterozygosity (LOH/AOH) and variants of uncertain significance are reported in Online Supplementary Table S2. Common copy number variations (CNV) or regions of LOH/AOH observed in the general population were deemed benign. Truly balanced rearrangements, some forms of polyploidy, low-level mosaicism, and point mutations were not detectable using this assay.
Definitions of outcomes
Overall survival was defined as the time from the day of transplantation to death from any cause. Patients who were alive at their last follow-up were censored. Death from causes other than relapse was considered non-relapse mortality. Relapse was defined as the time to onset of recurrent t-MDS, determined by morphological evidence in bone marrow or extramedullary sites. Relapse-free survival was defined as the time to relapse or death from any cause, whichever came first. Acute and chronic graft-versus-host disease were graded according to previously published criteria.25,26
Statistical analysis
The patients’ characteristics and disease- and transplant-related variables were summarized with descriptive statistics. Overall survival was computed using the Kaplan-Meier method. When calculating non-relapse mortality, relapse was counted as a competing risk, i.e. only patients who did not relapse and died were counted as an event. For patients who relapsed but were alive, the last contact was used as the latest follow-up. Similarly, the cumulative incidence of relapse was calculated with death before relapse as a competing risk factor. Gray method was used when calculating cumulative incidence of relapse and non-relapse mortality. Cox proportional hazard models were used for univariate and multivariate analyses and the hazard ratios are reported with a 95% confidence interval (CI). Stepwise variable selection with a backward Akaika information criterion was used for selecting variables. The full set of variables included age at transplant, sex combination of the donor and recipient, race, period of transplantation, time from diagnosis to transplantation, conditioning regimen, HLA transplant match, graft type, cytomegalovirus status for the donor and recipient and MDS classification. Due to correlations among t-MDS karyotype, marrow blast percentage before HCT and IPSS score (≤1 versus >1), these features were included in the model one at a time after model selection.
