Abstract
When a constraint is removed, confluent cells migrate directionally into the available space. How the migration directionality and speed increase are initiated at the leading edge and propagate into neighboring cells are not well understood. Using a quantitative visualization technique—Particle Image Velocimetry (PIV)—we revealed that migration directionality and speed had strikingly different dynamics. Migration directionality increases as a wave propagating from the leading edge into the cell sheet, while the increase in cell migration speed is maintained only at the leading edge. The overall directionality steadily increases with time as cells migrate into the cell-free space, but migration speed remains largely the same. A particle-based compass (PBC) model suggests cellular interplay (which depends on cell–cell distance) and migration speed are sufficient to capture the dynamics of migration directionality revealed experimentally. Extracellular Ca2+ regulated both migration speed and directionality, but in a significantly different way, suggested by the correlation between directionality and speed only in some dynamic ranges. Our experimental and modeling results reveal distinct directionality and speed dynamics in collective migration, and these factors can be regulated by extracellular Ca2+ through cellular interplay. Quantitative visualization using PIV and our PBC model thus provide a powerful approach to dissect the mechanisms of collective cell migration.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-017-2553-6) contains supplementary material, which is available to authorized users.
Keywords: Wound healing, Cell contractility, PDMS, Corneal epithelial cell, Cell communication, Blebbistatin
Introduction
Directionality and speed are two critical parameters in cell migration. When cells migrate in isolation, speed and directionality are regulated by different mechanisms, which orchestrate distinct cellular migratory behavior in different types of cells [1–7]. For example, Dictyostelium cells and neutrophil-like cells immobilized with latrunculin, which sequesters actin monomers and thus leads to degradation of actin filaments and decreases cell speed, are still capable of sensing chemoattractant gradients and establishing directionality [1].
Cells migrate collectively in wound healing, embryo development, tissue regeneration, and cancer metastasis [8]. How directionality and speed are regulated in collective migration is not well understood. Collective cell migration is not just simply the sum of the migration of a large group of individual cells. Collectively, cells migrate more efficiently in response to many directional cues than cells which migrate separately [9–12]. A cellular interplay has been proposed as the mechanism that underlies the increased efficiency in collective migration [8, 13]. This interplay may include biochemical and mechanical interactions such as propelling forces transmitted through cell–cell contacts [14, 15], contact-dependent cell polarity [11], adherens junction treadmilling [16], contact inhibition of locomotion [12, 17], and secreted molecules [18].
Monolayer wound healing assays are widely used in the study of collective cell migration. The barrier model allows cells to become confluent next to a barrier [19]. Cells migrate directionally toward the cell-free surface after removing the barrier. Advantages of the barrier-removal assay include that the cells at the edge are not damaged as in the scratch assay, and that the cells move over a surface on which the substratum is not affected by the scratching process [14, 15, 19–24]. Direction cues in this system may include space availability, population pressure, contact inhibition of locomotion, and activation of EGFR [25, 26].
Particle image velocimetry (PIV) is a cross-correlation technique initially developed in the field of hydrodynamics, which has been proven to be a useful tool for characterizing local displacements and has been used to study velocity dynamics in collective cell migration [27–30]. To investigate the transmission of directional movement signals from the free edge into a large sheet of corneal epithelial cells, we used PIV to quantitatively analyze and visualize collective cell migration with the detailed distinction between directionality and speed. Our results reveal remarkable distinctions between directionality and speed dynamics during collective migration of an epithelial cell confluent culture.
To investigate how cellular interplay may regulate migration directionality and speed in collective migration, we developed a particle-based compass (PBC) mathematical model. The key parameter for cellular interplay in this PBC model is the particle–particle distance (i.e., cell–cell distance). Following suggestions from the model, we experimentally tested the effects of extracellular Ca2+ on collective migration. We chose Ca2+ because early in the wound healing process, the concentration of Ca2+ in the wound fluid changes [31] and Ca2+ plays a significant role in membrane protrusion and cell–cell adhesion [32–34], which presumably underlie cellular interplay. Indeed, we find that Ca2+ plays different roles in regulating directionality and speed changes in collective migration of corneal epithelial cells.
Materials and methods
Reagents and cell line
Telomerase-immortalized human corneal epithelial cells (hTCEpi) were cultured at 37 °C, 5% CO2 in EpiLife medium containing 60 µM Ca2+ (Life Technologies, USA) supplemented with an EpiLife defined growth supplement (EDGS, Life Technologies, Grand Island, USA) and 1% (v/v) penicillin/streptomycin (Life Technologies). For Ca2+ intervention groups, 1 h before imaging, cell culture medium was switched to medium with high or low Ca2+. For low-Ca2+ experiments, 1 mM EGTA or 2 mM EGTA was added. For high-Ca2+ experiments, the medium was supplemented with an additional 60 µM or 120 µM Ca2+. For cell contractility intervention groups, 50 µM blebbistatin (a selective membrane-permeant inhibitor for non-muscle myosin II ATPase) was added 2 h before imaging. For the relative control group, DMSO (dimethyl sulfoxide) (1 µl/ml) was added. EGTA, CaCl2, DMSO, and blebbistatin were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Wound model
Before seeding the cells, PDMS strips were deposited on the surface of a six-well cell culture dish pre-coated with FNC (an aqueous solution of fibronectin and other cell adhesion proteins). 16–18 h after seeding, a confluent monolayer of cells was formed. Before image recording, the PDMS barrier was lifted with sterile tweezers to create a space next to the monolayer, and a clear and straight edge was left behind.
Time-lapse microscopy
Migration of cells from the edge of the wound was imaged by phase-contrast microscopy using an inverted microscope (Carl Zeiss, Oberkochen, Germany) equipped with a motorized stage, a specialized time-lapse imaging software (Metamorph NX; Molecular Device, Sunnyvale, USA) and a Carl Zeiss incubation system. A regular 10× objective lens was used for microscopy. Imaging began 10 min after wounding. The interval between image acquisitions was 5 min, and a typical experiment lasted for 6 h. To capture cell behavior over scales up to 0.5 mm behind the free edge, two images that covered the area of cell culture were acquired at each time point and were stitched together using Image J software from the National Institutes of Health (http://rsbweb.nih.gov/ij/).
Individual cell tracking
ImageJ software (MTrackJ) was used to quantify migration directionality and speed. Monolayer boundaries were computed by tracking ten points on the edge of the wound and computing an average to track the edge. For analysis of region dependence in cell migration during wound healing, we divided the first 15 rows of cells from the free edge into rows of 5 cells and defined them as the leading region, transition region, and trailing region, as illustrated in Supplementary Fig. S2a. The position of a cell was defined by its nuclear centroid. For each region, more than 70 cells in the monolayer were tracked at 10-min intervals. Directionality was defined as the cosine of the angle between cell trajectory and a line perpendicular to the free edge. A cell migrating directly toward the free surface (i.e., perpendicular to the free edge) would have a cosine equal to 1. Cells migrating directionless and randomly would have an average cosine equal to ~0. Cell migration speed is the accumulated migrated distance divided by time.
