Abstract
Native mass spectrometry (MS) is a rapidly advancing field in the analysis of proteins, protein complexes, and macromolecular species of various types. The majority of native MS experiments reported to-date has been conducted using direct infusion of purified analytes into a mass spectrometer. In this study, capillary zone electrophoresis (CZE) was coupled online to Orbitrap mass spectrometers using a commercial sheathless interface to enable high-performance separation, identification and structural characterization of limited amounts of purified proteins and protein complexes, the latter with preserved non-covalent associations under native conditions. The performance of both bare-fused silica and polyacrylamide-coated capillaries was assessed using mixtures of protein standards known to form non-covalent protein-protein and protein-ligand complexes. High-efficiency separation of native complexes is demonstrated using both capillary types, while the neutral-coated capillary showed better reproducibility and higher efficiency for more complex samples. The platform was then evaluated for the determination of monoclonal antibody aggregation and for analysis of proteomes of limited complexity using a ribosomal isolate from E. coli. Native CZE-MS, using accurate single stage and tandem-MS measurements, enabled identification of proteoforms and non-covalent complexes at femtomole levels. This study demonstrates that native CZE-MS can serve as an orthogonal and complementary technique to conventional native MS methodologies with the advantages of low sample consumption, minimal sample processing and losses, and high throughput and sensitivity. This study presents a novel platform for analysis of ribosomes and other macromolecular complexes and organelles, with the potential for discovery of novel structural features defining cellular phenotypes (e.g. specialized ribosomes).
Keywords: Capillary zone electrophoresis, Sheathless CZE-MS, Native mass spectrometry, Top-down mass spectrometry, Ultra-low flow separations, Protein complexes, Ribosome, Specialized ribosomes, Proteoform, Aggregation of biopharmaceutical proteins
Graphical abstract
Introduction
Native mass spectrometry (MS) enables the analysis of proteins, non-covalent assemblies of proteins, small ligand molecules and ions associated with proteins, and even macromolecular particles (e.g. viruses, ribonucleoprotein particles, exosomes) in their unaltered, biologically functional states under near-physiological conditions [1–6]. In native MS, non-covalent interactions are preserved, and tertiary and quaternary structures maintained [7, 8]. Recently, native MS has emerged as a powerful tool to provide information on topology, stoichiometry, function, dynamics, and composition of protein assemblies [8–10]. In more conventional top-down and intact MS characterization of proteins, non-covalent associations are lost as the protein is unfolded under denaturing conditions. Unfolding leads to an increased exposure of the protein to the aqueous buffer, thereby resulting in additional protonation. The resultant MS signal is then divided into a larger number of charge states in comparison to native MS conditions, resulting in complex spectra with poorly resolved ion species (especially those corresponding to molecular species with subtle structural differences), potential difficulties in data interpretation, and lower sensitivity (see Supplementary Figure 1). Importantly, the lower charge states under native conditions lead to the increased m/z spacing (resolution) of ion clusters corresponding to different charge states, potentially resulting in acquisition of structurally more informative spectra in analysis of proteoforms and similar Mr high molecular mass species. Additionally, experimental reproducibility is improved in native MS experiments in comparison to non-native approaches (i.e. middle-down, bottom-up, and top-down), as less sample handling and processing is required.
Subtle differences in protein structure and conformation may result in substantial differences in biological function of a protein or therapeutic properties of a biopharmaceutical. In biological systems, a protein encoded by a single gene may exist as a population of different proteoforms, e.g., charge isoforms, molecules with a variety of post-translational modifications (PTMs), sequence variants, products resulting from truncation of protein, or alternative mRNA splicing [11, 12]. Additionally, chemical modifications of proteins may occur at various stages of sample procurement, preparation, storage and analysis. As a result, comprehensive characterization of a single isolated protein – let alone a complex mixture – is highly challenging, requiring separation prior to high-resolution high mass accuracy MS analysis. We and others have recently demonstrated the power of CZE-MS in high-resolution separations of pharmaceutical glycoproteins [13–15]. In our study, a sheathless cross-linked polyethyleneimine-coated capillary coupled to the Orbitrap Elite MS was used to separate proteoforms of recombinant human interferon-β1 (Avonex), resulting in the detection of 138 proteoforms and the separation of deamidated, sialylated, truncated species, and positional glycoform isomers [15].
Nanoflow (<1 μL/min) and, especially, ultra-low flow (<25 nL/min) separation techniques coupled to nanoESI are highly desirable for native and top-down analyses of protein structures where an MS signal is divided into multiple channels corresponding to ion species of different charge states [16, 17]. In native MS, where physiological or near-physiological conditions are required, the task of separating non-denatured proteins and protein complexes at high resolution is further complicated. Conventional separation techniques compatible with native MS include size exclusion chromatography (SEC) [18], ion exchange chromatography (IEX) [19], hydrophobic interaction chromatography (HIC) [20], affinity chromatography [21], capillary isoelectric focusing (cIEF) [22, 23], native gel-eluted liquid fraction entrapment electrophoresis (GELFrEE) [24, 25], native polyacrylamide gel electrophoresis [26], capillary zone electrophoresis (CZE) [27, 28] and flow field-flow fractionation (F4) [29, 30]. However, for many of these methods, online interfacing to a mass spectrometer is challenging or yet impossible. SEC, which can allow separation under native conditions, provides insufficient resolution of species with subtle structural differences. cIEF may lead to unfolding of proteins and dissociation of protein complexes at their pIs and require the use of denaturing sheath flow buffers to maintain stable electrospray, which may limit native MS applications to relatively stable complexes. The native GELFrEE approach, although viable for the separation of native multimeric complexes and proteins in mixtures, has not yet been directly interfaced with MS or demonstrated the capability of high-resolution separations of charge-based proteoforms and species with very subtle structural differences [24, 25].
In contrast to the above techniques, capillary zone electrophoresis (CZE) can achieve high-resolution separation under native state conditions. CZE separates molecular species according to differences in their electrophoretic mobilities, which are dependent on analyte charge and hydrodynamic volume. Furthermore, negligible sample and buffer quantities are required, and sample losses and carryover are minimal due to the open tubular capillary geometry [31]. Currently, there are several commercial solutions for interfacing CZE online to MS that can potentially be used for native MS [28, 32–35]. The ability to separate non-covalent complexes from corresponding unbound complex-forming constituents can address a long-standing debate in the field of native ESI-MS on whether protein complexes and aggregates are formed in the gas phase during electrospray ionization, desolvation and ion manipulation in the mass spectrometer or the native MS data closely represent the structures of non-covalent complexes existing in aqueous solutions of biological systems [9, 36]. Such separation supports the capability of native MS to infer the structures of non-covalent macromolecular complexes existing in-solution from the results of gas-phase native ESI-MS measurements [37, 38].
Historically, native MS has been largely limited to the domain of TOF mass analyzers due to the high upper m/z thresholds of detection of these instruments (up to 100,000 Th) [2]. However, native MS analyses with TOF analyzers suffer from the limited resolving power of these instruments. Recent studies have demonstrated that high resolution Orbitrap mass analyzers can be used for the characterization of native proteins and large protein complexes [2, 39, 40]. For example, a modified Exactive Plus Orbitrap instrument was applied to distinguish between ATP- and ADP-bound states of the 0.8 MDa E. coli chaperonin GroEL, a fourteen subunit complex [2]. It was further demonstrated that a modified Exactive Plus Orbitrap instrument could be used for native MS experiments of substrates with molecular weights approaching 10.0 MDa while still possessing sufficient resolution and sensitivity to detect small changes to the large molecular mass of the multisubunit protein complex or macromolecular assembly [2, 41]. Moreover, recent studies have shown that an Orbitrap-based instrument can efficiently perform ion isolation of the complex and fragmentation into individual monomeric subunits (MS2) followed by subsequent isolation of subunit ions and fragmentation into backbone fragments (MS3) [39, 40].
Considering that a vast array of biological functions are conducted by protein assemblies rather than individual proteins, investigation of functional protein complexes, their composition, architecture, stoichiometry, and structural features is of great importance to understanding biological systems. The complexity of proteins and protein assemblies, as well as the heterogeneity of proteoforms in biomedical samples, necessitates both a high-efficiency separation component and a sensitive mass spectrometer of a high resolving power and an appropriate m/z range. In the present work, we survey the potential of an online sheathless CZE-Orbitrap MS platform to investigate model mixtures of intact proteins and protein complexes, heterogeneous biopharmaceutical proteins, and limited complexity proteomic samples. The approach is initially evaluated using model proteins with non-covalently associated multisubunit and small ligand entities, a biopharmaceutical monoclonal antibody, and then explores the E. coli ribosomal proteome. The platform facilitated the detection of numerous proteoforms and non-covalent complexes, with structures being confirmed by CZE-MS2 experiments. Particular molecular species could not be detected under denaturing conditions or in native direct infusion experiments. CZE separation, followed by higher energy collision-induced dissociation (HCD) MS2 resulted in the identification of 42 ribosomal protein groups, 144 proteoforms and 679 proteoform-spectrum matches (PfSMs) in a single analysis of the E. coli ribosomal proteome under native conditions. Characterization of the same sample by native CZE-MS1 and denaturing CZE-MS2 resulted in identification of 9 additional ribosomal protein groups, corresponding to a total of 51 identifications, which indicated the complementarity of techniques. Importantly, native CZE-MS2 resulted in a superior depth of proteomic profiling for the E. coli ribosomal proteome in comparison to denaturing CZE-MS2. While extraction of proteins from intact ribosomes involved a brief exposure to denaturing conditions prior to reconstitution into a buffer of a physiological pH, the resulting reconstituted protein extract was used as a model sample to assess the ability of native CZE-MS to survey complex biological samples that may contain non-covalent molecular assemblies that either survived the sample cleanup procedure or got re-associated and re-folded under native conditions. Several non-covalent associations of E. coli ribosomal proteins were elucidated using native CZE-MS analyses that could not otherwise be observed using conventional denaturing top-down or bottom-up approaches. Our work demonstrates the potential of CZE interfaced online with advanced mass spectrometry to enable native, high-resolution separations of proteins and protein complexes in their physiologically-relevant states and facilitate structurally-informative analyses via single stage and tandem MS under native non-denaturing conditions.
Experimental
Chemicals and Materials
All chemicals and materials used in this study were purchased from Sigma-Aldrich (St. Louis, MO) unless stated otherwise.
Preparation of the High-Purity Trastuzumab Standard
Trastuzumab (Herceptin, Genentech, South San Francisco, CA), kindly provided by Alain Beck (Centre d’Immunologie Pierre Fabre, St Julien-en-Genevois Cedex, France), was diluted in 25 mM ammonium acetate to a final concentration of 0.8 mg/mL and cleaned-up via Micro Bio-Spin size-exclusion columns with Bio-Gel P6 (SEC, Bio-Rad, Hercules, CA).
Preparation of Mixtures of Proteins and Protein Complexes
All protein standards were purchased from Sigma-Aldrich in lyophilized form. The following proteins were used: Enolase (P/N E6126; S. cerevisiae), carbonic anhydrase (P/N C3934; Bos taurus), alcohol dehydrogenase (P/N A8656; S. cerevisiae), protein disulfide isomerase (P/N P3818; Bos taurus), alpha crystallin (P/N C4163; Bos taurus), myoglobin (P/N M0630; E. caballus), cytochrome C (P/N C3131; Bos taurus), ribonuclease A (P/N R6513; Bos taurus), cytochrome C reductase type I (P/N C3381; S. scrofa), pyruvate kinase type III (P/N P9136; Oryctolagus caniculus), and fibrogen (P/N F8630; Bos taurus). All proteins were reconstituted in 25 mM ammonium acetate, pH 7.5 and mixed to the final concentrations specified below. Desalting was performed with Micro Bio-Spin 6 columns (Bio-Rad, Hercules, CA). A mixture of three proteins was prepared from enolase (9.18 μM), carbonic anhydrase (4.00 μM), and alcohol dehydrogenase (23.5 μM). A mixture of eleven proteins and protein complexes consisted of enolase (9.18 μM), carbonic anhydrase (4 μM), alcohol dehydrogenase (23.5 μM), pyruvate kinase type III (15.0 μM), alpha crystallin (21.6 μM), myoglobin (7.17 μM), cytochrome C (10.4 μM), ribonuclease A (17.8 μM), cytochrome C reductase type I (10.2 μM), protein disulfide isomerase (15.0 μM) and fibrogen (17.4 μM).
Preparation of the Ribosomal Protein Extract
The E. coli ribosomal protein extract was prepared as described [42, 43]. In brief, to remove RNA, 33 μL of cold, glacial acetic acid and 0.25 volumes (8.25 μL) of 100 mM magnesium chloride was added to 1 mg of the intact E. coli ribosomal isolate (13.3 μM; P0763S, New England Biolabs, Ipswich, MA). Next, the sample was spun-down at 10,000 RPM for 5 minutes; the resultant supernatant was collected and cleaned-up two times into 25 mM ammonium acetate, pH 7.5 using Micro Bio-Spin 6 SEC columns, according to manufacturer’s protocol (Bio-Rad). The pH of the mixture was closely monitored throughout the prep and brought close to neutral pH.
