Abstract
Vervet monkeys are amongst the most widely distributed nonhuman primates, show considerable phenotypic diversity, and have long been an important biomedical model for a variety of human diseases and in vaccine research. Using whole genome sequencing data from 163 vervets sampled from across Africa and the Caribbean, we find high diversity, within and between taxa, and clear evidence that taxonomic divergence was reticulate rather than following a simple branching pattern. A scan for diversifying selection across taxa reveals strong and highly polygenic selection signals affecting viral processes. Furthermore, selection scores are elevated in genes whose human orthologs interact with HIV, and in genes that show a response to experimental SIV infection in vervet monkeys but not in rhesus macaques, suggesting that part of the signal reflects taxon-specific adaptation to SIV.
Introduction
Vervet monkeys (genus Chlorocebus, also known as African green monkeys), are highly abundant in savannahs and riverine forests throughout sub-Saharan Africa, as well as on several Caribbean islands where they were introduced during the colonial era. There is a long history of research on vervet monkeys, ranging from studies of their social behavior1,2 to their use as an important model for a variety of human diseases3 and in vaccine research4–6. Vervet research colonies have been established, one of which is currently being genetically characterized7,8. Vervets are particularly interesting for HIV/AIDS research as they are the most abundant natural hosts of simian immunodeficiency virus (SIV), a close relative of HIV. SIV is highly prevalent across African vervets, but infected individuals typically avoid progression to immunodeficiency, despite high viral loads9,10. Elucidating the genetic mechanisms for host defense against this virus in vervets may identify new targets for preventive and therapeutic interventions for HIV/AIDS.
Here we follow the publication of the vervet reference genome11 by presenting a genus-wide survey of polymorphism. The genus Chlorocebus has alternatively been viewed as a single species (Ch. aethiops) with several subspecies or as 5–6 species with additional subspecies12. Our sampling strategy was intended to capture this diversity (Supplementary Data 1), by including a total of 163 individuals from nine countries in Africa and two countries in the West Indies, which harbor sizable feral populations (Fig. 1a, Supplementary Data 1). No previous study has conducted genome-wide resequencing in a non-human primate in such a large sample and over such a geographically extensive area.
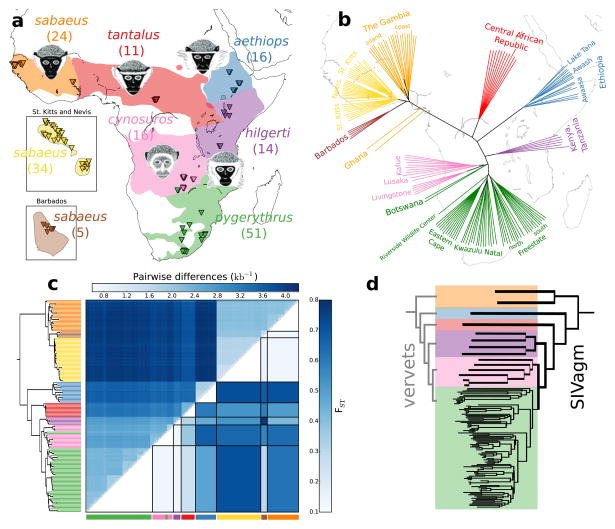
Fig. 1.
Sample information and genetic relatedness. (a) Taxon-distribution, approximate sampling locations (triangles) and representative drawings (where available: hilgerti and pygerythrus are morphologically very similar and have often not been considered separately). Number of whole-genome sequenced monkeys given in parentheses. (b) Neighbor-joining tree based on pairwise differences oriented to approximately fit geographic sampling locations. (c) Matrix-plot of pairwise genetic differences per callable site (above diagonal) and fixation index (FST) between groups (below diagonal). Rows and columns are sorted according to a hierarchical clustering (UPGMA) tree of vervet pairwise genetic differences. (d) Clustering tree of SIVagm pol gene sequences sampled from wild vervets (Africa only). Vervet taxonomic relationships are given on the left for comparison. The tree was constructed with sequences from both vervets included in this study and with sequences from the HIV Sequence Databases. Colors correspond to group labels in (a). Monkey heads in (a) are redrawn from Haus et al. (2013)16 adapted from Hill (1966)17. Distribution maps are adapted from IUCN (2017)18.
Vervets harbor extensive polymorphism, both within and between taxa, and we see clear evidence that taxonomic divergence involved gradual divergence and gene flow rather than following a simple branching pattern. A scan for diversifying selection across vervet taxa yields gene enrichments much stronger than in similar studies on humans13. In particular, we report strong and highly polygenic selection signals affecting viral processes — in line with recent evidence that proposes a driving role for viruses in protein evolution in mammals14. These signals are furthermore enriched in genes with known relevance to SIV: either their human orthologs interact with HIV, or they show a vervet-specific transcriptional response to SIV infection. Intriguingly, rather than affecting genes with antiviral and inflammatory-related functions15, selection in vervets appears to have primarily targeted genes involved in the transcriptional regulation of viruses, and in particular those that are harmful only under immunodeficiency, suggesting evolved tolerance of SIV rather than resistance against infection.
Results
Sequencing and polymorphism detection
100 bp paired-end Illumina data were generated for all samples. Coverage was relatively low (median 4.4X), but at least one member of each taxon was sequenced to 10X coverage or higher (Supplementary Table 1). Employing a standard pipeline for alignment to the reference Chlorocebus_sabaeus 1.111, derived from a St. Kitts-origin monkey, and joint variant detection across all samples, we discovered a total of over 97 million single nucleotide polymorphisms (SNPs), 61 million of which passed our quality filters (Online Methods, Supplementary Figs. 1 to 4).
Genetic relationships among vervet groups and SIV strains
Clustering of individuals based on pairwise genetic distance (Fig. 1, b and c) and principal component analysis (Supplementary Fig. 5) generally agrees with prior morphological and geographic classification, and led us to define six African and two Caribbean taxonomic groups: sabaeus (West Africa), aethiops, tantalus, hilgerti, cynosuros, pygerythrus, sabaeus (St.Kitts and Nevis) and sabaeus (Barbados). The genetic relatedness pattern also clearly confirms the status of sabaeus as an outgroup to other vervet taxa (Supplementary Note 1, Supplementary Figs. 6 and 7), and suggests isolation-by-distance within groups (Fig. 1b). Both geographic location and group identity contribute significantly to explaining the overall pattern of polymorphism (likelihood-ratio test p<2*10−3 and p<10−52, respectively, Supplementary Table 2). Our data agree with the morphology-based taxonomy12 in that sabaeus, aethiops and tantalus appear to be well-defined taxa, whereas hilgerti, cynosuros and pygerythrus are comparatively closer to each other, exhibiting substantial amounts of shared variation and strongly correlated allele frequencies (Fig. 1c, Supplementary Fig. 8). However, while morphological evidence groups hilgerti and pygerythrus as a single species (Ch. pygerythrus) distinct from cynosuros (Ch. cyonosuros), our data show that pygerythrus and cynosuros are closer to each other than either is to hilgerti. Indeed, two pygerythrus individuals from Botswana are more closely related to cynosuros than to other pygerythrus. This is probably due to admixture: as we note below, there is abundant evidence for admixture between these groups. Finally, the pattern of relatedness among SIV strains mirrors the pattern in vervets (Fig. 1d), suggesting that SIV existed in vervets prior to their initial divergence more than half a million years ago and has co-evolved with the taxa9,11.
