Abstract
The CRISPR-Cas9 system has revolutionized gene editing both on single genes and in multiplexed loss-of-function screens, enabling precise genome-scale identification of genes essential to proliferation and survival of cancer cells1,2. However, previous studies reported that a gene-independent anti-proliferative effect of Cas9-mediated DNA cleavage confounds such measurement of genetic dependency, leading to false positive results in copy number amplified regions3,4. We developed CERES, a computational method to estimate gene dependency levels from CRISPR-Cas9 essentiality screens while accounting for the copy-number-specific effect. As part of our efforts to define a cancer dependency map, we performed genome-scale CRISPR-Cas9 essentiality screens across 342 cancer cell lines and applied CERES to this dataset. We found that CERES reduced false positive results and estimated sgRNA activity for both this dataset and previously published screens performed with different sgRNA libraries. Here, we demonstrate the utility of this collection of screens, upon CERES correction, in revealing cancer-type-specific vulnerabilities.
Significant efforts using loss-of-function genetic screens to systematically identify genes essential to the proliferation and survival of cancer cells have been reported1–10. Genes identified by these approaches may represent specific genetic vulnerabilities of cancer cells, suggesting treatment strategies and directing the development of novel therapeutics. The CRISPR-Cas9 genome editing system has proven to be a powerful tool for multiplexed screening due to its relative ease of application and increased specificity compared to RNA interference technology11.
However, we and others have recently observed that measurements of cell proliferation in genome-scale CRISPR-Cas9 loss-of-function screens are influenced by the genomic copy number (CN) of the region targeted by the sgRNA-Cas9 complex1,3,4. Targeting Cas9 to DNA sequences within regions of high CN gain creates multiple DNA double-strand breaks (DSBs), inducing a gene-independent DNA damage response and a G2 cell-cycle arrest phenotype3. This systematic, sequence-independent effect due to DNA cleavage (copy-number effect) confounds the measurement of the consequences of gene deletion on cell viability (gene-knockout effect) and is detectable even among low-level CN amplifications and deletions. In particular, this phenomenon hinders interpretation of experiments performed in cancer cell lines that harbor many genomic amplifications since genes in these regions represent a major source of false positives3,4. Existing methods to handle the copy-number effect adopt filtering schemes9, which preclude examination of data from within amplified regions and ignore the effect at low-level alterations. Here, we present CERES, a method to estimate gene dependency from essentiality screens while computationally correcting the copy-number effect, enabling unbiased interpretation of gene dependency at all levels of CN.
As part of our efforts to define a Cancer Dependency Map, a catalog of cell line-specific genetic and chemical vulnerabilities10,12, we performed genome-scale CRISPR-Cas9 loss-of-function screens in 342 cancer cell lines representing 27 cell lineages (Supplementary Table 1, http://depmap.org/ceres) using the Avana sgRNA library13 (Supplementary Table 2) and assessed the effects of introducing each sgRNA on cell proliferation (Online Methods). After applying quality control measures, ROC analysis of sgRNAs targeting “gold standard” common core essential and nonessential genes14 demonstrated high screen quality in all cell lines (Fig. 1a). This collection of screens surpasses the scale of existing comparable datasets by roughly tenfold. To confirm the generalizability of our results in independent screens performed with different sgRNA libraries, we reanalyzed two published datasets derived from screens across 33 cancer cell lines of diverse cell lineage (GeCKOv2) 3 and 14 AML cell lines (Wang2017) 9 (Supplementary Fig. 1a).
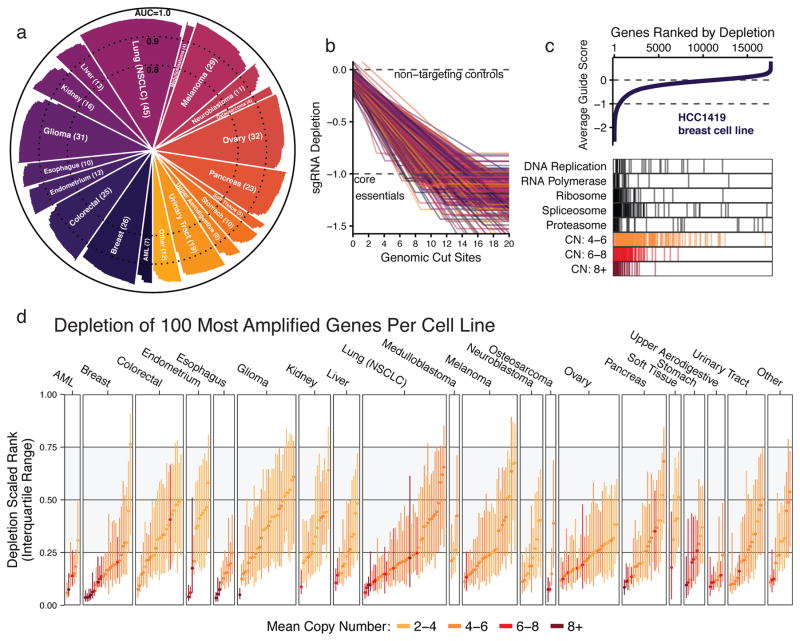
Figure 1. Genomic copy number confounds the interpretation of CRISPR-Cas9 loss-of-function proliferation screens of cancer cell lines.
(a) Screen quality for each cell line in the panel (n=342), as measured by area under the receiver operating characteristic curve (AUC) in discriminating between predefined sets of common core essential and nonessential genes. (b) The depletion of sgRNAs is regressed against the number of perfect-match genomic cut sites using a simple saturating linear fit, which is plotted for each cell line, colored by lineage, and scaled such that the median of sgRNAs targeting cell-essential genes is at −1, marked by a dashed line. (c) Genes are ranked by the mean depletion of targeting sgRNAs (average guide score) and plotted for an example cell line. Values of 0 and −1 represent the median scores of nonessential and cell-essential genes, respectively, indicated by dashed lines. Below, depletion ranks of genes involved in fundamental cell processes and genes at various ranges of CN amplification are shown. (d) The median and interquartile range (IQR) of depletion ranks for the 100 most amplified genes per cell line are plotted. Color indicates mean amplification level of these genes. The gray-shaded area indicates the IQR of all genes screened.
Using genomic copy number data from the Cancer Cell Line Encyclopedia (CCLE)15, we assessed the 342 cell lines screened in our Avana dataset for sensitivity to the copy-number effect as in Aguirre et al. 3. In consonance with previous observations, the relationship held in every cell line in our panel, where sgRNAs targeting more genomic loci were on average more depleted, frequently to levels at or below the depletion of sgRNAs targeting cell-essential genes (Fig. 1b, Supplementary Fig. 1b,c). In each of the three datasets, some of the observed variability in sensitivity was explained by the p53 mutational status of each line in CCLE (Supplementary Fig. 1d).
To quantify the extent to which this sgRNA-level effect translates into false positive gene dependencies, we ranked the genes in each cell line by the average depletion of their targeting sgRNAs (average guide score). In an example breast cancer cell line, HCC1419, high-ranking genes were enriched for both genes involved in fundamental cellular processes and genes with amplified CN (Fig. 1c). The depletion ranks of the 100 genes with the largest CN measurements were significantly higher than expected for the majority of cell lines (298/342 with p < 0.05, one-sample one-tailed K-S test; Fig. 1d, Supplementary Fig. 2a) and the extent of enrichment was significantly correlated with the average CN of these genes (Spearman ρ = 0.61, p < 10−15), consistent with previous studies (Supplementary Fig. 2b).
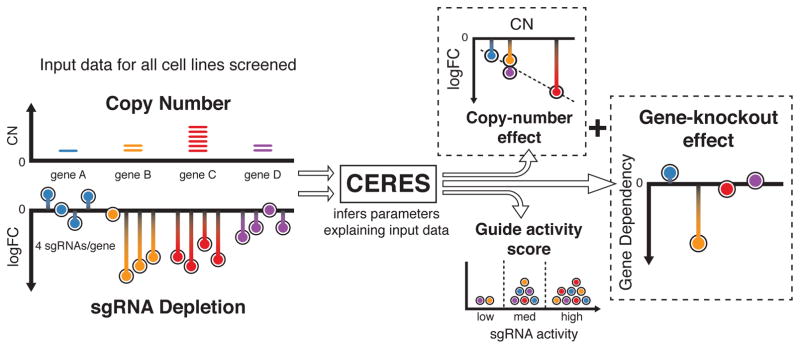
To decouple the gene-knockout effect from the copy-number effect, we developed CERES, which computationally models the measured sgRNA depletion (D) as a sum of these two effects (Fig. 2, Online Methods). Specifically, for each sgRNA i and cell line j, CERES assumes the following model (Equation 1):
where ε is a zero-mean, independent Gaussian noise term. The gene-knockout effect is a sum of cell line specific (gkj) and shared (hk) effects, which are aggregated across any gene targeted by sgRNA i (Gi). The copy-number effect is modeled by a piecewise linear spline, fj, evaluated at the number of genomic sites targeted, determined by the target loci (Li) and the CN at each locus (Clj) (Online Methods). The cumulative depletion effects are then scaled by a guide activity score (qi), restricted to values between 0 and 1, to capture and mitigate the influence of low-quality reagents13,16,17. The offset term oi accounts for noise in the measurement of sgRNA abundance in the reference pool (Online Methods). CERES infers the gene-knockout effects and all other parameters by fitting the model to the observed data via alternating least squares regression (Online Methods). The inferred gene-knockout effects are then scaled per cell line such that scores of 0 and −1 represent the median effects of nonessential genes and common core essential genes, respectively.
Figure 2. Schematic of the CERES computational model.
As input, CERES takes sgRNA depletion and CN data for all cell lines screened. During the inference procedure, CERES models the depletion values as a sum of gene-knockout and copy-number effects, multiplied by a guide activity score parameter. CERES then outputs the values of the parameters that produce the highest likelihood of the observed data under the model.