Results
Patients’ characteristics
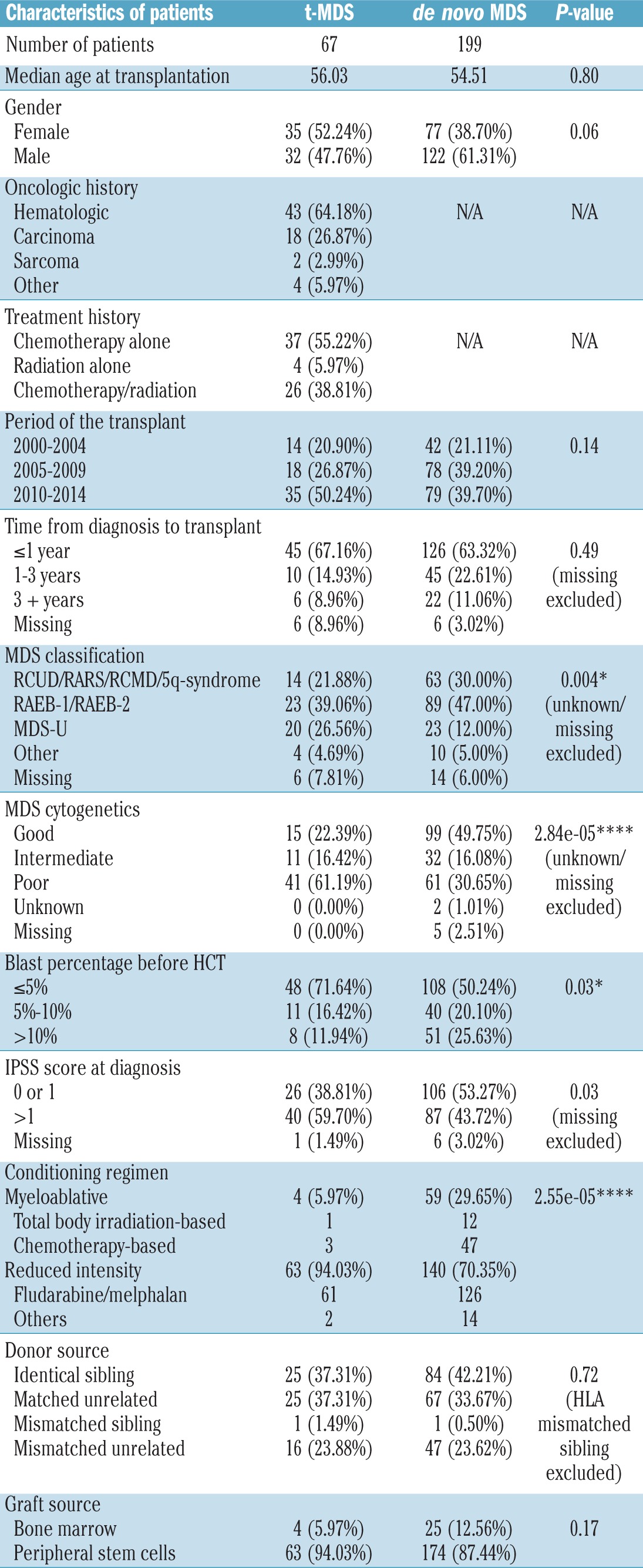
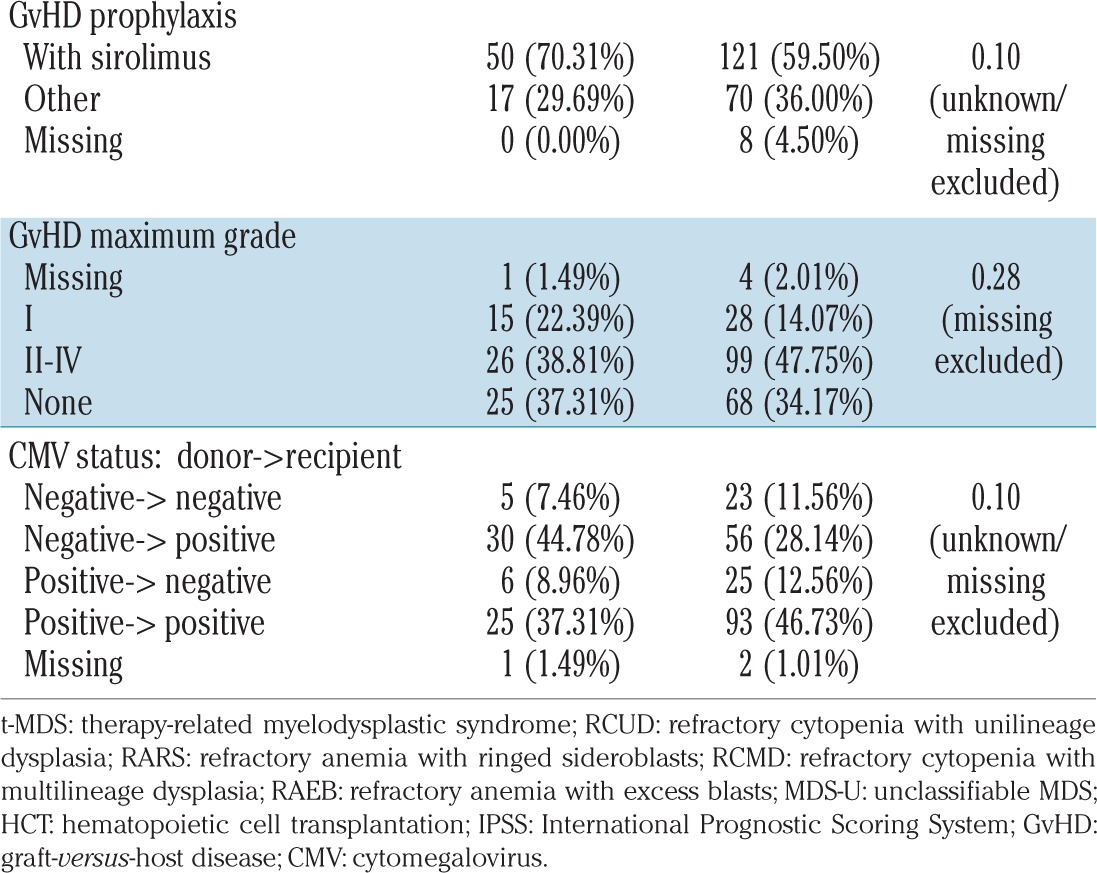
Of the 266 patients included in this analysis, 67 were classified as having t-MDS and 199 as having de novo MDS. The clinical characteristics of all included patients, and differences between t-MDS and de novo MDS are reported in Table 1. When compared to the de novo MDS cohort, t-MDS cases less frequently presented with refractory anemia with excess blasts-1/2 (39.1% versus 47.0%; P=0.004), had higher cytogenetic risk (P=0.00003), lower percentage of blasts at the time of HCT (P=0.03), higher IPSS score at diagnosis (P=0.03) and more frequently received reduced intensity conditioning (94.0% versus 70.4%; P=0.00003). There was no significant difference in median age at the time of HCT (P=0.80), gender (P=0.06), transplant era (P=0.14), time from diagnosis to HCT (P=0.49), donor type (P=0.72), graft source (P=0.17), graft-versus-host disease prophylaxis regimen (P=0.10) or donor-recipient cytomegalovirus status (P=0.10) between patients with t-MDS and those with de novo MDS.
Table 1.
Characteristics of the patients and their transplants.
Allogeneic hematopoietic cell transplantation outcomes
After a median follow-up of 4.8 years (range, 0.5–15.8) for surviving patients, the 5-year overall survival for the entire cohort was 52.8% (95% CI: 46.2–59.4%). There were no significant differences in the 5-year overall survival (49.9% versus 53.9%; P=0.61), relapse-free survival (47.2% versus 49.5%; P=0.68), non-relapse mortality (30.2% versus 26.8%; P=0.48) and relapse rate (22.6% versus 23.7%; P=0.81) between patients with t-MDS and those with de novo MDS (Figure 1A–C). There was also no significant difference in the incidence and severity of acute graft-versus-host disease between t-MDS and de novo MDS patients (grade II–IV: 38.8% versus 47.8%; P=0.28).
Figure 1.