Our data also confirm that, as surmised from the historical record, Caribbean vervets are derived from Western African sabaeus. Additionally, we are able to clarify the relationship between the vervet populations on different Caribbean islands. The fact that vervets from Barbados are nearly as different from vervets from St. Kitts and Nevis as they are from Gambian sabaeus (Fig. 1c) suggests that these two Caribbean populations were founded independently and experienced two independent bottlenecks (28% and 17% reduction in diversity relative to Gambia, respectively). The vervet population from Nevis, on the other hand, is genetically a subset of the St. Kitts population (17% reduction in diversity relative to St. Kitts), and likely was founded by individuals from this island, which is less than 4 km away. In human genetics there is currently great interest in sequencing studies of recently expanded bottlenecked populations, both for elucidating population genetic processes and for identifying deleterious variants with a strong impact on phenotypes that have reached high frequency through drift. However, while the site frequency spectrum in Caribbean vervets is generally biased towards higher allele frequencies, there is no evidence in our data that this effect is relatively stronger for putatively deleterious alleles (Supplementary Fig. 9).
Returning to Africa, variation within vervet taxa is much larger than in humans19 and other great apes20 but is typical for other primates (Fig. 1c, Supplementary Fig. 10)21 — which is perhaps surprising given the ubiquity of vervets. Divergence between vervet taxa is generally higher than between subspecies of other primates, with average pairwise sequence divergence between taxa of ~0.4%, compared to 0.2% to 0.32% across great ape subspecies20, and FST-values from 25% to 71% (Fig. 1c, Supplementary Fig. 11), compared to <15% across human populations22 or macaque subspecies23. However, maximum sequence divergence is substantially lower than between human and chimpanzee (~1.24%)24. This intermediate status is supported by the presence of substantial amounts of both shared variation and fixed differences between vervet taxa (Supplementary Fig. 8).
Evidence for genetic admixture
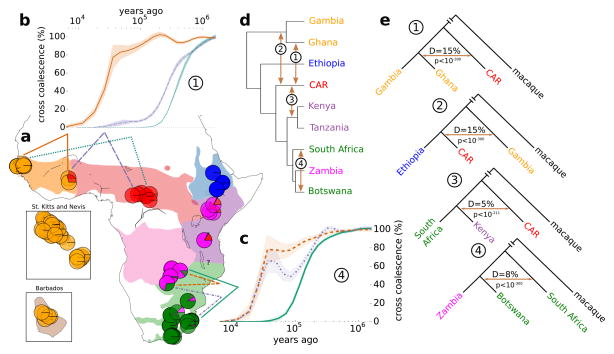
The process that gave rise to the current taxa was much more complex than a series of population splits. We used Admixture25 to cluster individuals into groups. This analysis generally resolves the above-mentioned taxa, and confirms the complicated relationships of south- and east-African vervets (Fig. 2a, Supplementary Figs. 12 and 13). However, there is evidence of admixture throughout. For example, Ghanaian sabaeus and Kenyan and Tanzanian hilgerti show substantial proportions of tantalus ancestry (Fig. 2a). D-statistic (ABBA-BABA test; Fig. 2, d and e, Supplementary Fig. 14, Supplementary Data 2)26,27 confirms that this shared ancestry represents gene flow between tantalus and both Ghanaian sabaeus and hilgerti (D = 15.2% and 5%, respectively; block jack-knifing p<10−300). The alternative explanation, ancestral population structure, seems less parsimonious, as the structure would have had to persist through multiple taxonomic splits. Using the multiple sequentially Markovian coalescent method (MSMC; Fig. 2, b and c, Supplementary Figs. 15 and 16, Supplementary Note 2)28 we show that the observed signatures of gene flow likely reflect ancient rather than recent admixture; for example, for tantalus, after initial divergence, gene flow ceased earlier with geographically more distant Gambian sabaeus than with geographically closer Ghanaian sabaeus (Fig. 2b). The lack of long shared haplotypes across taxa also supports the absence of recent admixture (Supplementary Table 3).
Fig. 2.
Evidence for gene flow across taxa. (a) Admixture clustering of individuals. Each pie-chart represents an individual and colours represent contributions from five assumed admixture clusters. The choice of five clusters is discussed in the legend of Supplementary Fig. 12. Full results are shown in Supplementary Fig. 13. Colored lines mark comparisons in panels b and c. (b) and (c) MSMC plots of cross-coalescence rate, a measure of gene flow, across time (on a log scale). Shaded areas correspond to +/− 3 block-jackknifing standard deviations. (d) UPGMA tree of pairwise distance matrix summarized by country. Arrows point to evidence of cross-taxon gene flow. (E) D-statistic (ABBA-BABA test) for instances of gene flow shown in (d). For full results see Supplementary Fig. 14 and Supplementary Data 2.
Turning to the east and south African cynosuros/hilgerti/pygerythrus complex, we find that, while simple clustering suggests that hilgerti is an outgroup to cynosuros and pygerythrus (Fig. 1, b and c), Admixture represents cynosuros individuals as a mixture of hilgerti and pygerythrus with a larger contribution of the former (Fig. 2a). MSMC suggests a complex history of varying gene flow between the three groups (Supplementary Fig. 16). We also investigated the status of pygerythrus from Botswana, which appear as sister-group to Zambian cynosuros in the clustering tree (Fig. 2d): D-statistic shows that they have an additional genetic contribution from South African pygerythrus (Fig. 2e, D = 7.6%, jack-knifing p<10−300) and MSMC confirms an intermediate status of Botswanian pygerythrus with comparable levels of genetic exchange with both South African pygerythrus and cynosuros until total separation from both groups ten thousand years ago (Fig. 2c), again compatible with isolation by distance. In summary, while inferred genetic relationships are generally consistent with current taxonomy, strong signals of excess allele sharing along geographic axes suggest that the evolutionary history of these taxa involved processes of gradual divergence, isolation, and secondary contact.
Strong signals of selection
Our data provide a rare opportunity to look for signals of adaptation on a continent-wide scale, across multiple taxa. To identify footprints of selection, we used an approach that incorporates information on both the distortion of allele frequency spectra within groups and the increase in differentiation among pairs of groups at loci close to a group-specific selective sweep (XP-CLR)29. To summarize the 30 XP-CLR-comparisons between African taxa (Supplementary Figs. 17 to 22), we calculated “selection scores” — the root mean square XP-CLR scores (across taxon comparisons) — on a 1000 base pair grid along the genome.