To analyze changes in cell density in collective cell migration, we quantified cell numbers in six regions of 160 µm width each, with the initial edge of cell sheets in the middle (Fig. S8a). Then we counted the cell numbers at 0, 3, and 6 h. Cell density was computed by dividing the cell number in each region by the area.
PIV migration analysis
Cell migration in the time-lapse images was quantified by PIV analysis using MATLAB (MathWorks) with a custom MATLAB code based on MatPIV1.6.1, a freeware distributed under the terms of the GNU general public license. The MATLAB code has been previously described in detail [35]. Kymographs were used to quantify and visualize spatiotemporal dynamics of horizontal velocity, directionality, and speed from the PIV measurements. For each data matrix from the PIV analysis, we computed the average value for each column parallel to the free edge and then derived a one-dimensional segment for each time point (Supplementary Fig. S4b). We defined the front of the directionality wave in the kymograph as the first grid of at least seven consecutive grids that reached a directionality threshold of cos(θ) ≥ 0.2588. Then we plotted the front of the directionality wave averaged over a defined time interval against each time point to obtain the best linear fit using the fitting equation y = ax + b, where x is the wave front position, y is the time, a is the slope of the linear fit, and b is the intercept of the linear fit. The propagation rate of the directionality wave was calculated as R = 1/a. (Supplementary Fig. S7) (please see the Supplementary Material for details).
PBC model
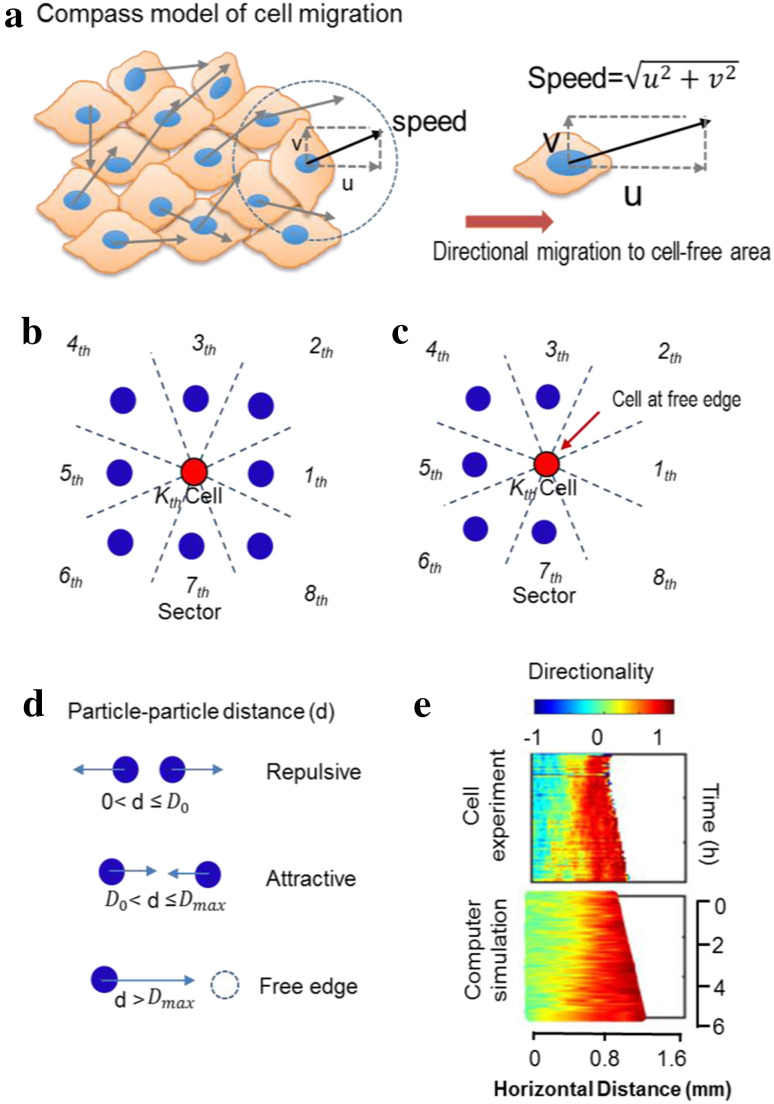
We developed a physical model based on wound healing assay experiments. All the length parameters were scaled such that 0.1 in model length units equaled 20 μm in real space (equivalent to the diameter of a single cell). The speed for the control group was set as 0.015 model length units (3 µm in real space) per simulation time step (4.8 min in real time), which is equivalent to the average speed of 37.3 µm/h measured in control group experiments. The space surrounding the center of a specific cell was divided into eight equal sectors (Fig. 2b, c). In our model, all cells were treated as particles with a diameter of 0.1. We assume that if the distance d between a specific cell and its nearest neighboring cell in a particular angular sector is less than the repulsive threshold distance D 0, the cell will have a repulsive interaction with the neighboring cell. The magnitude of this repulsive interaction is inversely proportional to d. If d is greater than the free edge threshold distance D max, this specific cell is considered a free edge cell and will gain a contribution to its motion direction toward the free space. If d is between D 0 and D max, a biased directionality of the specific cell toward the nearest neighboring cell is modeled. The magnitude of this biased directionality is directly proportional to d (Fig. 2d). This mechanism is applied to compute the cellular interplay in each of the eight angular sectors. By summing up the eight cell–cell interactions, the total cellular interplay vector for a specific cell can be obtained and the phase of this vector will be used to determine the cell migration angle. This algorithm is used to simulate collective cell migration in the experimental wound healing assay. In the control group simulations, D 0 is set at 0.12 (24 µm in real space) and D max is set at 0.30 (60 µm in real space) to best match the experimental data. More details of the model construction and numerical simulations are provided in the Supplementary Material.
Fig. 2.
PBC model to simulate collective migration. a Schematic of the model, which sets the speed of individual cells as particles and adjusts directionality depending on interactions with neighboring cells. b–d A particle representing a cell (red dot) at the free edge has three vacant neighbor sectors out of eight, which biases the direction of migration with the principles in d. The total cellular interplay for the kth cell is the sum of the eight interactions. Particle–particle distance d determines the cellular interactions, where D 0 and D max denote the threshold of repulsive and the threshold of the free surface effect. When 0 < d≤D 0, the particle is repulsed; when D 0 < d≤ D max, the particle is attracted; and when d > D max the cell feels a contribution from the free edge. e A kymograph from the model shows a pattern of directionality propagation similar to the experimental results. Computer simulation with repulsive threshold D 0 = 0.12 (24 µm) and average speed = 37.3 µm/h
Statistics
Data analyses, graphs, and statistical calculations were performed using Excel (Microsoft) and MATLAB (MathWorks). Data are presented as mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). Differences between conditions were compared using Student’s t test. The difference was considered statistically significant if p < 0.05.