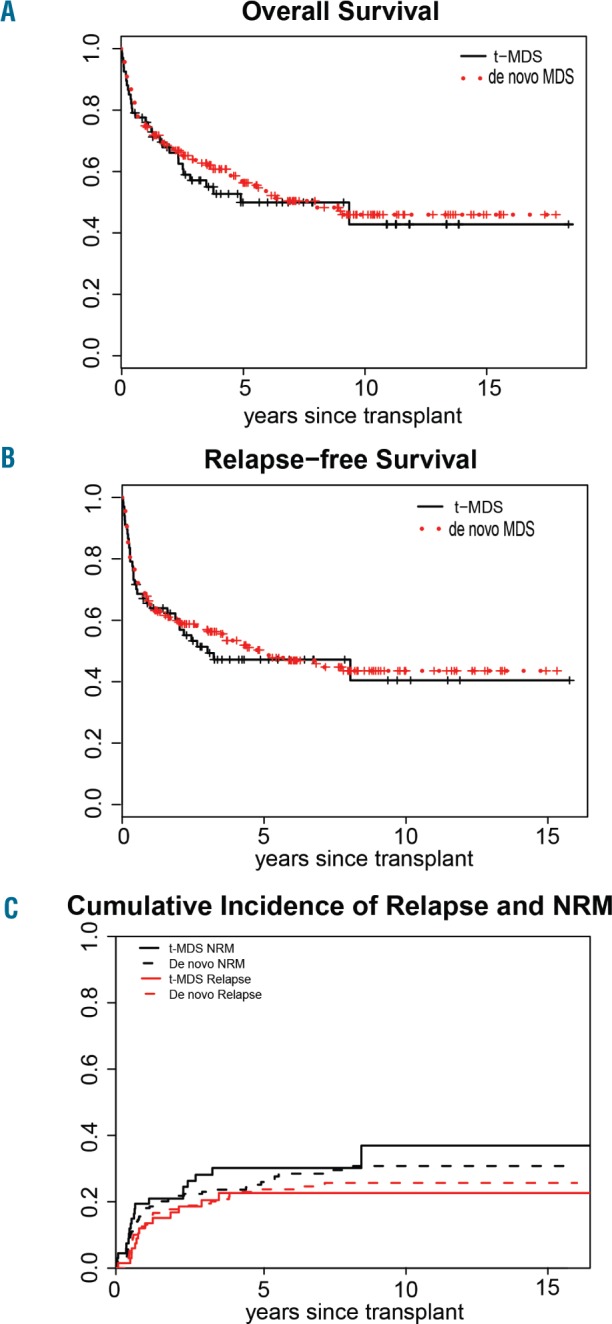
Transplant outcomes for patients with therapy-related (solid line) and de novo myelodysplastic syndrome (dotted line) following allogeneic hematopoietic cell transplantation. (A) Overall survival, (B) relapse free survival, (C) cumulative incidence of relapse and non-relapse mortality (NRM).
Prognostic factors for survival
On univariate analysis for patients with t-MDS, more recent era of HCT (2005–2009 and 2010–2014) was predictive of longer survival (HR=0.41; P=0.05) when compared with transplantation in the earlier era (2000–2004) (HR=0.32; P=0.01), while blast percentage at the time of HCT predicted inferior survival (HR=2.78; P=0.05). Within the group with de novo MDS, male patients and younger patients showed improved survival, whereas those with a poor-risk karyotype and those receiving stem cells from an unrelated donor had a worse survival.
In the multivariate model, prior cytotoxic therapy before MDS diagnosis (t-MDS) did not affect survival (P=0.7) in the whole cohort. For t-MDS (n=67), older age was associated with a trend toward a lower overall survival (HR: 1.04 for each year; P=0.06). Among t-MDS patients who underwent allogeneic HCT, compared with patients transplanted between 2000–2004, those transplanted during more recent periods had statistically superior overall survival (for 2005–2009: HR=0.27; P=0.02 and for 2010–2014: HR=0.21; P=0.002) and relapse-free survival (for 2005–2009: HR=0.28; P=0.16 and for 2010–2014: HR=0.20; P=0.002). Karyotypes, IPSS score and percentage of marrow blasts before allogeneic HCT were not independently associated with overall survival or relapse-free survival (Table 2).
Table 2.