Capillary Zone Electrophoresis Instrumentation and Methods
All capillary electrophoresis equipment, including bare fused silica (BFS) and neutral polyacrylamide-coated capillaries with hydrofluoric acid-etched emitters for sheathless nanoESI (aka CESI), were provided by SCIEX Separations (Brea, CA). A commercial CESI 8000 instrument was interfaced with either Exactive Plus extended mass range (EMR) or Q Exactive Plus mass spectrometers (Thermo Fisher Scientific) in all CZE-MS experiments. Both the BFS and polyacrylamide-coated capillaries were 30 μm inner diameter and 90 cm long, and contained an integrated sheathless etched porous nanospray tip.
For the analysis of the mixture of 3 standard proteins and complexes on the sheathless BFS – Exactive Plus EMR mass spectrometer, the capillary was first rinsed with 0.1 M sodium hydroxide (100 psi, 5 min), followed by 0.1 M hydrochloric acid (5 min) and nanopure water (5 min). Next, the separation capillary was conditioned with the BGE (25 mM ammonium acetate, pH 7.5) at 100 psi for 10 min. The conductive line capillary contained 3% acetic acid solution at 75 psi flow. Sample injection was performed at 2.5 psi for 15 seconds, corresponding to approximately 6 nL of sample (0.8% of the capillary volume). Separations were conducted for 60 minutes with a 20 kV potential applied across the capillary with a ramp time of one minute. At the end of the run, a ramp-down from 20 kV to 1 kV over five minutes was performed back to the start conditions for the next injections.
For the analysis of samples on the sheathless polyacrylamide-coated capillary, the column was first rinsed at 100 psi with 0.1 M hydrochloric acid (5 min), DDI water (5 min), followed by the BGE (40 mM ammonium acetate, pH 7.5) for 10 minutes; and the conductive line capillary was rinsed with a 3% acetic acid at 75 psi for 4 min. Injection was performed at 2.5 psi for 15 seconds, corresponding to approximately 6 nL of sample (0.8% of the capillary volume). Separations were performed for 60 minutes at 20 kV with a ramp time of one minute. Due to the significant suppression of EOF on polyacrylamide-coated capillaries, supplemental pressure was applied to generate flow of solvent and facilitate the migration of analytes towards the MS detector. For the analysis of the mixture of 11 proteins, 3 psi was used. For the ribosomal protein extract, separations were performed also at 3 psi supplemental pressure in CZE-MS1 experiments or with 1 psi supplemental pressure applied for the first 15 minutes of the separation, followed by a supplemental pressure of 3 psi for the remainder of the run (in CZE-MS2 experiments). A ramp-down from 20 kV to 1 kV over a five minute period was run after the conclusion of the separation.
Mass Spectrometry
All CZE-MS experiments performed on the Exactive Plus EMR (Thermo Fisher Scientific) used the following settings: a 1.5 kV spray potential, resolution of 35,000 at 200 Th, automatic gain control (AGC) of 3e6 and maximum injection time of 250 ms. In-source collision-induced dissociation (100 V × charge) was applied to provide sufficient desolvation without dissociation of non-covalently-bound subunits or fragmentation of covalent bonds. The instrument was operated in extended mass range mode (EMR). For AIF, fragmentation was performed with 150 V × charge in the HCD cell.
For the native CZE-MS1 analysis of the E. coli (strain B/BL21-DE3; UniProt) ribosomal protein extract, the Q Exactive Plus (Thermo Fisher Scientific) was operated with 1.3 – 1.5 kV spray potential, 100 eV for insource collision-induced dissociation, 1e6 for target AGC, 250 ms maximum injection time, and a 17,500 resolution at 200 Th. For CZE-MS2 experiments, the target AGC was set to 3e6, and the maximum injection time was 250 ms at a resolution of 140,000 at 200 Th. For native CZE-MS2, the normalized collision energy (NCE) was set at 33%. For both native CZE-MS1 and CZE-MS2, the instrument was operated in the standard mode with a HV pressure of approximately 3.6E-05 mbar.
For denaturing CZE-MS1, the MS detection settings were similar as the native experiments, with the exception of a lower in-source collision-induced dissociation energy (15 eV) and optimum NCE of 25% (for CZE-MS2).
For native MS infusion experiments of Trastuzumab, the polyacrylamide-coated capillary and a CESI 8000 instrument were used as an infusion platform into the Q Exative Plus MS. Infusion was achieved by filling the capillary with Trastuzumab (2 mg/mL, 20 mM ammonium acetate, pH 8.0) via an injection of 100 psi for 1 minute. Next, the separation capillary was inserted into the buffer vial at the anodic end containing 20 mM ammonium acetate, pH 8.0. A l supplemental pressure of 2.5 psi and if needed a 20 kV potential were applied to facilitate flow of liquid towards the MS inlet (cathodic end). These conditions were identical to parameters in CZE-MS experiments for this sample. A 1.5 kV voltage was applied to facilitate stable nESI. The inlet capillary temperature of the MS was 350 degrees Celsius and the rest of the MS parameters were set as defined above.
Data Analysis
Analysis of native CZE-MS1 runs from all system/instruments was performed using both XCalibur and Protein Deconvolution 4.0 (Thermo Fisher Scientific). Deconvolution was performed using the Sliding Window approach with either ReSpect for isotopically unresolved data, or Xtract for high resolution CZE-MS1 runs where isotopic resolution was achieved. Analysis of all-ion fragmentation (AIF) MS data acquired on an Exactive Plus EMR was performed using Byonic software (Version 2.5.6, Protein Metrics, San Carlos, CA) or manually, using fragment masses predicted by the Protein Prospector software (UCSF). It should be noted that for the analysis of AIF data acquired on an Exactive Plus EMR, which lacks a quadrupole and therefore the precursor ion selection capability for MS2, spectra in the regions of the CZE-MS1 run corresponding to migration times of separated or partially-separated proteins or protein complexes were averaged and analyzed individually. However, since baseline separation was not achieved for all mixture constituents, AIF-generated fragments from co-migrating species were often detected within the same AIF spectrum. For all CZE-MS2 experiments, Byonic (Version 2.5.6, Protein Metrics, San Carlos, CA) was employed for data interpretation. Byonic searches were performed with fully-specific digestion specificity and a maximum of two missed cleavages, as well as a maximum of one rare and one common modification for each proteoform-spectrum match (PfSM). Also, a 20 ppm fragment mass tolerance and a 10 ppm precursor mass tolerance were used in all Byonic searches. Common and expected modifications for the model protein mixtures and ribosomal extract were manually inputted into Byonic and used as dynamic modifications. Dynamic modifications included oxidation (methionine), deamidation (asparagine), phosphorylation (histidine, lysine, arginine, serine, threonine, and tyrosine), acetylation (N-terminal and lysine), N-terminal methionine loss, coordination of magnesium (2+) and zinc (2+) ions (C-terminal, aspartate, and glutamate), methylation (lysine and arginine), dimethylation (lysine), and N-terminal pyroglutamation (glutamate and glutamine). For analysis of ribosomal proteins and mixtures of protein standards, a database containing 208 protein sequences that combined the results of bottom-up proteomic profiling of the E. coli ribosomal isolate that included sequences of identified ribosomal proteins (UniProt, E. coli strain B/BL21-DE3), ribosome-associated proteins and sequences of common contaminants was used.
Bottom-Up NanoLC-MS-Based Proteomic Analysis
NanoLC-MS proteomic profiling was performed in a similar fashion to that described in [44]. A fused silica capillary (75 μm internal diameter, 20 cm long) (Polymicro Technologies, Phoenix, AZ) was packed with Magic C18AQ, 3 μm diameter, 200 Å pore size beads from Michrom Bioresources (Auburn, CA, USA). Liquid chromatography (LC) separation was performed on an Ultimate 3500 system from Thermo Fisher Scientific, and mass spectrometry data was acquired on an LTQ Orbitrap XL ETD (Tune Plus version 2.5.5) from Thermo Fisher Scientific. The sample was electrosprayed using a distal-coated, 20 μm internal diameter, 360 μm outer diameter tip with a 10 μm opening from New Objective (Woburn, MA, USA) connected directly to the column head using a Teflon sleeve.
Reversed-phase liquid chromatography was carried out using mobile phase A: 0.1% formic acid in water and mobile phase B: 0.1% formic acid in acetonitrile. Approximately 100 fmol of each trypsin digested protein was injected directly onto the analytical column with 2% B at 250 nL/min. The sample was loaded and desalted over 10 minutes in 2% B and separated at 250 nL/min flow rate using the following linear gradient: 2–37% B over 60 minutes, 37–95% B over 10 minutes, 95% B hold for 9 minutes, 95% to 2% B in 1 minute, 2% B hold for 10 minutes. Data were acquired on an LTQ Orbitrap XL mass spectrometer using parameters for conventional Top 10 analysis enabled through Xcalibur (v. 2.0.7). Full precursor scans were acquired over a 400–1700 Th range in the Orbitrap at 60k resolution (at 400 Th), with the AGC target set to 1e6 and 500 ms maximum ion accumulation time. The 10 highest intensity eligible precursors (Top 10) were sequentially selected in 2 Th wide isolation windows centered on the monoisotopic peak and activated for maximum 30 ms by CID at 35% normalized energy. In fragment ion accumulation, the AGC target was set to 200,000, the maximum injection time was 100 ms, and the ion trap was scanned at 0.5 Da resolution using dynamic exclusion of 60 seconds. Ions with a +1 charge state and lower than 500 intensity units were considered chemical noise.
The acquired data were analyzed in Proteome Discoverer 1.4 (Thermo Fisher Scientific) with Sequest HT peptide spectral match scoring and Percolator validation and filtering (q < 0.01) [45]. The search was conducted against a concatinated bovine, E. coli, equine, human, porcine, rabbit and S. cerevisiae (ATCC) UniProt databases containing canonical proteins and known variants from March 2015 appended with 47 common contaminants. The maximum precursor mass error was set to 20 ppm and the maximum fragment mass error was set to 0.6 Da. Carbamidomethylation of cysteine was set as a static modification, and oxidation of methionine, N-terminal acetylation and deamidation of glutamine and asparagine were set as dynamic modifications. Up to 2 missed tryptic cleavages were allowed. Amino acid sequences of all proteins identified in bottom-up experiments were consolidated into a single database, which was used for the identification of species in the intact CZE-MS1 and top-down CZE-MS2 approaches.
Results and Discussion
Analytical workflow
In this study, we demonstrate the power of CZE as an online high resolution separation technique compatible with native state MS analysis using: (1) mixtures of complex-forming protein standards, (2) a monoclonal antibody (Trastuzumab), and (3) an extract of the E. coli ribosomal proteome. Both bare fused silica (BFS) and polyacrylamide-coated capillaries were used for CZE separations under native conditions interfaced online via a SCIEX sheathless porous interface (CESI) for high sensitivity [28]. The CESI interface enabled the online integration of high resolution CZE separations with MS conducted at ultra-low flow rates (measured at 10 – 20 nL/min) and without analyte dilution prior to ionization [33, 46]. The flow rates used in our CZE-MS experiments are substantially lower than those typically utilized in more conventional sheath-liquid CZE platforms [28]. Ultra-low flow rate separations coupled to nanoESI-MS result in improved ionization efficiency, enhanced ion transfer into the mass spectrometer, and decreased ion suppression, collectively leading to the enhanced MS detection sensitivity [16, 17, 47]. We found that the acidic buffer used in the conductive line of the interface (3% acetic acid) did not significantly alter the pH of the electrosprayed BGE (e.g. 20–40 mM ammonium acetate, pH 7.5–8.5), which was important for maintaining native conformations of analytes. This is attributed largely to the design of the sheathless porous CESI interface (SCIEX), which allows for the limited passage of only small ions (i.e. not liquid) from the conductive line into the separation capillary at the CZE/ESI-MS porous junction without analyte dilution [28, 33, 46]. While the limited transport of hydroxonium ions from the conductive line into the separation capillary at the porous junction allowed for closing the CZE and ESI electric circuits and ionizing analyte molecules as well as for the improved sensitivity, stability and performance of CZE-ESI MS, the pH of the resultant solution measured at the outlet of the capillary emitter was found within the native range (pH 6.5–7.5). Additionally, the porous junction is approximately 2 cm in length at the outlet of the separation capillary, which makes the exposure of the analyte molecules to this minor change of the pH very short (seconds). Nearly identical results were found when 10% acetic acid was used in the conductive line. Identification of proteins and protein complexes under native conditions involved high mass accuracy single stage intact MS (CZE-MS1), followed by either all-ion fragmentation (AIF) on an Exactive Plus EMR or data-dependent high energy collision dissociation (DDA-HCD) CZE-MS2 on a Q Exactive Plus. Peptide maps of all proteins were obtained via conventional bottom-up nanoLC-MS.