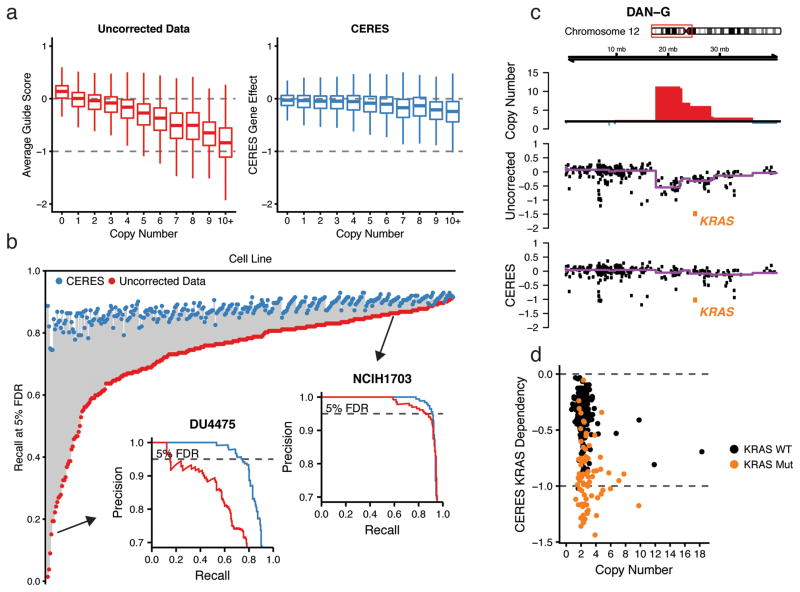
We applied CERES to the Avana dataset of 342 essentiality screens, as well as the GeCKOv2 and Wang2017 datasets, and analyzed the inferred gene-knockout effects (Supplementary Tables 3–5). As expected, CERES markedly reduced the relationship between CN and gene dependency found in the uncorrected average guide scores (Fig. 3a, Supplementary Fig. 3a) and removed it nearly entirely among unexpressed genes, determined using CCLE expression data (Supplementary Fig. 3b). For each gene, we correlated its CN measurements to its dependency scores before and after correction and found that CERES shifted the mean correlation to near zero (Supplementary Fig. 3c). CERES also improved the identification of essential genes in 339 out of 342 screens, as measured by the recall of common core essential genes at a 5% false discovery rate (FDR) of nonessential genes2, by an average of 13.8 percentage points (Fig. 3b, Supplementary Fig. 4a) (Online Methods). This improvement was substantially better than a simple linear model used to correct the relationship between average guide score and CN (Supplementary Fig. 4b) (Online Methods). Furthermore, CERES preserved an average of 134 genes per cell line that would have been removed using a simple filtering scheme. On average, six of these filtered genes per cell line scored as essential below a threshold of −0.6 after CERES correction (Supplementary Fig. 4c). Reassuringly, CERES preserved expected cancer-specific dependencies, even in amplified regions, such as KRAS in an example amplification on chromosome 12p of the DAN-G pancreatic cancer cell line (Fig. 3c, Supplementary Fig. 5). Additionally, KRAS-mutant cell lines remained substantially enriched over wild-type for KRAS gene dependency (Fig. 3d), which generalized to other known oncogenes (Supplementary Fig. 6).
Figure 3. CERES corrects the copy-number effect and improves the specificity of fCRISPR-Cas9 essentiality screens while preserving true gene dependencies.
(a) Boxplots of gene dependency scores are shown across CN for uncorrected average guide scores and CERES gene dependency scores. Data are scaled as in Fig. 1c. (b) The recall of cell-essential genes at a 5% FDR of nonessential genes is plotted for each cell line before (red) and after (blue) CERES correction. Precision-recall curves are inset for example cell lines with poor recall (bottom left) and good recall (top right) before CERES correction. (c) An example amplified region on chromosome 12p is shown for the DAN-G pancreatic cell line. The top track represents CN with amplifications shown in red. The middle track and bottom tracks show the average guide score and CERES score, respectively, for each gene in this region. The purple line is representing the median value in each CN segment. KRAS is highlighted in orange. (d) KRAS gene dependency and CN are shown for all cell lines after CERES correction, with mutant KRAS lines in orange.
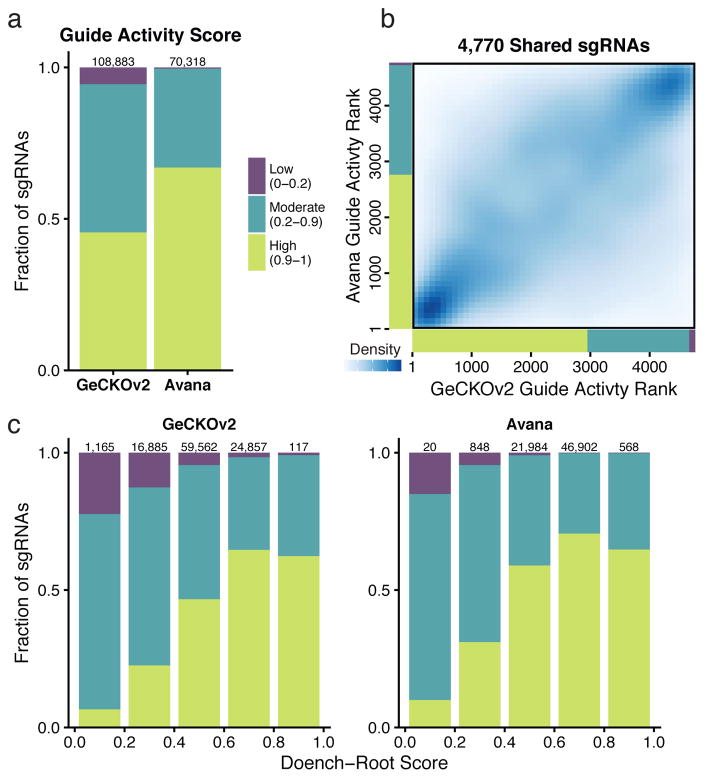
CERES estimates a guide activity score for each sgRNA used in the screens (Supplementary Tables 6–8). While it is infeasible to experimentally validate the activity of all, or even most, sgRNAs in a genome-scale library, sequence determinants have proven useful in the prediction of on-target activity13,18,19. The Avana sgRNA library was optimized using such predictions. Fittingly, CERES estimated higher guide activity scores on average for the Avana dataset relative to GeCKOv2, with a nearly twenty-fold increase in the ratio of high- to low-activity sgRNAs (161.3 to 1 and 8.3 to 1; Fig. 4a). The guide activity scores for the 4,770 sgRNAs common to both libraries showed substantial agreement (Spearman ρ = 0.53, p < 10−15; Fig. 4b), demonstrating that CERES captured a measure of sgRNA activity that is reproducible across independent collections of screens (Supplementary Fig. 7a,b). For both the GeCKOv2 and Avana libraries, we compared CERES guide activity scores to sequence-based predictions of sgRNA activity (Doench-Root scores) 13 and found significant correspondence (Avana: Pearson ρ = 0.21, p < 10−15; GeCKOv2: Pearson ρ = 0.37, p < 10−15; Fig. 4c). Taken together, these results demonstrate that the guide activity scores inferred by CERES are useful for estimating gene-knockout effects and, furthermore, suggests that they could assist in the selection of reagents for follow-up experiments.
Figure 4. CERES estimates guide activity scores for each sgRNA.
(a) sgRNAs are binned into groups with high (0.9–1), moderate (0.2–0.9), and low (0–0.2) guide activity scores. The compositions of guide activity scores are shown for the set of screens performed with the GeCKOv2 and the Avana sgRNA libraries. (b) For the set of 4,770 sgRNAs shared between the GeCKOv2 and Avana libraries, sgRNAs are ranked by guide activity scores in each dataset and are plotted against each other, with darker blue representing a greater density of sgRNAs. (c) sgRNAs are binned by predicted on-target activity using the Doench-Root score, and the composition of CERES-estimated guide activity scores is shown for each dataset.
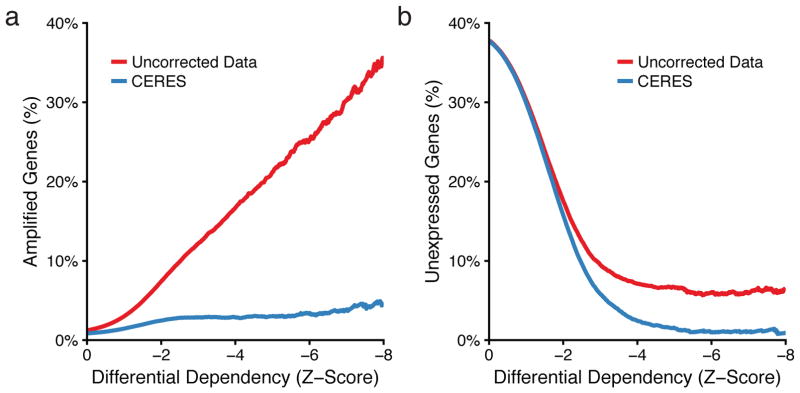
To identify cancer-specific genetic vulnerabilities, we used a metric of differential dependency representing the strength of dependency in a cell line relative to all other lines screened (Online Methods). We assessed an upper bound on the number of false positive differential dependencies due to CN amplifications by calculating the percentage of amplified genes at every possible threshold of differential dependency. In the uncorrected data, the percentage of amplified genes increased at stronger dependency thresholds, climbing above 30% at the highest levels of differential dependency, which CERES substantially reduces (Fig. 5a, Supplementary Fig. 8a). We next used a similar procedure to examine unexpressed genes, whose deletion or editing is not expected to induce phenotypic effects, and which represent an overt source of false positives if scored as differentially dependent. We found that for genes below a differential dependency of −8, CERES reduced the percentage of unexpressed genes from 6.6% to 0.9%, indicating a substantial improvement in specificity (Fig. 5b, Supplementary Fig. 8b).
Figure 5. CERES reduces false positive differential dependencies.
(a) The percentage of genes on amplified regions (CN > 4) below a given differential dependency threshold is plotted for the uncorrected average guide score in red and the CERES gene dependency score in blue. (b) The percentage of unexpressed genes (log2RPKM < −1) below a given differential dependency score is plotted as in (a).