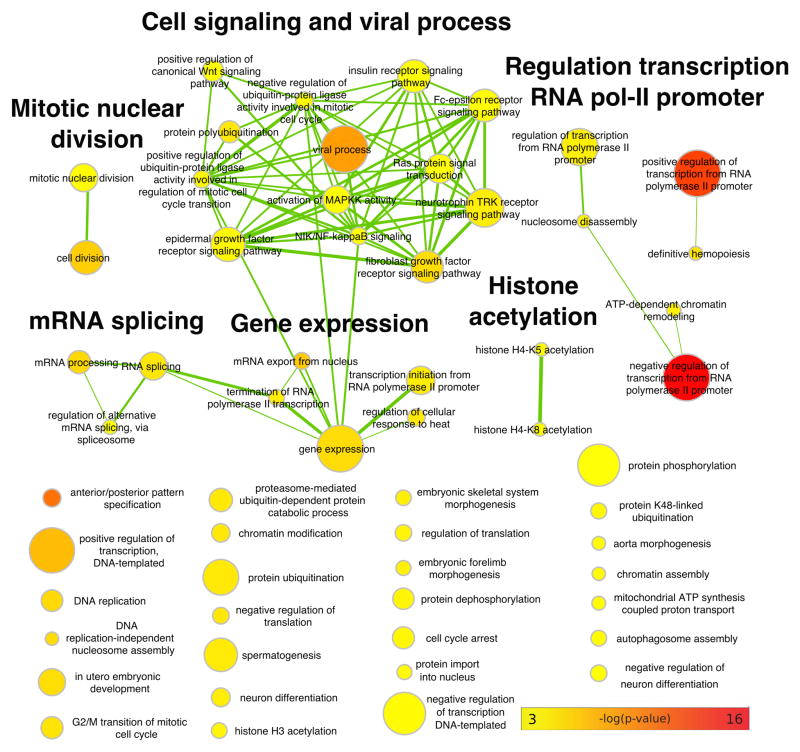
These scores clearly capture strong signals of selection, because they are significantly higher in genic than intergenic regions (one-sided Mann-Whitney U test p<10−300, Supplementary Fig. 23). To gain further insight, we compared the distribution of average selection scores for genes (Supplementary Data 3) across gene ontology (GO) terms with the R-package TopGO30. Testing for enrichment using the relative rank of all scores yielded stronger signals than testing genes with the highest scores against the background, suggesting that weaker, polygenic effects contribute strongly to the signal of selection (Supplementary Fig. 24). We found 157 significantly enriched GO terms, many of which are related to RNA transcription and cell signalling (Fig. 3, Supplementary Fig. 25, Supplementary Data 4). These GO enrichments show partial overlap with similar scores comparing human populations (Supplementary Fig. 26)13. However, there are many more significantly enriched GO terms for vervets than for humans and shared enrichments are generally much more significant in the current data set, suggesting that vervet taxa provide a powerful model to study diversifying selection across closely related primate taxa. The strongest selection scores are consistent with a dominant role of viral pathogens as selective agents in vervets. In particular, we note viral process (p=5*10−9), and positive and negative regulation of transcription from the polymerase II promoter (p=3*10−17, 5*10−14), which is known to interact with viral proteins (for example the HIV Tat protein during transcription elongation of HIV-1 LTP31). We note that these virus-related categories are not only enriched in the root mean square summary but also in many two-taxon XP-CLR comparisons (Supplementary Note 3). Furthermore, these categories do not show particularly large neutrality indices or significant enrichment for conserved elements32 (Supplementary Note 3; Supplementary Figs. 27–30), providing evidence that these signals are not predominantly driven by purifying selection (background selection), which can lead to confounding signals33.
Fig. 3.
Enrichment map network of Gene Ontology (GO) categories enriched for high average gene selection scores. Edges represent overlap in genes. Colors represent p-values on a log scale (red most highly significant, TopGO Kolmogorov-Smirnov weight01 p<0.001). Node size represents number of genes in a category (capped at 474). Terms are grouped using Cytoscape clustermaker34.
To test more specifically for virus-related selection signals, we looked for enrichment of signals among the orthologs of human HIV-interacting genes. Indeed, 43 out of 166 gene sets in the NCBI HIV-1 human interaction database35 show significant enrichment for high selection scores (Supplementary Fig. 31, Supplementary Data 5). However, we note that 21 and 71 partly overlapping gene categories from this database also show enrichment for human selection scores and conserved elements, respectively (Supplementary Figs. 31 and 32, Supplementary Note 3), suggesting that these gene sets are not very specific.
Selection signals linked to vervet specific response to SIV
SIVagm is prevalent in African vervets9,10 and has diverged into taxon-specific strains (Fig. 1d) 36,37. Furthermore, while SIVagm is highly pathogenic when used experimentally to infect pigtailed macaques that are not natural SIV hosts38,39, infected vervets generally do not progress to AIDS, suggesting coevolution of virus and host. We hypothesized that coevolution between taxon-specific SIV strains and vervet taxa could lead to an ongoing evolutionary arms race that would manifest itself as diversifying selection across taxa, specifically on genes involved in host defense (whereas adaptations shared across the genus would be very difficult to detect). To test this, we reanalyzed microarray data comparing the transcriptional response of vervets and macaques to infection with SIV40,41. Unlike vervets, macaques are not natural hosts of SIV and generally develop AIDS-like symptoms upon infection. If some of the selection signals reflect adaptation to SIV in vervets, we would expect selection scores to be elevated in genes that are differentially expressed in vervets — but not in macaque — as a response to infection. Indeed, selection scores are much higher in genes that show a significant expression difference before and after infection in vervets only, as compared to genes showing an expression difference in both species (one-sided Mann-Whitney U test p=5 10−9) or in macaque only (p=10−4) (Supplementary Fig. 33). Conversely, vervet-specific (but not shared or macaque-specific) differentially expressed genes are significantly enriched in high selection scores (p=0.003, p>0.99 and p>0.99, respectively).
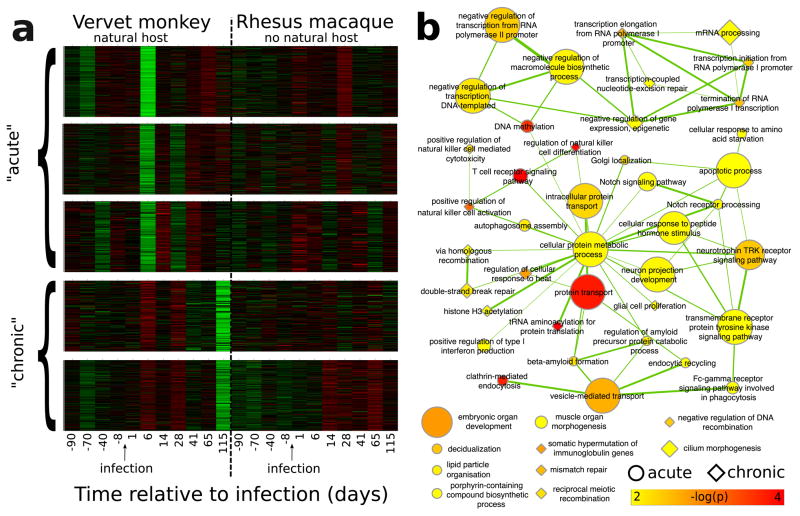
To further investigate the underlying mechanisms, we grouped differentially expressed genes by coexpression patterns using weighted gene co-expression network analysis (WGCNA, Supplementary Figs. 34 and 35)42. Five out of 33 gene co-expression modules show a significant enrichment for genes with high selection scores (FWER<0.05; Fig. 4a, Supplementary Fig. 35, Supplementary Data 6). Remarkably, the significant modules share similar expression patterns with strong changes in vervets post-infection, and weak, inconsistent signals in macaques. In particular, all the modules that are enriched for diversifying selection show changes in gene expression in vervets six days post-infection, which is around the time that the virus becomes detectable and activates early immune responses. Two modules also show expression differences in the chronic stage (115 days post infection), which is most relevant for progression to immunodeficiency. We ran GO enrichment analysis separately on the genes in the enriched WGCNA modules showing early (“acute”) and late (“chronic”) expression changes (Fig. 4). We found 30 and 20 significantly enriched GO categories, respectively, many of which are involved in response to HIV in humans (Supplementary Data 7 and 8). For example, for early expression response, enriched GO categories include clathrin-mediated endocytosis43, autophagosome assembly, positive regulation of type I interferon (IFN-I) production40,41 and innate immune response (marginally significant at p=0.01004). This is consistent with recent findings that in macaques the IFN-I response is delayed in response to SIV infection and inhibited during the first week of SIV infection44, while natural hosts mount a very early and transient IFN-I response40,41. Conversely, the three most highly enriched GO categories for genes in modules with late expression changes are positive regulation of natural killer (NK) cell activation, regulation of cellular response to heat45, and somatic hypermutation of immunoglobulin genes46, consistent with differences in NK cell responses during SIV infection in natural hosts as compared to non-natural hosts, the lack of viral replication in B cell follicles (Tfh cells) and preservation of lymph nodes immune function in natural hosts in contrast to macaques, as well as a better adaptation to the stress induced by the chronic infection47–50.