Native CZE-MS of mixtures of complex-forming proteins
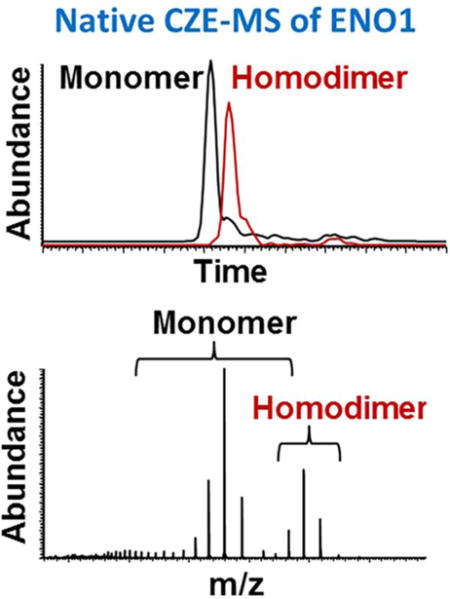
A mixture of three protein standards known to form non-covalent multi-subunit homomeric complexes and complexes with coordinated metal-ion ligands (alcohol dehydrogenase (S. cerevisiae), enolase (S. cerevisiae), and carbonic anhydrase (Bos taurus)) was analyzed using a BFS capillary coupled to an Exactive Plus EMR MS (Figure 1). This experiment was performed as a proof-of-principle for online native CZE-MS using a relatively simple mixture of three proteins and associated complexes. According to the product specifications, alcohol dehydrogenase 1 (ADH1) was expected to be observed as a homotetramer with the molecular mass of 141 – 151 kDa; enolase (ENO1) as a homodimer of 93 kDa with bound Mg2+ ions, and carbonic anhydrase 2 (CA2) as a 29 kDa complex with coordinated Zn2+ ions. Bottom-up proteomic analysis of the tryptic digested proteins revealed the presence of contaminants and isozymes, such as ENO1 and ENO2, as well as protein fragments, corresponding to truncated protein standards. Based on the results of bottom-up analysis and observed ion species in native CZE-MS experiments, theoretical molecular masses were calculated for monomers and complexes (see Supplementary Tables 1 and 2). CZE-MS1 was first conducted under native conditions using 25 mM ammonium acetate, pH 7.5 as the BGE, and the MS resolving power was set to 35,000 at 200 Th. Together, these CZE and MS conditions resulted in the separation and detection of the major components of the mixture between 12 and 32 minutes, including ENO1 homodimer and ADH1 tetramers (Figure 1A–B). ESI mass spectra of ENO1 monomer and homodimer, ENO2, CA2, and ADH1 tetramers are shown in Figure 1C. On the basis of accurate deconvoluted masses, the major mixture components, in addition to the most abundant proteoforms, were identified, as detailed next and in Supplementary Table 1.
Figure 1. Native CZE-MS analysis of a mixture of three protein complex-forming standards.
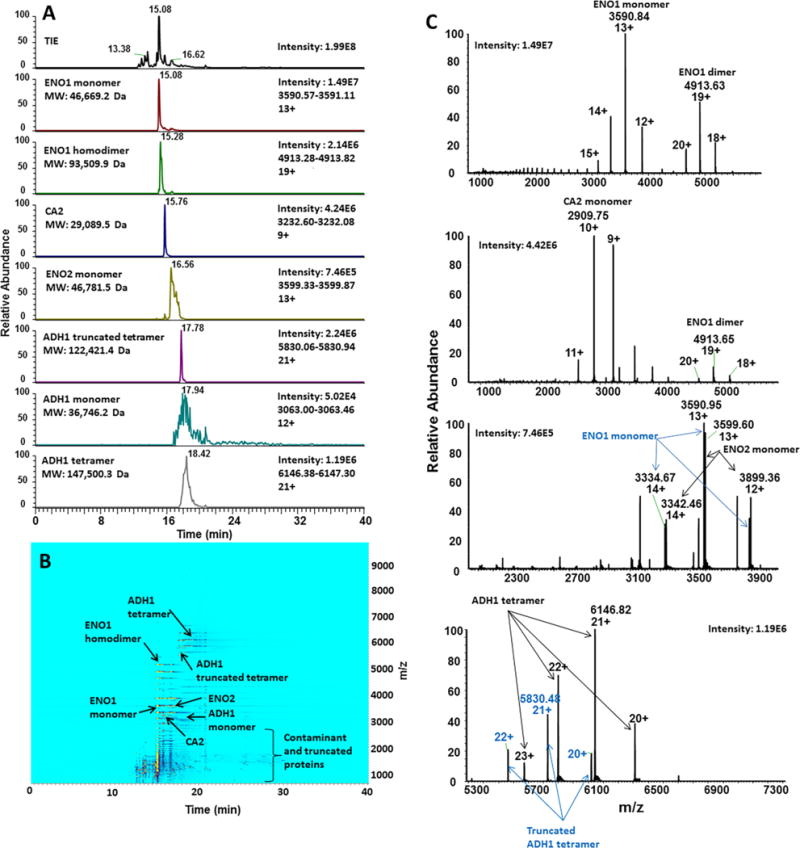
Monomers of ENO1 and ENO2, CA2, and ADH1 were detected and separated, as well as multimeric complexes for ENO1 (homodimer) and ADH1 (homotetramer and truncated homotetramer). For the separation performed on the sheathless BFS – Exactive Plus EMR system, extracted ion electropherograms (XIEs) for the most abundant ions of each protein/protein complex are shown (A). The separation and pattern and relative intensities can be observed on the ion density map (B). Examples of integrated MS spectra are shown for several major mixture components: ENO1 and ENO2, as well as truncated ADH1 and unmodified ADH1 tetramers (C).
CZE separation allowed structural analyses that could not be easily achieved using direct infusion. Partial separation was obtained between the ENO1 monomer (46,669.2 Da) and homodimer (93,509.9 Da), which were present at similar CZE-MS1 signal intensities (Figure 1A, C and Supplementary Figure 1). The separation yielded sufficient peak resolution between ENO1 monomer and dimer to enable accurate quantitative analysis and MS deconvolution (Figure 2). The latter is important for correct assignments of proteoforms and non-covalent binding (e.g. metal ions and multiple subunits) features that often cannot be analyzed by direct infusion. Both monomer and homodimer of ENO1 were also separated from ENO2 (46,781.5 Da), which has ~95% sequence homology with ENO1 and migrated as a broader peak (Figure 2A and Supplementary Table 1). Considering the high sequence homology of these isozymes, the peak width may correlate with the conformational stability of a protein under the employed separation conditions or the complexity of the mixture of proteoforms. The corresponding ENO2 homodimeric complex was not observed.
Figure 2. Native CZE-MS characterization of enolase 1.
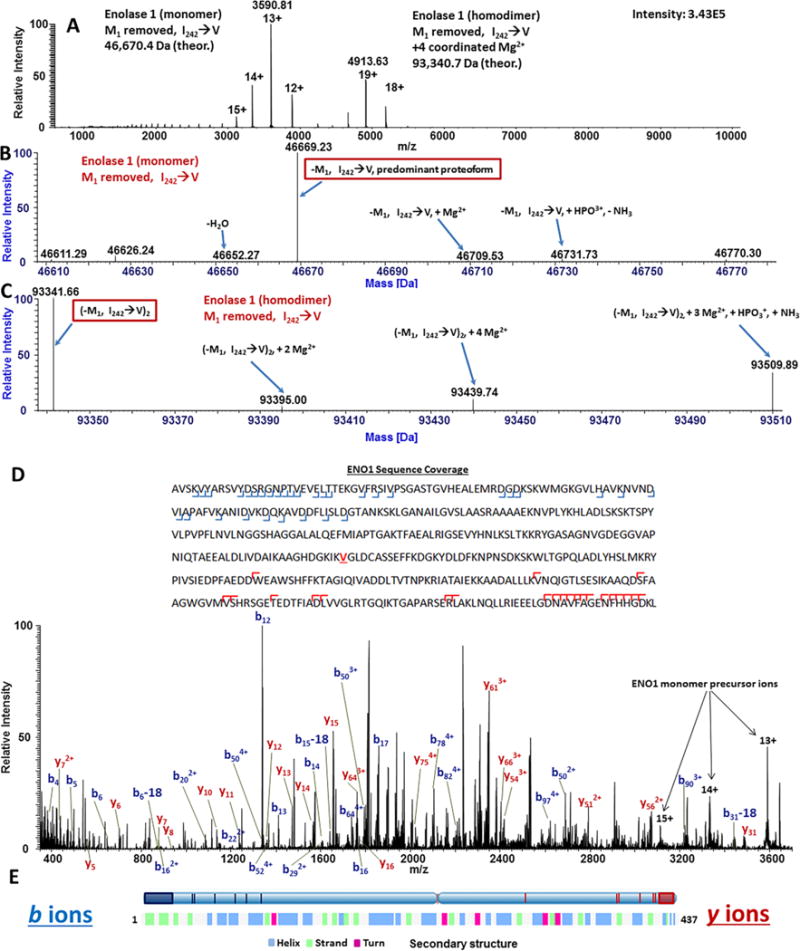
Averaged spectra across the entire migration time range corresponding to ENO1 electrophoretic peak resulted in the detection of both ENO1 monomer and homodimer (A). Deconvolution of the CZE-MS signal acquired during of the migration of the electrophoretic peak corresponding to ENO1 monomer resulted in the identification of major proteoforms of ENO1 (B). Deconvolution of the CZE-MS signal acquired during the migration time range corresponding mostly to ENO1 homodimer resulted in the identification of proteoforms of the dimer (C). Averaged AIF spectra of the entire migration time range corresponding to the ENO1 electrophoretic peak resulted in the generation of b- and y-ions characteristic to ENO1 (D). Fragmentation loci were mapped as blue (b-ions) and red (y-ions) vertical marks and shaded boxes on the secondary structure of ENO1, revealing that for the native globular protein ENO1, HCD fragmentation occurred predominantly on the N- and C-termini and turn loop regions (not colored on the lower bar representing the secondary structure of ENO1 (UniProt) (E).
For ADH1, monomer (36,746.2 Da) (8%) and homotetramer (147,500.3 Da) (92%) peaks were detected with slight shifts in migration times of peak maxima (Figure 3A). Based on these observations, information about the stability of complexes in biological samples and shifts in dissociation/association rates can be potentially generated based on CZE-MS1 quantitative characteristics of protein assemblies and corresponding monomeric subunits. In addition, another species of the ADH1 standard was also detected, likely corresponding to the homotetramer of truncated ADH1 (122,421.4 Da, experimental MW) (Figure 1A, C and Supplementary Table 1). Similarly, the combination of CZE separation and high mass accuracy MS detection under native conditions followed by MS deconvolution led to identification of CA2 (P00921, theoretical MW: 28,982.6 Da). Numerous contaminants, including intact and lower molecular mass truncated protein species were also detected in the mixture by both CZE-MS1 primarily within the 800 – 2,500 Th range (Figure 1B) and bottom-up nLC-MS analysis (see Supplementary Table 2).
Figure 3. Native CZE-MS analysis of alcohol dehydrogenase 1 and carbonic anhydrase 2.
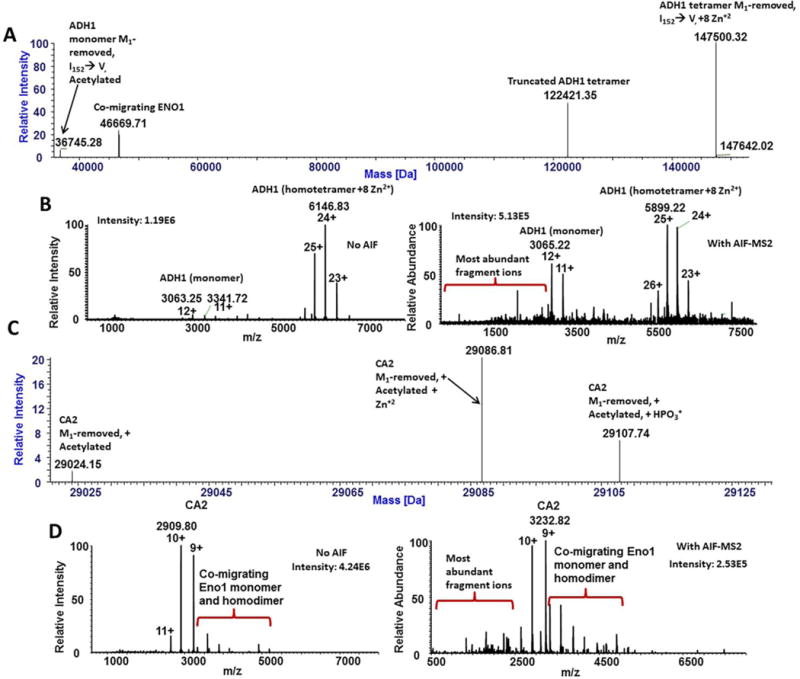
Deconvolution of the CZE-MS1 data in the electrophoregram region corresponding to ADH1 (17.5–22 min) resulted in the detection of peaks corresponding to monomer, tetramer, and truncated tetramer forms of ADH1 (A). MS fragmentation using HCD-AIF with a collision energy of 150 eV resulted in the fragmentation of ADH1 species, producing sequence-characteristic b- and y-ion fragments (B). Deconvolution of the electrophoretic peak corresponding to the CA2 (15.5–16.0 min) resulted in the identification of major proteoforms of CA2 (C). Fragmentation of the CA2 peak was achieved via HCD-AIF with collision energy of 150 eV, resulting in the generation of sequence-characteristic b- and y-ions (D).