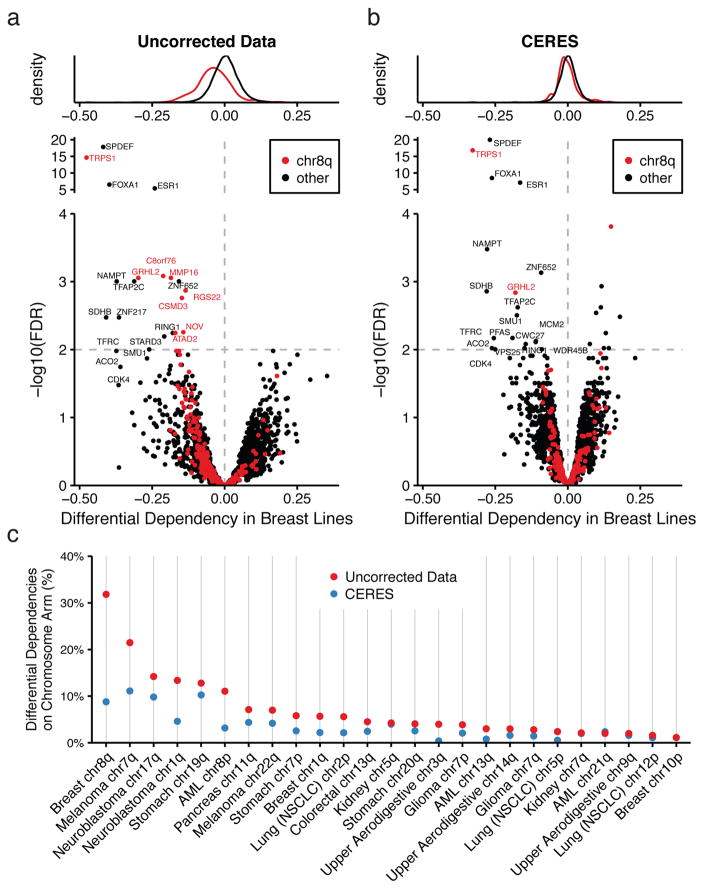
A dataset of this scale enables the discovery of genetic vulnerabilities specific to a subset of cancer cell lines defined by some cellular context, such as cell lineage. We hypothesized that in this setting, copy-number effects driven by recurrent CN alterations, even with small effect sizes, could introduce false positives. For each gene, we compared average guide scores in 26 breast cancer cell lines to those of all other cell lines (Online methods). Indeed, we found several differential dependencies resident on chromosome 8q, which is recurrently amplified in breast tumors (Fig. 6a). However, when we used CERES-corrected dependency scores, we found that only two of the original chr8q genes, TRPS1 and GRHL2, remained (Fig. 6b). To confirm this finding using a complementary assay, we analyzed this set of genes in a dataset derived from genome-scale RNAi screens across 501 cancer cell lines20. We found that these were the only two genes on chr8q that scored as differentially dependent in the 34 breast lines, while most genes in other regions validated (Supplementary Fig. 9a,b). Previous studies have implicated these transcription factors in breast cancer progression21,22, and the high expression levels of these and other transcription factors in breast lines identified suggest that they are likely to be true differential dependencies (Supplementary Fig. 9c). We extended this analysis to all cell lineages with recurrently amplified chromosome arms and quantified the enrichment of differential dependencies before and after CERES correction in each context. We observed that CERES reduced the fraction of differential dependencies on the recurrently amplified chromosome arm in 24 out of 25 such cases (Fig. 6c) (Online Methods).
Figure 6. CERES reduces false positives among lineage-specific differential dependencies due to recurrently amplified chromosome arms.
(a) The distributions of differential dependencies in breast lines are plotted red for genes on chromosome 8q (commonly gained in breast tumors) and black for all other genes. Below, the differential dependency of each gene is plotted against the FDR-corrected p-value, calculated from a student’s t-test, with colors as above. The dashed line represents an FDR of 5%. (b) Data are shown for CERES-inferred gene effects as in (a). (c) The percentages of lineage-specific differential dependencies (FDR < 0.05) that are on recurrently amplified chromosome arms are shown, before and after CERES correction.
While CERES leverages data across many cell lines to infer guide activity scores, we confirmed that this approach can be applied to datasets of any size - even a screen of a single cell line - given predetermined guide activity scores. These may be pre-computed from a larger set of screens, predicted using available tools, or assumed uniform. In random sub-samplings of cell lines from the Avana dataset, CERES performed nearly as well as when applied to the full set. Furthermore, we tested CERES on single cell lines, using fixed uniform guide activities, and found that the median improvement per cell line was over 97% that of the run on all 342 cell lines (Supplementary Fig 10) (Online Methods).
In summary, we introduce a large set of uniformly performed CRISPR-Cas9 essentiality screens of cancer cell lines, propose a methodology to estimate gene dependency while removing false positives due to copy-number effects, and demonstrate the power of these two resources in revealing genetic vulnerabilities of cancer. To facilitate the use of the Avana dataset and CERES, we make the software available as an R package at https://depmap.org/ceres, along with all data and analysis scripts used in this study.
Online Methods
CRISPR-Cas9 essentiality screening assay
Cancer cell lines were transduced with a lentiviral vector expressing the Cas9 nuclease under blasticidin selection (pXPR-311Cas9). Each Cas9-expressing cell line was subjected to a Cas9 activity assay3 to characterize the efficacy of CRISPR/Cas9 in these cell lines (Supplementary Table 1). Cell lines with less than 45% measured Cas9 activity were considered ineligible for screening. Stable polyclonal Cas9+ cell lines were then infected in replicate (n = 3) at low multiplicity of infection (MOI < 1) with a library of 76,106 unique sgRNAs (Avana), which after filtering out sex chromosomes was composed of 70,086 targeting 17,670 genes (~4 sgRNAs per gene) annotated in the consensus coding sequence (CCDS) database, and 995 non-targeting control sgRNAs (Supplementary Table 2). Cells were selected in puromycin and blasticidin for 7 days and then passaged without selection while maintaining a representation of 500 cells per sgRNA until 21 days after infection. Genomic DNA was purified from endpoint cell pellets, the sgRNA barcodes are PCR amplified with sufficient gDNA to maintain representation, and the PCR products are sequenced using standard Illumina machines and protocols.
Preprocessing and quality control
After sequencing the sgRNA barcodes, raw barcode counts are deconvoluted from sequence data using PoolQ software (http://portals.broadinstitute.org/gpp/public/dir/download?dirpath=protocols/screening&filename=Pooled_Screening_Deconvolution_using_PoolQ.pdf) and are summed across sequencing lanes. Samples were removed if they failed to reach 15 million reads. We calculated normalized read counts for each sample according to the procedure in Cowley et al.7. We then calculated pairwise Pearson correlation coefficients between replicate samples from the same cell line to identify and remove poor quality replicates using a threshold of 0.7. All sample read counts were then divided by their representation in the starting plasmid DNA library (pDNA) to compute a fold-change (FC). We computed robust Strictly Standardized Mean Difference (SSMD)23 statistics for the replicates using FCs between non-targeting control sgRNAs and FCs from sgRNAs targeting the spliceosomal, ribosomal, or proteasomal genes in KEGG genesets24–26. We remove replicates with SSMDs that fail to reach −0.5. We also followed standard fingerprinting procedures to remove mismatched cell lines7. logFC data were then normalized within each cell line replicate by subtracting the median logFC value and dividing by the median average deviation (MAD) before input to CERES.
Copy number data
Copy number data for all cancer cell lines were obtained from the Cancer Cell Line Encyclopedia (CCLE)15 data portal (https://portals.broadinstitute.org/ccle). CN data were derived from Affymetrix SNP6.0 arrays. Segmentation of normalized log2 ratios was performed using the circular binary segmentation (CBS) algorithm. The dataset is available at (https://data.broadinstitute.org/ccle_legacy_data/dna_copy_number/CCLE_copynumber_2013-12-03.seg.txt).
Gene expression and mutation data
Gene expression and mutation data for all cell lines were obtained from CCLE data portal. These datasets are available at https://data.broadinstitute.org/ccle/CCLE_RNAseq_081117.rpkm.gct and https://data.broadinstitute.org/ccle/ccle2maf_081117.txt.
sgRNA genome mapping
sgRNA sequences are mapped to the hg19 reference genome using the bowtie short read aligner, version 1.1.227. Bowtie was run using the options “-a -v 0” in order to find all perfect matches in the genome. Only sgRNAs with fewer than 100 alignments were included and alignments were filtered to include an NGG protospacer-adjacent motif (PAM). Alignments were then mapped to gene coding sequences using the consensus coding sequence (CCDS) database.
Model fitting
To fit CERES to input data, we solve the following optimization problem:
Where D̂ij is computed according to Equation (1). The constants M, N, and K in the objective function are, respectively, the total number of sgRNAs, cell lines, and genes in the dataset. The right-hand term in the objective function acts as a regularizer on the cell-line specific deviation from the shared gene-knockout effect, where the hyperparameter λg modulates the strength of the regularization. The first constraint on the model parameters ensures that the guide activity scores are between 0 and 1. The second constraint guarantees that the copy-number effect functions are monotonically decreasing in their arguments. As the objective function is not jointly convex in the model parameters, we fit CERES using alternating least squares, first solving for the gene essentiality scores and copy-number effect parameters with the guide activity scores and offsets held constant, then solving for the guide activity scores and offsets as follows:
Algorithm 1.1.
CERES alternating minimization.
| given ε > 0 | |
| initialize | |
| 1. | gene-knockout and copy-number effect coefficients [g, f] := [0,0] |
| 2. | guide activity scores and offsets [q, o] := [1,0] |
| repeat | |
| 1. | Solve for gene-knockout and copy-number effects. Compute optimal parameters [g*, f*] |
| 2. | Update. [g*, f*] := [g*, f*] |
| 3. | Solve for guide activity scores and offsets. Compute optimal parameters [q*, o*] |
| 4. | Update. [q*, o*] := [q*, o*] |
| 5. | Evaluate mean squared error (mse). mset := ||D − D̂||2/MN |
| 6. | Evaluate decrease in error. Δmse := mset − mset−1 |
| 7. | Stopping criterion. quit if Δmse < ε |
Due to the presence of constraints, we use numerical optimization techniques to solve for the optimal parameters [g*, f*] and [q*, o*] in steps 1 and 328. Note that we use the bracket notation [g, f] to indicate that the enclosed parameters are inferred simultaneously as variables in a system of constrained linear equations.
