Abstract
In acute myeloid leukemia, there is growing evidence for splicing pattern deregulation, including differential expression of linear splice isoforms of the commonly mutated gene nucleophosmin (NPM1). In this study, we detect circular RNAs of NPM1 and quantify circRNA hsa_circ_0075001 in a cohort of NPM1 wild-type and mutated acute myeloid leukemia (n=46). Hsa_circ_0075001 expression correlates positively with total NPM1 expression, but is independent of the NPM1 mutational status. High versus low hsa_circ_0075001 expression defines patient subgroups characterized by distinct gene expression patterns, such as lower expression of components of the Toll-like receptor signaling pathway in high hsa_circ_0075001 expression cases. Global evaluation of circRNA expression in sorted healthy hematopoietic controls (n=10) and acute myeloid leukemia (n=10) reveals circRNA transcripts for 47.9% of all highly expressed genes. While circRNA expression correlates globally with parental gene expression, we identify hematopoietic differentiation-associated as well as acute myeloid leukemia subgroup-specific circRNA signatures.
Introduction
Acute myeloid leukemia (AML) is the most common acute leukemia in adults and despite recent progress in understanding leukemia biology, many patients relapse and ultimately die of the disease.1 While AML is a genetically heterogeneous disease, it has become clear that not only changes at the genome level contribute to AML pathogenesis. During the past few years, next-generation sequencing (NGS) has revealed an accumulation of mutations in genes regulating the splicing process in approximately 10% of AML patients,2 and a large comprehensive NGS study in AML determined AML patients with spliceosome mutations as a clinically relevant distinct AML subgroup.3 This places a novel focus on alternative and aberrant splicing events that seem to play a general role in AML, and an improved understanding of these aberrations might harbor the potential for novel treatment strategies.4
Aberrant splicing events might also impact the large subset of AML patients carrying a mutation in the nucleophosmin (NPM1) gene, which encodes a multifunctional chaperone protein involved in ribosomal biogenesis, apoptosis, and cell proliferation.5 In NPM1-mutated AML patients, an insertion into exon 12 of NPM1 leads to an aberrant localization of the protein in the cytoplasm, which contributes to the leukemogenic phenotype and is considered an AML-defining mutation.6,7 Moreover, there is evidence that both the impairment of NPM1 function due to deletion or dislocation, and the enhancement of NPM1 function due to overexpression, can confer a tumorigenic effect.8,9 As NPM1 bears both proto-oncogene and tumor-suppressor properties, its deregulation by means other than gene mutation might also play a pathogenic role.
Recently, it has been shown that, in AML, several alternatively spliced linear isoforms are produced from the NPM1 gene, of which a short R2 variant (NCBI Reference Sequence: NM_001037738.2) was shown to be differentially expressed in a large cohort of AML patients.10 Notably, there was also an association between high NPM1 R2 expression and better outcome in CN-AML patients, especially in patients without concomitant FLT3-ITD mutation, thereby providing further evidence that deregulated splicing patterns might contribute to AML pathogenesis.
In addition to splicing events affecting protein coding genes, recent transcriptome studies have revealed that more than 60% of the human genome is transcribed and reproducibly detected as RNA transcripts, while only 2% of the genome codes for protein sequences.11 This implies an important biological function and a need for more comprehensive analyses of the non-coding transcriptome. In accordance with this, several studies have also pointed to an important role for non-coding RNAs, such as microRNAs (miRNA) and long non-coding RNAs (lncRNA), in AML pathogenesis.12–14
Recently, a new class of non-coding RNAs termed circular RNAs (circRNAs) was discovered.15 CircRNAs are abundant, highly conserved, and their expression is specifically regulated, making them promising candidates for biomarker research.15–17 CircRNAs are formed through a ligation of the 5′ and the 3′ end of a transcript (so-called backsplicing), which leads to a non-canonical order of exons. Different mechanisms have been suggested for the biogenesis of circRNAs, which are able to bring downstream donors into close proximity with upstream splice acceptors, including binding sites for RNA-binding proteins and reverse complementary sequences in flanking introns.18,19 Given the general deregulation in splicing mechanisms in AML, expression of circRNAs might also be impaired in leukemia cells, and altered circRNAs could contribute to leukemogenesis.20
In the current study, based on the relevance of NPM1 in AML, we investigate the expression of circular NPM1 transcripts in both healthy hematopoietic and leukemic cells. In addition to known NPM1 circRNAs (annotated in the database circBase21), we provide evidence for novel variants as well as NPM1 circRNAs differentially expressed in AML, such as hsa_circ_0075001. Quantifying the expression of hsa_circ_0075001 in a cohort of 46 AML patients, we reveal NPM1 mutation-independent expression groups characterized by distinct gene expression profiles. This further suggests a potential functional relevance of circular NPM1 transcripts and adds another level of complexity to the multifaceted gene, NPM1. Via a more comprehensive and unbiased RNA-Seq-based transcriptome analysis, we gain additional insights into the circular RNAome of the hematopoietic system, and determine changes in the circRNA repertoire throughout myeloid differentiation, as well as non-physiological patterns associated with leukemic transformation.