Fig. 4.
Gene co-expression modules with differential expression pre- and post-SIV-infection which are also significantly enriched for high selection scores. Genes that were differentially expressed in CD4+ blood cells in vervet or macaque as response to SIV infection were grouped into 36 co-expression modules using WGCNA (shown in Supplementary Fig. 35). (a) Expression pattern of the five co-expression modules which are significantly enriched for high selection scores (p<0.01, FWER <0.05). (b) Joint enrichment map network of GO enrichments of the genes in the top three panels of (a) (“acute”, circles) and the bottom two panels of (a) (“chronic”, diamonds). GO enrichment was tested using TopGO Fisher’s exact test with weight01 algorithm. Edges represent overlap in genes. Node size represents number of genes in a category (capped at 474).
Candidate targets of selection
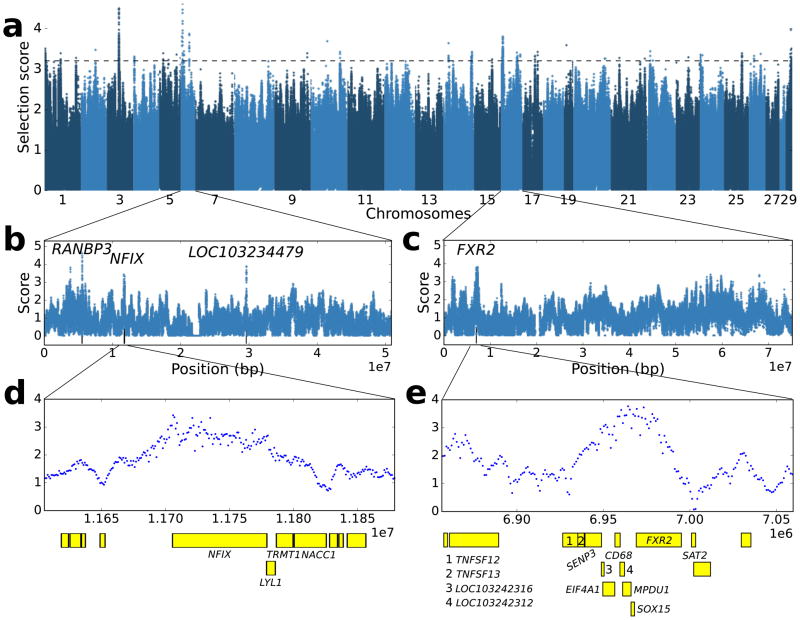
While enrichment analysis identifies categories of genes under selection, and is likely driven by large numbers of genes with moderate effects, the highest selection scores identify candidate regions for strong selection (Fig. 5). The highest score is for an uncharacterized gene on chromosome 6 (Fig. 5b) with 97% sequence identity to the human gene encoding for RAN binding protein 3 (RANBP3), a protein connected to influenza A virus replication51 and that is involved in nucleocytoplasmic export of RNAs from human T-cell leukemia virus type 1 (HTLV-I)52 and HIV53,54. Another gene that displays among the highest selection scores is NFIX nuclear factor I/X (Fig. 5d), encoding for a transcription factor that binds the palindromic sequence 5′-TTGGCNNNNNGCCAA-3 in viral and cellular promoters. Nuclear factor I proteins can serve as a transcription selectivity factor for RNA polymerase II, and play a critical role in transcription and regulation of JC virus in humans55 and Simian virus 40 in vervet cells56. Remarkably, these closely related viruses are usually harmless but cause disease under immunodeficiency, specifically in SIV/HIV infection in macaque57 and human58. However, the lack of common genetic variants in the coding sequence of this gene suggests that selection is more likely to have targeted regulatory variants.
Fig. 5.
Selection scores across the genome and candidate genes with strong selection signals. (a) Manhattan plot of selection scores across all chromosomes. (b–c) Selection scores along chromosomes 6 and 16, respectively (d) Magnification of the region containing NFIX. (e) Magnification of a peak containing multiple candidates, among them CD68 (Cluster of Differentiation 68), a glycoprotein highly expressed on monocytes/macrophages, and FXR2 (Fragile X mental retardation, autosomal homolog 2) that interacts with HIV-1 Tat gene. Slightly downstream of the shown region, we note the highly scoring gene KDM6B (lysine-specific demethylase 6B), which is upregulated by HIV-1 gp120 in human B-cells.
Discussion
We have genetically characterized Chlorocebus, vervet monkeys, a genus of African primates with continent-wide distribution and substantial, recently established, populations on three Caribbean islands. We find vervets to be genetically diverse with an average nucleotide diversity (heterozygosity) within taxonomic subgroups about twice that of humans (~0.2% in vervets)19. A recent study59, based on unpublished data from a subset of the samples described here, infers more than five times lower diversity; less than half the lowest synonymous diversity reported in a study of 76 species of all phyla60. We suggest that the extremely low diversity values reported in the vervet study mentioned above are due to biases in GATK HaplotypeCaller leading to an excess of homozygous reference calls (Online Methods) and/or an overestimate of the accessible genome size.
There has been considerable debate about vervet taxonomy, both concerning taxonomic levels of different groups (species or subspecies) and relationships between groups. While taxonomic assignment can reflect a variety of morphological, genetic and behavioral information, our results suggest that — despite evidence for substantial genetic exchange — Chlorocebus includes both genetically well-separated taxa (sabaeus, aethiops, tantalus, and pygerythrus) as well as more closely related groups (pygerythrus, cynosurus, and hilgerti). The latter groups would naturally fall at a taxonomic level below the former.
Different phylogenetic relationships between vervet taxa have been proposed. Using two different clustering algorithms we find that West African sabaeus split off the common ancestor of other vervets first, followed by aethiops, with the last split separating tantalus from pygerythrus (the latter including cynosuros and hilgerti). This result is consistent with Warren et al. (2015)11, who inferred the same branching pattern using whole genome sequencing data of a single representative per taxonomic group. It contradicts Pfeifer (2017)59, who suggested that aethiops (rather than sabaeus) constitutes the outgroup of vervet taxa. Pfeifer attributed this difference to Warren et al. having neglected to distinguish fixed from shared polymorphism. However, Pfeifer apparently used fixed differences only to infer taxon relatedness. This procedure leads to erroneous conclusions, because inferred branch lengths for taxa with comparatively high rates of drift (low effective population size), such as aethiops, will be biased upward, and branch lengths for taxa that have been exchanging genes, such as sabaeus and tantalus, will be biased downward. By aligning a subset of the data against the macaque reference genome and computing genome-wide summaries of the coalescent histories of the samples, we strongly confirm the outgroup status of sabaeus (Supplementary Fig. 7, Supplementary Note 1).
These results notwithstanding, our analyses also clearly show that a simple phylogenetic tree cannot fully capture the pattern of relatedness across vervets. As in several recent studies61,62, the process of speciation must have been gradual and involved gene flow, leading to a fundamentally reticulate pattern of relatedness. Although we see no evidence for current gene flow between vervet taxa, definite conclusions will require more appropriate sampling (especially in putative hybrid zones, as these taxa are expected to readily hybridize63,64) and sequencing strategies that are better able to resolve haplotypes.
Finally, we carry out a screen for diversifying selection across vervet taxa, our primary goal being to look for signs of adaption to SIV. To this end, we use a method that tests whether the change in allele frequency between taxa at a locus occurred too quickly to be due to random drift. This approach is expected to be sensitive to both recent and relatively older selection events, and to pick up signals of recurrent adaptation29.