Spline functions
The piecewise linear spline functions fj in the CERES model equations allow for flexible modeling of the characteristic saturation of the copy-number effect at high numbers of cuts. They are implemented with B-spline regression methods and are each parameterized by 25 slope coefficients plus a single intercept parameter. These are inferred directly in the regression that determines the gene-knockout effects. Each spline has an initial knot point at CN = 0. The additional knot points are determined by running average linkage clustering on the CN data for each cell line.
Hyperparameter optimization and test set evaluation
To improve the generalizability of our model and minimize overfitting of the training data, we regularized the cell line specific gene effects. To find the best value of λg, we evaluated the mean squared error (MSE) obtained on a randomly selected held-out validation set (one-tenth of all observations) for each of 25 values of λg sampled log-uniformly from the interval [0.01, 1]. After the 25 models were evaluated, the value of λg yielding the lowest MSE was used to fit the final model on the full set of observations (Supplementary Fig. 11). The optimized value of λg was 0.562, 0.681, and 0.681 for the Avana, GeCKOv2, and Wang2017 datasets, respectively.
Model complexity
Given a collection of CRISPR screening data, let N be the number of sgRNAs, M be the number of cell lines, and K be the number of targeted genes in the dataset. CERES fits KM cell line specific gene effect parameters and an additional K parameters for the shared gene effects. The model also fits M(S + 1) copy-number effect parameters, where S is the number of CN segments in each piecewise linear spline, and 2N parameters for the guide activity scores and offsets. Ignoring the degrees of freedom lost by regularization and constraints, CERES takes in MN data points and fits MN(1/N + S/N + 2/M + K/N + K/MN) parameters.
Software and implementation
Matrix operations for the optimization procedure were implemented using the open source C++ linear algebra library Eigen, version 3.3, available at http://eigen.tuxfamily.org. These operations were then wrapped into the R statistical software using the ‘RcppEigen’ package, downloaded from http://cran.r-project.org. The optimization routine and final fit for each dataset were run using Google Cloud Platform services.
Precision-recall analysis
Precision-recall curves were generated using the sets of common core essential and nonessential genes defined in Hart et al. 14. The best threshold for which greater than 95% of hits are essential genes is calculated for an FDR of 5%. The percentage of all essential genes that score as hits at this threshold is calculated as the recall at 5% FDR.
Comparison with linear regression
For each cell line, average guide scores were regressed against gene-level copy number data using a linear model. The fit residuals are taken as the LM-corrected gene dependency scores. Precision-recall analysis was performed as above.
Subsampling analysis
We simulated CERES performance generalization to other dataset sizes by downsampling from the Avana dataset. Specifically, for each number p in the set {1, 2, 4, 8, 16, 32, 64} we ran trials (up to rounding), such that each cell line appeared once in each run of size p. For each p and each cell line, we evaluated the harmonic mean of precision and recall (referred to as the F1-measure) at the point of equiprobability between the essential and nonessential gene classes. We then compared this number to the F1-measure obtained by running CERES on the full Avana dataset. For p < 5 we fixed all guide activity scores to a value of 1.
Differential dependency
Differential dependency is calculated as the difference between a single cell line’s dependency score for a given gene and the mean score for that gene across all lines screened, and then z-score normalized to that cell line’s entire set of differential dependencies to reduce the influence of noisy cell lines. For calculating the fraction of differential dependencies that are amplified or unexpressed, only genes with a negative dependency score in at least one cell line are considered.
Recurrent chromosome arm amplifications
We called recurrent chromosome arm amplifications for a lineage across the entire CCLE CN dataset. A chromosome arm was called as amplified if the weighted median of copy number segments on that arm was greater than 2.8. Recurrently amplified chromosome arms for a lineage were then defined using a one-tailed Fisher’s exact test to test for enrichment of amplified arms in that lineage, at an FDR-corrected p-value of 0.05.
Lineage-specific differential dependencies
For every lineage in our dataset with at least five cell lines, we calculate the difference in means in gene dependency between cell lines of that lineage and the rest of the dataset, and assess significance with a two-tailed student’s t-test (df=340), for each gene screened. Differential dependencies are called with a negative effect size at a significance of FDR-corrected p-value < 0.05. For each chromosome arm that was recurrently amplified for that lineage, we calculate the fraction of significant differential dependencies on that chromosome arm before and after CERES correction.
Supplementary Material
Supplementary Table 1. Sample information for the 342 cancer cell lines used in this study.
Supplementary Table 2. sgRNA barcode sequences included in the Avana library with genome and coding sequence mappings.
Supplementary Table 3. CERES-estimated gene-knockout effects for 342 cancer cell lines screened with the Avana sgRNA library.
Supplementary Table 4. CERES-estimated gene-knockout effects for 33 cancer cell lines screened with the GeCKOv2 sgRNA library published in Aguirre et al. (2016).
Supplementary Table 5. CERES-estimated gene-knockout effects for 14 AML cell lines screened with the Wang2017 sgRNA library published in Wang et al. (2017).
Supplementary Table 6. CERES-estimated guide activity scores for sgRNAs in the Avana dataset.
Supplementary Table 7. CERES-estimated guide activity scores for sgRNAs in the GeCKOv2 dataset.
Supplementary Table 8. CERES-estimated guide activity scores for sgRNAs in the Wang2017 dataset.
Table 1.
| Name | Source | Lineage | Histology | Gender | Age | Primary/Metastasis | Achilles culture medium |
|---|---|---|---|---|---|---|---|
| 143B_BONE | CCLE | Osteosarcoma | osteosarcoma | female | 13 | primary | EMEM; 10% FBS; 0.015 mg/ml 5-bromo-2′-seoxyuridine |
| 42MGBA_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | RPMI 1640 + EMEM (1:1): 80.0% |
| 5637_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | RPMI-1640: 90.0% |
| 59M_OVARY | CCLE | Ovary | carcinoma | NA | NA | NA | “DMEM; 10% FBS + 2 mM Glutamine, sodium pyruvate, ITS” |
| 639V_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | DMEM; 10% FBS |
| 647V_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | “DMEM; 15% FBS, 2mMGlutamax-1” |
| 769P_KIDNEY | CCLE | Kidney | carcinoma | female | 63 | primary | RPMI; 10% FBS |
| 786O_KIDNEY | CCLE | Kidney | carcinoma | male | 58 | primary | RPMI; 10% FBS |
| 8305C_THYROID | CCLE | Thyroid | carcinoma | NA | NA | NA | RPMI-1640: 85.0% |
| 8MGBA_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | EMEM: 80.0% |
| A2058_SKIN | CCLE | Melanoma | malignant_melanoma | male | 43 | metastasis | DMEM; 10% FBS |
| A2780_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | RPMI; 10% FBS |
| A549_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 58 | primary | DMEM; 10% FBS |
| ABC1_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 47 | primary | EMEM; 10% FBS |
| AGS_STOMACH | CCLE | Stomach | carcinoma | female | 54 | primary | F12K; 10% FBS |
| ASPC1_PANCREAS | CCLE | Pancreas | carcinoma | female | 62 | metastasis | RPMI; 10% FBS |
| AU565_BREAST | CCLE | Breast | carcinoma | female | 43 | metastasis | DMEM; 10% FBS |
| BC3C_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | M10 |
| BFTC905_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | DMEM: 90.0% |
| BFTC909_KIDNEY | CCLE | Kidney | carcinoma | male | 64 | primary | DMEM; 10% FBS |
| BHY_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | male | NA | NA | DMEM; 10% FBS |
| BICR22_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | male | NA | primary | DMEM; 10% FBS; 2mM Glutamine; 0.4ug/ml hydrocortisone |
| BICR6_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | male | NA | NA | DMEM; 10% FBS |
| BT549_BREAST | CCLE | Breast | carcinoma | female | 72 | primary | RPMI; 10% FBS; 10 ug/ml insulin |
| C2BBE1_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | 72 | primary | DMEM; 10% FBS; 0.01mg/ml transferrin; 2 mM glutamine |
| C32_SKIN | CCLE | Melanoma | malignant_melanoma | male | 53 | primary | EMEM; 10% FBS; 0.1mM NEAA |
| CAKI1_KIDNEY | CCLE | Kidney | carcinoma | male | 49 | metastasis | McCoy’s 5A; 10% FBS |
| CAKI2_KIDNEY | CCLE | Kidney | carcinoma | male | 69 | primary | McCoy’s 5A; 10% FBS |
| CAL27_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | male | 56 | primary | DMEM; 10% FBS |