Following the CZE-MS1 experiments, all-ion fragmentation (AIF) scans of all detected precursors were conducted in the HCD cell of the Exactive Plus EMR. The AIF fragmentation resulted in the generation of HCD-characteristic b- and y-ions that validated the major components of the mixture identified by CZE-MS1. In spite of the sample complexity, separation, combined with accurate intact mass measurements and AIF, allowed the characterization of proteoforms and protein complexes of the mixture. The majority of observed molecular masses of proteins and protein complexes in this mixture (see below) were mapped to theoretically expected masses, based on protein sequences reported in UniProt within a 30 ppm mass deviation (see Supplementary Table 1). In a few cases, the mass deviation was slightly higher, which we attribute to incomplete desolvation or non-specific binding of adducts.
Using the ReSpect and Sliding Window algorithms of the Protein Deconvolution 4.0 software suite, identities of proteoforms of the ENO1 monomer and homodimer were determined (Figure 3B, C). Fragment ion m/z values were mapped using a tolerance of 20 ppm (FDR<1%). In Figure 3B, the major form of ENO1 is seen to be a previously reported proteoform with an N-terminal Met-loss and a likely Ile → Val substitution at position 242 of the canonical protein sequence (P00924) [48, 49]. However, confirmation of the exact location of this substitution was not possible via AIF-MS. It should be noted that the observed variants of ENO1 are consistent with a previously reported primary structure and known modifications [48, 49]. Minor ion species (below 5%) likely corresponded to other reported ENO1-associated structural features, such as phosphorylation, oxidation, and magnesium ion coordination [50] (Supplementary Table 1). Additionally, products of incomplete ion desolvation and in-source decay were observed (Figure 2B). For the ENO1 homodimer, in addition to Met1-truncation and the Ile242→Val substitution, coordination of two magnesium ions per subunit was observed (Figure 2C), in agreement with the literature [51]. Confirmation of the identity of the Met-removed, Ile242 → Val variant as the most abundant ENO1 proteoform was achieved via mapping of the m/z values of fragment ions to the sequence with the above substitution and a fragment mass tolerance of 20 ppm (Figure 2D, Supplementary Figure 2). However, localization of the exact substitution site was not achieved. Similar to previously-reported findings for other proteins studied under native conditions, as shown in Figure 2E, mapping of the most abundant b- and y-fragment ions onto the ENO1 secondary structure revealed by X-ray crystallography [52], expectedly demonstrated that the majority of identified fragments corresponded to N-and C-termini and turn loop regions, which are typically exposed to the surrounding aqueous environment and tend to contain polar and charged amino acid residues. Supplementary Figure 2 shows the CZE-AIF-MS spectrum for ENO1 homodimer. As expected, the AIF fragmentation patterns for ENO1 homodimer and monomer were very similar, likely because the majority of fragment ions originated from the monomer that was released from the homodimer in the process of HCD activation.
The combination of native CZE separation with MS1 and AIF was also important for the characterization of ADH1 (Figure 1). Deconvolution of the ADH1 peak resulted in the detection of the ADH1 monomer with a MW of 36,745.3 Da, which corresponds to the N-terminal methionine truncated form, and the more prevalent homotetramer (12-fold higher) with a MW of 147,500.3 Da, which corresponds to the N-terminal methionine loss on each subunit and a total of eight coordinated Zn2+ ions (two Zn2+ ions per subunit are required for appropriate folding and functional activity of ADH1) (Figure 3A) [53]. Additionally, a likely truncated form of the ADH1 tetramer (122,421.4 Da) was observed in the mixture, as well as the ADH1 standard. The experimental mass of the monomer closely matched a previously reported ADH1 variant (P00330) with Met1-truncation and acetylation at the N-terminal Ser residue, along with a single substitution of Ile with Val at position 152 or 338 [53, 54]. This match resulted in a mass difference of 9 ppm between the experimentally determined and calculated average mass values (Supplementary Table 1). Similar to enolase, AIF of ADH1 led to the validation of the major forms of ADH1 determined from intact mass measurements (Figure 3B and Supplementary Figure 3A).
A previous study of ADH1 tetramers by direct infusion native MS on a Fourier transform ion cyclotron resonance (FTICR)-MS with electron capture dissociation (ECD) reported on the fragmentation of residues exclusively at the N-terminus of the protein [10]. The authors correlated this fragmentation pattern with the X-ray crystal structure of the ADH tetramer that showed that the N terminus of the protein is exposed and available for fragmentation, while the C terminus is hidden and involved in the interface of the subunits. Fragmentation coverage was also limited as a result of the formation of charge-reduced ADH1 tetramer species by ECD, resulting in no dissociation of ADH1 subunits from the tetrameric complex [10]. In our study, in addition to N-terminal fragments we also observed C-terminal fragment ions (Supplementary Figure 3A). A different mechanism of fragmentation (HCD AIF accompanied with in-source CID) was used in our experiments, in addition to a different mass analyzer type (Orbitrap) and MS instrument, which could be responsible for these differences in fragmentation patterns. Also, in our study, the combination of in-source CID used for desolvation and AIF possibly promoted partial dissociation of ADH1 tetramers into monomers, which made both protein termini accessible for fragmentation. In a different study, a variety of fragmentation techniques, including collisionally activated dissociation (CAD), infrared multiphoton dissociation (IRMPD), in-source dissociation, and ECD were used for the top-down characterization of native ADH using an FTICR-MS instrument [55]. CAD and, especially IRMPD, resulted in the significant generation of sequence-informative N- and C-terminal fragment ions. These results, and particularly CAD results, are similar to those obtained in our experiments using HCD AIF accompanied with in-source CID. However, expectedly, sequence coverage was considerably more comprehensive using CAD and IRMPD in [55] than our approach, involving HCD AIF. In our study, similar to the previous reports [10, 55], the sequence origin of the detected fragment ions can be correlated to the outer surface regions of the complex and protein termini.
The characterization of proteoforms and complexes of CA2 (P00921, theoretical MW: 28,982.6 Da) was achieved; deconvolution revealed three major proteoforms of CA2 (Figure 3C). The most abundant species (experimental MW: 29,086.8 Da) closely matched the proteoform with a loss of the N-terminal methionine, N-terminal acetylation, and a coordinated Zn2+ ion (calculated MW: 29,089.7 Da). Both Met1-truncation and coordination of Zn2+-ions by three histidyl residues of CA2 were previously reported [56]. The second most abundant form was tentatively identified to contain N-terminal Met-loss, N-terminal acetylation, and phosphorylation (experimental MW: 29,107.7 Da) (Supplementary Table 1). The third detected form of CA2 was determined to possess N-terminal Met-loss and acetylation (experimental MW: 29,024.2 Da), with an abundance of less than 4% relative to the Zn2+-coordinated species (Figure 3C, Supplementary Table 1). From CZE-MS utilizing AIF, proteoforms of CA2 were fragmented into corresponding b- and y-ions (Figure 3D). Analysis of fragment ion spectra resulted in the confirmation of the identity of the most abundant proteoform (Supplementary Figure 3B). Coordination of one Zn2+ ion by monomeric CA2 under native conditions and the primary structure of the most abundant form of CA2 – confirmed by the AIF MS – fragmentation pattern are also in good agreement with the crystal structure of this protein and previous reports [56].
Following the characterization of a simple mixture of three proteins, a more complex mixture of eleven protein standards known to form non-covalent multisubunit protein complexes and complexes with coordinated metal ions was evaluated using the sheathless CZE-MS system. However, first, the components were analyzed separately using a conventional bottom-up nanoLC-MS2 approach (FDR<1%). The sample heterogeneity extended well beyond the 11 commercial standards used to prepare the mixture, considering the number of contaminant proteins detected in the bottom-up approach (Supplementary Table 2). Sequences of these proteins were assembled into a protein database to facilitate identification of proteins and protein complexes (including identification of predominant proteoforms and sequence variants) in subsequent native CZE-MS1 experiments.
The BFS capillary demonstrated high separation performance in the analysis of the three protein mixture under native conditions because the choice of ammonium acetate, pH 7.0–8.5 as the BGE effectively suppressed adsorption of these specific proteins on the capillary surface. However, in CZE separations of intact proteins and protein complexes, the performance of BFS capillaries is often suboptimal due to the large number of surface charges – both on the protein and on the inner capillary surface – thereby leading to non-specific adsorption to the capillary wall and hence, potentially resulting in peak tailing, lower separation efficiency, and carryover issues [28, 57–59]. Thus, the separation performance of the BFS capillary was found inadequate for the mixture of 11 proteins and associated complexes as well as for higher complexity samples and high molecular mass proteins (e.g. mAbs). Presumably, ionic strength and salt concentration of the ammonium acetate BGE used in these experiments was insufficient for effectively preventing adsorption for a number of proteins of the mixture. Therefore, a recently-commercialized capillary with an uncharged polyacrylamide coating preventing protein adsorption [28] was used for analysis of a higher complexity samples.
The components of the mixture of 11 proteins and associated complexes migrated between 15 and 35 minutes and, as expected, many of them were detected in native CZE-MS but not in direct infusion analyses (Figure 4, Supplementary Figure 4). Successful characterization of the mixture of 11 proteins and other higher complexity samples via direct infusion native MS was precluded by considerable overlapping of m/z’s of different proteins, resulting in difficult to interpret, less informative spectra. Moreover, in direct infusion native MS mode, difference in abundances and ionization efficiencies between individual analytes resulted in considerable signal suppression of lower abundance and lower ionization efficiency species.
Figure 4. Characterization of the complex mixture on the sheathless cPA capillary-Exactive EMR system.
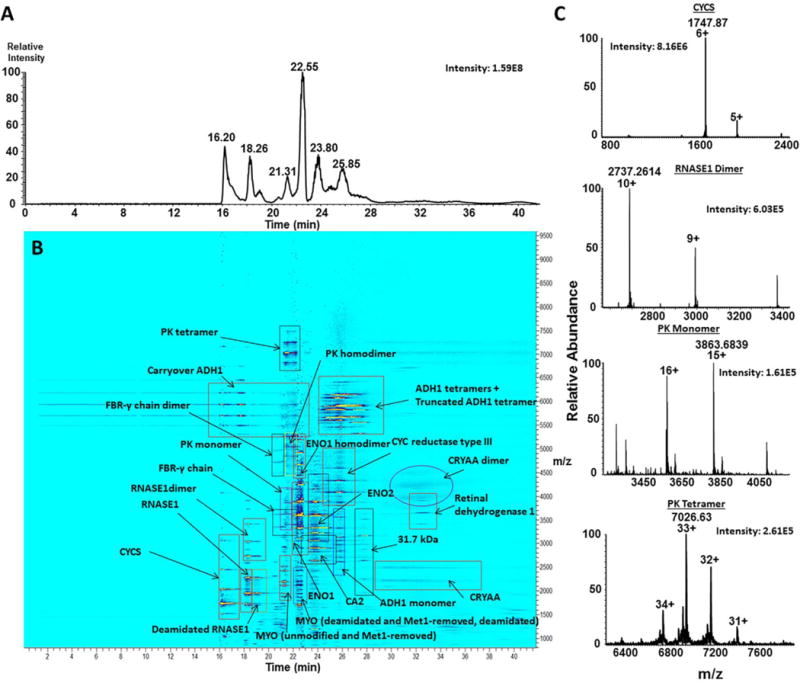
The mixture of 11 complex-forming proteins was separated and characterized on the sheathless polyacrylamide-coated – Exactive EMR system and resulted in the detection of all 11 protein standards in their monomeric forms, in addition to multisubunit complexes for PK, ADH1, RNASE1, ENO1, CRYAA, and truncated ADH1 as well as contaminant proteins and truncated species. Panel (A) represents the total ion electropherogram for the separation, (B) represents the annotated ion density map, showing migration times (horizontal axis) of detected proteins vs. their corresponding m/z values (vertical axis), and (C) represents native MS spectra for CYCS monomer, RNASE1 homodimer, PK monomer and PK homotetramer detected on the Exactive Plus EMR.
Accurate mass measurements from the native CZE-MS1 run were used for the identification of major forms of individual proteins. A higher molarity BGE (40 mM, relative to the 25 mM ammonium acetate we used previously), was utilized in these experiments to improve the separation and more-closely mimic physiological conditions, thereby improving the stability of non-covalent protein complexes in the mixture during the analysis. It should be noted that the molarity of the ammonium acetate BGE used in this study was limited by the performance and the long-term stability of the porous sheathless emitter. The 40 mM ammonium acetate BGE did not result in any changes in spray stability and the lifetime of the emitter. Separation efficiency exceeding 150,000 theoretical plates per meter were readily observed for selected components of the mixture using the employed native conditions.