Multivariate analysis for overall survival and relapse-free survival.
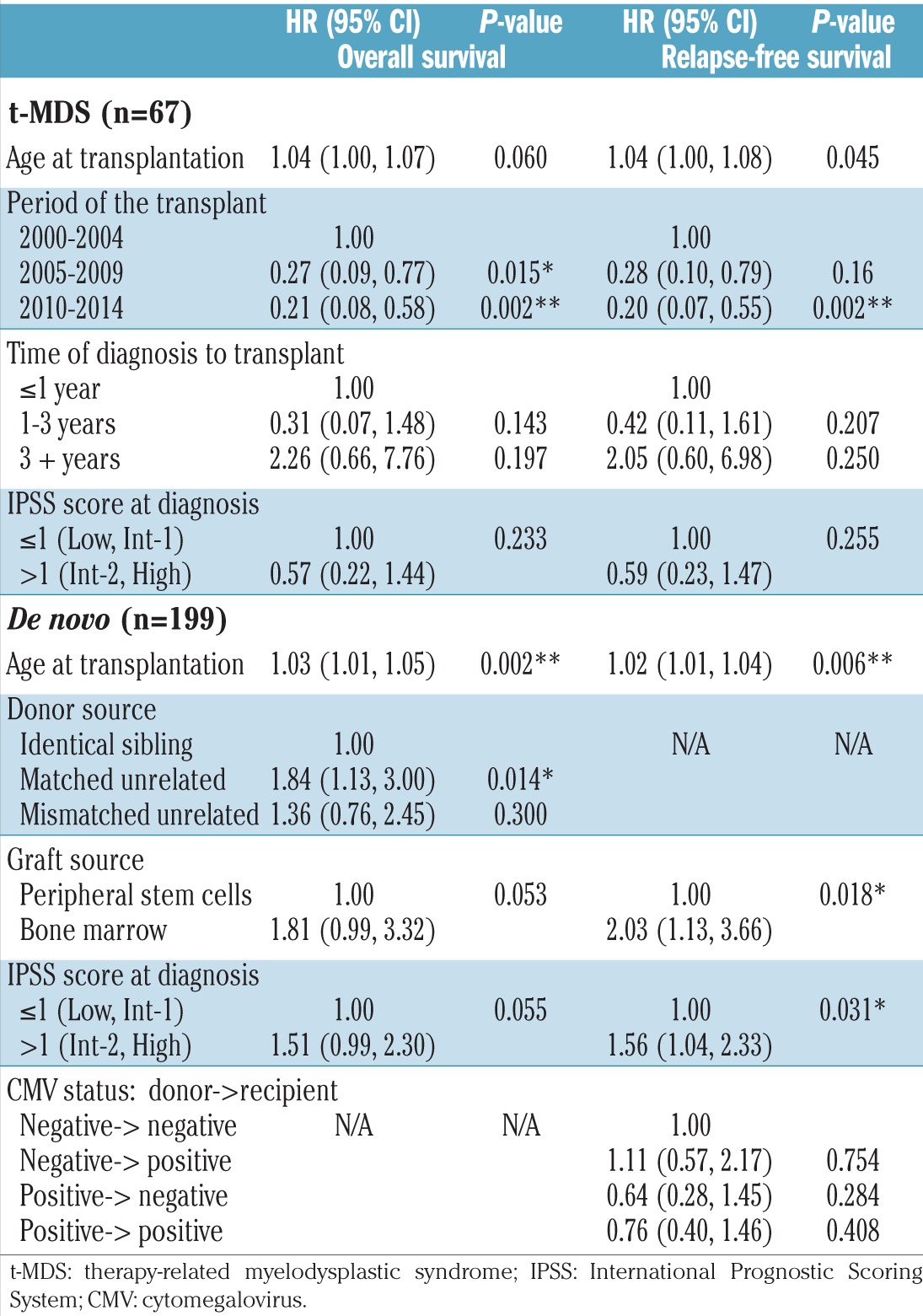
For de novo MDS (n=199), older age (HR=1.03 for each year; P=0.002), unrelated donor (HR=1.84; P=0.01) and IPSS Intermediate-2/High classification (HR=1.51; P=0.06) were the only independent predictors of overall survival (Table 2). Younger age (HR=1.02 for each year; P=0.006), bone marrow as the graft source (HR=2.03; P=0.02) and Intermediate-2/High IPSS score (HR=1.56; P=0.03) were independently associated with shorter relapse-free survival.
Somatic mutations in therapy-related myelodysplasia
We focused our molecular analysis on t-MDS patients only. Of the 67 t-MDS patients, 60 had available pre-HCT DNA samples, of which 43 (72%) had at least one detectable gene mutation. Among patients with detectable mutations, the median number of mutations per case was 2 (range, 1–6) (Figure 2). The most common mutated gene was TP53, which was present in 18 (30%) cases. Other more common mutations (observed in ≥4 cases) were RUNX1 (12%), TET2 (8%), U2AF1 (8%), ASXL1 (8%), DNMT3A (7%) and SETBP1 (7%). Of 18 cases with somatic mutations involving the TP53 gene, five (28%) had multiple distinct TP53 mutations (two mutations: n=4; three mutations: n=1). Most TP53 mutations were localized at the DNA binding domain (Online Supplementary Figure S1). Additionally, non-TP53 mutations (U2AF1, RUNX1, ASXL1, TET2, and STAG2) were observed in 33% of TP53-mutated cases (Online Supplementary Table S2).
Figure 2.
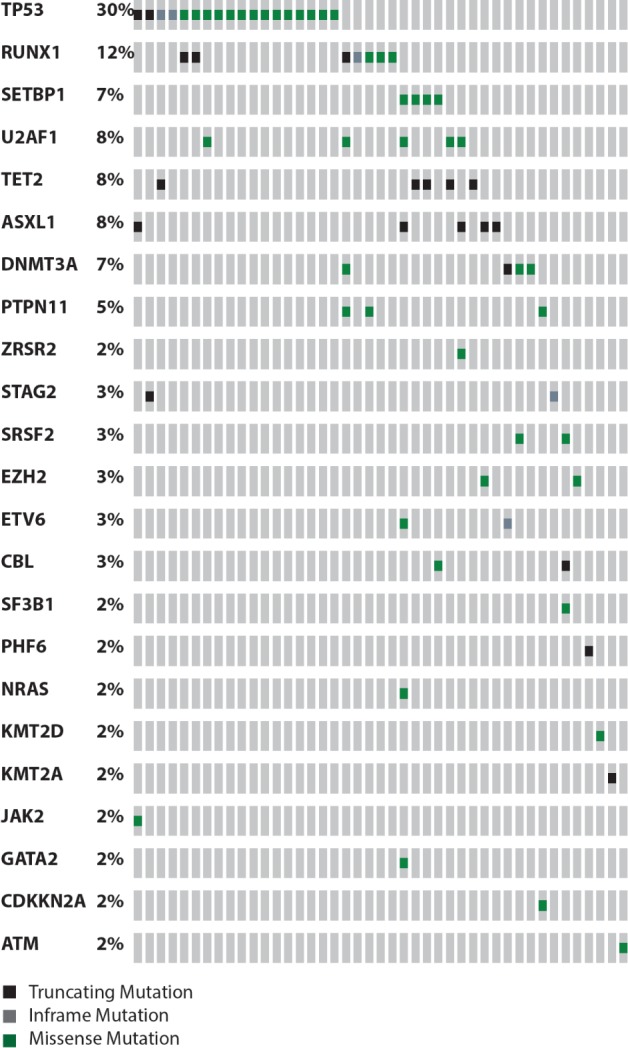
The distribution and frequency of mutations detected by next-generation sequencing in 43 out of 60 patients with therapy-related myelodysplastic syndrome with detected mutation(s).