Results
Outward collective migration was initiated from the free edge
To induce directional collective migration, we cultured human corneal epithelial cells against a polydimethylsiloxane (PDMS) barrier to confluence at ~18 h after seeding the cells. Upon removal of the barrier, cells migrated into the cell-free areas as previously reported with increases in the number of cells in the imaging field, decreases in cell-free area, and advancement of the leading edge cells into the cell-free area (Supplementary Fig. S1, Supplementary Video S1) [36]. Because of the short time period of imaging (3–6 h), increased cell numbers in the cell-free area were mainly due to cell migration and not cell proliferation. Examination of time-lapse videos showed that less than 2% of cells divided during this period.
Directionality waves propagated into the cell sheet
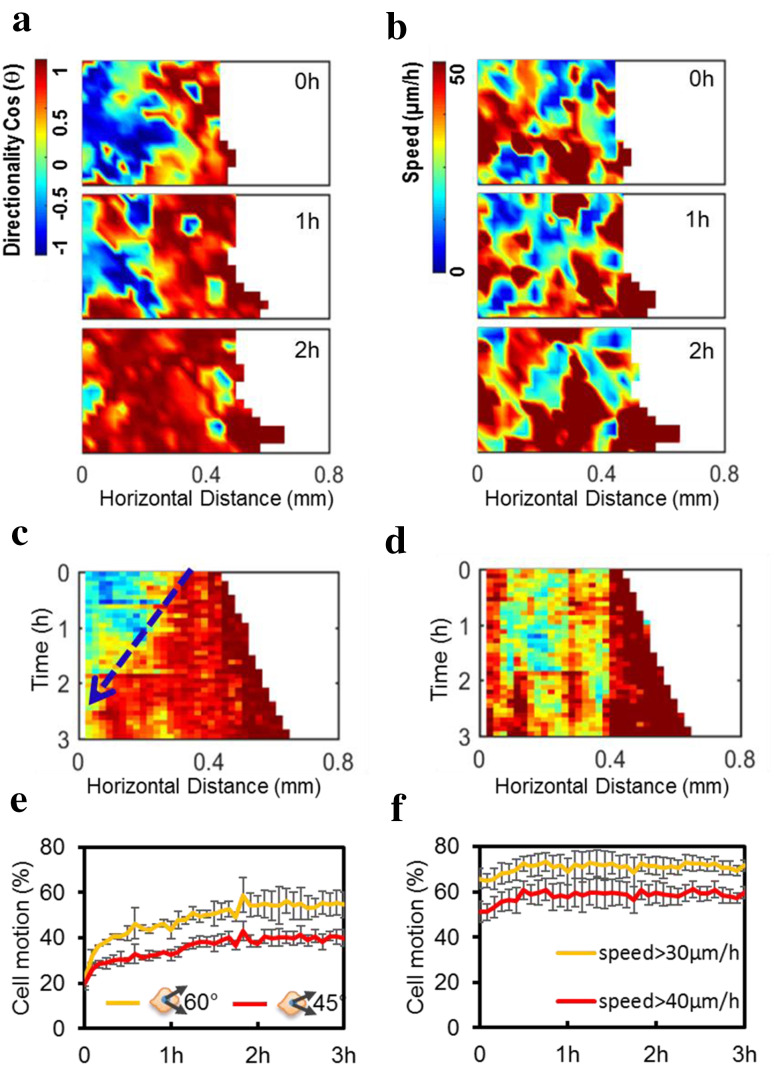
To visualize changes in directionality and speed of cell migration following free edge generation, we took advantage of PIV, which provides quantitative visualization of directionality and speed of the mass movement of whole cell groups. Remarkably, the directionality of cell movement into the cell-free area (X-directionality) first increased at the leading edge and then propagated into neighboring confluent cells (Fig. 1a; Supplementary Video S2), while changes in the speed of cell movement (i.e., cell migration speed) did not show propagation from the free edge into the following cells (Fig. 1b; Supplementary Video S2).
Fig. 1.
Free edge induced a wave-like propagation of directionality but not the speed in confluent epithelial cells. a Heat maps of cell migration directionality at different time points show the increase in directionality as a wave propagating from the leading cells into the following cells after removal of the PDMS barrier. “Hot” colors like red show migration to the right, into the free space. “Cold” colors like blue show migration to the left, away from the free space. b Heat maps of cell migration speed, however, lack a similar wave propagation from the leading cells into the following cells. c Kymograph showing an increase in directionality propagating from the leading edge into the cell sheet along time (blue arrow); by 3 h, all the cells migrated directionally to the right (coded red). d Kymograph coding migration speed shows that speed increased at the leading edge, but did not propagate into the cell sheet. e, f After PDMS barrier removal, cells showed increased directionality: the proportion of cell motion directed to the free edge within a bias angle less than 60° or 45° increased over time (e), while the fraction of cells with a speed over 30 or 40 µm/h remained stable (f). Directionality and speed of cell migration were calculated using PIV (see “Materials and methods”). Color indicates the value of cosine theta (directionality) from −1 to 1 (a) or the cell speed from 0 to 50 µm/h (b). Data are presented as mean ± SD from three independent experiments
Kymographs of directionality and speed of migration confirmed the contrast between these two migration parameters. Increases in directionality at the free edge propagated into the cell sheet over time as a wave (blue arrow in Fig. 1c). Changes in cell migration speed, however, did not show a similar propagation (Fig. 1d). The fraction of cell motion directed to the free edge within a bias angle less than 60° or 45° increased over time (Fig. 1e), while the speed remained stable (Fig. 1f). The directionality wave propagated into the cell sheet with a speed significantly higher than the speed at which the free edge advanced into the cell-free area (49.74 ± 33 vs. 34.66 ± 6.55 µm/h, p < 0.05).
Tracking migration of individual cells validated PIV results
We then manually tracked cells to validate the results with those obtained using PIV analysis (Supplementary Fig. S2). We divided the first 15 rows of cells from the free edge into three regions which are 0–160, 160–320, and 320–480 µm from the free edge (Supplementary Fig. S2a). Migration trajectories of individual cells in each group were plotted with the initial position at the origin (Supplementary Fig. S2b–d). Cell migration showed gradually increased directionality starting from the leading edge: cells in the 0–160 µm region responded first with steady displacement into the cell-free area while cells in the 160–320 µm region (about five cell rows from the free edge) took 1–2 h to show directional displacement. Cells in the 320–480-μm region took even longer (2–3 h) to show directional cell migration (Supplementary Fig. S2b–e).
From the trajectory data, we further analyzed migration directionality and speed in each region. The directionality of the 0–160-, 160–320-, and 320–480-µm regions are significantly different in the first hour after barrier removal. Cells in the 0–160-µm region have significantly higher directionality into the cell-free area (perpendicular to the free edge) compared to cells in the other two regions. This difference gradually reduced as the cells in the 160–320- and 320–480-µm regions began to follow the leading edge (Supplementary Fig. S3a, b). The mean speed of the cells in all three regions remained the same (Supplementary Fig. S3c, d). Together with the speed and directionality measurements, these data indicated that directional guidance cues from the free edge extended gradually into the cell sheet in a spatial–temporal-dependent manner, but that the speed remained relatively stable. Manual tracking analysis confirmed the results from PIV analysis. A similar propagation pattern in the directional velocity was evident when the velocity of the movement into the cell-free area was visualized over time (Supplemental Fig. S4a, c), which was similar to previous reports of the leading edge movement in studies without detailed and separate analysis of directionality and speed.