| CAL29_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | DMEM; 10% FBS |
| CAL51_BREAST | CCLE | Breast | carcinoma | female | 45 | metastasis | DMEM; 20% FBS |
| CAL78_BONE | CCLE | Chondrosarcoma | chondrosarcoma | NA | NA | NA | “RPMI-1640, 20% FBS” |
| CALU6_LUNG | CCLE | Lung (NSCLC) | carcinoma | female | 61 | primary | EMEM; 10% FBS |
| CAMA1_BREAST | CCLE | Breast | carcinoma | female | 51 | metastasis | EMEM; 10% FBS |
| CAOV3_OVARY | CCLE | Ovary | carcinoma | female | 54 | primary | DMEM; 10% FBS |
| CAS1_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 63 | primary | DMEM; 10% FBS |
| CCFSTTG1_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | RPMI; 10% FBS |
| CCK81_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | NA | “EMEM, 10% FBS” |
| CFPAC1_PANCREAS | CCLE | Pancreas | carcinoma | male | 26 | primary | DMEM; 10% FBS |
| CHAGOK1_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 45 | primary | RPMI; 10% FBS; 2mM glutamine |
| CHP212_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | NA | NA | NA | EMEM:F12 (1:1); 10% FBS |
| CJM_SKIN | CCLE | Melanoma | malignant_melanoma | NA | NA | metastasis | Hams F-12; 10% FBS |
| CL40_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | NA | “DMEM/F-12 (1:1), 20% FBS” |
| COLO678_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | NA | NA | RPMI; 10% FBS |
| COLO679_SKIN | CCLE | Melanoma | malignant_melanoma | female | 47 | metastasis | RPMI; 10% FBS |
| COLO792_SKIN | CCLE | Melanoma | malignant_melanoma | male | 62 | metastasis | RPMI; 10% FBS |
| COLO800_SKIN | CCLE | Melanoma | malignant_melanoma | male | 14 | primary | RPMI-1640; 10%FBS |
| CORL279_LUNG | CCLE | Lung (SCL) | carcinoma | male | 63 | metastasis | RPMI; 10% FBS; 2mM glutamine |
| CORL47_LUNG | CCLE | Lung (SCL) | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| COV318_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | EMEM; 10% FBS |
| COV362_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | DMEM; 10% FBS; 2mM L-glutamine |
| COV434_OVARY | CCLE | Ovary | sex_cord-stromal_tumour | female | NA | primary | DMEM; 10% FBS; 2mM L-glutamine |
| COV504_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | DMEM; 10% FBS; 2mM L-glutamine |
| COV644_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | DMEM; 10% FBS; 2mM L-glutamine |
| D283MED_CENTRAL_NERVOUS_SYSTEM | CCLE | Medulloblastoma | primitive_neuroectodermal_tumour-medulloblastoma | male | NA | NA | “DMEM; 10% FBS; 2mM L-glutamine, 2mM sodium pyruvate” |
| D341MED_CENTRAL_NERVOUS_SYSTEM | CCLE | Medulloblastoma | primitive_neuroectodermal_tumour-medulloblastoma | male | NA | NA | DMEM:F12 (1:1); 15% FBS |
| DANG_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | RPMI-1640: 90.0% |
| DAOY_CENTRAL_NERVOUS_SYSTEM | CCLE | Medulloblastoma | primitive_neuroectodermal_tumour-medulloblastoma | NA | NA | NA | EMEM: 90.0% 10%FBS |
| DETROIT562_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | female | NA | NA | EMEM; 10% FBS |
| DKMG_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | female | 67 | primary | RPMI; 10% FBS; 2mM glutamine |
| DLD1_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | NA | primary | RPMI; 10% FBS |
| DU4475_BREAST | CCLE | Breast | carcinoma | female | 70 | metastasis | RPMI; 10% FBS |
| EFM19_BREAST | CCLE | Breast | carcinoma | female | 50 | metastasis | RPMI; 10% FBS |
| EFO21_OVARY | CCLE | Ovary | carcinoma | female | 56 | metastasis | RPMI; 20% FBS; 0.1mM NEAA; 1mM Sodium Pyruvate |
| EFO27_OVARY | CCLE | Ovary | carcinoma | female | 36 | metastasis | RPMI; 20% FBS; 0.1mM NEAA; 1mM Sodium Pyruvate |
| EKVX_LUNG | CCLE | Lung (NSCLC) | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| EPLC272H_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 57 | primary | RPMI; 20% FBS |
| ES2_OVARY | CCLE | Ovary | carcinoma | female | 47 | primary | RPMI; 10% FBS |
| ESS1_ENDOMETRIUM | CCLE | Endometrium | carcinoma | female | 76 | primary | RPMI; 20% FBS |
| F5_CENTRAL_NERVOUS_SYSTEM | “Dunn Lab, Harvard” | Meningioma | meningioma | male | NA | NA | RPMI; 10% FBS |
| FADU_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | male | NA | NA | EMEM; 10% FBS |
| FU97_STOMACH | CCLE | Stomach | carcinoma | female | NA | NA | DMEM;10% FBS; Human Insulin: 0.01 mg/mL |
| G292CLONEA141B1_BONE | CCLE | Osteosarcoma | osteosarcoma | NA | NA | NA | McCoy’s 5A; 10% FBS |
| G401_SOFT_TISSUE | CCLE | Soft Tissue | rhabdoid_tumour | male | 0.25 | primary | McCoy’s 5A; 10% FBS |
| GAMG_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | “DMEM, 10%FBS, 2mM Glutamax-1” |
| GB1_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 35 | primary | EMEM; 10% FBS |
| GCIY_STOMACH | CCLE | Stomach | carcinoma | female | NA | primary | MEM; 15% FBS |
| GI1_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | DMEM: 90.0% |
| GSS_STOMACH | CCLE | Stomach | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| H4_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | DMEM: 90.0% |
| HARA_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 57 | primary | RPMI; 10% FBS |
| HCC1143_BREAST | CCLE | Breast | carcinoma | female | 52 | primary | RPMI; 10% FBS |
| HCC1359_LUNG | CCLE | Lung (NSCLC) | carcinoma | female | 55 | primary | RPMI; 10% FBS |
| HCC1395_BREAST | CCLE | Breast | carcinoma | female | 43 | primary | RPMI; 10% FBS |
| HCC1419_BREAST | CCLE | Breast | carcinoma | NA | NA | NA | RPMI-1640: 90.0% |
| HCC1428_BREAST | CCLE | Breast | carcinoma | female | 49 | metastasis | RPMI; 10% FBS |
| HCC15_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 47 | primary | RPMI; 10% FBS |
| HCC1806_BREAST | CCLE | Breast | carcinoma | female | 60 | primary | RPMI; 10% FBS |
| HCC1937_BREAST | CCLE | Breast | carcinoma | female | 24 | primary | RPMI; 10% FBS |
| HCC1954_BREAST | CCLE | Breast | carcinoma | female | 61 | primary | RPMI; 10% FBS |
| HCC202_BREAST | CCLE | Breast | carcinoma | female | 82 | primary | RPMI; 10% FBS |
| HCC56_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | NA | EMEM; 10% FBS |
| HCC827_LUNG | CCLE | Lung (NSCLC) | carcinoma | female | 39 | primary | RPMI; 10% FBS |
| HCC95_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 65 | primary | RPMI; 10% FBS |
| HEC1A_ENDOMETRIUM | CCLE | Endometrium | carcinoma | female | 71 | primary | McCoy’s 5A; 10% FBS |
| HEC1B_ENDOMETRIUM | CCLE | Endometrium | carcinoma | NA | NA | NA | EMEM; 10%FBS |
| HEC251_ENDOMETRIUM | CCLE | Endometrium | carcinoma | female | NA | primary | EMEM; 0.15% FBS |
| HEC50B_ENDOMETRIUM | CCLE | Endometrium | carcinoma | female | NA | primary | EMEM; 15% FBS |
| HEC59_ENDOMETRIUM | CCLE | Endometrium | carcinoma | female | NA | primary | EMEM; 0.15% FBS |
| HEC6_ENDOMETRIUM | CCLE | Endometrium | carcinoma | NA | NA | NA | EMEM; 15% FBS |
| HEYA8_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | RPMI; 10% FBS |
| HGC27_STOMACH | CCLE | Stomach | carcinoma | NA | NA | NA | Minimum Essential Media (MEM); 10% FBS; NEAA( Non-essential Amino Acids): 5.0 ml; L-glutamine: 2.0 mM |
| HLF_LIVER | CCLE | Liver | carcinoma | male | 69 | primary | EMEM; 10% FBS |
| HMC18_BREAST | CCLE | Breast | carcinoma | NA | NA | NA | RPMI-1640: 10%FBS |
| HOP62_LUNG | CCLE | Lung (NSCLC) | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| HS294T_SKIN | CCLE | Melanoma | malignant_melanoma | male | 56 | metastasis | DMEM; 10% FBS |
| HS578T_BREAST | CCLE | Breast | carcinoma | female | 74 | primary | DMEM; 10% FBS |
| HS683_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 76 | primary | DMEM; 10% FBS |
| HS695T_SKIN | CCLE | Melanoma | malignant_melanoma | male | NA | NA | EMEM: 10% FBS |
| HS729_SOFT_TISSUE | CCLE | Soft Tissue | rhabdomyosarcoma | NA | NA | NA | DMEM; 5% FBS |
| HS766T_PANCREAS | CCLE | Pancreas | carcinoma | male | 46 | primary | DMEM; 10% FBS |
| HS944T_SKIN | CCLE | Melanoma | malignant_melanoma | male | 51 | metastasis | DMEM; 10% FBS |
| HSC3_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | male | 64 | primary | EMEM; 10% FBS |
| HT1080_SOFT_TISSUE | CCLE | Soft Tissue | fibrosarcoma | male | 35 | metastasis | EMEM; 10% FBS |
| HT115_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | NA | DMEM; 15% FBS; 2mM Glutamax-1 |
| HT1197_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | male | 44 | primary | EMEM; 10% FBS |
| HT1376_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | EMEM; 10% FBS |
| HT144_SKIN | CCLE | Melanoma | malignant_melanoma | male | NA | NA | McCoy’s 5A; 10% FBS |
| HT55_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | primary | EMEM; 20% FBS; 2mM L-glutamine; 0.1mM NEAA |
| HUH1_LIVER | CCLE | Liver | carcinoma | male | 35 | primary | DMEM; 10% FBS |
| HUH6_LIVER | CCLE | Liver | other | NA | NA | NA | DMEM; 10% FBS |
| HUH7_LIVER | CCLE | Liver | carcinoma | male | 57 | primary | DMEM; 10% FBS |
| HUPT3_PANCREAS | CCLE | Pancreas | carcinoma | male | 66 | primary | MEM; 10% FBS |
| IGR1_SKIN | CCLE | Melanoma | malignant_melanoma | male | 42 | metastasis | DMEM; 10% FBS |
| IGR39_SKIN | CCLE | Melanoma | malignant_melanoma | male | 26 | primary | DMEM; 15% FBS |
| IMR32_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | male | NA | NA | EMEM; 10% FBS |