TP53 mutations were more frequently associated with complex and/or monosomal karyotype compared to TP53-wild type cases (78% versus 38%; P=0.02). No statistically significant differences in age, sex, prior therapy, prior malignancies, latency from prior diagnosis to MDS diagnosis, marrow blasts, number of cytopenias or IPSS score were observed between patients with mutated TP53 and those with the wild-type gene (Table 3). There was also no difference in the number of patients who had received hypomethylating agents before allogeneic HCT between the TP53-mutated and TP53 wild-type groups (39% versus 36%). Interestingly, a TP53 mutation did not adversely affect post-transplant overall survival (HR=1.12; P=0.79) or relapse-free survival (HR=1.42; P=0.37) (Figure 3A,B). The 3-year overall survival and relapse-free survival rates for TP53-mutated t-MDS cases were 51.3% and 41.2%, respectively. The presence of more than one TP53 mutation also did not affect either overall survival or relapse-free survival (Online Supplementary Figure S2A,B).
Table 3.
Characteristics of t-MDS based on TP53 and high-risk mutations.
Figure 3.
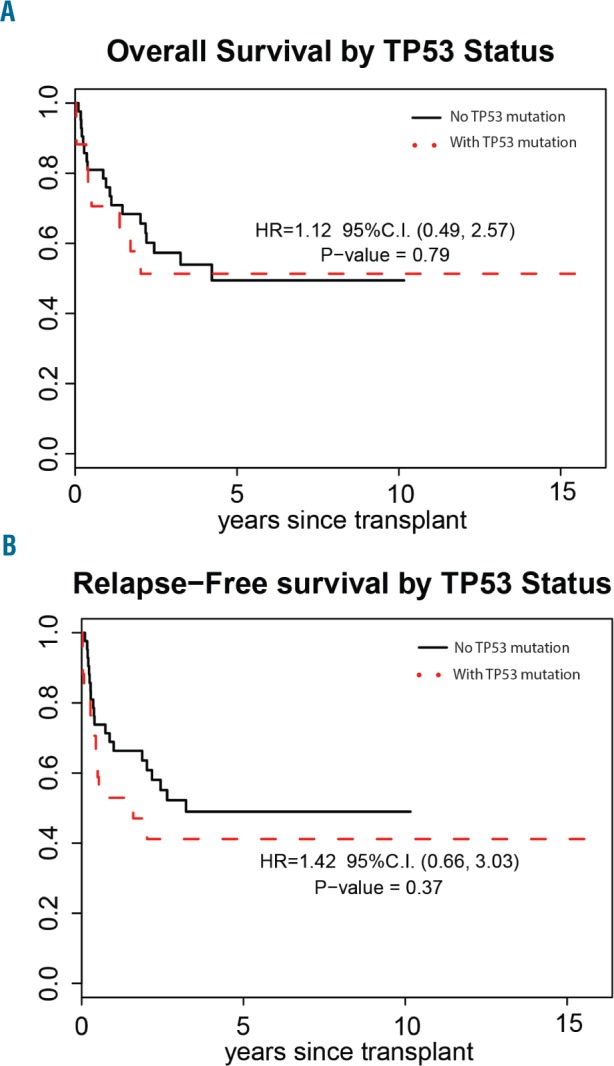
Transplant outcomes for patients with therapy-related myelodysplastic syndrome with (dashed line) and without (solid line) TP53 mutation who underwent allogeneic hematopoietic cell transplantation. (A) Overall survival, and (B) relapse-free survival.
Investigating previously proposed high-risk genes (TP53, EZH2, ETV6, RUNX1 and ASXL1) in MDS,19 we identified that at least one of these genes was mutated in 29 (48%) of our t-MDS cases. t-MDS cases with high-risk mutations were more common after hematologic malignancies (P=0.005) and had additional mutations (55% versus 23%; P=0.02) compared to t-MDS with non-high-risk mutations, but no differences were observed in cytogenetics (P=0.15) or other features (Table 3). Contrary to previous reports,19 the presence of one of the high-risk mutations did not adversely influence overall survival (HR=1.29; P=0.52) or relapse-free survival (HR=1.58; P=0.22) among t-MDS patients who underwent allogeneic HCT in our cohort (Figure 4A,B). While there was a trend toward worse relapse-free survival in patients carrying high-risk genes, this trend did not reach statistical significance, possibly due to the lack of power of the study.
Figure 4.