Gene ontology analysis yields strong enrichment of selection scores in multiple biological processes, generally driven by polygenic signals. Our data have the potential to yield insights into taxon-specific adaptations (e.g., altitude adaptation in aethiops). In the present study, we focus on loci that show signals of repeated (but differential) adaptations across multiple taxa, consistent with host-pathogen coevolution. As hypothesized, our screen revealed a strong excess of signal in genes that interact with viruses, consistent with findings in other organisms14,29. While there is potential for coevolution with different types of viruses in vervets, integration of our selection study with gene expression analysis of SIV-infected monkeys provides evidence that part of the signal results from vervet co-evolution with SIV. Interestingly, the genes identified do not include the virus (co-)receptor genes involved in the virus docking mechanism, but rather genes involved in cell signalling and transcriptional regulation, consistent with recent results suggesting that natural selection has shaped primate CD4+ T-cell transcription65, and suggesting adaptation to living with the virus rather than avoidance of infection. Indeed, one of the highest scoring genes controls the expression of a virus known to cause disease under SIV-induced immunodeficiency.
While there is no doubt that our analysis is picking up real signals of selection in virus-related genes, it is difficult to determine the mode of selection conclusively. For example, we carried out several tests to confirm that our results are not primarily driven by purifying (rather than diversifying) selection, but further orthogonal approaches and functional validation will be necessary to ultimately understand the evolutionary dynamics of vervets and their pathogens. The data and results presented here should aid this endeavor, and may prove useful in the quest for antiviral vaccinations and therapies.
Online Methods
Sample collection and sequencing
All blood samples were collected under approved country specific permits that meet standardized bioprospecting regulations. DNA samples were obtained from blood (PaxGene DNA tubes, ACD tubes or archival blood cell pellet collection) except for sample AG23 that was obtained from a B lymphoblastoid cell line transformed with herpesvirus papio. Individuals were sequenced at variable coverage (Supplementary Data 1) on a Illumina HiSeq2000 platform obtaining 100bp paired-end reads.
Alignment, variant detection and filtering
Sequences were aligned against the ChlSab1.1 reference11 using bwa-mem66 with a total coverage of 798X and a median coverage of 4.4X. On average, more than 98% of the reads mapped for all taxa, suggesting that reference bias is weak. Following the GATK recommended workflow67,68, alignments against ChlSab1.1 (see URLs) were locally realigned, base quality scores were recalibrated using a first round of variant calling and variants were detected using GATK UnifiedGenotyper. We also called variants using GATK HaplotypeCaller but found these calls to have a strong bias towards homozygous reference alleles in the low coverage samples. We hence only used UnifiedGenotyper variant calls for further analysis. Biallelic SNP calls were hard filtered with a combination of GATK best practices68 and custom filters to yield the data set used for further analysis (Supplementary Table 1, Supplementary Fig. 1). We used VCFtools diff-indv-discordance to compare the genotype calls from Warren et al. 201511 and the current dataset for the five individuals that are shared between the studies. For non-filtered SNP the discordance rate was 0.12% – 0.26% and for all non-filtered sites it was 0.006% – 0.012%.
Given the large differences in coverage between individuals (2X–45X), a stringent control on false positive rate would have led to strong a bias towards lower diversity (and especially a lower number of singletons) in low coverage samples. We suggest that for population genomic analysis it is conservative to reduce bias at the cost of increased noise. Our dataset does show correlation between coverage and individual heterozygosity (Pearson’s r=0.48, p=10−10, Supplementary Fig. 3), especially for individuals with less than 4X coverage, but cross-individual sequence divergence within and between taxa is not strongly affected by coverage (Pearson’s r=0.009 and 0.026, respectively, Supplementary Fig. 3). The ancestral state for each SNP was determined by aligning the macaque reference genome, rheMac2, against ChlSab1.1 using nucmer69, only considering one-to-one mappings with a minimum length of 200bp.
To test whether our results could be affected by coverage bias, we repeated some of the analysis with a subset of the sequencing reads aligned to the Rhesus macaque genome, Mmul 8.0.1 (Supplementary Note 1, Supplementary Figs. 6 and 36).
Minor in-silico contamination of the original read-files of 18 South African individuals with RNA-seq reads was detected at a late stage (after review). While the effect of this on variant calls was very small (median genotype concordance between the original call-set and an updated version with contamination removed was 99.91%), it led to some highly expressed genes being masked by the high coverage filter (~2% additional PASS SNPs in the recall). We therefore repeated the complete selection analysis (Figs. 3–5, Supplementary Figs. 15–31) using the updated call-set.
Accessible genome size
To compare levels of polymorphism and divergence across individuals and to previous studies, we measured the proportion of the genome accessible to our variant detection process. In particular, we excluded all sites that did not pass our quality filters, and, for each individual, all sites for which UnifiedGenotyper could not make a genotype call (Ns) (Supplementary Fig. 2).
Diversity and divergence
Nucleotide diversity was calculated by computing the number of pairwise differences for each comparison divided by the accessible genome size for each pair as derived above. For each group, nucleotide diversity was estimated as the average of within-group comparisons. We found 16–27 million SNPs segregating within taxa, corresponding to an average number of pairwise differences per site (nucleotide diversity) of 0.17–0.22% (Fig. 1c, Supplementary Fig. 7) and effective population sizes generally above 35,000 (except for aethiops for which we estimate ~29,000). Site frequency spectra within taxa (Supplementary Fig. 4) generally agree with neutral expectations, except for a general lack of low frequency variants and an excess of high frequency derived variants, most likely as a consequence of low power to call low frequency variants and erroneous inference of the ancestral state, respectively.
Divergence was calculated as the average of pairwise differences across all comparisons of two groups. Two taxon site frequency spectra (Supplementary Fig. 9) generally showed fixed differences as well as shared and private variation, except for hilgerty/cynosorus/pygerythrus which showed few fixed differences and highly correlated allele frequencies.
To assess the relative contribution of geography and taxon label to explaining the genetic relatedness among vervets, we calculated principal components (PCs) from autosomal SNPs using PCAdapt version 05/26/14 in mode fast70 setting K=6, which gave the best fit to our data. Next, for each of the six PCs we performed likelihood ratio tests in R (function anova with option test=‘LRT’) to test whether a linear model “PC ~ latitude + longitude + taxon label” gave a significantly better fit than a model using either only geography or only taxon (Supplementary Table 4). The outgroup branch of the vervet phylogeny was confirmed using genome wide summaries of the average time to the most recent common ancestor among samples from different taxa (Supplementary Note 1, Supplementary Fig. 7).
FST was calculated using the Weir-Cockerham estimator71 using vcftools72 for all autosomal SNPs. For each pairwise comparison, we summarised FST-values in minor allele frequency (MAF) bins (Supplementary Fig. 8; the maximum across MAFs in Fig. 1c).
SIVagm phylogenetic analyses
A 602 bp pol integrase fragment of SIVagm, obtained as described previously9, was used for phylogenetic analyses of a large sample of SIVagm strains from the different subtaxa of vervet with different origin. pol nucleotide sequence alignments were obtained from the Los Alamos National Laboratory HIV Sequence Database (see URLs). Newly derived SIV sequences were aligned using MUSCLE73 and alignments were edited manually where necessary. Regions of ambiguous alignment and all gap-containing sites were excluded.