A PBC model reproduced the directionality propagation
To study how the migration directionality and speed are differentially regulated in collective cell migration, we developed a two-dimensional PBC model (please see Supplementary Material for details). Briefly, cells in a group tend to migrate into cell-free space (Fig. 2a). As cells at the free edge migrate, they move away from the cells behind, and experience reduced cell–cell contact inhibition (modeled as a repulsive interaction), as well as biased migration of the cells toward the free space (modeled as an attractive interaction). Cells farther behind are then biased to migrate toward the cell-free area. Thus, in this model, we simplified cellular interplay to one factor, i.e., the particle–particle distance, which is mathematically defined as the distance between a specific cell and its immediately adjacent cells in each of the eight equally spaced angular sectors in a 2D plane (centered at the location of the specific cell) (Fig. 2b, c). Varying the threshold cell–cell distance parameters, i.e., D 0 for the transition from repulsive to attractive interaction and D max for the transition from the distance-dependent attractive interaction to the free edge biased migration, mediates the dynamics of cell migration directionality (Fig. 2d). We set cell speed to be a constant plus noise in the model, because our experiments showed stable migration speeds (Fig. 1f).
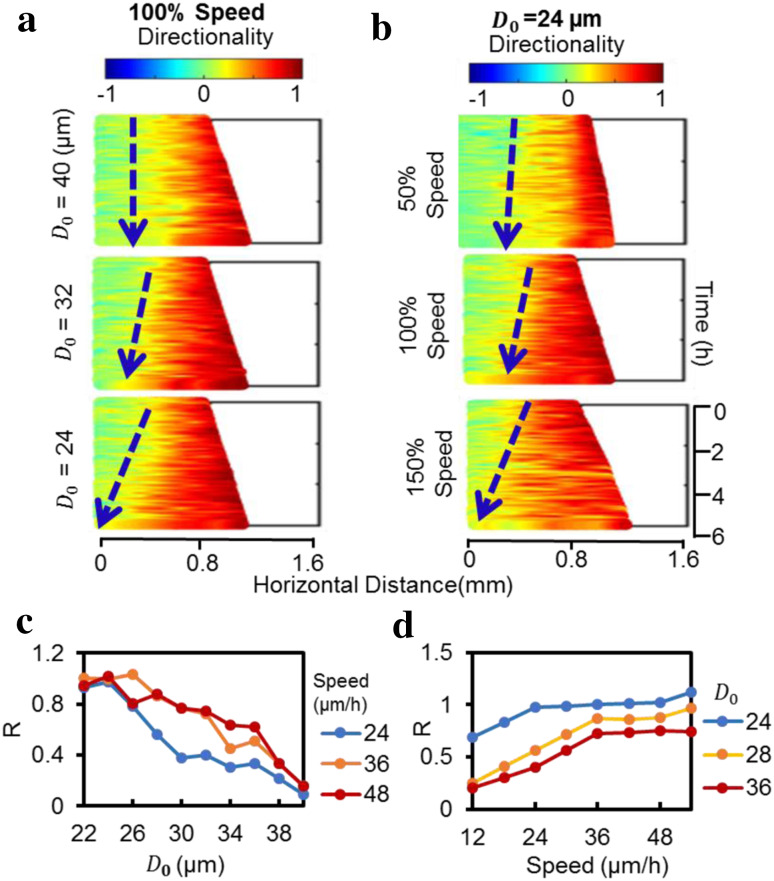
Computer simulations based on this PBC model showed similar pattern of directional collective migration as cell culture experiments. Cells close to the free edge migrate directionally first and the cells behind follow in a time-dependent manner (Supplementary Video S3). Kymographs of directionality from the simulation show similar patterns to that from the experiment (Fig. 2e). We used D 0 and migration speed as two major model parameters. In initial simulations, we set the migration speed constant according to cell culture experiments (Fig. 1f) and varied the repulsive interaction threshold D 0. D 0 is crucial to determine the propagation rate of the directionality wave (Fig. 3a, c). Holding D 0 constant and varying migration speed also affected directionality wave propagation (Fig. 3b, d). However, it is worth noting that the directionality propagation rate is less sensitive to the speed when the speed is over a certain threshold value at a defined D 0 (Fig. 3d).
Fig. 3.
Computer simulations suggested D 0 and speed as two major parameters that affect directionality wave propagation. a Effect of repulsive threshold D 0 (24, 36 or 40 µm) on directionality wave propagation. Speed is set at 37.3 µm/h. D 0 is the threshold of repulsive interactions. The value ranges from 24 to 40 µm. Decreasing D 0 accelerated wave propagation. b Effect of speed (50, 100, 150% of the control speed of 37.3 µm/h) on directionality wave propagation. Repulsive threshold D 0 = 24 µm. The value ranges from 0 to 150% (12–55 µm/h). Increasing speed accelerates wave propagation. c, d The rate of directionality wave propagation depends on D 0 and particle speed. R is the normalized rate of directionality wave propagation. R was set as 1 when D 0 = 24 µm and speed = 37.3 µm/h. D 0 negatively correlated with the rate, whereas the rate was positively correlated with speed
Different roles of Ca2+ in propagation of speed and directionality waves
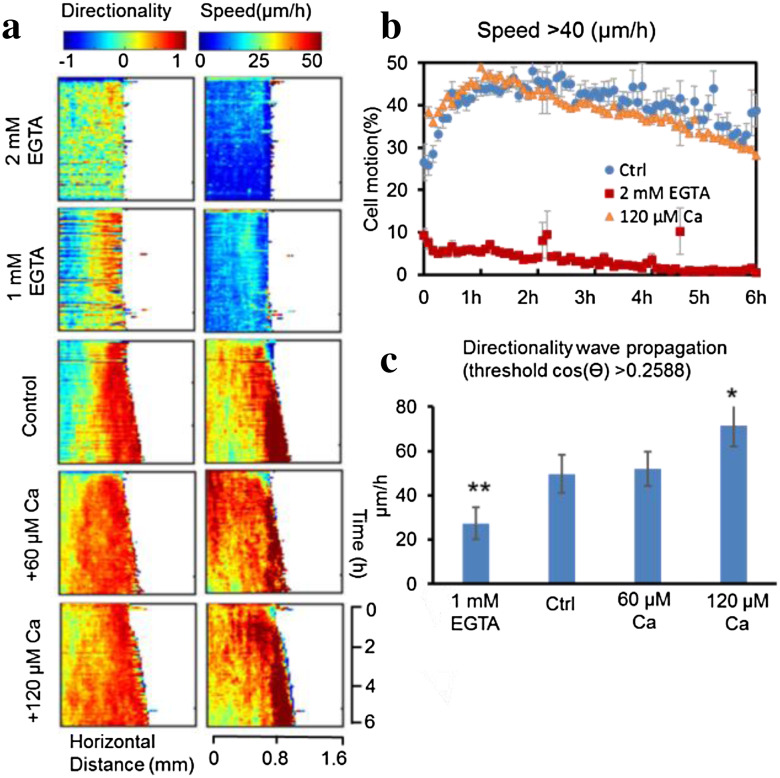
To examine mechanisms of directionality wave propagation, we manipulated extracellular Ca2+. EGTA, a Ca2+ chelator, significantly slowed down cell migration (Fig. 4a, b; Supplementary Fig. S5a, b, Video S4, 5). EGTA treatment at both 1 mM and 2 mM reduced migration speed to a similar level (Fig. 4a, b). However, the effects of 1 and 2 mM EGTA on directionality were different in that 2 mM EGTA almost abolished migration directionality, while cells in 1 mM EGTA maintained obvious directionality (Fig. 4a).