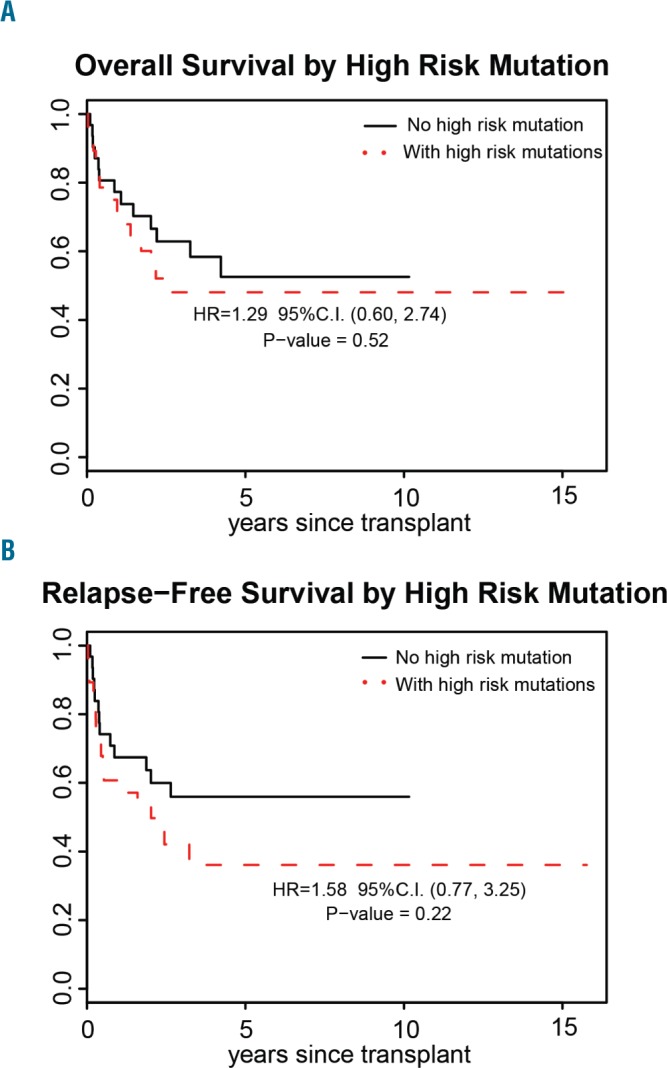
Transplant outcomes for patients with therapy-related myelodysplastic syndrome with (dashed line) and without (solid line) one of high-risk mutations who underwent allogeneic hematopoietic cell transplantation. (A) Overall survival, and (B) relapse-free survival.
Cytogenomic studies in therapy-related myelodysplasia
Of 34 cases with t-MDS with available pre-HCT genomic DNA, cytogenomic microarray analysis provided novel CNV/LOH findings in 24 cases (71%) (Online Supplementary Table S3). These novel findings included the identification of additional abnormalities or modified/clarified previously reported observations. More precisely, 15/34 cases (44%) had LOH not otherwise detectable by conventional cytogenetics or fluorescence in situ hybridization (FISH). The most common CNV occurred in chromosomes 5, 7, 12, 13 and 17, consistent with typical cytogenetic changes reported in MDS. Additional CNV were detected in chromosomes 2, 6, 11, and 18, which were not detectable by standard MDS FISH panels. Importantly, we found four cases of MDS in which TP53 alterations were found only by cytogenomic microarray or FISH and not by NGS. The overall survival of the TP53 mutated and TP53 wild-type groups remained similar when these additional del17p cases were re-classified into the TP53 mutated group (data not shown).
Discussion
Here we report one of the largest single center series of t-MDS patients undergoing allogeneic HCT. Our results, in agreement with previous reports,9,12 indicated that allogeneic HCT is a curative therapeutic option for patients with t-MDS, and approximately half of transplanted patients could achieve long-term survival and perhaps a cure. Importantly, our study illustrates comparable outcomes between patients with t-MDS and those with de novo MDS despite the former having a greater prevalence of high-risk cytogenetic features and higher IPSS scores. Hence, having been previously exposed to cytotoxic therapy was not associated with worse survival (P=0.7) in our cohort. Interestingly, the non-relapse mortality rate was also not higher in t-MDS patients than in de novo MDS ones, despite the former having a history of prior exposure to cytotoxic therapy. The significance of these results also validated the feasibility of reduced intensity conditioning, which is a critical finding as patients with t-MDS are frequently ineligible for myeloablative conditioning regimens due to comorbidities and prior exposure to cytotoxic therapy.
In our cohort, we found that cytogenetics, IPSS score and marrow blast percentage at the time of transplantation did not adversely affect survival following allogeneic HCT in t-MDS patients, suggesting that allogeneic HCT may attenuate or abrogate the adverse prognostic impact that these factors were found to have in non-transplanted and/or mixed cohorts of MDS patients. Nonetheless, we acknowledge that our cohort may not have had the statistical power to rule out this prognostic association. In our multivariate models for t-MDS patients, younger age and more recent era of allogeneic HCT were independently associated with longer overall survival and longer relapse-free survival. The positive impact of younger age and a more recent transplant were consistent with prior publications.9,12
Using NGS, we showed that the majority of t-MDS cases in our cohort carried somatic mutations involved in multiple mechanisms and pathways including transcription factors (TP53, RUNX1, GATA, ETV6), splicing (U2AF1, SF3B1, ZRSR2), epigenetics (TET2, ASXL1, DNMT3A, EZH2, IDH), kinase signaling (PTPN11, NRAS, CBL) and others (ATM, STAG2), in accordance with previous reports.5,15,25 Furthermore, a TP53 mutation was the most common molecular alteration among t-MDS patients in this cohort, consistent with prior reports.5,16,17 Comprehensive analysis of TP53 function requires characterization of single nucleotide variations, small indels, copy number alterations and LOH of the 17p locus. Our results demonstrated that 27% of t-MDS cases positive by NGS for TP53 alterations had more than one mutation. Although we could not confirm whether these mutations were present in cis or trans, it is likely that they were compound heterozygotes. CNV/LOH was examined by cytogenomic microarray and FISH in 76% of cases assessed by NGS. We found that 10% of cases that were negative for TP53 alterations by NGS were positive for CNV/LOH by cytogenomic microarray or FISH studies and that 11% of cases with TP53 mutations detected by NGS had concurrent CNV/LOH TP53 abnormalities. In our study, however, TP53 mutation did not adversely affect the overall survival of transplanted t-MDS patients, regardless of whether they had received hypomethylating agents prior to allogeneic HCT, or had more than one TP53 mutation. These findings are clinically very relevant, since previous reports showed a very limited survival (3-year overall survival of 20%)5 for TP53-mutated MDS patients, including those undergoing transplantation.5,16,18,20,22 In these studies, the majority of TP53-mutated patients succumbed to either relapse or non-relapse mortality following allogeneic HCT,20,21 thereby leading to the conclusion that alternative therapies other than such transplants should be explored. Furthermore, while we did observe the occurrence of TP53 mutations and/or at least one other mutated gene among those incorporated in a recent high-risk five-gene panel for MDS19 in at least half of our patients, these molecular aberrations did not adversely affect transplant outcomes. Thus, we concluded that allogeneic HCT should be considered as the primary approach for t-MDS, regardless of the patient’s genetic features including, but not limited to, TP53 mutation.