Phylogenetic trees were inferred from the nucleotide sequence alignments by the neighbor-joining method using the HKY85 model of nucleotide substitution73,74 implemented using PAUP*75. The reliability of branching order was assessed by performing 1,000 bootstrap replicates, again using neighbor-joining and the HKY85 model. Phylogenetic trees were also inferred by maximum-likelihood using PAUP* with models inferred from the alignment using Modeltest76. The neighbor-joining tree topology was used as the starting tree in a heuristic search using TBR branch swapping.
Admixture analysis
The software Admixture 1.2325 was run on autosomal SNPs filtered for minor allele frequency > 5%, converted to binary .bed format using GATK VariantToBinaryPed and LD pruned using the plink flag --indep 50 10 2 (Fig. 3a and Supplementary Fig. 11).
Cross-taxon gene flow across time
Missing genotypes in our autosomal SNP calls were imputed using beagle 477, version 03Oct15.284:
java -jar beagle.jar gl=biallelic_pass_snps.vcf out=beagle_out.vcf ibd=false
Samples were phased using shapeit.v2.r837.GLIBCv2.12.Linux78:
shapeit -phase --input-vcf beagle_out.vcf --window 0.1 -O phased.tmp
shapeit -output-vcf --input-haps phased.tmp -O phased.vcf
For each geographic sample group of interest, we chose the individual with the highest coverage for further analysis. For pairs of individuals from different groups, we extracted the alleles that segregate within or between the two individuals and their phase as needed as input to MSMC. The number of informative sites between two segregating variants was determined for each pair of individuals separately from the all-sites VCF (the whole genome including non-variant sites) by counting the number of non-filtered sites for which both individuals had genotype calls.
Three runs of MSMC2 (see URLs) were produced for each pair, two inferring coalescent rates across time within each of the two samples
msmc2 -I 0,1 -o within_1 input_chrom1 ... input_chrom29
msmc2 -I 2,3 -o within_2 input_chrom1 ... input_chrom29
and one run for inferring coalescent rates across time between the two samples
msmc2 -P 0,0,1,1 -o between input_chrom1 ... input_chrom29
The outputs of these runs were combined by interpolating the mid-point of each time interval in the former two on the mid-points in the latter run. Cross-coalescent rate was calculated as 2*between/(within_1+within_2) (Figs. 2, b and c, Supplementary Figs. 12 and 13, Supplementary Note 2). Evolutionary time was scaled to years using a mutation rate of 1.5*10−8 and a generation time of 8.5 years11. Each MSMC2 analysis was re-run 29 times leaving one chromosomes out at a time and block-jackknifing variance was calculated.
D-statistic
D-statistic was calculated from autosomal SNPs using Admixtools 3.027, treating samples from each country as a populations and performing all tests that were consistent with the country UPGMA tree shown in Fig. 2d. Macaque was used as an outgroup and the analysis was restricted to sites where the macaque allele could be inferred. Due to limitations in Admixtools the analysis was restricted to vervet chromosomes 1–24. Block-jackknifing was performed with Admixtools standard settings.
Diversifying selection scan
Autosomal biallelic PASS SNP genotypes were converted to XP-CLR input format. For each comparison of two groups, we excluded SNPs if they were not segregating within or between the groups or if they had more than 20% missing genotypes across the two groups. The genetic map from Huang et al.7 was interpolated to our SNP positions to obtain genetic distance in Morgan. A handful of extremely flat regions in the genetic map lead to numeric errors in XPCLR. The problematic markers were removed and map distance was instead interpolated from the adjacent markers left and right.
XP-CLR was run on all 30 possible comparison of the 6 African taxa (for each comparison, using each taxon once as objective and once as reference population). Parameters supplied to XP-CLR were -w1 0.001 500 1000 -p0 0, meaning that a set of grid points as the putative selected allele positions are placed along the chromosome with a spacing of 1 kb, the sliding window size was 0.1 cM around the grid points and if the number of SNPs within a window is beyond 500, some SNPs were randomly dropped to control for SNP number. Alleles were assumed unphased (-p0) and SNPs in high LD were not down-weighted (final 0).
To find loci that were repeatedly under diversifying selection across several group comparisons, we calculated for each grid point the root mean square selection score across all 30 comparisons (Fig. 5a). To test whether these scores capture biological signal, we confirmed that scores are significantly higher in genic (introns + exons) than in intergenic regions (Supplementary Fig. 21, one sided Mann-Whitney U test, p<10−300). Since the Mann-Whitney U test assumes independence of scores, a condition which is not totally met in our data due to linkage disequilibrium, we also calculated the average of the mean selection score across each gene and compared the resulting value to a background distribution. The background distribution was obtained by first concatenating all chromosomes in a circle and randomly shifting (rotating) the scores against their genomic positions, then calculating mean gene scores from these rotated data. We again found that genic scores are significantly larger (p<10−5).
Gene set enrichment analysis
z-score transformed selection scores across genes (exons and introns) were used for gene enrichment analysis. Gene locations were extracted from NCBI Chlorocebus sabaeus Annotation Release 100 (see URLs). To test for enrichment in gene ontology (GO) terms, we first used the the R-package TopGO30 with the “weight01” algorithm which allows to account for the hierarchical structure (and thus overlap) of GO terms when testing significance and thereby implicitly corrects for multiple testing. GO annotations were obtained from the R package org.Hs.eg.db (Bioconductor 3.2). We restricted the analysis to 5777 GO terms with more than ten genes in our annotation. Note that our gene scores are not biased by gene length because we are calculating the average score across genes rather than taking the maximum score. However, enrichment results are qualitatively similar if the maximum is taken. Results are also similar if only exons are used (rather than exons + introns). We also note that the most significantly enriched categories contain many genes (Supplementary Data 4) and do not show strong clustering in particular genomic regions.
HIV-1 human interaction categories
The NCBI HIV-1 Human Interaction Database35 was downloaded from (see URLs). We only kept categories from the database that had ten or more genes in our annotation. We implemented the sumstat statistic13 to compare observed and expected gene-averaged selection scores in HIV-1 human interaction categories to random sets of known genes.
Gene expression analysis
We conducted Weighted Gene Co-expression Network Analysis (WGCNA) on gene expression data of vervets and macaques essayed at different time points pre- and post-infection with SIVagm and SIVmac, respectively40, using the WGCNA R package as previously described42,79. We used as a starting point the list of genes with differentially expressed transcripts in CD4+ cells before and after SIV infection in either vervet or rhesus40. Correlation coefficients were constructed between expression levels of genes, and a connectivity measure (topological overlap, TO) was calculated for each gene by summing the connection strength with other genes. Genes were then clustered based on their TO, and groups of coexpressed genes (modules) were identified. Each module was assigned a color, and the first principal component (eigengene) of a module was extracted from the module and considered to be representative of the gene expression profiles in a module. We identified 36 modules (Supplementary Figs. 34 and 35, Supplementary Data 6), 33 of which contained more than 10 genes in our annotation and were used for enrichment testing.
Data availability
All genomic data for the vervets sequenced in this study are available through the NCBI SRA public repositories under NCBI BioProject numbers PRJNA168521, PRJNA168472, PRJNA168520, PRJNA168527, PRJNA168522. Variant Call Format (VCF) files are available from the Dryad Digital Repository (see URLs).