Detection of large, multimeric protein complexes, such as ADH1 (~148 kDa) and pyruvate kinase (PK) (~231 kDa), was enabled by the Exactive Plus EMR. The enhanced gas flow into the HCD cell of the instrument allowed for collisional cooling and trapping of large proteins and complexes. The combination of advanced CZE- and MS-based technologies enabled the separation of individual proteins and detection of corresponding complexes (Figure 4B and Supplementary Figure 4). Baseline CZE separation was achieved between isozymes of enolase – ENO1 and ENO2; however, ENO1 monomer and homodimer were not fully resolved. Similarly, ADH1 monomer, homodimer and homotetramer were not well-resolved electrophoretically, although the corresponding peaks were of different shapes and demonstrated noticeable offsets in migration times, which in combination with MS detection made the corresponding CZE-MS1 migration patterns easy to distinguish. Also, the ADH1 monomer peak was considerably sharper relative to the experiments conducted on the BFS capillary. The peak intensity for the tetramer was approximately 90 and 80 fold higher than for the monomer and the dimer, respectively, which reflects the abundance ratios of these forms in solution. Examination of the extracted ion electropherograms corresponding to the most abundant charge states of these forms indicates small levels of in-source dissociation of the ADH1 complexes (the tetramer into the dimer and both the tetramer and the dimer into the monomer). Monomer (~58.0 kDa), homodimer (~116 kDa) and homotetramer (~232 kDa) PK species were detected as peaks of distinct shapes and with visible offsets in migration time of their peak maxima. In the case of the PK species, the monomeric species showed signal intensity approximately 2–3-fold higher relative to the dimer and tetramer peaks. Low levels of in-source dissociation of the multisubunit PK complex were also evident from the comparative examination of the extracted ion electropherograms for all three detected PK species. Other detected multisubunit complexes included RNASE1 dimer, alpha-crystallin (CRYAA) dimer, and fibrinogen-γ (FBR-γ) dimer. Baseline resolution was achieved between unmodified and deamidated myoglobin (MYO), as well as Met1-removed and the deamidated form of Met1-removed MYO. Similarly, deamidated RNASE1 was separated from the unmodified proteoform (Figure 4, Supplementary Figure 4). The CZE separation allowed us to distinguish and reliably identify these near-isobaric species with a mass difference of close to 1 Da, and showed the potential for relative quantitation of such proteoforms and complexes, which would be considerably more challenging in direct infusion experiments.
Native CZE-MS of a monoclonal antibody (mAb)
A recombinant, humanized monoclonal anti-Her2 IgG1 antibody, Trastuzumab, was examined under native conditions as a means to assess the heterogeneity of a biopharmaceutical protein and determine the potential presence of aggregates. To our knowledge, this experiment represents the first attempt of the characterization of an intact mAb using online native CZE-MS. A neutral polyacrylamide-coated capillary was coupled to the Exactive EMR Plus mass spectrometer and used for this experiment. Figure 5A shows the separation of the antibody sample components in a BGE of 20 mM ammonium acetate pH 8.0. The total ion electropherogram is shown in the top panel, followed by extracted ion electropherograms of major identified species. Predominant forms of the intact mAb, corresponding to 2X-glycosylated, 1X-glycosylated (hemi-glycosylated), and dimeric structures co-migrated at 24.4 min. Molecular species of approximately 101 kDa, presumably corresponding to the mAb with a loss of two light chains, were detected at 26.62 min and separated from the major forms of Trastuzumab. The corresponding dissociated light chains were detected as monomeric forms at 24.41 min and as dimeric forms at 22.83 min (Figure 5A). This supports previously reported observations that in some mAbs, individual chains are held together by non-covalent interactions instead of disulfide bonds [60, 61]. As a result, these chains may dissociate, especially if exposed to conditions other than the formulation buffer. Since these structures were separated by CZE, the dissociation of chains did not occur inside the mass spectrometer.
Figure 5. Native CZE-MS analysis of Trastuzumab.
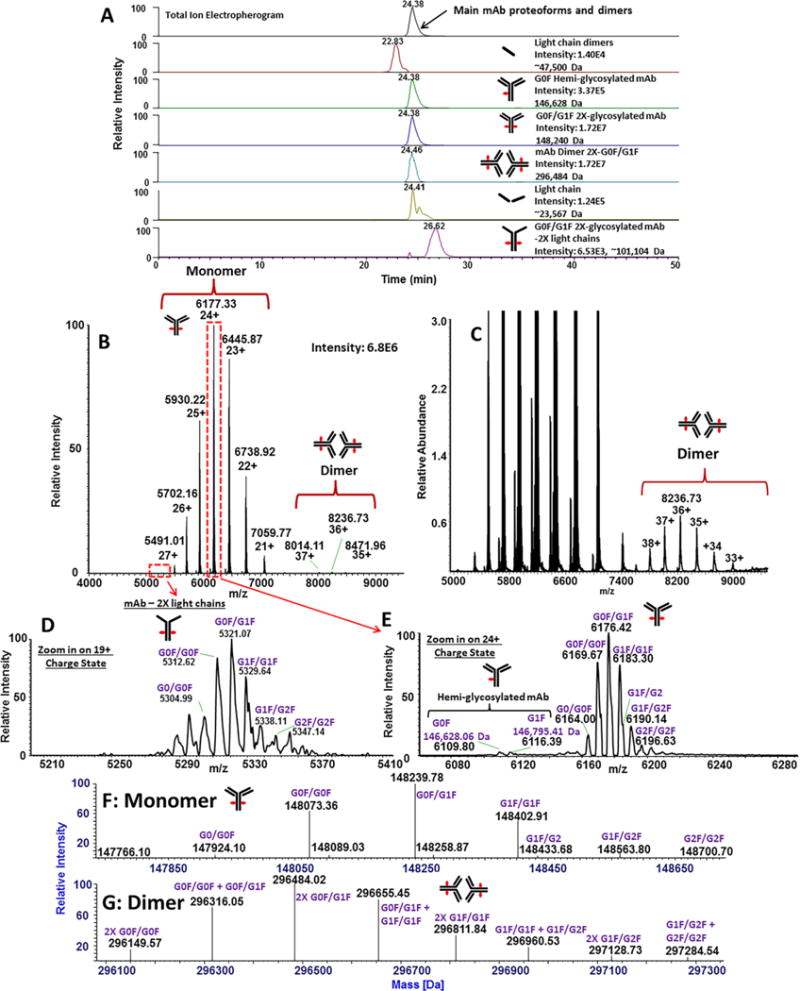
The total ion electropherogram is shown in the top panel, as well as identified components of the mAb represented by extracted ion electropherograms. An averaged spectrum across the entire electrophoretic peak resulted in the identification of charge states corresponding to monomeric and dimeric forms of Trastuzumab (B). A zoomed-in spectrum revealed the m/z distribution of Trastuzumab dimers (C). A zoom-in of the 19+ charge state of species corresponding to the mAb with depleted two light chains is shown (D). A zoomed-in view of the 24+ charge state shows 2X-glycosylated and 1X-glycosylated (hemi-glycosylated) mAb structures (E). Deconvolution of Trastuzumab monomer peaks resulted in the identification of major glycoforms of the protein (F). Deconvolution also resulted in the characterization of dimeric forms, corresponding to combinations of glycoforms of Trastuzumab (G).
The spectrum integrated across the main electrophoretic peak (24.38 min) revealed the presence of the monomer and a trace (<1%) of a dimeric species (Figure 5B, C). As stated above, the mAb depleted of two light chains was also detected. As an illustration of this component, a zoomed-in view of the 19+ charge state of these forms is shown Figure 5D. A zoomed-in spectrum of the 24+ charge state of the main forms is shown in Figure 5E, revealing 1X- and 2X-glycosylated structures of the intact mAb. For the 2X-glycosylated forms, deconvolution of the MS signal from the entire electrophoretic migration window of the mAb sample constituents resulted in the identification of several biantennary glycoforms (Figure 5F). In addition, several dimers (assumed to be non-covalently associated) were also observed (Figure 5G). It is important to note that dimers of Trastuzumab could not be observed in denaturing CZE-MS (or LC-MS) experiments conducted by us and others [62].
The most prominent charge states, 24+ and 36+, were observed for the monomer and the dimeric (aggregated) mAb species, respectively (Figures 5B, C). Deconvolution of native CZE-MS1 of Trastuzumab was performed using ReSpect with the Sliding Window algorithm. Depending on the target mass range (e.g. 100 – 300 kDa, 140 – 150 kDa, 280 – 300 kDa) and m/z range (e.g. 5,000 – 9,000 Th, 7,500 – 9,000 Th, 5,000 – 6,500 Th), the intensities of the resultant peaks corresponding to Trastuzumab dimers were ~140 – 250 fold lower than the intensity of the predominant peak corresponding to the monomer. Relative quantitation of the monomer and the aggregate can therefore be achieved using the native CZE-MS approach.
The average experimental molecular mass of the predominant form of the antibody is 148,073.36 Da (Figure 5F), which is a close match (15.81 Da greater) to the theoretical molecular mass of 148,057.55 Da calculated from the sequence reported in reference [63] with two G0F moieties and 16 disulfide bonds that corresponds to the molecular formula C6560H10132N1728O2090S44. The difference between the theoretical and experimental molecular masses of the dimer was 34.48 Da, possibly due to strong coordination of one water molecule or replacement of a proton with an ammonium ion per monomer. It is worth noting that DrugBank (www.drugbank.ca/drugs/DB00072) has a different sequence for the heavy chain (HC) of the innovator version of Trastuzumab (Genentech), including DELTK for residues 360–364 of the HC instead of the EEMTK, and an extra P in the sequence with VEPPK instead of VEPK at residues 218–222 of the HC, as also reported by others [63, 64]. Additionally, the absence of the C-terminal K is yet another difference between the DrugBank sequence and the one reported in [63], which also agrees with our experimental findings.
From deconvolution of the MS1 spectrum corresponding to the monomer, several combinations of biantennary glycans in the FC region of the monomer were detected under native CZE-MS experiments (Figure 5G). These structures represent glycosylation at both FC/2 domains corresponding to the expected G0/G0F, G0F/G0F, G0F/G1F, G1F/G1F, G1F/G2, G1F/G2F, and G2F/G2F forms. The presence of asymmetric glycosylation, characterized by different glycan compositions for both FC/2 domains in monoclonal antibodies, has also been reported previously for mAbs, including Trastuzumab [62, 65]. Deconvolution of the MS1 spectra of dimeric Trastuzumab, presented in Figure 5G, shows the composition of dimeric proteoforms. In addition to homodimers, characterized by symmetric Trastuzumab monomers (2X G0F/G0F, 2X G0F/G1F, 2X G1F/G1F, 2X G1F/G2F), heterodimers characterized by asymmetric monomers (G0F/G0F + G0F/G1F, G0F/G1F + G1F/G1F, G1F/G1F + G1F/G2F, G1F/G2F + G2F/G2F) were also detected (Figure 5G). Interestingly, dimers were determined to be combinations of 2X-glycosylated Trastuzumab forms, with no dimeric species containing partially deglycosylated subunits (i.e. 1X-glycosylated).
While identifications of the mAb components presented above could be also achieved by direct infusion native MS conducted using the same capillary and the CESI-MS platform at the identical flow rate and MS conditions, we expect that the CZE separation alleviates the ion suppression effects that are still present in the direct infusion mode experiments even at such low flow rates as 20–40 nL/min and improves the detection (and potentially relative quantitation) of minor sample components when combined with native MS.
It should be noted that a variety of other approaches exist for the identification of protein aggregation, including SEC coupled to multi-angle light scattering (MALS) [66] and NMR [67]. However, for SEC-MALS, detailed characterization of subunit composition may be not achieved. For example, individual glycosylation states of Trastuzumab dimers, as identified in this study, could not be characterized by SEC-MALS. For current NMR methodologies, while smaller protein dimers may be identified, mAbs such as Trastuzumab are too large for their structure to be resolved with these approaches [68]. Thus, techniques such as native CZE-MS, enabling characterization of protein sequence and identification of higher order structure, can potentially provide important structural information. The ability to detect non-covalent assemblies of mAb molecules can be used to explore the pathways of aggregation (native and non-native) [69, 70] and assess the extent of protein aggregation, which is precluded by most other separation techniques that could not be performed online with native MS. The native CZE-MS approach can be potentially further extended to the relative quantitative or semi-quantitative analysis of aggregates in mAb-based immunotherapeutics, antibody-drug conjugates or other biopharmaceutical proteins, as well as different types of protein complexes in heterogeneous biological samples.
Native CZE-MS of the E. coli ribosomal extract
In this study, we also assessed the capabilities of native CZE-MS in the characterization of biological samples and proteomes of low complexity, using the protein isolate from the E. coli ribosomes. Ribosomes are non-membranous organelles that synthesize proteins from mRNA. These complex molecular machines are typically composed of over 50 distinct proteins with one to three rRNA molecules, depending on the organism. Ribosomes have traditionally been viewed as complex reproducibly-assembled ribozymes of a virtually fixed protein composition, whose function is to catalyze protein synthesis with a limited to non-existent ability to regulate this process [71]. Recent studies have revealed that ribosomes may be highly heterogeneous in their composition among various cell and tissue types [71–73]. These studies also indicated the possibility of variable protein-protein interactions within ribosomes that may alter their functional activities and specializations [71–75]. Detailed proteomic profiling of ribosomes and analysis of protein-protein interactions are necessary for elucidating possible mechanisms of specialized ribosomal activities. Native infusion MS analysis of the non-lysed ribosome and ribosomal subunits was recently demonstrated [41]. To our knowledge, however, no MS-based studies of ribosomal proteins performed with online separations under native conditions have been reported. In this work, we evaluated the capabilities of sheathless CZE with a commercial polyacrylamide-coated capillary coupled to a Q Exactive Plus mass spectrometer for characterization of individual intact proteoforms and non-covalent assemblies from a model sample of the E. coli ribosome protein extract under native conditions (Figure 6A–B). Data dependent acquisition (Top N) was used to generate MS/MS spectra. The results were then compared to those of CZE-MS analysis performed under denaturing conditions (Figures 6C–D).