Methods
Patient material and healthy controls
Acute myeloid leukemia patients were selected from a larger cohort enrolled in the AMLSG_07-04 study (clinicaltrials.gov identifier: 00151242). Informed patient consent was obtained for the study and concomitant scientific investigations, and the research was conducted after approval by the local ethical committee and according to the principles of the Declaration of Helsinki. Cytogenetically normal AML cases were selected for the quantification of hsa_circ_0075001 (n=46) and for RNA-Seq (n=10). For more detailed information please refer to the Online Supplementary Methods and Online Supplementary Table S1.
Healthy control samples were collected from 6 individuals after informed consent. For RNA-Seq, bone marrow samples of 3 healthy controls were FACS-sorted as previously described.22
Oxford Nanopore sequencing
PCR products for Oxford Nanopore sequencing were generated with outward facing, so-called divergent primers specific for NPM1 (Online Supplementary Table S2), with product sizes ranging from 150bp to 2500bp. 1 mg purified PCR product was subjected to library preparation using the SQK-NSK007 Nanopore Sequencing Kit (v.R9, Oxford Nanopore Technologies, Oxford, UK), following the standard protocol for Amplicon sequencing for the MinION™ device. 135 ng of end-prepped DNA was subjected to adapter ligation. Oxford Nanopore reads were aligned using BWA-MEM23 and NCBI BLAST.24
Integration of gene expression data
The 46 AML patients were dichotomized based on the following criteria: NPM1 mutational status [wild-type (wt) vs. mutated], circNPM1 hsa_circ_0075001 expression (high vs. low, relative to the median expression), or total NPM1 expression (high vs. low). Principal component analysis (PCA) of microarray data was conducted using the Partek® Genomics Suite® software (Partek, St. Louis, MO, USA). One-way ANOVA was used to examine differences between the respective groups. Differentially expressed genes were defined as log2FC>|0.6| and P<0.05 after controlling the false discovery rate (FDR) using the Benjamini–Hochberg procedure. Pathway analysis was performed using the iPathwayGuide web application (Advaita, Plymouth, MI, USA). Statistical thresholds were set at P<0.05 and log2FC>|0.6|. Microarray data are available at Gene Expression Omnibus (GEO; accession number GSE104099).
RNA-Seq
Libraries were prepared from 1 μg of input total RNA using the TruSeq Stranded Total RNA Kit with Ribo-Zero Human/Mouse/Rat (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. The pooled RNA libraries were sequenced on an Illumina HiSeq2000 with 100bp paired-end reads and an average coverage of 62 (±24)×106 reads per sample. RNA-Seq data were aligned and quantified using STAR.25 Reads derived from circRNAs mapping to a backsplice junction were identified using an in-house analysis pipeline detecting exons in a shuffled order. A more detailed description of the pipeline is provided in the Online Supplementary Methods. Circular reads were then normalized and transformed to the logarithmic scale using DESeq226 and PCA was performed based on those 500 genes with the highest variance of circRNA expression across all samples. Genes differentially expressing circRNAs were tested for log2FC>|0.6|, and FDR<0.1 was set for correction of multiple comparisons. Gene set enrichment analysis was performed using GSEA and the Molecular Signatures Database (MsigDB).27 To calculate the percentage of genes which also produce circRNAs, the top 10,794 genes with a mean read count over 100 were considered as “highly expressed” and the top 17,755 genes with a mean read count over 10 were considered “markedly expressed” (Online Supplementary Figure S1). Normalized RNA-Seq data are provided in Online Supplementary Tables S3 and S4.