While we could not identify definitive factors explaining the difference between the outcomes of our cohort and patients in previous studies, it is possible that the difference in conditioning regimen may have played a role. In a recent study reported by the Dana Farber Cancer Institute,19 the majority of TP53 cases were conditioned with a busulfan/fludarabine reduced intensity conditioning regimen (14/18), while the majority of our TP53 mutated patients (16/18) received fludarabine/melphalan as their reduced intensity conditioning in combination with tacrolimus/sirolimus-based graft-versus-host disease prophylaxis.27,28 In fact recent retrospective studies comparing busulfan/fludarabine with fludarabine/melphalan as reduced intensity conditioning for HCT in patients with acute leukemia and MDS have demonstrated an association between busulfan/fludarabine and higher cumulative incidence of relapse and lower progression-free survival;29–31 while a large MDS study analyzing 1514 patients enrolled in the Center for International Blood and Marrow Transplant Research Repository found that the survival rates of patients with TP53 mutations were similar between those conditioned with myeloablative regimens or reduced intensity regimens.5 In addition, CV/LOH was not assessed in the Dana Farber Cancer Institute study, which may have affected their analysis.
Conventional cytogenetics remains the leading prognostic factor across different prognostic scores in MDS. However, the limitations of classical cytogenetics lie in the difficulty of identifying cryptic structural abnormalities and the inability to identify copy neutral LOH. These abnormalities could consequently lead to inactivation of tumor suppressor genes and activation of oncogenes, which are encountered not uncommonly in myeloid malignancies. Therefore, detection of these abnormalities could potentially be used as prognostic tools, which may improve the classification of MDS scores. In our study, microarray analysis identified additional CNV abnormalities and LOH regions in addition to those that could be found by conventional cytogenetic analysis in 24/34 cases. Of note, several LOH regions detected solely by single nucleotide polymorphism array contained genes previously reported to be mutated in MDS, including FLT3, ASXL1, CBL1, FANCC and TP53. In addition, single nucleotide polymorphism array analysis helped to clarify observations from metaphase cytogenetics in several cases. These findings underscore the significant advantage of using genome-wide single nucleotide polymorphism microarray analyses to identify cryptic pathogenic changes in t-MDS. Abnormalities missed by cytogenomic microarray were either below the detection limit of this assay, or could be attributed to the fact that cytogenomic microarray was conducted on peripheral blood specimens and not the corresponding bone marrow, due to lack of availability.
In conclusion, despite limitations inherent to the retrospective nature of the analysis and potential for selection bias related to the decisions regarding prior transplant treatment and timing of transplantation, which were not made uniformly for all t-MDS patients, our study shows that allogeneic HCT is curative for patients with t-MDS. These transplanted patients have outcomes similar to those of transplanted patients with de novo MDS, despite more frequently having cytogenetic, molecular and clinical higher-risk features prior to allogeneic HCT. Furthermore, we demonstrated that there was no increase in non-relapse mortality in t-MDS patients despite prior exposure to cytotoxic therapy. Importantly, although TP53 mutations were highly prevalent in t-MDS patients, they did not adversely affect transplant outcome, leading us to conclude, in contrast to prior reports, that TP53-mutated t-MDS can also be cured with allogeneic HCT. Our study is limited by the lack of TP53 mutation analysis in patients with de novo MDS to serve as a control cohort for genomic comparison; however, this analysis has been conducted by other groups, showing lower rates of TP53 mutations overall in de novo MDS.5,16 The present findings warrant future prospective studies focusing on the heterogeneity of TP53 mutated t-MDS cases to identify which subsets could potentially be cured with allogeneic HCT and which cases require alternative novel therapies.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Sally Mokhtari for assistance with editing this manuscript. This work was supported by a Hematological Malignancies Cancer Center Seed Grant, City of Hope.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/12/2030
References
- 1.Larson RA. Therapy-related myeloid neoplasms. Haematologica. 2009;94(4):454–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 3.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. [DOI] [PubMed] [Google Scholar]
- 4.Kayser S, Dohner K, Krauter J, et al. The impact of therapy-related acute myeloid leukemia (AML) on outcome in 2853 adult patients with newly diagnosed AML. Blood. 2011;117(7):2137–2145. [DOI] [PubMed] [Google Scholar]
- 5.Lindsley RC, Saber W, Mar BG, et al. Prognostic mutations in myelodysplastic syndrome after stem-cell transplantation. N Engl J Med. 2017;376(6):536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith SM, Le Beau MM, Huo D, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003;102(1):43–52. [DOI] [PubMed] [Google Scholar]