Fig. 4.
Calcium regulates the propagation of directionality waves. a Extracellular Ca2+ significantly affects directionality and speed for cell migration. Kymographs show that Ca2+ had different effects on directionality wave propagation and speed. b High Ca2+ did not affect migration speed. The graph shows the time course of the proportion of cells in the sheet reaching a speed >40 µm/h for control, EGTA 2 mM, calcium 120 µM groups. c High extracellular Ca2+ increased, while depletion of Ca2+ decreased, the rate of directionality wave propagation. *p < 0.05, **p < 0.01 when compared with the control value using a Student’s t test. Data of the proportion of cells reaching a certain threshold over time are mean ± SEM from three independent experiments. Data on wave propagation rates are mean ± SEM from four independent wounds
Addition of higher Ca2+ to 60 or 120 µM did not increase the average speed of cell migration (Fig. 4a, b; Supplementary Fig. S6a, b, Video S4, 6), however, the rate of directionality wave propagation was significantly elevated (Fig. 4a, c). The fraction of cell motion in the sheet with a bias angle less than 60° in the control, EGTA 2 mM, Ca2+ 120 µM groups confirmed these differences in directionality propagation (Supplementary Fig. S6c). The time course of the fraction of cell motion in the sheet with migration speed 40 µm/h above for control, EGTA 2 mM, and high-Ca2+ groups also confirmed the effect on migration speed (Fig. 4b). These same experiments also showed that the directionality wave propagated faster across the monolayer when Ca2+ was increased (Fig. 4c).
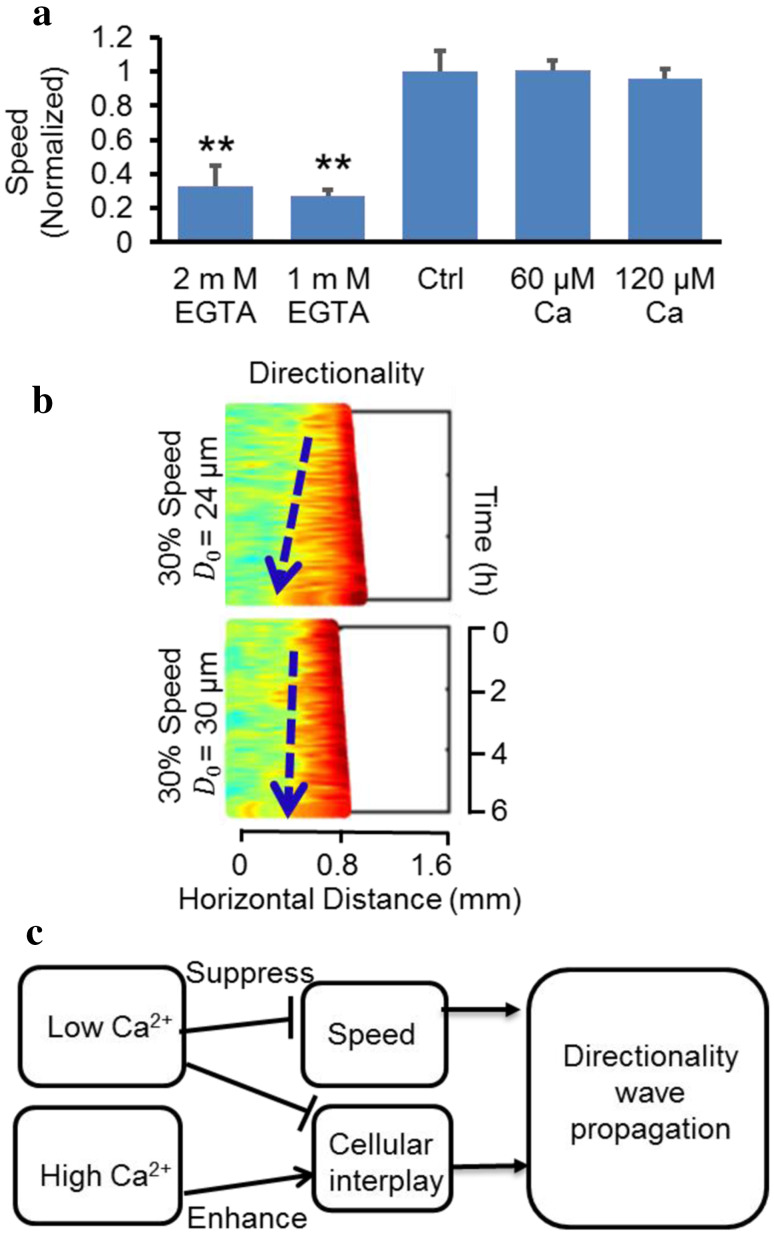
We then investigated how Ca2+ regulates propagation of the directionality wave—whether it is through its effect on migration speed, on the interaction threshold (D 0), or on both. In the PBC model, two major parameters that affected directionality wave propagation were speed and the repulsive action threshold, D 0. In high-Ca2+ experiments, there was no apparent change in cell speed (Fig. 5a), thus D 0 must be decreased to cause an increase in the propagation rate of directionality wave (Fig. 5c). EGTA treatment decreased the speed of cell migration, which reduced the directionality wave propagation. The average speed of cell migration was significantly reduced after EGTA treatment: 27.47 and 32.92% of that in the control value in 1 and 2 mM EGTA, respectively (Fig. 5a). We simulated cells using 30% of the control group cell speed but with the same D 0 as in the control group. The simulation results suggested that simply reducing the cell speed to 30% was insufficient to inhibit the directionality wave propagation (Fig. 5b). An increase in D 0 along with reduced cell speed produced a directionality wave pattern consistent with the experimental results (Fig. 5b), suggesting that EGTA treatment affected both cell speed and cellular interplay, which in combination inhibited directionality wave propagation. In high-Ca2+ simulations, a change in D 0 alone was able to recapitulate experimental observations of directionality wave propagation (Fig. 5c).
Fig. 5.