Results
Circular NPM1 transcripts in leukemic and healthy cells
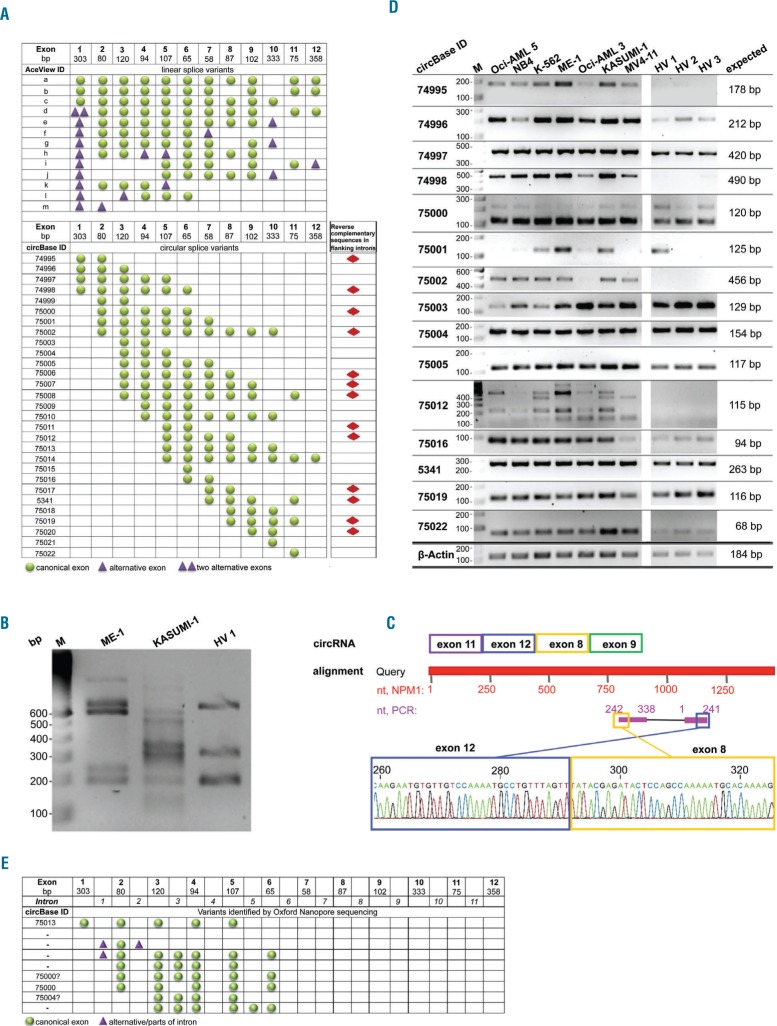
Several linear and circular transcripts are known to be transcribed from the NPM1 gene, but circRNA biogenesis is only partly understood. An overview of variants as annotated in the databases AceView28 for linear NPM1 transcripts and circBase21 for circRNAs is given in Figure 1A. Since reverse complementary intronic sequences, such as inverted Alu repeats, are thought to be of importance for the formation of the circle,16,29 we investigated the presence of such sequences in introns of the NPM1 gene (ENSG00000181163) using NCBI BLAST.24 Moreover, we assessed a possible correlation of the presence of reverse complementary intronic sequences with exon combinations that take part in backsplicing events. Out of 29 known NPM1 variants, we found reverse complementary sequences in flanking introns of 13 (44.8%) annotated circular transcripts, possibly promoting their biogenesis (Online Supplementary Table S5). However, we also found inverse complementary sequences in introns that are not known to take part in backsplicing events to form a circRNA. Thus, reverse complementary intronic sequences are not the only mechanism promoting circRNA formation.
Figure 1.
Characterization of circular NPM1 splice variants. (A) Predicted exon composition of linear and circular NPM1 splice variants, based on information provided in the AceView and circBase databases. Presence of reverse complementary sequences in the flanking introns of a particular circRNA was evaluated and is indicated by red diamonds. (B) A PCR using divergent NPM1-specific primers was performed with cDNA of two AML cell lines and 1 healthy volunteer sample. (C) Sanger sequencing of a KASUMI-1 cell line PCR product and NCBI BLAST alignment confirmed the existence of a backsplice junction and non-canonical exon order of a circular RNA-derived PCR product. (D) Detection of circNPM1 transcripts in leukemia cell lines (n=7) and healthy volunteers (HV, n=3) with backsplice-specific primers. β-actin (ACTB) served as an internal control. IDs according to circBase and length of the expected amplicon are indicated. (E) Exon and intron composition of circNPM1 splice variants detected via Oxford Nanopore long-read sequencing of PCR products.
As a first experimental approach, we performed PCR with divergent primers in two NPM1 wt (NPM1wt) leukemia cell lines, ME-1 and KASUMI-1, and one sample derived from a healthy volunteer. Primers were designed for exons of NPM1 which are located in the majority of annotated circular NPM1 transcripts. This resulted in multiple PCR products of circular origin in all samples, reflecting various circRNAs produced from the NPM1 gene (Figure 1B). Validation by Sanger sequencing confirmed the specificity for NPM1, the non-canonical exon order, and the backsplice sequence of these circRNAs. A representative example of a previously unknown circNPM1 variant detected in the KASUMI-1 cell line consists of exons 8, 9, 11, and 12 with a backsplice junction located between exons 12 and 8 of NPM1 (Figure 1C).
Next, for the specific detection of individual circNPM1 transcripts, backsplice sequence-specific PCR primers were designed, and the screening of cell lines was extended. In total, we analyzed six AML cell lines (including the NPM1 mutant cell line OCI-AML3), one CML cell line in terminal myeloid blast crisis, and 3 samples derived from the peripheral blood mononuclear cell fraction of healthy volunteers, and we were able to specifically amplify 15 circNPM1 variants. While some variants were uniformly expressed among the control and leukemia samples, including hsa_circ_0074997, _0075004, _0075005, _0005341, _0075019 and _0075022, other variants showed lower expression in healthy volunteers compared to leukemia cell lines, such as hsa_circ_0074995, _0074998, _0075001, _0075002 and _0075012 (Figure 1D).