- 7.Zeidan AM, Al Ali N, Barnard J, et al. Comparison of clinical outcomes and prognostic utility of risk stratification tools in patients with therapy-related vs de novo myelodysplastic syndromes: a report on behalf of the MDS Clinical Research Consortium. Leukemia. 2017;31(6):1391–1397. [DOI] [PubMed] [Google Scholar]
- 8.Granfeldt Ostgard LS, Medeiros BC, Sengelov H, et al. Epidemiology and clinical significance of secondary and therapy-related acute myeloid leukemia: a national population-based cohort study. J Clin Oncol. 2015;33(31):3641–3649. [DOI] [PubMed] [Google Scholar]
- 9.Kroger N, Brand R, van Biezen A, et al. Risk factors for therapy-related myelodysplastic syndrome and acute myeloid leukemia treated with allogeneic stem cell transplantation. Haematologica. 2009;94(4):542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Litzow MR, Tarima S, Perez WS, et al. Allogeneic transplantation for therapy-related myelodysplastic syndrome and acute myeloid leukemia. Blood. 2010;115(9):1850–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nevill TJ, Hogge DE, Toze CL, et al. Predictors of outcome following myeloablative allo-SCT for therapy-related myelodysplastic syndrome and AML. Bone Marrow Transplant. 2008;42(10):659–666. [DOI] [PubMed] [Google Scholar]
- 12.Yakoub-Agha I, de La Salmoniere P, Ribaud P, et al. Allogeneic bone marrow transplantation for therapy-related myelodysplastic syndrome and acute myeloid leukemia: a long-term study of 70 patients-report of the French society of bone marrow transplantation. J Clin Oncol. 2000;18(5):963–971. [DOI] [PubMed] [Google Scholar]
- 13.Gibson CJ, Lindsley RC, Tchekmedyian V, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol. 2017;35(14):1598–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Wang F, Kantarjian H, et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 2017;18(1):100–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ok CY, Patel KP, Garcia-Manero G, et al. Mutational profiling of therapy-related myelodysplastic syndromes and acute myeloid leukemia by next generation sequencing, a comparison with de novo diseases. Leuk Res. 2015;39(3):348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ok CY, Patel KP, Garcia-Manero G, et al. TP53 mutation characteristics in therapy-related myelodysplastic syndromes and acute myeloid leukemia is similar to de novo diseases. J Hematol Oncol. 2015;8:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540):552–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bally C, Ades L, Renneville A, et al. Prognostic value of TP53 gene mutations in myelodysplastic syndromes and acute myeloid leukemia treated with azacitidine. Leuk Res. 2014;38(7):751–755. [DOI] [PubMed] [Google Scholar]
- 19.Bejar R, Stevenson K, Abdel-Wahab O, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bejar R, Stevenson KE, Caughey B, et al. Somatic mutations predict poor outcome in patients with myelodysplastic syndrome after hematopoietic stem-cell transplantation. J Clin Oncol. 2014;32(25):2691–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Della Porta MG, Galli A, Bacigalupo A, et al. Clinical effects of driver somatic mutations on the outcomes of patients with myelodysplastic syndromes treated with allogeneic hematopoietic stem-cell transplantation. J Clin Oncol. 2016. September 6 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: a joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19(1):4–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bejar R. Implications of molecular genetic diversity in myelodysplastic syndromes. Curr Opin Hematol. 2017;24(2):73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Przepiorka D, Weisdorf D, Martin P, et al. 1994 Consensus conference on acute GVHD grading. Bone Marrow Transplant. 1995;15(6):825–828. [PubMed] [Google Scholar]
- 27.Nakamura R, Palmer JM, O’Donnell MR, et al. Reduced intensity allogeneic hematopoietic stem cell transplantation for MDS using tacrolimus/sirolimus-based GVHD prophylaxis. Leuk Res. 2012;36(9):1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura R, Rodriguez R, Palmer J, et al. Reduced-intensity conditioning for allogeneic hematopoietic stem cell transplantation with fludarabine and melphalan is associated with durable disease control in myelodysplastic syndrome. Bone Marrow Transplant. 2007;40(9):843–850. [DOI] [PubMed] [Google Scholar]
- 29.Damlaj M, Alkhateeb HB, Hefazi M, et al. Fludarabine-busulfan reduced-intensity conditioning in comparison with fludarabine-melphalan is associated with increased relapse risk in spite of pharmacokinetic dosing. Biol Blood Marrow Transplant. 2016;22(8):1431–1439. [DOI] [PubMed] [Google Scholar]
- 30.Kawamura K, Shuichi B, Ishiyama K, et al. Comparison of reduced-intensity conditioning with fludarabine/busulfan and fludarabine/melphalan in patients 50 years or older. Blood. 2016;128(22):3414. [Google Scholar]
- 31.Caddell RJ, Ma Z, Dimaggio E, et al. Fludarabine and melphalan results in fewer relapses compared to fludarabine and targeted busulfan in patients receiving reduced intensity conditioning for AML and MDS. Blood. 2016;128(22):3486. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