Supplementary Material
Acknowledgments
Samples were collected through the UCLA Systems Biology Sample Repository funded by NIH grants R01RR016300 and R01OD010980 to N.F. For permits allowing us to collect samples, we thank the Gambia Department of Parks & Wildlife Management; Botswana Ministry of Environment & Wildlife and Tourism; Ghana Wildlife Division, Forestry Commission; Zambia Wildlife Authority; Ethiopian Wildlife Conservation Authority; Ministry of Forestry & the Environment, Department of Environmental Affairs, South Africa; Department of Economic Development and Environmental Affairs, Eastern Cape; Department of Tourism, Environmental and Economic Affairs, Free State Province; the Ezemvelo KZN Wildlife in KwaZulu Natal Province; and the Department of Economic Development, Environment and Tourism, Limpopo Province. We also thank Dr. Gene Redmond and St. Kitts Biomedical Research Foundation for facilitating sample collection in St. Kitts and Nevis. We thank Drs. Jason Brenchley, Keith Reimann (R24OD010976) and Jean Baulu and the Barbados Primate Research Center and Wildlife Reserve for providing samples from Tanzania origin and Barbados vervets. For help with sample collection and processing we thank Dr. Jennifer Danzy-Cramer, Ms. Yoon Jung, Mr. Oliver Pess Morton, and Mr. Jake Freimer. We thank Jack Kamm for discussion, Ümit Seren, Jessica Wasserscheid, and Nikoleta Juretic for IT support, and Reena Halai for help with figure design. H.S. has been supported by a travel grant of the Austrian Ministry of Science and Research. CA is supported by RO1 AI119346 from NIAID. We acknowledge the support of the NINDS Informatics Center for Neurogenetics and Neurogenomics (P30 NS062691). We would like to thank Ms. Fuying Gao for assistance with microarray data analysis.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare that there are no competing financial interests.
URLs
Vervet reference genome
https://www.ncbi.nlm.nih.gov/assembly/GCF_000409795.2
Los Alamos National Laboratory HIV Sequence Database
Vervet reference gene annotation
https://www.ncbi.nlm.nih.gov/genome/annotation_euk/Chlorocebus_sabaeus/100/
MSMC2
https://github.com/stschiff/msmc2
Human-HIV1 interaction database
https://www.ncbi.nlm.nih.gov/refseq/HIVInteractions (last access January 2016)
Vervet variant call data from this study
Author contributions
N.B.F., T.R.T., M.N., A.J.J., K.D., W.C.W., and R.K.W. conceived the study. M.N. and H.S. designed the analysis strategy. H.S. analysed the data and prepared tables and figures. C.A. contributed the SIVagm sequence analysis. G.C. contributed the WGCNA analysis. B.J. and M.M-T. provided expertise on the expression data analysis and SIV. Y.H. and V.R. provided bioinformatic support. C.A.S, J.P.G., M.A., and T.R.T collected samples and obtained permits. N.B.F., G.W., R.K.W., K.D. and W.C.W. oversaw sequencing. M.N., H.S., N.F., and A.J.J. wrote the manuscript. All authors read and approved the manuscript.
Main text references
- 1.Cheney DL, Seyfarth RM. The recognition of social alliances by vervet monkeys. Anim Behav. 1986;34:1722–1731. [Google Scholar]
- 2.Seyfarth RM, Cheney DL, Marler P. Vervet monkey alarm calls: Semantic communication in a free-ranging primate. Anim Behav. 1980;28:1070–1094. [Google Scholar]
- 3.Jasinska AJ, et al. Systems Biology of the Vervet Monkey. ILAR J. 2013;54:122–143. doi: 10.1093/ilar/ilt049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Briggs CM, et al. Live attenuated tetravalent dengue virus host range vaccine is immunogenic in African green monkeys following a single vaccination. J Virol. 2014;88:6729–6742. doi: 10.1128/JVI.00541-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsuoka Y, et al. African green monkeys recapitulate the clinical experience with replication of live attenuated pandemic influenza virus vaccine candidates. J Virol. 2014;88:8139–8152. doi: 10.1128/JVI.00425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pripuzova NS, et al. Exploring of primate models of tick-borne flaviviruses infection for evaluation of vaccines and drugs efficacy. PLoS One. 2013;8:e61094. doi: 10.1371/journal.pone.0061094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang YS, et al. Sequencing strategies and characterization of 721 vervet monkey genomes for future genetic analyses of medically relevant traits. BMC Biol. 2015;13:41. doi: 10.1186/s12915-015-0152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jasinska AJ, et al. Genetic variation and gene expression across multiple tissues and developmental stages in a non-human primate. Nature Genetics. 2017 doi: 10.1038/ng.3959. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma D, et al. SIVagm infection in wild African green monkeys from South Africa: epidemiology, natural history, and evolutionary considerations. PLoS Pathog. 2013;9:e1003011. doi: 10.1371/journal.ppat.1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma D, et al. Factors Associated with Simian Immunodeficiency Virus Transmission in a Natural African Nonhuman Primate Host in the Wild. J Virol. 2014;88:5687–5705. doi: 10.1128/JVI.03606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren WC, et al. The genome of the vervet ( Chlorocebus aethiops sabaeus ) Genome Res. 2015;4 doi: 10.1101/gr.192922.115. gr.192922.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enstam KL, Isbell LA. The guenons (genus Cercopithecus) and their allies. In: Campbell CJ, editor. Primates in Perspective. Oxford University Press; USA: 2007. pp. 252–274. [Google Scholar]
- 13.Daub JT, et al. Evidence for Polygenic Adaptation to Pathogens in the Human Genome. Mol Biol Evol. 2013;30:1544–1558. doi: 10.1093/molbev/mst080. [DOI] [PubMed] [Google Scholar]
- 14.Enard D, David E, Le C, Carina G, Petrov DA. Viruses are a dominant driver of protein adaptation in mammals. Elife. 2016:5. doi: 10.7554/eLife.12469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quach H, et al. Genetic Adaptation and Neandertal Admixture Shaped the Immune System of Human Populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haus T, et al. Mitochondrial diversity and distribution of African green monkeys (chlorocebus gray, 1870) Am J Primatol. 2013;75:350–360. doi: 10.1002/ajp.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill WCO. Primates, comparative anatomy and taxonomy. Edinburgh: Edinburgh University press; 1966. 6. Catarrhini, Cercopithecoidea, Cercopithecinae; pp. 533–581. [Google Scholar]
- 18.IUCN. [Accessed: 09/2017];IUCN Red List of threatened species. 2017 (2017). Available at: http://www.iucnredlist.org.