To gain insight into the internal structure of circRNAs, we made use of the long-read Oxford Nanopore technology to sequence circular-derived PCR products created with NPM1-specific divergent primers. We detected already annotated circNPM1 variants, including hsa_circ_0074997, _0075000 and _0075004 (Figure 1E). Moreover, we found novel variants that retained complete introns, especially the complete intronic sequences between exons 3 and 4 of NPM1, and between exons 5 and 6. Furthermore, parts of intron 1 were shown to take part in backsplicing events, for example, with exon 6 and parts of intron 2 (Figure 1E).
Hsa_circ_0075001 correlates with distinct gene expression patterns
As circNPM1 75001 (circBase ID hsa_circ_0075001) exhibited a highly differential expression pattern in the AML cell lines, it was selected for quantitative screening in a cohort of 46 AML patients in parallel to the expression of total NPM1. No considerable difference could be detected regarding the mean, median, and the range of expression between NPM1wt (n=23) and NPM1 mutated (NPM1mut, n=23) patients (Online Supplementary Figure S2A and B). However, we observed high expression of hsa_circ_0075001 in patients with no or minimal maturation of the predominant blast population [subtypes M0 or M1 according to the French-American-British (FAB) classification system] (Online Supplementary Figure S3). Expression of hsa_circ_0075001 in patients with a more mature blast population (M2, M4 and M5) was significantly lower (median fold change=3; P<0.001, unpaired t-test).
Across all AML samples, there was a 4-fold difference in expression (P<0.0001, unpaired t-test) and patients were allocated to a low and a high circNPM1 75001 expression cohort based on the median (Figure 2A). Furthermore, expression of hsa_circ_0075001 correlated strongly with total NPM1 expression (Pearson correlation coefficient R=0.72) (Figure 2B), raising the question of whether the expression of circRNAs might generally be correlated with that of the respective parental gene.
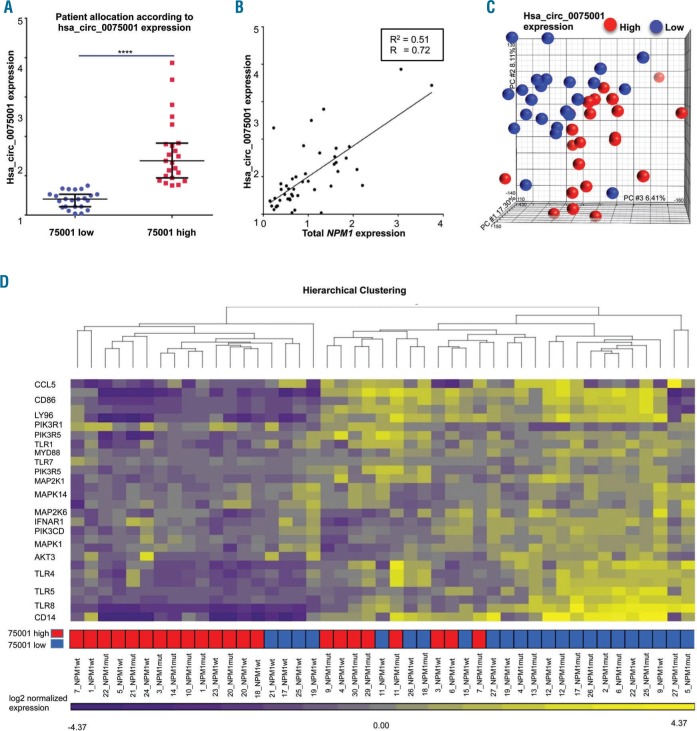
Figure 2.
Expression of circular NPM1 transcript hsa_circ_0075001 and impacts on gene expression in a cohort of 46 acute myeloid leukemia (AML) patients. Expression of hsa_circ_0075001 and total NPM1 was measured in 46 AML patients by qPCR. All values are normalized to β-actin (ACTB) and are relative to the respective mean expression. (A) Difference in hsa_circ_0075001 expression in AML patients allocated to the high (red) or low (blue) expression group (dichotomized at the median; P<0.001, unpaired t-test). Black lines indicate the 25% percentiles. (B) Expression of hsa_circ_0075001 in relation to total NPM1 in the AML cohort. (C) Principal component analysis of the distribution of global gene expression differences in AML patients allocated to the high (red) or low (blue) hsa_circ_0075001 expression group. (D) Key components of the Toll-like receptor (TLR) signaling pathway were down-regulated in patients with high hsa_circ_0075001 expression (red). The heatmap illustrates gene expression of the top 20 genes of the TLR pathway with low expression colored purple and high expression colored yellow. Hierarchical clustering of AML patients with high (red) or low (blue) hsa_circ_0075001 expression based on these genes. R: Pearson correlation coefficient; R2: coefficient of determination.