Figure 6. Comparison of native and denaturing CZE-MS analysis of ribosomal proteins.
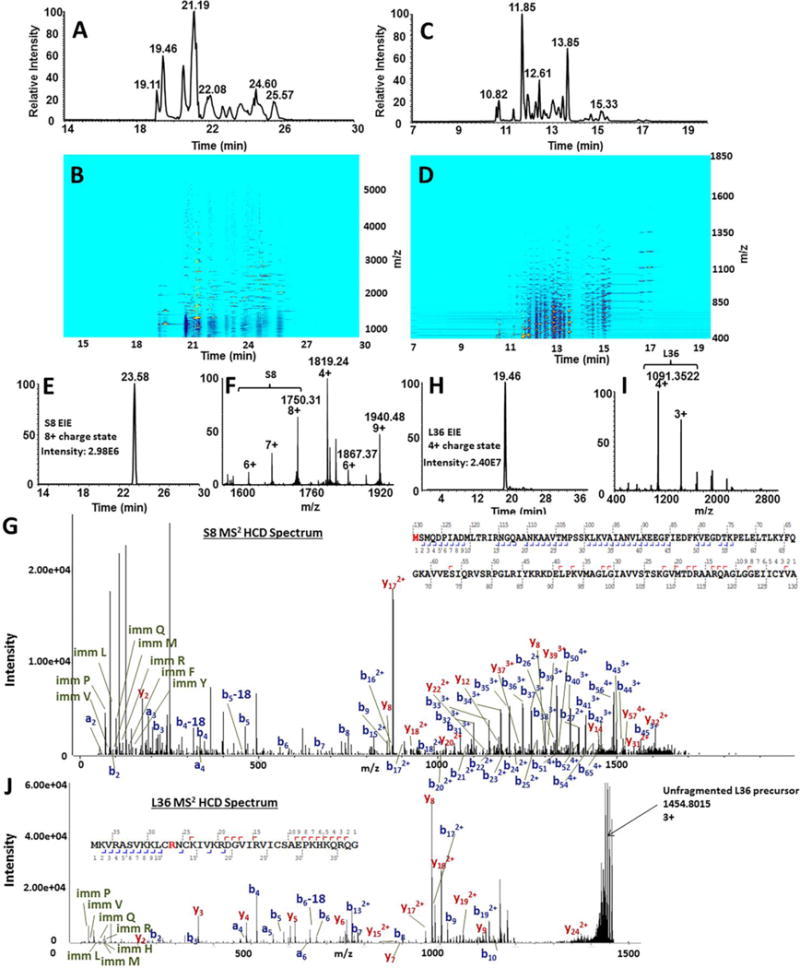
Combined results of native and denaturing CZE-MS analyses of ribosomal proteins on the polyacrylamide-coated capillary resulted in the total identification of 51 RPs. Panel (A) shows the total ion electropherogram for the experiment performed under native conditions. The ion density map for the native experiment, showing the migration time (horizontal axis) vs. m/z (vertical axis), is shown in (B). The total ion electropherogram for the denaturing separation of RPs separation with a 3% acetic acid BGE (pH 2.5) is shown in (C) and the ion density map is shown in (D). An example of RP characterization using the developed workflow is demonstarated for S8 (14.0 kDa). Native CZE-MS1 analysis resulted in the migration of S8 at 23.58 minutes, represented by the extracted ion electropherogram for the most prominent charge state (+8) (E). The ESI mass spectrum for the entire S8-corresponding electrophoretic peak is revealed in (F). Fragmentation of S8 via DDA-HCD resulted in unambiguous identification of S8 with a N-terminal Met-loss. Detected b- and y-ion distributions are shown in (G). Another example is shown for RP L36. The extracted ion electropherogram for L36 is shown in (H), followed by the corresponding ESI mass spectrum (I) and the CZE-MS2 spectrum with identified fragment ion series (J).
The compiled sequence database of ribosomal proteins (RPs), in combination with native CZE-tandem MS, allowed the identification of 42 ribosomal protein groups, 603 proteoform-spectrum matches (PfSMs), and 137 proteoforms in a single experiment at <1% FDR (Supplementary Tables 3, 4). An additional six RP identifications, corresponding to 48 in total, were made from deconvolution of native CZE-MS1 data and manual matching of experimental and theoretical molecular masses (Supplementary Table 5). The combination of native CZE-MS1 and CZE-MS2 resulted in identification of close to 90% of the expected E. coli RPs. The migration order of RPs was closely-related to hydrodynamic volume, as smaller proteins (e.g. L36, L33) were detected first and larger proteins (e.g. S8, S9) last (Figure 7A–D, Supplementary Figure 5).
Figure 7. Comparison of ribosomal protein identificaiton between native and denaturing CZE-MS analyses.
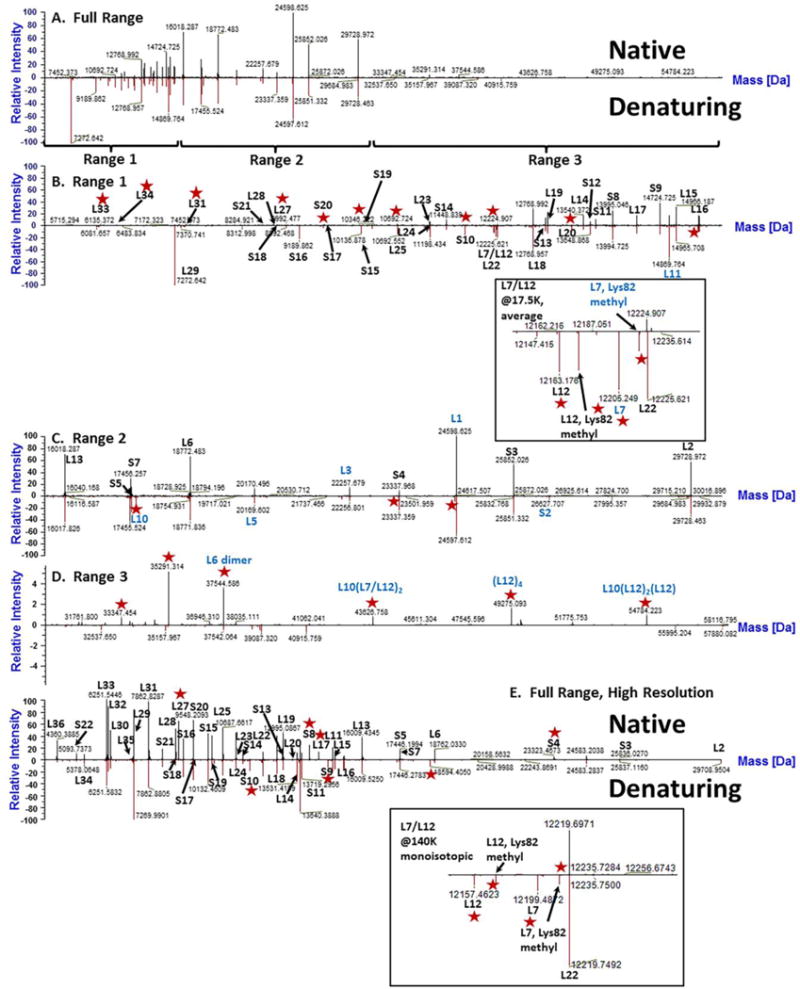
CZE-MS1 analyses of proteins extracted from the E. coli ribosomal isolate were conducted under native and denaturing conditions using ammonium acetate (pH 7.5) and 3% acetic acid (pH 2.5) BGEs, respectively. MS data acquisition was performed using either low or high resolving power (17,500 or 140,000 at 200 Th, respectively) on a Q Exactive Plus instrument. (A). A comparison of deconvolution results between native (all top panels) and denaturing (all bottom panels) CZE-MS1 analyses resulted in a similar distribution and quantity of identified proteins in mass Ranges 1 and 2 (4,000 – 15,000 Da and 15,000 – 31,000 Da) (B–C). However, for higher molecular weight species, native separations resulted in the identification of more peaks than the corresponding denaturing run (D). For CZE separations performed with high resolution MS data acquisition, numerous low MW proteins were identified that were not observed through deconvolution of low resolution spectra (E). Specifically, L36, S22, and L34 – all below 5,550 Da – were readily identified. Identifications of specific ribosomal proteins were achieved either via Byonic analysis of CZE-MS2 data (black labels) or via manual interpretation of CZE-MS1 deconvolution data (blue labels). Peaks elucidated by one set of conditions (native or denaturing) and not the other are annotated with red stars. For both low resolution and high resolution experiments, zoom-in views of the region corresponding to proteins L7/L12 and L22 (12,150 Da – 12,270 Da) are shown as separate inserts.
Next, we conducted conventional CZE-MS characterization under denaturing conditions (Figure 6C–D), and a comparison of RP species identified through deconvolution of native and denaturing CZE-MS1 data is shown in Figure 7. The number of molecular features detected at the low molecular weight portion of the deconvoluted spectrum (<15,000 Da) under both conditions CZE-MS1 experiments was comparable (Figure 7 A, B). However, in the higher molecular weight region (>30,000 Da), the native condition resulted in the detection of a larger number of molecular ion species, potentially corresponding to non-covalent protein complexes (Figure 7 A and C). Protein-protein interactions, especially in the stalk complex of the ribosome, are not well-characterized, which made assignment of high molecular weight species difficult (Figure 7D) [76]. These results were then supplemented with a denaturing CZE-MS2 experiment (Supplementary Table 4) where a lower number of species were found. We explain this by the “dilution” of the analyte signal across more charge states and the reduced spacing between protein ion species of different charges as well as between species with subtle mass differences, resulting from higher net charges under denaturing conditions. Nevertheless, the denaturing CZE-MS2 experiments resulted in identification of three additional RPs not found under native conditions, demonstrating the complementarity of the approaches. In total, the combination of native and denaturing CZE-MS approaches resulted in the identification of 51 out of 55 known proteins of the E. coli ribosome, close to 200 different proteoforms, and 679 PfSMs, which was unachievable in direct infusion experiments.
The above identification of RPs from native and denaturing CZE-MS2 experiments was achieved using Byonic software (Protein Metrics). In our searches, we used a fragment mass tolerance of 20 ppm and an FDR<1%. As an example of RP identifications, in Figure 6, we show the extracted ion electropherogram (Figure 6E) and specific precursor ions for the ribosomal protein S8 (MW: 14.1 kDa) (Figure 6F). The corresponding CZE-MS2 spectrum is then matched to theoretical peaks in situ, resulting in the identification of the protein with an N-terminal Met-truncation, which matched the corresponding experimental mass with an error of ~5 ppm (Figure 6G). Another example involves the ribosomal protein L36 (MW: 4.37 kDa). For L36, the extracted ion electropherogram is shown in Figure 6H, followed by the detected precursor ions, shown in Figure 6I. The CZE-MS2 spectrum used to identify an oxidation with high confidence at Arg11 is then shown in Figure 6J. Other ribosomal and ribosome-associated proteins and their specific PTMs, as well as non-covalent complexes, were determined in an analogous manner and are shown in Supplementary Table 5.
We also processed the CZE-MS2 data using ProSightPC in addition to the analysis via Byonic. While the re-analysis of native experiments did not yield additional identifications, searches of the denaturing data resulted in the identification of an additional five ribosomal proteins (L24, L6, L14, L34, S10). However, the ProSightPC searches did not yield any additional RP identifications through the combination of native and denaturing approaches. Nevertheless, several ribosome-associated proteins and additional PfSMs corresponding to RPs, not previously found by Byonic, were identified by ProSightPC. The former included stationary-phase-induced ribosome-associated protein and cold shock protein A that were likely co-purified along with other ribosomal proteins during the isolation and protein extraction steps.
Identification of ribosomal extract components by both software tools was achieved using database searching of CZE-DDA-MS2 against a database constructed on the basis of the results from the bottom-up LC-MS2 analysis (i.e. bottom-up guided native MS analysis) (Supplementary Table 3). Identification of individual proteins and protein complexes from a complex mixture would be considerably more challenging without a target database (e.g. limited samples) and may lead to a lower profiling depth and an increased FDR. Higher efficiency fragmentation techniques and more advanced bioinformatics approaches (both database- and spectral library-based in combination with MS deconvolution) are needed to address these limitations.
It should be mentioned that in this study, glacial acetic acid was used to precipitate ribosomal RNA for the extraction of RPs, similar to the approach described in [42]. A brief exposure to denaturing conditions was then followed by reconstitution of the protein extract into physiological pH buffer (ammonium acetate, pH 7.5). Even a brief exposure to denaturing conditions is expected to result in protein unfolding and dissociation of non-covalent interactions for at least some complexes. However, certain non-covalent interactions, including coordination of metal ions and multisubunit protein-protein interactions, can be stable at such conditions [77–79]. We anticipated that the reconstituted RP extracts would contain non-covalent complexes that were (a) stable enough to survive the denaturing conditions and (b) re-assembled after the buffer exchange to native conditions. While the latter reconstituted complexes may not exactly represent structural features characteristic to naïve ribosomes due to protein misfolding and scrambled interactions, we used the ribosomal protein extract as a model sample to assess the capabilities of native CZE-MS in analysis of biological samples containing non-covalent protein complexes.