| IPC298_SKIN | CCLE | Melanoma | malignant_melanoma | female | 64 | primary | RPMI; 10% FBS |
| JHH1_LIVER | CCLE | Liver | carcinoma | male | 50 | primary | William’s E Medium; 10% FBS |
| JHH4_LIVER | CCLE | Liver | carcinoma | NA | NA | NA | EMEM; 10% FBS |
| JHH5_LIVER | CCLE | Liver | carcinoma | male | 50 | primary | William’s E Medium: 90.0% |
| JHH7_LIVER | CCLE | Liver | carcinoma | NA | NA | NA | William’s E medium with 10% FCS |
| JHOC5_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | DMEM:F12 (1:1); 10% FBS; 0.1mM NEAA |
| JHOM1_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | DMEM:F12 (1:1); 10% FBS; 0.1mM NEAA |
| JHOS2_OVARY | CCLE | Ovary | carcinoma | female | 45 | primary | DMEM/F12 (1:1);10 % FBS; |
| JHOS4_OVARY | CCLE | Ovary | carcinoma | female | 44 | primary | DMEM:F12 (1:1); 10% FBS |
| JIMT1_BREAST | CCLE | Breast | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| JMSU1_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | RPMI; 10% FBS; 2mM Glutamax-1 |
| K029AX_SKIN | CCLE | Melanoma | malignant_melanoma | NA | NA | primary | RPMI; 10% FBS |
| KALS1_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | female | NA | primary | RPMI; 5% FBS |
| KARPAS299_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | “Weinstock Lab, DFCI” | T-cell Lymphoma (ALCL) | lymphoid_neoplasm | male | NA | NA | “RPMI, 20% FBS, 2 mM L-glutamine” |
| KELLY_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | NA | NA | NA | RPMI; 10% FBS |
| KIJK_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | “Weinstock Lab, DFCI” | T-cell Lymphoma (ALCL) | lymphoid_neoplasm | male | NA | NA | RPMI; 20% FBS |
| KLE_ENDOMETRIUM | CCLE | Endometrium | carcinoma | NA | NA | NA | “DMEM/F-12 (1:1), 10%FBS” |
| KM12_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| KMBC2_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | DMEM; 10% FBS |
| KMRC1_KIDNEY | CCLE | Kidney | carcinoma | male | NA | primary | DMEM; 10% FBS |
| KMRC20_KIDNEY | CCLE | Kidney | carcinoma | NA | NA | primary | DMEM; 10% FBS |
| KNS42_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | EMEM; 5% FBS |
| KNS60_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 55 | primary | DMEM; 0.05% FBS |
| KNS62_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 49 | primary | EMEM; 20% FBS |
| KNS81_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 65 | primary | DMEM; 5% FBS |
| KP2_PANCREAS | CCLE | Pancreas | carcinoma | female | 65 | primary | RPMI; 10% FBS |
| KP3_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| KP4_PANCREAS | CCLE | Pancreas | carcinoma | male | 50 | metastasis | DMEM:F12 (1:1); 10% FBS |
| KPL1_BREAST | CCLE | Breast | carcinoma | female | 50 | metastasis | DMEM; 10% FBS |
| KPNYN_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | NA | NA | NA | RPMI; 10% FBS |
| KS1_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | NA | NA | NA | MEM; 10% FBS; 2mMGlutamax-1 |
| KU1919_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | RPMI; 10%heat inactive FBS |
| KURAMOCHI_OVARY | CCLE | Ovary | carcinoma | female | NA | primary | RPMI; 10% FBS |
| KYSE180_OESOPHAGUS | CCLE | Esophagus | carcinoma | male | NA | NA | RPMI; 10% FBS |
| KYSE270_OESOPHAGUS | CCLE | Esophagus | carcinoma | NA | NA | NA | RPMI 1640:F12 (1:1): 90.0% |
| KYSE30_OESOPHAGUS | CCLE | Esophagus | carcinoma | male | 64 | primary | RPMI:F12 (1:1); 20% FBS |
| KYSE410_OESOPHAGUS | CCLE | Esophagus | carcinoma | NA | NA | NA | RPMI-1640: 90.0% |
| KYSE450_OESOPHAGUS | CCLE | Esophagus | carcinoma | male | 59 | primary | RPMI:HamsF-12(1:1) (RPMI-1640 (Hyclone Cat.# SH30027.02):Hams F-12 (Hyclone Cat.# SH30026.01)); 10% FBS |
| KYSE70_OESOPHAGUS | CCLE | Esophagus | carcinoma | male | 77 | primary | RPMI; 10% FBS |
| LCLC103H_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 61 | metastasis | RPMI; 10% FBS |
| LI7_LIVER | CCLE | Liver | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| LK2_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | primary | DMEM; 10% FBS |
| LN18_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 65 | primary | DMEM; 5% FBS |
| LN235_CENTRAL_NERVOUS_SYSTEM | “Lynda Chin, MD Anderson” | Glioma | glioma | male | NA | NA | DMEM; 10% FBS |
| LN382_CENTRAL_NERVOUS_SYSTEM | “Lynda Chin, MD Anderson” | Glioma | glioma | male | NA | NA | DMEM; 10% FBS |
| LN443_CENTRAL_NERVOUS_SYSTEM | “Mikael Rinne, DFCI” | Glioma | glioma | male | NA | NA | DMEM; 10% FBS |
| LNZ308_CENTRAL_NERVOUS_SYSTEM | “Lynda Chin, MD Anderson” | Glioma | glioma | female | NA | NA | DMEM; 10% FBS |
| LOVO_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | 56 | metastasis | F12K; 10% FBS |
| LS1034_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | NA | NA | RPMI; 10% FBS |
| LS180_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | female | NA | NA | EMEM; 10% FBS |
| LS513_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | 63 | primary | RPMI; 10% FBS |
| LUDLU1_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 72 | primary | RPMI; 10% FBS |
| LXF289_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 63 | primary | Hams F-12; 10% FBS |
| M059K_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 33 | primary | DMEM/F12 (1:1); 10 % FBS |
| MALME3M_SKIN | CCLE | Melanoma | malignant_melanoma | male | 43 | metastasis | RPMI; 10% FBS |
| MCAS_OVARY | CCLE | Ovary | carcinoma | NA | NA | NA | EMEM:15%FBS |
| MDAMB157_BREAST | CCLE | Breast | carcinoma | female | 44 | metastasis | RPMI; 10% FBS |
| MDAMB231_BREAST | CCLE | Breast | carcinoma | female | 51 | metastasis | RPMI; 10% FBS |
| MDAMB415_BREAST | CCLE | Breast | carcinoma | female | 38 | metastasis | L-15; 15% FBS; 2mM glutamine; 10mcg/mL Insulin; 10mcg/mL Glutathione |
| MDAMB435S_SKIN | CCLE | Melanoma | malignant_melanoma | female | NA | NA | RPMI; 10% FBS |
| MDAMB436_BREAST | CCLE | Breast | carcinoma | female | 43 | metastasis | RPMI; 10% FBS; 16ug/ml glutathione |
| MDAMB453_BREAST | CCLE | Breast | carcinoma | female | 48 | metastasis | RPMI; 10% FBS |
| MDAMB468_BREAST | CCLE | Breast | carcinoma | female | 51 | metastasis | DMEM; 10% FBS |
| MDST8_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | NA | DMEM; 10% FBS; 2mM Glutamine |
| MELHO_SKIN | CCLE | Melanoma | malignant_melanoma | female | NA | primary | RPMI; 10% FBS |
| MELJUSO_SKIN | CCLE | Melanoma | malignant_melanoma | female | NA | NA | RPMI; 10% FBS |
| MFE319_ENDOMETRIUM | CCLE | Endometrium | carcinoma | NA | NA | NA | 40% RPMI 1640 + 40% MEM (with Earle’s salts) + 20% h.i. FBS |
| MHHNB11_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | male | NA | NA | RPMI; 10% FBS |
| MIAPACA2_PANCREAS | CCLE | Pancreas | carcinoma | male | 65 | primary | DMEM; 10% FBS |
| MKN45_STOMACH | CCLE | Stomach | carcinoma | female | 62 | metastasis | RPMI; 10% FBS |
| MOLM13_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | CCLE | AML | haematopoietic_neoplasm | male | 20 | primary | RPMI; 20% FBS |
| MORCPR_LUNG | CCLE | Lung (NSCLC) | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| MV411_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | CCLE | AML | haematopoietic_neoplasm | male | 10 | primary | IMDM; 10% FBS |
| NB1_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | male | NA | NA | RPMI; 10% FBS |
| NB4_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | CCLE | AML | haematopoietic_neoplasm | female | 23 | primary | RPMI; 10% FBS |
| NCIH1299_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 43 | metastasis | RPMI; 10% FBS |
| NCIH1437_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 6 | metastasis | RPMI; 10% FBS |
| NCIH1581_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 44 | primary | DMEM:F12 (1:1); 10% FBS |
| NCIH1650_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 27 | metastasis | RPMI; 10% FBS |
| NCIH1693_LUNG | CCLE | Lung (NSCLC) | carcinoma | female | 55 | metastasis | RPMI; 10% FBS |
| NCIH1703_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 54 | primary | RPMI; 10% FBS |
| NCIH1792_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 50 | metastasis | RPMI; 10% FBS |
| NCIH1944_LUNG | CCLE | Lung (NSCLC) | carcinoma | female | 62 | metastasis | RPMI; 10% FBS |
| NCIH2023_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 26 | metastasis | “DMEM:HAM’s F12 (1:1); 5% FBS; .005 mg/ml insulin, .01 mg/ml transferrin, 30nM sodium selenite, 10 nM hydrocortisone, 10 nM beta estradiol, 10 mM HEPES, 2 mM L-glutamine” |
| NCIH2030_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | metastasis | RPMI; 10% FBS |
| NCIH2087_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 69 | metastasis | RPMI; 5% FBS |
| NCIH2110_LUNG | CCLE | Lung (NSCLC) | carcinoma | NA | NA | metastasis | RPMI; 10% FBS |
| NCIH2122_LUNG | CCLE | Lung (NSCLC) | carcinoma | female | 46 | metastasis | RPMI; 10% FBS |
| NCIH2126_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 65 | metastasis | “DMEM:HAM’s F12 (1:1); 5% FBS; .005 mg/ml insulin, .01 mg/ml transferrin, 30nM sodium selenite, 10 nM hydrocortisone, 10 nM beta estradiol, 10 mM HEPES, 2 mM L-glutamine” |
| NCIH2170_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | primary | RPMI; 10% FBS |