Gene expression profiling for the high versus low hsa_circ_0075001 expression groups identified 2292 differentially expressed (DE) genes between patients using an FDR-corrected P<0.05 and a threshold for log2 fold change (log2FC)>|0.6|. Unsupervised principal component analysis (PCA) illustrates that the hsa_circ_0075001 expression status defined distinct subgroups of patients (Figure 2C). One-way ANOVA confirmed significant differences in gene expression between the high versus low hsa_circ_0075001 expression groups, and a comparative gene expression pathway analysis (statistical thresholds for differential gene expression set at P<0.05 and log2FC>|0.6|) revealed significant differences in the expression of genes involved in the Toll-like receptor (TLR) signaling pathway as an example. This pathway was significantly down-regulated in patients with high hsa_circ_0075001 expression compared to patients with low hsa_circ_0075001 expression (FDR-corrected P<0.0001) (Online Supplementary Table S6). A total of 94 genes were perturbed in this pathway, with the main dysregulated components of the pathway being several TLR genes, such as TLR1, TRL4, TLR5, and TLR7/8, as well as levels of co-receptors like CD14, and downstream adaptor proteins like MYD88 (Figure 2D). Moreover, 185 differentially expressed genes, whose expression was decreased in patients with high hsa_circ_0075001 expression, are known target genes of miR-181, such as Caspase recruitment domain-containing protein 8 (CARD8), Caspase 1 (CASP1) Macrophage scavenger receptor 1 (MSR1) solute carrier family 11 member 1 (SLC11A1), and TLR4. It is known that miR-181 is commonly deregulated in cytogenetically normal AML.30
In contrast, a comparison of AML cases with high versus low total NPM1 expression did not reveal an enrichment of TLR signaling pathway genes among the top 40 pathways, but rather ribosomal protein genes being up-regulated in patients with high total NPM1 expression (FDR-corrected P<0.0001) (Online Supplementary Table S7).
Unbiased evaluation of the circular RNAome in AML
Given the differential expression of NPM1 circRNA in AML, we aimed for a more global insight into circRNA expression in hematopoietic cells. To this end, we performed ribosomal RNA-depleted RNA-Seq of 10 AML patients (n=5 NPM1mut and n=5 NPM1wt cases), and 10 FACS-sorted healthy control samples [n=4 immature myeloid differentiation stages (myeloblasts and promyelocytes), n=6 more mature myeloid differentiation stages (metamyelocytes and neutrophils)]. Using an in-house script for circRNA identification, in total, we detected circRNAs for 31.7% (n=5635) out of 17,755 markedly expressed genes (mean normalized read count >10 across all 20 RNA-Seq samples) and for 47.9% (n=5173) out of the 10,794 highly expressed genes (mean normalized read count >100) (Online Supplementary Figure S1). While circular expression of many genes was comparable between AML patients and healthy control samples (Pearson correlation coefficient R=0.749) (Figure 3A), some genes particularly showed high circular expression in AML but not in healthy samples, and vice versa.
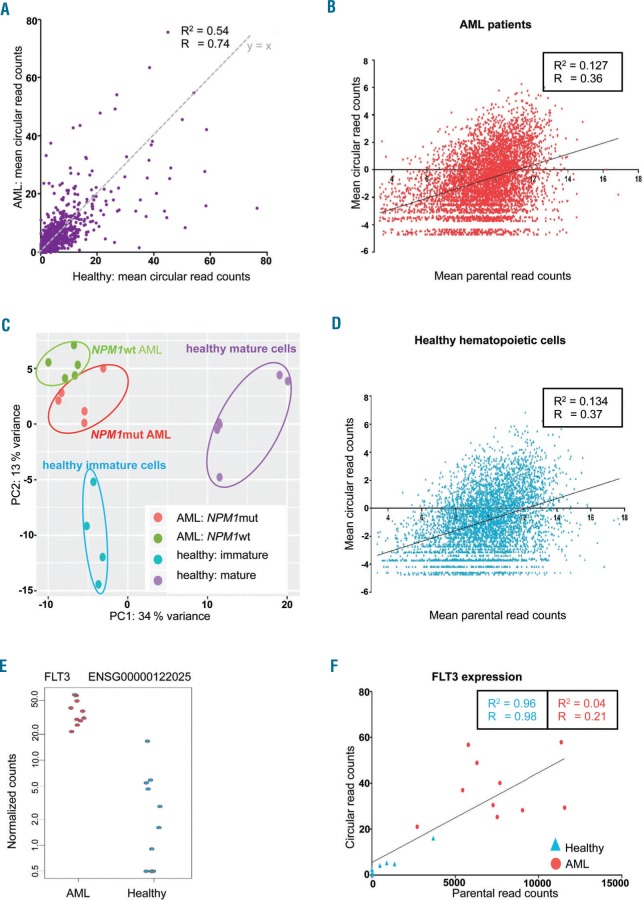
Figure 3.