Hypothetical effects of Ca2+ on the propagation of directionality wave through speed and cellular interplay. a Depletion of Ca2+ reduced migration speed to 30% of the control. The average speed of the control group is normalized to 1, and average speed of EGTA groups is about 0.3. b Simulation results from the simulation model indicate that when cell speed is tuned down to 30% of the control value with other parameters unchanged, the directionality wave keeps propagating for a considerable distance. A combination adjustment of speed and D 0 reproduces a kymograph similar to the EGTA experimental data (Fig. 4a). c The schematic diagram shows how Ca2+ affects directionality waves through cell speed and cellular interplay. Low Ca2+ reduces speed and cellular interplay (increasing D 0), and thus decreases directionality wave propagation. High Ca2+ increases cellular interplay (decreasing D 0) and enhances directionality wave propagation with little effect on the speed. Data are shown as mean ± SEM from three independent experiments. *p < 0.05, **p < 0.01
Directionality wave propagation is independent of cell contractility
To investigate the role of myosin in directionality wave propagation, we used blebbistatin, an inhibitor for non-muscle myosin II ATPase. After peeling off the PDMS barrier, both the blebbistatin-treated group and the control group migrated directionally toward the cell-free area. Blebbistatin-treated cells became elongated with an extend cell body dragging behind as previously reported [37–39] (Supplementary Video S7). No significant difference was found in the distance between cells (cell density) along the x-axis (Fig. S8b), advancement of the leading edge into the cell-free area (Fig. S8c), directionality wave propagation (Fig. S8d), time evolution of directionality (Fig. S8e), and cell migration speed (Fig. S8f) between the blebbistatin-treated group and the control group.
Discussion
In this study, we used PIV-based techniques to reveal different dynamics in directionality and speed in collective cell migration. In confluent human corneal epithelial cells, a directionality wave was initiated at the leading edge when constraint was removed, and the directionality wave propagated backward into the cell sheet while migration speed remained unchanged. PBC modeling suggested two parameters—cell–cell distance and migration speed—could control the rate of the directionality wave propagation. Experimental tests of this prediction using Ca2+ showed that changes in “cellular interplay” and cell migration speed can regulate directionality wave propagation.
PIV analysis of cell movement relies on basic pattern matching instead of tracking individual cells to quantify cell movement. No labeling is required and many types of microscope images meet the requirements for the analysis. PIV provides significantly detailed parameters for collective cell migration, including directionality and speed in all directions over the whole cell area. These parameters can be analyzed and, importantly, visualized with high spatial and temporal resolution over a large size of field [40]. Traditional manual tracking of individual cells confirmed the reliability of PIV analysis. Thus, PIV has been widely used in quantify and visualize collective cell migration [14, 27–30]. Visualization of cell sheet movement using PIV revealed striking propagation of a directionality wave from the leading edge cells to the inside of the cell sheet (Fig. 1a, c), whereas the increase in migration speed was localized to cells at the leading edge (Fig. 1b, d). In other words, the directionality signal can be transmitted independently of changes in cell migration speed. In both chemotaxis and galvanotaxis of isolated cells, speed and directionality of migration have been suggested to have different control mechanisms [5–7]. Systematic screening of the signaling pathways involved in the collective migration of endothelial cells has revealed the molecular basis for independent functional modules, such as proliferation, cell migration speed, directed migration, and cell–cell coordination [41]. Quantitative visualization of directionality and speed in collective cell migration will facilitate understanding of these mechanisms. Some aspects have been reported before in different context [24, 39, 42]. For example, previous work found that directionality waves keep propagating for some time, even after the two advancing edges of cell groups meet, which suggests that the free edge is necessary for directional migration of the cells at edge, but not essential for propagation of the directionality wave [24].
Multiple models of collective cell migration have been reported to simulate force transmission between cells [14], the generation of finger cells [43, 44], the advancement of free edges [45] and other features of collective migration, but little has been done on cell migration velocity evolution in wound healing. Similar particle-based modeling has been used to simulate collective migration [43, 46]. Sepulveda et al. have elegantly demonstrated cell motion at early stages after barrier removal, as well as the formation of leader cells and fingers at the free edge [43]. Long-term effects of barrier removal and the cell migration dynamics behind free edge, however, have not been investigated. On the other hand, compass modeling has been used for simulating cell polarity/asymmetry and planar cell polarity signaling in isolated individual cells [47–50]. Our PBC model for collective cell migration integrates directional cues from neighboring angular sectors. This model simplifies the complex cellular interactions and provides an approximated approach to capture directionality wave propagation. It offers a quick in silico test for the key parameters that may affect collective cell migration (Figs. 2, 3).
For this particular corneal epithelial cell line, we observed that some cells at the edge migrated away from the group. We also noticed that neighboring cells did not attach by forming tight cell–cell junctions. Nonetheless, the robust transition of directionality persists both experimentally and in simulations, which suggests that cell–cell mechanical coupling is not essential for transmission of directional signals, although it has been suggested to be required for collective migration of MDCK cells [14].
Extracellular Ca2+ regulates many aspects of cell migration including polarity [34, 51–54], migration speed [34, 55–57], cell–cell adhesion (e.g., cadherin-based adhesions) [51, 58], and membrane protrusion. In collective cell migration, additional Ca2+ promoted directionality wave propagation. The propagation rate of directionality waves was significantly higher than that of the control group (Fig. 4a, c), whereas migration speed in high-Ca2+ conditions was the same as the control group, suggesting that high-Ca2+-accelerated directionality wave propagation was independent of cell migration speed. Simulations indicated that the reduction in speed could not fully account for inhibition of the directionality wave propagation (Fig. 5b, c). This is supported by experiments which showed that only 120 µM Ca2+ significantly increased the directionality wave propagation, whereas cells in both 60 and 120 µM Ca2+ had the same speed (Fig. 5a). Low-Ca2+ levels affected cell migration speed (Fig. 4b) and also affected the collective directional response, presumably through cellular interplay. These results provide further evidence for the separation of directionality and speed in collective cell migration and suggest regulation by Ca2+, which warrants further investigation to elucidate downstream mechanisms underlying different regulation of directionality and speed of collective migration by Ca2+.
In summary, PIV analysis revealed that a free edge in corneal epithelial monolayer induced a directionality wave of cell migration propagating from the leading edge over several hundred microns into the following confluent cells, whereas the increase in cell migration speed was limited to the leading edge cells. The directionality wave was differentially regulated by extracellular Ca2+, presumably through cellular interplay and migration speed.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by NIH EY019101(to. M.Z.) and AFOSR FA9550-16-1-0052 (to M.Z.). This study was supported in part by the Major Program Grant of Zhejiang Provincial Science and Technology (No. 2012C03007-6) (to Z.X.), NIH GM 68952 (to A.M.), an Unrestricted Grant from Research to Prevent Blindness, Inc. (to M.Z.), and an NEI core grant (to M.Z.). We thank Dr. James Jester (UC Irvine), Dr. Vijay Krishna Raghunathan and Dr. Christopher J. Murphy (UC Davis) for the generous gift of the hTCEpi cell, Brian Reid (UC Davis) for English editing and proofreading. Y.Z. is supported by a fellowship from the China Scholarship Council. F. Lin thanks the support from a Collaborative Research Travel Grant provided by the Burroughs Wellcome Fund.