| NCIH2172_LUNG | CCLE | Lung (NSCLC) | carcinoma | female | NA | primary | RPMI; 10% FBS |
| NCIH2291_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | metastasis | RPMI; 10% FBS |
| NCIH23_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 51 | primary | RPMI; 10% FBS |
| NCIH322_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 52 | primary | RPMI; 10% FBS; 2mM glutamine |
| NCIH441_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | metastasis | RPMI; 10% FBS |
| NCIH460_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | metastasis | RPMI; 10% FBS |
| NCIH520_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | primary | RPMI; 10% FBS |
| NCIH716_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | 33 | metastasis | RPMI; 10% FBS |
| NCIH747_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | 69 | metastasis | RPMI; 10% FBS |
| NCIH838_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 59 | metastasis | RPMI; 10% FBS |
| NCIN87_STOMACH | CCLE | Stomach | carcinoma | male | NA | metastasis | RPMI; 10% FBS |
| NOMO1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | CCLE | AML | haematopoietic_neoplasm | female | 31 | primary | RPMI; 10% FBS |
| NUGC3_STOMACH | CCLE | Stomach | carcinoma | male | 72 | primary | RPMI; 10% FBS |
| OAW28_OVARY | CCLE | Ovary | carcinoma | NA | NA | NA | DMEM; 10% FBS |
| OE21_OESOPHAGUS | CCLE | Esophagus | carcinoma | NA | NA | NA | RPMI-1640: 90.0% |
| OE33_OESOPHAGUS | CCLE | Esophagus | other | female | 73 | primary | RPMI; 10% FBS |
| ONS76_CENTRAL_NERVOUS_SYSTEM | CCLE | Medulloblastoma | primitive_neuroectodermal_tumour-medulloblastoma | NA | NA | NA | RPMI; 10% FBS |
| OSRC2_KIDNEY | CCLE | Kidney | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| OUMS23_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | NA | DMEM; 10% FBS |
| OV7_OVARY | CCLE | Ovary | carcinoma | female | 78 | primary | DMEM:F12 (1:1); 5% FBS; 2mM L-glutamine; 0.5ug/ml hydrocortisone; 10ug/ml insulin |
| OV90_OVARY | CCLE | Ovary | carcinoma | female | 64 | metastasis | DMEM; 10% FBS(Tony) [1:1 mixture of MCDB 105 medium with 1.5 g/L sodium bicarbonate added and Medium 199; 10% FBS] |
| OVCAR8_OVARY | CCLE | Ovary | carcinoma | female | 64 | primary | RPMI; 10% FBS |
| OVISE_OVARY | CCLE | Ovary | carcinoma | female | 40 | primary | RPMI; 10% FBS |
| OVK18_OVARY | CCLE | Ovary | carcinoma | NA | NA | NA | MEM10 |
| OVMANA_OVARY | CCLE | Ovary | carcinoma | female | 51 | primary | RPMI; 10% FBS |
| OVTOKO_OVARY | CCLE | Ovary | carcinoma | female | 78 | metastasis | RPMI; 10% FBS |
| P31FUJ_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | “Ebert Lab, DFCI” | AML | haematopoietic_neoplasm | male | NA | NA | RPMI; 10% FBS |
| PANC0203_PANCREAS | CCLE | Pancreas | carcinoma | female | NA | NA | RPMI; 10% FBS; 1mM sodium pyruvate |
| PANC0403_PANCREAS | CCLE | Pancreas | carcinoma | male | NA | NA | RPMI; 15% FBS; 20ug/ml human insulin |
| PANC1005_PANCREAS | CCLE | Pancreas | carcinoma | male | NA | primary | RPMI; 15% FBS; 2mM glutamine; 1.5 g/L Sodium bicarbonate; 4.5g/L glucose; 10mM HEPES; 1mM Sodium Pyruvate; 10 units/mL Insulin |
| PATU8988S_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | “DMEM:;10%FBS, 2mMGlutamax” |
| PC14_LUNG | CCLE | Lung (NSCLC) | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| PECAPJ34CLONEC12_UPPER_AERODIGESTIVE_TRACT | CCLE | Upper Aerodigestive | carcinoma | male | 60 | primary | IMDM; 10% FBS; 2mM Glutamine |
| PF382_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | “Steigmaier Lab, DFCI” | T-cell ALL | lymphoid_neoplasm | NA | NA | NA | RPMI; 10% FBS |
| PK1_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | RPMI-1640:10%FBS |
| PK45H_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| PK59_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| PLCPRF5_LIVER | CCLE | Liver | carcinoma | male | 24 | primary | DMEM; 10% FBS |
| PSN1_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | primary | RPMI; 10% FBS; 2mM glutamine |
| RCC10RGB_KIDNEY | CCLE | Kidney | carcinoma | male | NA | primary | DMEM; 10% FBS |
| RD_SOFT_TISSUE | CCLE | Soft Tissue | rhabdomyosarcoma | female | 7 | primary | “DMEM:HAM’s F12 (1:1); 5% FBS; .005 mg/ml insulin, .01 mg/ml transferrin, 30nM sodium selenite, 10 nM hydrocortisone, 10 nM beta estradiol, 10 mM HEPES, 2 mM L-glutamine” |
| RERFLCAD1_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 70 | primary | RPMI; 10% FBS |
| RERFLCAI_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | NA | primary | EMEM; 10% FBS |
| RH30_SOFT_TISSUE | CCLE | Soft Tissue | rhabdomyosarcoma | male | 17 | metastasis | RPMI; 10% FBS |
| RKN_SOFT_TISSUE | CCLE | Ovary | leiomyosarcoma | female | 45 | primary | Hams F-12; 10% FBS |
| RKO_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | NA | NA | primary | MEM; 10% FBS |
| RMUGS_OVARY | CCLE | Ovary | carcinoma | female | 62 | primary | Hams F-12; 10% FBS |
| RPMI7951_SKIN | CCLE | Melanoma | malignant_melanoma | female | 18 | metastasis | RPMI; 10% FBS |
| RT112_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | female | NA | primary | RPMI; 10% FBS |
| RT11284_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | EMEM; 10% FBS;2mMGlutamax; 1%NEAA |
| RT4_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | M10 |
| RVH421_SKIN | CCLE | Melanoma | malignant_melanoma | male | NA | NA | RPMI; 10% FBS |
| SCABER_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | “E10+ L-glutamine: 2.0 mM, NEAA( Non-essential Amino Acids): 0.1 mM, Sodium Pyruvate: 0.1 mM” |
| SF295_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | female | 67 | primary | RPMI; 10% FBS |
| SF767_CENTRAL_NERVOUS_SYSTEM | “Lynda Chin, MD Anderson” | Glioma | glioma | female | NA | NA | DMEM; 10% FBS |
| SH10TC_STOMACH | CCLE | Stomach | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| SIMA_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | male | NA | NA | RPMI; 10% FBS |
| SJSA1_BONE | CCLE | Osteosarcoma | osteosarcoma | male | 19 | primary | RPMI; 10% FBS |
| SKBR3_BREAST | CCLE | Breast | carcinoma | female | 43 | metastasis | McCoy’s 5A; 10% FBS |
| SKHEP1_LIVER | CCLE | Liver | carcinoma | male | 52 | metastasis | EMEM; 10% FBS |
| SKMEL24_SKIN | CCLE | Melanoma | malignant_melanoma | male | 67 | metastasis | EMEM; 10% FBS |
| SKMEL30_SKIN | CCLE | Melanoma | malignant_melanoma | male | 67 | metastasis | RPMI; 10% FBS |
| SKMES1_LUNG | CCLE | Lung (NSCLC) | carcinoma | male | 65 | metastasis | DMEM; 10% FBS |
| SKNAS_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | female | NA | NA | DMEM; 10% FBS; NEAA |
| SKNBE2_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | male | NA | NA | EMEM:F12 (1:1); 10% FBS |
| SKNDZ_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | female | NA | NA | DMEM; 10% FBS; NEAA |
| SKNFI_AUTONOMIC_GANGLIA | CCLE | Neuroblastoma | neuroblastoma | male | NA | NA | DMEM; 10% FBS; NEAA |
| SKNMC_BONE | CCLE | Ewing Sarcoma | Ewings_sarcoma-peripheral_primitive_neuroectodermal_tumour | female | NA | NA | EMEM; 10% FBS |
| SKOV3_OVARY | CCLE | Ovary | carcinoma | female | 64 | metastasis | McCoy’s 5A; 10% FBS |
| SLR20_KIDNEY | “Kaelin Lab, DFCI” | Kidney | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| SLR23_KIDNEY | “Kaelin Lab, DFCI” | Kidney | carcinoma | NA | NA | primary | RPMI;10% FBS w/kanamycin |
| SLR26_KIDNEY | “Kaelin Lab, DFCI” | Kidney | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| SNGM_ENDOMETRIUM | CCLE | Endometrium | carcinoma | NA | NA | NA | Ham F-12: 80.0% |
| SNU1_STOMACH | CCLE | Stomach | carcinoma | NA | NA | NA | RPMI-1640: 90.0% |
| SNU201_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 58 | primary | RPMI; 10% FBS |
| SNU213_PANCREAS | CCLE | Pancreas | carcinoma | male | NA | NA | “RPMI; 10% FBS, 2mM L-glutamine” |
| SNU349_KIDNEY | CCLE | Kidney | carcinoma | male | 68 | primary | RPMI; 10% FBS |
| SNU398_LIVER | CCLE | Liver | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| SNU410_PANCREAS | CCLE | Pancreas | carcinoma | male | NA | NA | RPMI; 10% FBS |
| SNU449_LIVER | CCLE | Liver | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| SNU503_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | NA | NA | RPMI; 10% FBS |
| SNU685_ENDOMETRIUM | CCLE | Endometrium | carcinoma | female | NA | NA | RPMI; 10% FBS |
| SNU8_OVARY | CCLE | Ovary | carcinoma | female | 55 | primary | RPMI; 10% FBS |
| SNU840_OVARY | CCLE | Ovary | carcinoma | female | 45 | primary | RPMI; 10% FBS |
| SUIT2_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| SUPM2_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | “Weinstock Lab, DFCI” | T-cell Lymphoma (ALCL) | lymphoid_neoplasm | female | NA | NA | RPMI; 20% FBS |
| SUPT1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | “Steigmaier Lab, DFCI” | T-cell ALL | lymphoid_neoplasm | male | 8 | metastasis | RPMI; 10% FBS |
| SW1463_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | female | NA | NA | RPMI; 10% FBS |
| SW403_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | female | 51 | primary | Leibovitz’s L-15; 10%FBS |