Circular RNAome in acute myeloid leukemia (AML) patients and healthy hematopoietic cells. Ribosomal RNA-depleted RNA-Seq of 5 NPM1mut, 5 NPM1wt AML, and 10 healthy hematopoietic control samples. (A) Global circular gene expression in 10 AML patients compared to expression in 10 healthy controls. Circular-derived reads were detected for 5694 genes. The x-value of each dot is the mean circular read count for the respective gene in 10 healthy samples; y-value represents the mean circular read count in 10 AML patient samples. y=x is plotted in gray. (B) Mean expression of circRNA transcripts relative to parental gene expression of “markedly expressed” genes in 10 AML patients. The log2 of the mean normalized read counts across all 10 samples is shown. The linear regression function is indicated. (C) Mean expression of circRNA transcripts relative to parental gene expression of “markedly expressed” genes in 10 healthy hematopoietic control samples. (D) CircRNA expression in AML patients compared to healthy control samples. Principal component analysis (PCA) of circRNA expression data of 5 NPM1mut patients (red) and 5 NPM1wt patients (green), and 10 healthy control samples, of which 4 were derived from immature myeloid differentiation stages (blue: myeloblasts and promyelocytes) and 6 from more mature myeloid differentiation stages (purple: metamyelocytes and neutrophils). PCA was performed based on the 500 genes with the highest variance across all samples. (E) Normalized circular read counts in 10 AML patients and 10 healthy control samples are shown for the fms like tyrosine kinase 3 (FLT3) gene which is one of the genes differentially expressing circRNAs. (F) Correlation of circFLT3 expression with parental gene expression in 10 AML patients (red circles) and 10 healthy control samples (blue triangles). Values are shown as normalized read counts. R: Pearson correlation coefficient; R2: coefficient of determination.
In accordance with our previous finding for NPM1, we again observed a tendency towards higher circRNA expression in genes with higher parental gene expression in both leukemia cells (Figure 3B) and healthy control cells (Figure 3C). Nevertheless, there are genes that produce only few or no circRNA transcripts despite high gene expression levels, while others with lower parental gene expression show comparatively high circRNA expression. Furthermore, there was a slight, but distinct difference between AML and healthy control cells with circRNAs for 46.7% of the highly expressed genes in AML and only 43.8% in normal cells (P=0.009, χ2 test).
AML patients and healthy controls differ in circRNA signature
We performed PCA on the circular-derived RNA-Seq data comparing AML with healthy control samples, and at the same time contrasting NPM1mut and NPM1wt patients and healthy myeloid cell samples of more mature myeloblasts and promyelocytes, and more immature metamyelocytes and neutrophils (Figure 3D). This analysis showed that circRNA signatures are associated with myeloid differentiation and are distinct in leukemic cells, which is reflected in the fact that AML samples do not merely resemble immature myeloid cells. Although there was no difference in circNPM1 expression, NPM1mut patients could be distinguished from NPM1wt patients based on their global circRNA expression.
In total, differentially expressed circRNAs from 27 genes were found comparing AML to healthy control samples (P<0.05) (Online Supplementary Table S8). Notably, these genes were significantly enriched for the RADMACHER_AML_PROGNOSIS gene set (FDR-corrected P<0.05, GSEA) based on the differential expression of angiopoietin 1 (ANGPT1; log2FC=3.3), UDP-glucose ceramide glucosyltransferase (UGCG; log2FC=−3.5), and fms related tyrosine kinase 3 (FLT3; log2FC=3.4) circRNAs. Among 14 genes with higher circRNA expression in AML (log2FC range 2.1–4.7), the most significantly deregulated circRNAs were produced from genetic suppressor element 1 (GSE1; log2FC=4.3) and FLT3 (see above). Circular-derived FLT3 reads mainly mapped to three different variants: two already annotated variants (hsa_circ_0100163, hsa_circ_0100164) and one novel variant with a backsplice sequence between exons 19 and 16 of FLT3. Since FLT3 is often mutated and constitutively activated in AML,3 we were particularly interested in circFLT3 expression in AML patients compared to healthy cells (Figure 3E). The correlation of total circFLT3 expression with parental gene expression was high in healthy samples (Pearson correlation coefficient R=0.98), whereas this was not observed in AML samples (Pearson correlation coefficient R=0.21) (Figure 3F).
Discussion
Here, we investigated circRNA expression of one of the most frequently mutated genes in AML. In agreement with others who detected alternative splicing events in circRNAs,31 we found that there are many different circRNA variants for NPM1 that comprise non-canonical exon and intron sequences. Previously, circRNAs were predicted based on non-canonical exon-exon junctions found by RNA-Seq.15,17 Using a new technology for long-read sequencing, Oxford Nanopore, we elucidated the internal structure of several circNPM1 variants for the first time in AML and demonstrated the power of this novel approach. We could show that, in addition to canonical exons, some variants retained full introns. Moreover, we found parts of intronic sequences taking part in backsplicing events. Since these sequences have not been taken into account in many circRNA studies so far, backsplice events between canonical exons and intronic sequences have probably been under-estimated.