In contrast to analytical approaches requiring denaturing conditions, CZE-MS2 characterization of the E. coli RP extract under native conditions resulted in the detection of multiple non-covalent protein-metal ion complexes. Metal ions (e.g. Mg2+, Zn2+) coordinated with specific ribosomal proteins are essential to the functional integrity of the ribosome [80]. CZE-MS2 under native conditions allowed for the detection of ribosomal proteins L13, L17, and L22 with coordinated Mg2+ cofactors and L16, L29, L35, and S18 with coordinated Zn2+ cofactors. This was evident from both accurate intact mass measurements and fragmentation spectra, even though unambiguous localization of the coordination sites was often not possible from the acquired HCD spectra (Supplementary Figure 6A–C). Several PTMs were readily detected including oxidation, N-terminal methylation and acetylation, internal methylation and acetylation, N-terminal methionine truncation, phosphorylation, and deamidation. Methylation at the N-terminal α-amino group is an important PTM commonly found on large macromolecular assemblies, such as ribosomes [81]. N-terminal methylation of S11, L16 and L33 was unambiguously elucidated via native CZE-MS2 analyses (Supplementary Figure 7A–C), which are in agreement with findings reported in a previous study [82]. N-terminal acetylation is also a common protein PTM that confers stability to proteins, in addition to being crucial for cellular localization, regulation, and function. N-terminal acetylation at S5 and S18 was reliably confirmed via native CZE-MS2 (Supplementary Figure 8A–B), in agreement with previous reports [82–85]. In both cases, acetylation was observed on N-terminal Ala residues of the Met1-truncated proteoforms, as expected. Numerous oxidation sites, N-terminally deamidated, and Met1-truncated proteoforms of RPs were also observed. Methylation of Lys and Arg, as well as di-methylation of Lys, were also found on various internal residues of RPs (L25, S8, S3, S16, S12, L2, L16, L35, L24, L20, L17, and S4). However, localization of the aforementioned PTMs at specific residues was not possible in some cases due to incomplete sequence coverage from DDA-HCD MS2. Similarly, a phosphorylated variant of L36 was also detected, with the corresponding PTM likely at Ser6. All proteoforms identified from CZE-MS2, as well as their match scores, determined and expected molecular masses, and sequence coverage values are summarized in Supplementary Table 4. These results show that the native CZE-MS approach can be effectively used for detailed structural characterization of ribosomes and other macromolecular complexes to better understand their functional specialization and biology. However, alternative, gentler protein extraction techniques that do not require denaturing conditions should be used to more closely characterize biologically relevant complexes. In comparison, direct infusion conducted under native conditions does not readily allow for the identification of lower abundance proteoforms in such complex biological samples.
The native MS approach to protein analysis requires sophisticated and specialized MS instrumentation. For example, selection of precursor ions for MS2 fragmentation is limited by a 2,500 Th upper selection threshold on the quadrupole of the Q Exactive Plus mass spectrometer. A considerable number of analytes were above the 2,500 Th threshold, thereby precluding their selection for MS2 analysis. However, under denaturing acidic conditions, due to the overall increase in protein protonation and the corresponding decrease in m/z distributions of precursor ions, MS2 analysis of larger ionized molecules was possible. In our case, denaturing CZE-MS2 experiments resulted in additional identifications of RPs and their PTMs. For example, proteins S21, L7/L12, and L11 were detected in the intact CZE-MS2 experiment using a denaturing BGE but not observed with the native BGE; presumably these proteins were present as multi-subunit RP complexes under native conditions.
Detected exclusively under denaturing CZE-MS conditions, protein L7/L12 is an extensively-studied ribosomal protein that is biologically active as a tetramer bound to the protein L10. Proteins L7 and L12 possess identical primary structures and differ by a single PTM at their N-termini. L7 is known to be acetylated at the N-terminal Ser residue, while L12 is known to possess methylation instead of acetylation at the same residue [85, 86]. Additionally, proteins L7 and L12 are singly-methylated at their ε-amino groups of residues Lys81 [82]. L7/L12 forms a pentameric complex with L10 (L10(L7/L12)2(L7/L12)2) as a part of the ribosomal stalk called L8 [76, 87]. The L8 complex interacts with the translation factors IF2, EF-Tu, EF-G, and RF3, which were not identified in native or denaturing analyses; however, several of these were detected in bottom-up experiments (Supplementary Table 3). The top-down CZE-MS2 approach with the denaturing BGE employed in this work resulted in the identification of internal methylation at either Lys84 or Lys81 of L7/L12. While the exact localization of the PTM was not achieved via DDA-HCD MS2, previous studies reported on internal methylation at Lys81 [82] (Supplementary Figure 9A). Additionally, N-terminally acetylated (L7) (Supplementary Figure 9B) and non-acetylated (L12) variants were also detected (Supplementary Figure 9C).
A potentially important application of native CZE-MS is the detection of protein-protein interactions. However, as stated previously, due to the sample preparation technique used in this study to extract RPs, protein-protein and protein-metal ion complexes detected after sample reconstitution may not represent native confirmations. Here, we demonstrate proof-of-principle results to show the potential of native CZE-MS in characterization of non-covalent protein complexes and interactions. For the ribosomal protein extract, several complexes were detected in native CZE-MS analyses. Several peaks detected exclusively under native conditions were interpreted as potential non-covalent complexes, including such constituents of L8 as L10(L7/L12)2, (L12)4 tetramer, L10(L12)2(L12) tetramer [87] (Figure 7D), while both L7 and L12 were revealed only under denaturing analysis (Insert in Figure 7B and Figure 7C). Also, species corresponding to the molecular weight of 37,554 Da were detected under native conditions and presumed to be a homodimer of L6, which was identified in several MS2 spectra in its monomeric form (18,772 Da) (Supplementary Figure 10A, B). These non-covalent associations were not detected in our denaturing CZE-MS experiments, native MS direct infusion experiments, or in the previously-reported analyses of the E. coli ribosomal protein extract via nanoLC-MS conducted under denaturing separation conditions [43].
The combined advantages of the polyacrylamide-coated capillary and the Q Exactive Plus, coupled via the commercial sheathless CESI interface, enabled analysis of the protein isolate of the E. coli ribosomal extract with confirmation of protein identities with DDA-HCD-MS2. Out of the total 55 unique E. coli ribosomal proteins, a single native CZE-MS2 experiment resulted in the identification of 42 proteins of the ribosomal proteome. Additional identifications made from native CZE-MS1 data (6) and the top-down CZE-MS2 experiment performed under denaturing conditions (3) resulted in a total of 51 identifications, corresponding to approximately 93% of the expected proteins present in the E. coli ribosomal proteome. The depth of RP profiling is similar to the one reported in a recent study (46 RPs identified) that used the top-down reversed-phase LC-MS approach [43]. Additionally, our native CZE-MS approach resulted in identification of several non-covalent protein-protein and protein-metal ion RP complexes in the reconstituted ribosomal protein extract, which demonstrates the potential of the technique to characterize protein interactions in biological and clinical samples. In our hands, direct infusion native MS experiments were far less informative and failed to provide the similar depth of RP profiling, which highlights the need for high resolution and high sensitivity native separation coupled online to MS. Separation efficiency exceeding 150,000 theoretical plates per meter were observed for selected analyte species under native CZE-MS conditions. Both high resolution and high sensitivity are among the important attributes of native CZE-MS, which were critical for enabling deep profiling of the ribosomal proteome using limited sample amounts.
Conclusions
This work represents one of the first examples of online high performance liquid-phase separation directly interfaced with MS under native conditions. The paper demonstrates the potential of CZE for high resolution separation of proteins, protein complexes and organelle proteome-level samples under native conditions coupled online to mass spectrometry. Native CZE-MS using commercially-available instrumentation enabled detailed structural characterization of such analytes in their native, biologically active states with unaltered non-covalent interactions and spatial molecular configurations. Attributes of the online CZE-MS platform used in this study such as minimal sample consumption, ultralow flow rate separation, sheathless CZE-MS without analyte dilution, the open tubular geometry, and integration with advanced MS enabled the high sensitivity (low fmol level) for characterization of intact and functionally active proteins and non-covalent protein complexes. The platform was evaluated using a high purity monoclonal antibody, mixtures of complex-forming protein standards, and a limited proteome-level isolate of E. coli ribosomes. Analysis of the E. coli ribosomal isolate on the sheathless polyacrylamide-coated capillary CZE – Q Exactive Plus system resulted in the identification of 51 intact proteins, close to 200 proteoforms, and several non-covalent protein complexes from a complementary analysis involving both native and denaturing CZE-MS, which can be instrumental in exploring ribosome specialization.
Considering that most native MS experiments have thus far been performed either with direct infusion into the MS or with a prior offline fractionation, our efforts demonstrate that the high performance separation under non-denaturing conditions can be performed online using native CZE-MS. Such an approach could serve as an effective technique in a wide variety of potential biomedical applications requiring the analysis of stable and transient protein-interactions and proteins in their native states. The high performance separation capability added to native mass spectrometry will allow one to address challenges of intrinsic heterogeneity of protein-based biomedical samples and improve the accuracy of quantitative analysis. Enabled separation of non-covalent multisubunit complexes from their monomeric and lower order multisubunit constituents could minimize the possible ambiguity of native MS-based structural characterization and quantitation. The native CZE-MS approach seems also promising for effective characterization of whole complex proteomes if combined with up-front fractionation.
The platform could be further improved via modification of MS instrumentation to enable selection of higher m/z precursors for MS2 fragmentation than the instruments used in this study allowed for. Alternative fragmentation techniques, such as UVPD, ETD, and EThcD and their combined use would also aid in fragmentation of internal backbone residues of larger molecular weight proteins. Lastly, availability of MS3 or MSn analysis on a modified Q Exactive instrument as in [39, 40], coupled with online CESI CZE with a neutral polyacrylamide-coated capillary, is expected to aid in the characterization of individual subunits of protein complexes and elucidate locations of potential PTMs down to the level of individual residues.
Supplementary Material
Acknowledgments
The authors would like to thank SCIEX for generously providing the CESI 8000 instrument and consumables used for all CZE experiments. The authors also thank Chitra Ratnayake and Anna Lou (SCIEX) for providing the prototype neutral coated OptiMS capillaries initially used in this study and Alain Beck at Centre D’Immunollogie Pierre Fabre for providing the Trastuzumab samples. We thank Thermo Fisher Scientific for providing access to an Exactive EMR instrument and Drs. Neil Kelleher and Ioanna Ntai (Northwestern University) for assistance with ProSightPC searches. We also thank Drs. Nikolai Slavov (Northeastern University) and St John Skilton (Protein Metrics) for helpful discussions. This work was supported by the ASMS 2015 Research Award (ARI), NIH 1R01GM120272 award (ARI) and the Barnett Institute of the Chemical and Biological Analysis at Northeastern University.