| SW48_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | female | 82 | primary | RPMI; 10% FBS |
| SW620_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | 51 | metastasis | L-15; 10% FBS |
| SW837_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | 53 | primary | RPMI; 10% FBS |
| T24_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | McCoy 5A: 90.0% |
| T3M4_PANCREAS | CCLE | Pancreas | carcinoma | NA | NA | NA | Ham F-10: 90.0%;10%FBS |
| T84_LARGE_INTESTINE | CCLE | Colorectal | carcinoma | male | NA | NA | DMEM:F12(1:1); 5% FBS; 2mM Glutamine |
| T98G_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | 61 | primary | EMEM; 10% FBS |
| TCCPAN2_PANCREAS | CCLE | Pancreas | carcinoma | female | NA | NA | RPMI; 10% FBS |
| TCCSUP_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | female | 67 | primary | EMEM; 10% FBS; 1mM NEAA; 1mM Sodium Pyruvate |
| TE1_OESOPHAGUS | CCLE | Esophagus | carcinoma | male | NA | NA | RPMI; 10% FBS |
| TE5_OESOPHAGUS | CCLE | Esophagus | carcinoma | NA | NA | NA | RPMI; 10% FBS |
| TEN_ENDOMETRIUM | CCLE | Endometrium | carcinoma | NA | NA | NA | MEM;10%FBS |
| TF1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | CCLE | AML | haematopoietic_neoplasm | male | NA | NA | RPMI-1640: 10%FBS; 2ng/ml GM-CSF |
| THP1_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | CCLE | AML | haematopoietic_neoplasm | male | 1 | primary | RPMI; 10% FBS; 50uM B-mercaptoethanol |
| TOV21G_OVARY | CCLE | Ovary | carcinoma | female | 62 | primary | MCDB 105:Medium 199 (1:1); 15% FBS |
| TUHR10TKB_KIDNEY | CCLE | Kidney | carcinoma | NA | NA | primary | RPMI; 10% FBS |
| TUHR4TKB_KIDNEY | CCLE | Kidney | carcinoma | NA | NA | primary | DMEM; 10% FBS |
| U118MG_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | NA | primary | DMEM; 10% FBS |
| U178_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | NA | NA | DMEM; 10% FBS |
| U251MG_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | male | NA | primary | DMEM; 10% FBS |
| U2OS_BONE | CCLE | Osteosarcoma | osteosarcoma | female | 15 | primary | McCoy’s 5A; 10% FBS |
| U343_CENTRAL_NERVOUS_SYSTEM | “Lynda Chin, MD Anderson” | Glioma | glioma | NA | NA | NA | DMEM; 10% FBS |
| U87MG_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | female | 44 | primary | EMEM; 10% FBS |
| U937_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE | “Ebert Lab, DFCI” | Lymphoma (DLBCL) | lymphoid_neoplasm | male | 37 | metastasis | RPMI; 10% FBS |
| UACC257_SKIN | CCLE | Melanoma | malignt_melanoma | NA | NA | primary | RPMI; 10% FBS |
| UACC62_SKIN | CCLE | Melanoma | malignant_melanoma | NA | NA | NA | RPMI; 10% FBS |
| UMUC3_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | EMEM; 10% FBS |
| UOK101_KIDNEY | “Kaelin Lab, DFCI” | Kidney | carcinoma | female | NA | NA | DMEM; 10% FBS |
| VMCUB1_URINARY_TRACT | CCLE | Urinary Tract | carcinoma | NA | NA | NA | DMEM; 10%FBS |
| WM115_SKIN | CCLE | Melanoma | malignant_melanoma | female | NA | NA | EMEM; 10% FBS |
| WM1799_SKIN | CCLE | Melanoma | malignant_melanoma | NA | NA | NA | RPMI; 10% FBS |
| WM2664_SKIN | CCLE | Melanoma | malignant_melanoma | female | NA | NA | DMEM; 10% FBS |
| WM793_SKIN | CCLE | Melanoma | malignant_melanoma | NA | NA | NA | RPMI; 10% FBS |
| WM983B_SKIN | CCLE | Melanoma | malignant_melanoma | NA | NA | NA | RPMI; 10% FBS |
| YAPC_PANCREAS | CCLE | Pancreas | carcinoma | male | NA | NA | RPMI; 10% FBS |
| YKG1_CENTRAL_NERVOUS_SYSTEM | CCLE | Glioma | glioma | female | NA | primary | DMEM; 10% FBS |
| ZR751_BREAST | CCLE | Breast | carcinoma | female | NA | NA | RPMI; 10% FBS |
Acknowledgments
This work was supported by U01 CA176058, U01 CA199253, and P01 CA154303 grants (W.C.Hahn) and by the Slim Initiative for Genomic Medicine, a project funded by the Carlos Slim Foundation and the H.L. Snyder Foundation.
Footnotes
Code availability
We have made the CERES software and documentation available as an R package at https://depmap.org/ceres. We have deposited a code repository of scripts for regenerating analyses and figures here as well.
Data availability
We have made all CRISPR-Cas9 screening data presented here available at https://depmap.org/ceres. We also have posted these data and all other datasets used for analysis in a Figshare record, available at https://doi.org/10.6084/m9.figshare.5319388.
Author contributions
R.M.M, J.G.B, and A.T. conceived of and designed the study. R.M.M., J.G.B., and J.M.M. performed computational analysis and interpretation of results. J.G.B. wrote and implemented the modeling software. R.M.M., B.A.W., and A.E.S. processed and managed data. H.X., and N.V.D. assisted with computational analysis. P.G.M. provided computational tools. G.S.C., S.P., and F.V. provided project management. A.G., Y.L., L.D.A., G.J., R.L., W.F.H., M.S., T.W., D.C.H., V.A.Z., M.R.W., Z.K., J.J.C., and M.O. assisted with data generation. R.M.M., J.G.B., J.M.M, W.C.H., and A.T. wrote and/or revised the manuscript with assistance from other authors. K.S., T.R.G., J.S.B., F.V., D.E.R., W.C.H., and A.T. supervised the study and performed an advisory role.
Competing financial interests
W.C. Hahn reports receiving a commercial research grant from Novartis and is a consultant/advisory board member for the same as well as for KSQ Therapeutics. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hart T, et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Aguirre AJ, et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 2016;6:914–929. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munoz DM, et al. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov. 2016;6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- 5.Cheung HW, et al. Systematic investigation of genetic vulnerabilities across cancer cell lines reveals lineage-specific dependencies in ovarian cancer. Proc Natl Acad Sci USA. 2011;108:12372–12377. doi: 10.1073/pnas.1109363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcotte R, et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012;2:172–189. doi: 10.1158/2159-8290.CD-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowley GS, et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Scientific Data. 2014;1:140035. doi: 10.1038/sdata.2014.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzelepis K, et al. A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Reports. 2016;17:1193–1205. doi: 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang T, et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 2017 doi: 10.1016/j.cell.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsherniak A, et al. Defining a Cancer Dependency Map. Cell. 2017;170:564–570.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellmann C, Gowen BG, Lin P-C, Doudna JA, Corn JE. Cornerstones of CRISPR-Cas in drug discovery and therapy. Nat Rev Drug Discov. 2016 doi: 10.1038/nrd.2016.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corsello SM, et al. The Drug Repurposing Hub: a next-generation drug library and information resource. Nature Medicine. 2017;23:405–408. doi: 10.1038/nm.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart T, Brown KR, Sircoulomb F, Rottapel R, Moffat J. Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol. 2014;10:733–733. doi: 10.15252/msb.20145216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hart T, Moffat J. BAGEL: a computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics. 2016;17:164. doi: 10.1186/s12859-016-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu H, et al. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsherniak A, Vazquez F, Golub TR, Boehm JS, Hahn WC. Defining a Cancer Dependency Map. Cell. 2017:1–31. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiang X, et al. Grhl2 Determines the Epithelial Phenotype of Breast Cancers and Promotes Tumor Progression. PLoS ONE. 2012;7:e50781. doi: 10.1371/journal.pone.0050781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Werner S, et al. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem. 2013;288:22993–23008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89:552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyd S, Vandenberghe L. Convex Optimization. Cambridge University Press; 2004. pp. 1–730. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Sample information for the 342 cancer cell lines used in this study.
Supplementary Table 2. sgRNA barcode sequences included in the Avana library with genome and coding sequence mappings.
Supplementary Table 3. CERES-estimated gene-knockout effects for 342 cancer cell lines screened with the Avana sgRNA library.
Supplementary Table 4. CERES-estimated gene-knockout effects for 33 cancer cell lines screened with the GeCKOv2 sgRNA library published in Aguirre et al. (2016).
Supplementary Table 5. CERES-estimated gene-knockout effects for 14 AML cell lines screened with the Wang2017 sgRNA library published in Wang et al. (2017).
Supplementary Table 6. CERES-estimated guide activity scores for sgRNAs in the Avana dataset.
Supplementary Table 7. CERES-estimated guide activity scores for sgRNAs in the GeCKOv2 dataset.
Supplementary Table 8. CERES-estimated guide activity scores for sgRNAs in the Wang2017 dataset.