Besides intronic sequences, we also detected truncated alternative exons of NPM1, which participated in forming the fusion site (truncated versions of exons 2, 5, 7, 8 and 12). In this context, it would be of interest to study the possible effects of splicing mutations in AML on the formation and the internal structure of circRNAs. Moreover, future elucidation of circRNA function will help to understand the potential necessity of the large circRNA variety and their role in AML.
Quantification of the circNPM1 variant hsa_circ_0075001 and integration of global gene expression data revealed a distinct hsa_circ_0075001-associated gene expression signature, pointing to a biological relevance of this circRNA. High hsa_circ_0075001 expression was highly associated with a significantly lower expression of genes involved in the TLR signaling pathway. While TLRs form a critical part of the innate immune response, they have recently been implicated in the differentiation of normal hematopoietic cells. Furthermore, TLR1 expression has been linked to leukemic stem cell survival in AML, as well as enhanced TLR1/TLR2 activation in leukemic stem cell differentiation.32–34 This is in line with our observation that high hsa_circ_0075001 expression is associated with low TLR gene expression and a more immature phenotype of the AML blasts. Future functional experiments will be needed to show whether in AML, the TLR pathway deregulation is in part caused by aberrant expression of circRNAs, such as hsa_circ_0075001, or whether the respective circRNA expression changes are in turn secondary to TLR-deregulation, for example, as a marker for differentiation.
Interestingly, Marcucci et al. have shown that the expression of TLR signaling pathway genes is associated with prognostically relevant microRNA signatures in AML.35 In particular, the expression levels of miRNA-181 family members were inversely correlated with the expression levels of the TLR signaling pathway genes. In our study, we found that the expression of various known miR-181 target genes was significantly decreased in patients with high hsa_circ_0075001 expression. Since the NPM1 gene contains binding sites for miR-181,36 it would be of great interest to investigate whether circular NPM1 transcripts interact with members of the miR-181 family and thereby influence the expression of genes involved in the TLR signaling pathway.
In contrast, the TLR signaling pathway was not among the top 40 pathways associated with total NPM1 expression, although its deregulation could be detected at a weaker level considering the positive correlation of total NPM1 and hsa_circ_75001 expression. However, the fact that the signature is more pronounced in patients with high hsa_circ_75001 expression supports the view that circRNA expression is the decisive factor correlating with TLR pathway deregulation.
Among the linear-driven NPM1 pathways, we found that ribosomal protein genes were strongly up-regulated in patients with high total NPM1 expression. Since NPM1 is closely involved in the biogenesis of ribosomes, this finding supports the accuracy and fidelity of our pathway analysis. Moreover, these findings underline the conceivably different functions and impacts of circular and linear NPM1 transcripts. While we could show correlation of circNPM1 variant expression and distinct gene expression patterns, the significance of the respective NPM1 variants is still unknown, and further studies are warranted to study the impact of circular NPM1 RNAs on regulation of gene expression.
With regard to circRNA biogenesis, complementary intronic sequences are most likely not the only key mechanism. This is consistent with findings of such sequences in only around half of the circNPM1 variants. Whether circRNA expression is directly linked to the transcription of the linear isoforms of a gene is still an issue of current research and might not be consistent for all genes. The much longer half life of circular transcripts compared to their linear counterparts, as well as impairment of the splicing machinery, might also impact findings. In our study, a positive correlation was found for hsa_circ_0075001 and total NPM1 expression. In line with this, RNA-Seq data revealed a general correlation of circRNA and parental gene expression. However, we also found many exceptions. Not all highly expressed genes produce circRNAs, and those circRNAs with the highest expression level are generally not derived from genes with highest mRNA expression. Moreover, circFLT3 expression did correlate with parental gene expression only in healthy samples, but not in AML cells, thereby further pointing towards a non-random deregulation of circRNAs in AML, independent of mRNA expression.
Circular RNAs have been shown to be tightly regulated in the course of differentiation. Hundreds of circRNAs are, for example, differentially expressed during epithelial-mesenchymal transition,37 and circRNAs are globally up-regulated during neuronal differentiation.38 For the first time, we have shown that the expression of circRNAs, in line with mRNA, miRNA and lncRNA expression, is deregulated in AML, a disease characterized by inadequate differentiation of hematopoietic progenitor cells. We detected AML-associated and differentiation-independent differences between healthy hematopoietic cells and AML cells, and could also distinguish different AML subgroups based on their circRNA expression profile. These differences might in part be explained by the different degree of differentiation of the leukemia-initiating cells, but also by the genomic alterations influencing splicing factors and epigenetic modulators. As a consequence, the determination of the circular RNAome provides additional valuable insights into the biology of a leukemic cell. Furthermore, circRNAs are more stable than their linear mRNA counterparts,39,40 and thus could serve as potential biomarkers for classification and risk stratification in AML. In the future, additional studies will be required: i) to further determine the biological function of circRNA candidates in both healthy and malignant tissue; and ii) to define the prognostic and predictive impact of circRNA signatures in AML.