Abbreviations
- PBC
Particle-based compass
- PDMS
Polydimethylsiloxane
- PIV
Particle image velocimetry
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Francis Lin, Email: flin@physics.umanitoba.ca.
Zhengping Xu, Email: zpxu@zju.edu.cn.
Min Zhao, Email: minzhao@ucdavis.edu.
References
- 1.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278(23):20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 2.Servant G, Weiner OD, Herzmark P, Balla T, Sedat JW, Bourne HR. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287(5455):1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parente CA, Blacklock BJ, Froehlich WM, Murphy DB, Devreotes PN. G protein signaling events are activated at the leading edge of chemotactic cells. Cell. 1998;95(1):81–91. doi: 10.1016/S0092-8674(00)81784-5. [DOI] [PubMed] [Google Scholar]
- 4.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 5.Bültmann BD, Gruler H. Analysis of the directed and nondirected movement of human granulocytes: influence of temperature and ECHO 9 virus on N-formylmethionylleucylphenylalanine-induced chemokinesis and chemotaxis. J Cell Biol. 1983;96(6):1708–1716. doi: 10.1083/jcb.96.6.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruler H, Franke K. Automatic control and directed cell movement. Novel approach for understanding chemotaxis, galvanotaxis, galvanotropism. J Biosci. 1990;45(11–12):1241–1249. doi: 10.1515/znc-1990-11-1226. [DOI] [PubMed] [Google Scholar]
- 7.Gruler H, Nuccitelli R. The galvanotaxis response mechanism of keratinocytes can be modeled as a proportional controller. Cell Biochem Biophys. 2000;33(1):33–51. doi: 10.1385/CBB:33:1:33. [DOI] [PubMed] [Google Scholar]
- 8.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10(7):445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 9.Li L, Hartley R, Reiss B, Sun Y, Pu J, Wu D, Lin F, Hoang T, Yamada S, Jiang J, Zhao M. E-cadherin plays an essential role in collective directional migration of large epithelial sheets. Cell Mol Life Sci. 2012;69(16):2779–2789. doi: 10.1007/s00018-012-0951-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M, Agius-Fernandez A, Forrester JV, McCaig CD. Directed migration of corneal epithelial sheets in physiological electric fields. Invest Ophthalmol Vis Sci. 1996;37(13):2548–2558. [PubMed] [Google Scholar]
- 11.Theveneau E, Marchant L, Kuriyama S, Gull M, Moepps B, Parsons M, Mayor R. Collective chemotaxis requires contact-dependent cell polarity. Dev Cell. 2010;19(1):39–53. doi: 10.1016/j.devcel.2010.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20(6):319–328. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayor R, Etienne-Manneville S. The front and rear of collective cell migration. Nat Rev Mol Cell Biol. 2016;17(2):97–109. doi: 10.1038/nrm.2015.14. [DOI] [PubMed] [Google Scholar]
- 14.Serra-Picamal X, Conte V, Vincent R, Anon E, Tambe DT, Bazellieres E, Butler JP, Fredberg JJ, Trepat X. Mechanical waves during tissue expansion. Nat Phys. 2012;8(8):628–634. doi: 10.1038/nphys2355. [DOI] [Google Scholar]
- 15.Das T, Safferling K, Rausch S, Grabe N, Boehm H, Spatz JP. A molecular mechanotransduction pathway regulates collective migration of epithelial cells. Nat Cell Biol. 2015;17(3):276–287. doi: 10.1038/ncb3115. [DOI] [PubMed] [Google Scholar]
- 16.Peglion F, Llense F, Etienne-Manneville S. Adherens junction treadmilling during collective migration. Nat Cell Biol. 2014;16(7):639–651. doi: 10.1038/ncb2985. [DOI] [PubMed] [Google Scholar]
- 17.Cheng G, Youssef BB, Markenscoff P, Zygourakis K. Cell population dynamics modulate the rates of tissue growth processes. Biophys J. 2006;90(3):713–724. doi: 10.1529/biophysj.105.063701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weijer CJ. Dictyostelium morphogenesis. Curr Opin Genet Dev. 2004;14(4):392–398. doi: 10.1016/j.gde.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Das AM, Eggermont AM, ten Hagen TL. A ring barrier-based migration assay to assess cell migration in vitro. Nat Protoc. 2015;10(6):904–915. doi: 10.1038/nprot.2015.056. [DOI] [PubMed] [Google Scholar]
- 20.Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106(4):489–498. doi: 10.1016/S0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- 21.Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421(6924):753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- 22.Raftopoulou M, Etienne-Manneville S, Self A, Nicholls S, Hall A. Regulation of cell migration by the C2 domain of the tumor suppressor PTEN. Science. 2004;303(5661):1179–1181. doi: 10.1126/science.1092089. [DOI] [PubMed] [Google Scholar]
- 23.Klarlund JK. Dual modes of motility at the leading edge of migrating epithelial cell sheets. Proc Natl Acad Sci USA. 2012;109(39):15799–15804. doi: 10.1073/pnas.1210992109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaritsky A, Kaplan D, Hecht I, Natan S, Wolf L, Gov NS, Ben-Jacob E, Tsarfaty I. Propagating waves of directionality and coordination orchestrate collective cell migration. PLoS Comput Biol. 2014;10(7):e1003747. doi: 10.1371/journal.pcbi.1003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klarlund JK, Block ER. Free edges in epithelia as cues for motility. Cell Adhes Migr. 2011;5(2):106–110. doi: 10.4161/cam.5.2.13728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Block ER, Tolino MA, Lozano JS, Lathrop KL, Sullenberger RS, Mazie AR, Klarlund JK. Free edges in epithelial cell sheets stimulate epidermal growth factor receptor signaling. Mol Biol Cell. 2010;21(13):2172–2181. doi: 10.1091/mbc.E09-12-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chepizhko O, Giampietro C, Mastrapasqua E, Nourazar M, Ascagni M, Sugni M, Fascio U, Leggio L, Malinverno C, Scita G, Santucci S, Alava MJ, Zapperi S, La Porta CA. Bursts of activity in collective cell migration. Proc Natl Acad Sci USA. 2016;113(41):11408–11413. doi: 10.1073/pnas.1600503113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milde F, Franco D, Ferrari A, Kurtcuoglu V, Poulikakos D, Koumoutsakos P. Cell image velocimetry (CIV): boosting the automated quantification of cell migration in wound healing assays. Integr Biol. 2012;4(11):1437–1447. doi: 10.1039/c2ib20113e. [DOI] [PubMed] [Google Scholar]