- 19.Leffler EM, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry GH, et al. Comparative RNA sequencing reveals substantial genetic variation in endangered primates. Genome Res. 2012;22:602–610. doi: 10.1101/gr.130468.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1, 092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez RD, et al. Demographic histories and patterns of linkage disequilibrium in Chinese and Indian rhesus macaques. Science. 2007;316:240–243. doi: 10.1126/science.1140462. [DOI] [PubMed] [Google Scholar]
- 24.Ebersberger I, Ingo E, Dirk M, Carsten S, Svante P. Genomewide Comparison of DNA Sequences between Humans and Chimpanzees. Am J Hum Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Mol Biol Evol. 2011;28:2239–2252. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patterson N, et al. Ancient Admixture in Human History. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H, Patterson N, Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. 2016. [Google Scholar]
- 31.Zhou C, Rana TM. A bimolecular mechanism of HIV-1 Tat protein interaction with RNA polymerase II transcription elongation complexes. J Mol Biol. 2002;320:925–942. doi: 10.1016/s0022-2836(02)00556-9. [DOI] [PubMed] [Google Scholar]
- 32.Siepel A, et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng K, Corcoran P. The Effects of Background and Interference Selection on Patterns of Genetic Variation in Subdivided Populations. Genetics. 2015;201:1539–1554. doi: 10.1534/genetics.115.178558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Demchak B, et al. Cytoscape: the network visualization tool for GenomeSpace workflows. F1000Res. 2014 doi: 10.12688/f1000research.4492.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ako-Adjei D, et al. HIV-1, human interaction database: current status and new features. Nucleic Acids Res. 2015;43:D566–70. doi: 10.1093/nar/gku1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapusinszky B, et al. Local Virus Extinctions following a Host Population Bottleneck. J Virol. 2015;89:8152–8161. doi: 10.1128/JVI.00671-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Müller MC, et al. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J Virol. 1993;67:1227–1235. doi: 10.1128/jvi.67.3.1227-1235.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goldstein S, et al. Plateau Levels of Viremia Correlate with the Degree of CD4 -T-Cell Loss in Simian Immunodeficiency Virus SIVagm-Infected Pigtailed Macaques: Variable Pathogenicity of Natural SIVagm Isolates. J Virol. 2005;79:5153–5162. doi: 10.1128/JVI.79.8.5153-5162.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandell DT, et al. Pathogenic features associated with increased virulence upon Simian immunodeficiency virus cross-species transmission from natural hosts. J Virol. 2014;88:6778–6792. doi: 10.1128/JVI.03785-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jacquelin B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009 doi: 10.1172/jci40093. [DOI] [PMC free article] [PubMed]
- 41.Jacquelin B, et al. Innate immune responses and rapid control of inflammation in African green monkeys treated or not with interferon-alpha during primary SIVagm infection. PLoS Pathog. 2014;10:e1004241. doi: 10.1371/journal.ppat.1004241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miyauchi K, Kim Y, Latinovic O, Morozov V, Melikyan GB. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell. 2009;137:433–444. doi: 10.1016/j.cell.2009.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barouch DH, et al. Rapid Inflammasome Activation following Mucosal SIV Infection of Rhesus Monkeys. Cell. 2016;165:656–667. doi: 10.1016/j.cell.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan XY, et al. Heat Shock Factor 1 Mediates Latent HIV Reactivation. Sci Rep. 2016;6:26294. doi: 10.1038/srep26294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klein F, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pereira LE, Johnson RP, Ansari AA. Sooty mangabeys and rhesus macaques exhibit significant divergent natural killer cell responses during both acute and chronic phases of SIV infection. Cell Immunol. 2008;254:10–19. doi: 10.1016/j.cellimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Meythaler M, et al. Early induction of polyfunctional simian immunodeficiency virus (SIV)-specific T lymphocytes and rapid disappearance of SIV from lymph nodes of sooty mangabeys during primary infection. J Immunol. 2011;186:5151–5161. doi: 10.4049/jimmunol.1004110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brenchley JM, et al. Differential infection patterns of CD4+ T cells and lymphoid tissue viral burden distinguish progressive and nonprogressive lentiviral infections. Blood. 2012;120:4172–4181. doi: 10.1182/blood-2012-06-437608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang R, et al. Envelope-specific B-cell populations in African green monkeys chronically infected with simian immunodeficiency virus. Nat Commun. 2016;7:12131. doi: 10.1038/ncomms12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Predicala R, Zhou Y. The role of Ran-binding protein 3 during influenza A virus replication. J Gen Virol. 2013;94:977–984. doi: 10.1099/vir.0.049395-0. [DOI] [PubMed] [Google Scholar]
- 52.Hakata Y, Yamada M, Shida H. A multifunctional domain in human CRM1 (exportin 1) mediates RanBP3 binding and multimerization of human T-cell leukemia virus type 1 Rex protein. Mol Cell Biol. 2003;23:8751–8761. doi: 10.1128/MCB.23.23.8751-8761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langer K, et al. Insights into the Function of the CRM1 Cofactor RanBP3 from the Structure of Its Ran-Binding Domain. PLoS One. 2011;6:e17011. doi: 10.1371/journal.pone.0017011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shida H, Hisatoshi S. Role of Nucleocytoplasmic RNA Transport during the Life Cycle of Retroviruses. Front Microbiol. 2012:3. doi: 10.3389/fmicb.2012.00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Messam CA, Jean H, Gronostajski RM, Major EO. Lineage pathway of human brain progenitor cells identified by JC virus susceptibility. Ann Neurol. 2003;53:636–646. doi: 10.1002/ana.10523. [DOI] [PubMed] [Google Scholar]
- 56.Traut W, Fanning E. Sequence-specific interactions between a cellular DNA-binding protein and the simian virus 40 origin of DNA replication. Mol Cell Biol. 1988;8:903–911. doi: 10.1128/mcb.8.2.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaliyaperumal S, et al. Frequent infection of neurons by SV40 virus in SIV-infected macaque monkeys with progressive multifocal leukoencephalopathy and meningoencephalitis. Am J Pathol. 2013;183:1910–1917. doi: 10.1016/j.ajpath.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferenczy MW, et al. Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus-induced demyelinating disease of the human brain. Clin Microbiol Rev. 2012;25:471–506. doi: 10.1128/CMR.05031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pfeifer SP. The Demographic and Adaptive History of the African Green Monkey. Mol Biol Evol. 2017;34:1055–1065. doi: 10.1093/molbev/msx056. [DOI] [PubMed] [Google Scholar]
- 60.Romiguier J, et al. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature. 2014;515:261–263. doi: 10.1038/nature13685. [DOI] [PubMed] [Google Scholar]
- 61.Novikova PY, et al. Sequencing of the genus Arabidopsis identifies a complex history of nonbifurcating speciation and abundant trans-specific polymorphism. Nat Genet. 2016;48:1077–1082. doi: 10.1038/ng.3617. [DOI] [PubMed] [Google Scholar]
- 62.Mallet J, Besansky N, Hahn MW. How reticulated are species? Bioessays. 2016;38:140–149. doi: 10.1002/bies.201500149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Matsubayashi K, Hirai M, Watanabe T, Ohkura Y, Nozawa K. A case of patas-vervet hybrid in captivity. Primates. 1978;19:785–793. [Google Scholar]
- 64.de Jong YA, Butynski TM. Three Sykes’s Monkey Cercopithecus mitis × Vervet Monkey Chlorocebus pygerythrus Hybrids in Kenya. Primate Conserv. 2010;25:43–56. [Google Scholar]
- 65.Danko CG, et al. Natural Selection has Shaped Coding and Non-coding Transcription in Primate CD4+ T-cells. 2016 doi: 10.1101/083212. [DOI] [Google Scholar]
- 66.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013 arXiv [q-bio.GN]
- 67.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Auwera GA, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kurtz S, et al. Versatile and open software for comparing large genomes. Genome Biol. 2004;5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duforet-Frebourg N, Bazin E, Blum MGB. Genome scans for detecting footprints of local adaptation using a Bayesian factor model. Mol Biol Evol. 2014;31:2483–2495. doi: 10.1093/molbev/msu182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weir BS, Clark Cockerham C. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 72.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 75.Swofford DL, Jack S. Phylogeny inference based on parsimony and other methods using PAUP. The Phylogenetic Handbook. :267–312.
- 76.Posada D, Crandall KA. Selecting the best-fit model of nucleotide substitution. Syst Biol. 2001;50:580–601. [PubMed] [Google Scholar]
- 77.Browning BL, Browning SR. Genotype Imputation with Millions of Reference Samples. Am J Hum Genet. 2016;98:116–126. doi: 10.1016/j.ajhg.2015.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Delaneau O, Howie B, Cox AJ, Zagury JF, Marchini J. Haplotype estimation using sequencing reads. Am J Hum Genet. 2013;93:687–696. doi: 10.1016/j.ajhg.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All genomic data for the vervets sequenced in this study are available through the NCBI SRA public repositories under NCBI BioProject numbers PRJNA168521, PRJNA168472, PRJNA168520, PRJNA168527, PRJNA168522. Variant Call Format (VCF) files are available from the Dryad Digital Repository (see URLs).