Supplementary Material
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/102/12/2039
Funding
The authors would like to thank the Deutsche Forschungsgemeinschaft (SFB 1074 project B3 to KD and LB and Heisenberg-Professur BU 1339/8-1 to LB) and the Studienstiftung des deutschen Volkes which supported SH for funding this project.
References
- 1.Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. [DOI] [PubMed] [Google Scholar]
- 2.Dolnik A, Engelmann JC, Scharfenberger-Schmeer M, et al. Commonly altered genomic regions in acute myeloid leukemia are enriched for somatic mutations involved in chromatin remodeling and splicing. Blood. 2012;120(18):e83–e92. [DOI] [PubMed] [Google Scholar]
- 3.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic Classification and Prognosis in Acute Myeloid Leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SC-W, Dvinge H, Kim E, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med. 2016;22(6):672–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falini B, Nicoletti I, Bolli N, et al. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007;92(4):519–532. [DOI] [PubMed] [Google Scholar]
- 6.Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 8.Nozawa Y, Van Belzen N, Van der Made AC, Dinjens WN, Bosman FT. Expression of nucleophosmin/B23 in normal and neoplastic colorectal mucosa. J Pathol. 1996;178(1):48–52. [DOI] [PubMed] [Google Scholar]
- 9.Grisendi S, Mecucci C, Falini B, Pandolfi PP. Nucleophosmin and cancer. Nat Rev Cancer. 2006;6(7):493–505. [DOI] [PubMed] [Google Scholar]
- 10.Zajac M, Dolnik A, Stasiak G, et al. Analysis of NPM1 splice variants reveals differential expression patterns of prognostic value in acute myeloid leukemia. Oncotarget. 2017. August 3 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rücker F, Russ A, Cocciardi S, et al. Altered miRNA and gene expression in acute myeloid leukemia with complex karyotype identify networks of prognostic relevance. Leukemia. 2013;27(2):353–361. [DOI] [PubMed] [Google Scholar]
- 13.Russ AC, Sander S, Lück SC, et al. Integrative nucleophosmin mutation-associated microRNA and gene expression pattern analysis identifies novel microRNA-target gene interactions in acute myeloid leukemia. Haematologica. 2011; 96(12):1783–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garzon R, Volinia S, Papaioannou D, et al. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci USA. 2014;111(52):18679–18684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- 18.Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56(1):55–66. [DOI] [PubMed] [Google Scholar]
- 19.Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177. [DOI] [PubMed] [Google Scholar]
- 20.Guarnerio J, Bezzi M, Jeong JC, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165(2):289–302. [DOI] [PubMed] [Google Scholar]
- 21.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun SM, Dijkstra MK, Bijkerk AC, et al. Transition of highly specific microRNA expression patterns in association with discrete maturation stages of human granulopoiesis. Br J Haematol. 2011;155(3):395–398. [DOI] [PubMed] [Google Scholar]
- 23.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013;arzXiv:1303.3997v2. [Google Scholar]
- 24.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(Suppl 2):W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thierry-Mieg D, Thierry-Mieg J. AceView: a comprehensive cDNA-supported gene and transcripts annotation. Genome Biol. 2006;7(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28(20):2233–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwind S, Maharry K, Radmacher MD, et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(36):5257–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun. 2016;7:12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai Y, Garrett KP, Ohta S, et al. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24(6):801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okamoto M, Hirai H, Taniguchi K, et al. Toll-like Receptors (TLRs) are expressed by myeloid leukaemia cell lines, but fail to trigger differentiation in response to the respective TLR ligands. Br J Haematol. 2009;147(4):585–587. [DOI] [PubMed] [Google Scholar]
- 34.Eriksson M, Peña P, Chapellier M, et al. Toll-like Receptor 1 Is a Candidate Therapeutic Target in Acute Myeloid Leukemia. Blood. 2014;124(21):5782–5782. [Google Scholar]
- 35.Marcucci G, Radmacher MD, Maharry K, et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1919–1928. [DOI] [PubMed] [Google Scholar]
- 36.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA. org resource: targets and expression. Nucleic Acids Res. 2008;36(Suppl 1):D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015; 160(6):1125–1134. [DOI] [PubMed] [Google Scholar]
- 38.Rybak-Wolf A, Stottmeister C, Glažar P, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015; 58(5):870–885. [DOI] [PubMed] [Google Scholar]
- 39.Bahn JH, Zhang Q, Li F, et al. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem. 2015;61(1):221–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PloS One. 2015;10(10):e0141214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.