- 29.Poujade M, Grasland-Mongrain E, Hertzog A, Jouanneau J, Chavrier P, Ladoux B, Buguin A, Silberzan P. Collective migration of an epithelial monolayer in response to a model wound. Proc Natl Acad Sci USA. 2007;104(41):15988–15993. doi: 10.1073/pnas.0705062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petitjean L, Reffay M, Graslandmongrain E, Poujade M, Ladoux B, Buguin A, Silberzan P. Velocity fields in a collectively migrating epithelium. Biophys J. 2010;98(9):1790–1800. doi: 10.1016/j.bpj.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grzesiak JJ, Pierschbacher MD. Shifts in the concentrations of magnesium and calcium in early porcine and rat wound fluids activate the cell migratory response. J Clin Investig. 1995;95(1):227. doi: 10.1172/JCI117644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lagunowich LA, Grunwald GB. Expression of calcium-dependent cell adhesion during ocular development: a biochemical, histochemical and functional analysis. Dev Biol. 1989;135(1):158–171. doi: 10.1016/0012-1606(89)90166-8. [DOI] [PubMed] [Google Scholar]
- 33.Gipson IK. Adhesive mechanisms of the corneal epithelium. Acta Ophthalmol. 1992;70(S202):13–17. doi: 10.1111/j.1755-3768.1992.tb02162.x. [DOI] [PubMed] [Google Scholar]
- 34.Lawson MA, Maxfield FR. Ca2+-and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature. 1995;377(6544):75–79. doi: 10.1038/377075a0. [DOI] [PubMed] [Google Scholar]
- 35.Lee RM, Kelley DH, Nordstrom KN, Ouellette NT, Losert W. Quantifying stretching and rearrangement in epithelial sheet migration. New J Phys. 2013;15(2):025036. doi: 10.1088/1367-2630/15/2/025036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang CC, Park AY, Guan JL. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2007;2(2):329–333. doi: 10.1038/nprot.2007.30. [DOI] [PubMed] [Google Scholar]
- 37.Kolega J. The role of myosin II motor activity in distributing myosin asymmetrically and coupling protrusive activity to cell translocation. Mol Biol Cell. 2006;17(10):4435–4445. doi: 10.1091/mbc.E06-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mironmendoza M, Graham E, Kivanany PB, Quiring J, Petroll WM. The role of thrombin and cell contractility in regulating clustering and collective migration of corneal fibroblasts in different ECM environments. Invest Ophthalmol Vis Sci. 2015;56(3):2079–2090. doi: 10.1167/iovs.15-16388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng MR, Besser A, Danuser G, Brugge JS. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J Cell Biol. 2012;199(3):545–563. doi: 10.1083/jcb.201207148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raffel M, Willert CE, Kompenhans J. Particle image velocimetry: a practical guide. New York: Springer; 2013. [Google Scholar]
- 41.Vitorino P, Meyer T. Modular control of endothelial sheet migration. Genes Dev. 2008;22(23):3268–3281. doi: 10.1101/gad.1725808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nnetu KD, Knorr M, Kas JA, Zink M. The impact of jamming on boundaries of collectively moving weak-interacting cells. New J Phys. 2012;14(11):115012. doi: 10.1088/1367-2630/14/11/115012. [DOI] [Google Scholar]
- 43.Sepulveda N, Petitjean L, Cochet O, Graslandmongrain E, Silberzan P, Hakim V. Collective cell motion in an epithelial sheet can be quantitatively described by a stochastic interacting particle model. PLoS Comput Biol. 2013;9(3):e1002944. doi: 10.1371/journal.pcbi.1002944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masuzzo P, Van Troys M, Ampe C, Martens L. Taking aim at moving targets in computational cell migration. Trends Cell Biol. 2016;26(2):88–110. doi: 10.1016/j.tcb.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Vitorino P, Hammer MM, Kim J, Meyer T. A steering model of endothelial sheet migration recapitulates monolayer integrity and directed collective migration. Mol Cell Biol. 2011;31(2):342–350. doi: 10.1128/MCB.00800-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szabo B, Szollosi GJ, Gonci B, Juranyi Z, Selmeczi D, Vicsek T. Phase transition in the collective migration of tissue cells: experiment and model. Phys Rev E. 2006;74(6):061908. doi: 10.1103/PhysRevE.74.061908. [DOI] [PubMed] [Google Scholar]
- 47.Kusch J, Liakopoulos D, Barral Y. Spindle asymmetry: a compass for the cell. Trends Cell Biol. 2003;13(11):562–569. doi: 10.1016/j.tcb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 48.Vladar EK, Antic D, Axelrod JD. Planar cell polarity signaling: the developing cell’s compass. Cold Spring Harbor Perspect Biol. 2009 doi: 10.1101/cshperspect.a002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rickert P, Weiner OD, Wang F, Bourne HR, Servant G. Leukocytes navigate by compass: roles of PI3Kγ and its lipid products. Trends Cell Biol. 2000;10(11):466–473. doi: 10.1016/S0962-8924(00)01841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arrieumerlou C, Meyer T. A local coupling model and compass parameter for eukaryotic chemotaxis. Dev Cell. 2005;8(2):215–227. doi: 10.1016/j.devcel.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 51.Tsai F-C, Seki A, Yang H, Hayer A, Carrasco S, Malmersjö S, Meyer T. A polarized Ca2+, diacylglycerol and STIM1 signalling system regulates directed cell migration. Nat Cell Biol. 2014;16(2):133–144. doi: 10.1038/ncb2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brundage RA, Fogarty KE, Tuft RA, Fay FS. Calcium gradients underlying polarization and chemotaxis of eosinophils. Science. 1991;254(5032):703–706. doi: 10.1126/science.1948048. [DOI] [PubMed] [Google Scholar]
- 53.Lee J, Ishihara A, Oxford G, Johnson B, Jacobson K. Regulation of cell movement is mediated by stretch-activated calcium channels. Nature. 1999;400(6742):382–386. doi: 10.1038/22578. [DOI] [PubMed] [Google Scholar]
- 54.Hahn K, DeBiasio R, Taylor DL. Patterns of elevated free calcium and calmodulin activation in living cells. Nature. 1992;359(6397):736–738. doi: 10.1038/359736a0. [DOI] [PubMed] [Google Scholar]
- 55.Janmey PA. Phosphoinositides and calcium as regulators of cellular actin assembly and disassembly. Annu Rev Physiol. 1994;56(1):169–191. doi: 10.1146/annurev.ph.56.030194.001125. [DOI] [PubMed] [Google Scholar]
- 56.Mandeville J, Ghosh RN, Maxfield FR. Intracellular calcium levels correlate with speed and persistent forward motion in migrating neutrophils. Biophys J. 1995;68(4):1207. doi: 10.1016/S0006-3495(95)80336-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tran POT, Hinman LE, Unger GM, Sammak PJ. A wound-induced [Ca2+]i increase and its transcriptional activation of immediate early genes is important in the regulation of motility. Exp Cell Res. 1999;246(2):319–326. doi: 10.1006/excr.1998.4239. [DOI] [PubMed] [Google Scholar]
- 58.Kohn EC, Alessandro R, Spoonster J, Wersto RP, Liotta LA. Angiogenesis: role of calcium-mediated signal transduction. Proc Natl Acad Sci. 1995;92(5):1307–1311. doi: 10.1073/pnas.92.5.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.