References
- 1.Boeri Erba E, Petosa C. The emerging role of native mass spectrometry in characterizing the structure and dynamics of macromolecular complexes. Protein Sci. 2015;24:1176–1192. doi: 10.1002/pro.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rose RJ, Damoc E, Denisov E, Makarov A, Heck AJ. High-sensitivity Orbitrap mass analysis of intact macromolecular assemblies. Nat Methods. 2012;9:1084–1086. doi: 10.1038/nmeth.2208. [DOI] [PubMed] [Google Scholar]
- 3.Heck AJ. Native mass spectrometry: a bridge between interactomics and structural biology. Nat Methods. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 4.Weiss VU, Bereszcazk JZ, Havlik M, Kallinger P, Gosler I, Kumar M, et al. Analysis of a common cold virus and its subviral particles by gas-phase electrophoretic mobility molecular analysis and native mass spectrometry. Analytical Chemistry. 2015;87:8709–8717. doi: 10.1021/acs.analchem.5b01450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Waterbeemd M, Snijder J, Tsvetkova IB, Dragnea BG, Cornelissen JJ, Heck AJ. Examining the Heterogeneous Genome Content of Multipartite Viruses BMV and CCMV by Native Mass Spectrometry. J Am Soc Mass Spectrom. 2016;27:1000–1009. doi: 10.1007/s13361-016-1348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saliou JM, Manival X, Tillault AS, Atmanene C, Bobo C, Branlant C, et al. Combining native MS approaches to decipher archaeal box H/ACA ribonucleoprotein particle structure and activity. Proteomics. 2015;15:2851–2861. doi: 10.1002/pmic.201400529. [DOI] [PubMed] [Google Scholar]
- 7.Blackwell AE, Dodds ED, Bandarian V, Wysocki VH. Revealing the quaternary structure of a heterogeneous noncovalent protein complex through surface-induced dissociation. Anal Chem. 2011;83:2862–2865. doi: 10.1021/ac200452b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia BA. What does the future hold for Top Down mass spectrometry? J Am Soc Mass Spectrom. 2010;21:193–202. doi: 10.1016/j.jasms.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Leney AC, Heck AJR. Native Mass Spectrometry: What is in the Name? J Am Soc Mass Spectr. 2017;28:5–13. doi: 10.1021/jasms.8b05378. [DOI] [PubMed] [Google Scholar]
- 10.Zhang H, Cui WD, Wen JZ, Blankenship RE, Gross ML. Native Electrospray and Electron-Capture Dissociation in FTICR Mass Spectrometry Provide Top-Down Sequencing of a Protein Component in an Intact Protein Assembly. J Am Soc Mass Spectr. 2010;21:1966–1968. doi: 10.1016/j.jasms.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith LM, Kelleher NL, Consortium for Top Down, P Proteoform: a single term describing protein complexity. Nat Methods. 2013;10:186–187. doi: 10.1038/nmeth.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khawli LA, Goswami S, Hutchinson R, Kwong ZW, Yang J, Wang X, et al. Charge variants in IgG1: Isolation, characterization, in vitro binding properties and pharmacokinetics in rats. mAbs. 2010;2:613–624. doi: 10.4161/mabs.2.6.13333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haselberg R, Brinks V, Hawe A, de Jong GJ, Somsen GW. Capillary electrophoresis-mass spectrometry using noncovalently coated capillaries for the analysis of biopharmaceuticals. Anal Bioanal Chem. 2011;400:295–303. doi: 10.1007/s00216-011-4738-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haselberg R, de Jong GJ, Somsen GW. Low-flow sheathless capillary electrophoresis-mass spectrometry for sensitive glycoform profiling of intact pharmaceutical proteins. Anal Chem. 2013;85:2289–2296. doi: 10.1021/ac303158f. [DOI] [PubMed] [Google Scholar]
- 15.Bush DR, Zang L, Belov AM, Ivanov AR, Karger BL. High Resolution CZE-MS Quantitative Characterization of Intact Biopharmaceutical Proteins: Proteoforms of Interferon-beta1. Anal Chem. 2015 doi: 10.1021/acs.analchem.5b03218. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Plouffe BD, Belov AM, Ray S, Wang X, Murthy SK, et al. An Integrated Platform for Isolation, Processing and Mass Spectrometry-based Proteomic Profiling of Rare Cells in Whole Blood. Mol Cell Proteomics. 2015 doi: 10.1074/mcp.M114.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov AR, Zang L, Karger BL. Low-attomole electrospray ionization MS and MS/MS analysis of protein tryptic digests using 20-microm-i.d. polystyrene-divinylbenzene monolithic capillary columns. Anal Chem. 2003;75:5306–5316. doi: 10.1021/ac030163g. [DOI] [PubMed] [Google Scholar]
- 18.Kukrer B, Filipe V, van Duijn E, Kasper PT, Vreeken RJ, Heck AJ, et al. Mass spectrometric analysis of intact human monoclonal antibody aggregates fractionated by size-exclusion chromatography. Pharmaceutical research. 2010;27:2197–2204. doi: 10.1007/s11095-010-0224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muneeruddin K, Nazzaro M, Kaltashov IA. Characterization of intact protein conjugates and biopharmaceuticals using ion-exchange chromatography with online detection by native electrospray ionization mass spectrometry and top-down tandem mass spectrometry. Anal Chem. 2015;87:10138–10145. doi: 10.1021/acs.analchem.5b02982. [DOI] [PubMed] [Google Scholar]
- 20.Debaene F, Boeuf A, Wagner-Rousset E, Colas O, Ayoub D, Corvaia N, et al. Innovative native MS methodologies for antibody drug conjugate characterization: High resolution native MS and IM-MS for average DAR and DAR distribution assessment. Anal Chem. 2014;86:10674–10683. doi: 10.1021/ac502593n. [DOI] [PubMed] [Google Scholar]
- 21.Gahoual R, Heidenreich AK, Somsen GW, Bulau P, Reusch D, Wuhrer M, et al. Detailed characterization of monoclonal antibody receptor interaction using affinity liquid chromatography hyphenated to native mass spectrometry. Anal Chem. 2017 doi: 10.1021/acs.analchem.7b00211. [DOI] [PubMed] [Google Scholar]
- 22.Martinovic S, Berger SJ, Pasa-Tolic L, Smith RD. Separation and detection of intact noncovalent protein complexes from mixtures by on-line capillary isoelectric focusing-mass spectrometry. Analytical Chemistry. 2000;72:5356–5360. doi: 10.1021/ac0004557. [DOI] [PubMed] [Google Scholar]
- 23.Fonslow BR, Kang SA, Gestaut DR, Graczyk B, Davis TN, Sabatini DM, et al. Native capillary isoelectric focusing for the separation of protein complex isoforms and subcomplexes. Anal Chem. 2010;82:6643–6651. doi: 10.1021/ac101235k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skinner OS, Do Vale LH, Catherman AD, Havugimana PC, de Sousa MV, Compton PD, et al. Native GELFrEE: a new separation technique for biomolecular assemblies. Anal Chem. 2015;87:3032–3038. doi: 10.1021/ac504678d. [DOI] [PubMed] [Google Scholar]
- 25.Tran JC, Doucette AA. Gel-eluted liquid fraction entrapment electrophoresis: an electrophoretic method for broad molecular weight range proteome separation. Anal Chem. 2008;80:1568–1573. doi: 10.1021/ac702197w. [DOI] [PubMed] [Google Scholar]
- 26.Zheng J, Wei C, Zhao L, Liu L, Leng W, Li W, et al. Combining blue native polyacrylamide gel electrophoresis with liquid chromatography tandem mass spectrometry as an effective strategy for analyzing potential membrane protein complexes of Mycobacterium bovis bacillus Calmette-Guerin. BMC genomics. 2011;12:40. doi: 10.1186/1471-2164-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han X, Wang Y, Aslanian A, Fonslow B, Graczyk B, Davis TN, et al. In-line separation by capillary electrophoresis prior to analysis by top-down mass spectrometry enables sensitive characterization of protein complexes. J Proteome Res. 2014;13:6078–6086. doi: 10.1021/pr500971h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busnel JM, Schoenmaker B, Ramautar R, Carrasco-Pancorbo A, Ratnayake C, Feitelson JS, et al. High capacity capillary electrophoresis-electrospray ionization mass spectrometry: coupling a porous sheathless interface with transient-isotachophoresis. Analytical chemistry. 2010;82:9476–9483. doi: 10.1021/ac102159d. [DOI] [PubMed] [Google Scholar]
- 29.Rambaldi DC, Reschiglian P, Zattoni A. Flow field-flow fractionation: recent trends in protein analysis. Anal Bioanal Chem. 2011;399:1439–1447. doi: 10.1007/s00216-010-4312-5. [DOI] [PubMed] [Google Scholar]
- 30.Reschiglian P, Zattoni A, Roda B, Roda A, Parisi D, Moon MH, et al. Hollow-fiber flow field-flow fractionation: a gentle separation method for mass spectrometry of native proteins. Annali di chimica. 2006;96:253–257. doi: 10.1002/adic.200690026. [DOI] [PubMed] [Google Scholar]
- 31.Issaq HJ, Janini GM, Atamna IZ, Muschik GM. Separations by High-Performance Liquid-Chromatography and Capillary Zone Electrophoresis - a Comparative-Study. Journal of Liquid Chromatography. 1991;14:817–845. [Google Scholar]
- 32.Maxwell EJ, Chen DD. Twenty years of interface development for capillary electrophoresis-electrospray ionization-mass spectrometry. Anal Chim Acta. 2008;627:25–33. doi: 10.1016/j.aca.2008.06.034. [DOI] [PubMed] [Google Scholar]
- 33.Moini M. Simplifying CE-MS operation. 2 Interfacing low-flow separation techniques to mass spectrometry using a porous tip. Analytical chemistry. 2007;79:4241–4246. doi: 10.1021/ac0704560. [DOI] [PubMed] [Google Scholar]
- 34.Maxwell EJ, Zhong X, Zhang H, van Zeijl N, Chen DD. Decoupling CE and ESI for a more robust interface with MS. Electrophoresis. 2010;31:1130–1137. doi: 10.1002/elps.200900517. [DOI] [PubMed] [Google Scholar]
- 35.Wojcik R, Dada OO, Sadilek M, Dovichi NJ. Simplified capillary electrophoresis nanospray sheath-flow interface for high efficiency and sensitive peptide analysis. Rapid Commun Mass Spectrom. 2010;24:2554–2560. doi: 10.1002/rcm.4672. [DOI] [PubMed] [Google Scholar]
- 36.Breuker K, McLafferty FW. Stepwise evolution of protein native structure with electrospray into the gas phase, 10(−12) to 10(2) s. Proc Natl Acad Sci U S A. 2008;105:18145–18152. doi: 10.1073/pnas.0807005105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hewitt D, Marklund E, Scott DJ, Robinson CV, Borysik AJ. A hydrodynamic comparison of solution and gas phase proteins and their complexes. The journal of physical chemistry B. 2014;118:8489–8495. doi: 10.1021/jp501950d. [DOI] [PubMed] [Google Scholar]
- 38.Meyer T, Gabelica V, Grubmüler H, Orozco M. Proteins in the gas phase. WIREs Comput Mol Sci. 2013:408–425. [Google Scholar]
- 39.Ben-Nissan G, Belov ME, Morgenstern D, Levin Y, Dym O, Arkind G, et al. Triple-Stage Mass Spectrometry Unravels the Heterogeneity of an Endogenous Protein Complex. Analytical Chemistry. 2017;89:4708–4715. doi: 10.1021/acs.analchem.7b00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Belov ME, Damoc E, Denisov E, Compton PD, Horning S, Makarov AA, et al. From protein complexes to subunit backbone fragments: a multi-stage approach to native mass spectrometry. Anal Chem. 2013;85:11163–11173. doi: 10.1021/ac4029328. [DOI] [PubMed] [Google Scholar]
- 41.van de Waterbeemd M, Fort KL, Boll D, Reinhardt-Szyba M, Routh A, Makarov A, et al. High-fidelity mass analysis unveils heterogeneity in intact ribosomal particles. Nat Methods. 2017 doi: 10.1038/nmeth.4147. [DOI] [PubMed] [Google Scholar]
- 42.Hardy SJ, Kurland CG, Voynow P, Mora G. The ribosomal proteins of Escherichia coli. I. Purification of the 30S ribosomal proteins. Biochemistry. 1969;8:2897–2905. doi: 10.1021/bi00835a031. [DOI] [PubMed] [Google Scholar]
- 43.Cannon JR, Cammarata MB, Robotham SA, Cotham VC, Shaw JB, Fellers RT, et al. Ultraviolet photodissociation for characterization of whole proteins on a chromatographic time scale. Analytical Chemistry. 2014;86:2185–2192. doi: 10.1021/ac403859a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kreimer S, Belov ME, Danielson WF, Levitsky LI, Gorshkov MV, Karger BL, et al. Advanced Precursor Ion Selection Algorithms for Increased Depth of Bottom-Up Proteomic Profiling. J Proteome Res. 2016;15:3563–3573. doi: 10.1021/acs.jproteome.6b00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 46.Whitt JT, Moini M. Capillary electrophoresis to mass spectrometry interface using a porous junction. Anal Chem. 2003;75:2188–2191. doi: 10.1021/ac026380j. [DOI] [PubMed] [Google Scholar]
- 47.Valaskovic GA, Kelleher NL, McLafferty FW. Attomole protein characterization by capillary electrophoresis-mass spectrometry. Science. 1996;273:1199–1202. doi: 10.1126/science.273.5279.1199. [DOI] [PubMed] [Google Scholar]
- 48.Holland MJ, Holland JP, Thill GP, Jackson KA. The primary structures of two yeast enolase genes. Homology between the 5′ noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981;256:1385–1395. [PubMed] [Google Scholar]
- 49.Chin CC, Brewer JM, Wold F. The amino acid sequence of yeast enolase. J Biol Chem. 1981;256:1377–1384. [PubMed] [Google Scholar]
- 50.Poyner RR, Laughlin LT, Sowa GA, Reed GH. Toward identification of acid/base catalysts in the active site of enolase: comparison of the properties of K345A, E168Q, and E211Q variants. Biochemistry. 1996;35:1692–1699. doi: 10.1021/bi952186y. [DOI] [PubMed] [Google Scholar]
- 51.Brewer JM. Specificity and Mechanism of Action of Metal-Ions in Yeast Enolase. FEBS Lett. 1985;182:8–14. [Google Scholar]
- 52.Lebioda L, Stec B. Crystal structure of enolase indicates that enolase and pyruvate kinase evolved from a common ancestor. Nature. 1988;333:683–686. doi: 10.1038/333683a0. [DOI] [PubMed] [Google Scholar]
- 53.Leskovac V, Trivic S, Pericin D. The three zinc-containing alcohol dehydrogenases from baker’s yeast, Saccharomyces cerevisiae. FEMS Yeast Res. 2002;2:481–494. doi: 10.1111/j.1567-1364.2002.tb00116.x. [DOI] [PubMed] [Google Scholar]
- 54.Jornvall H. The primary structure of yeast alcohol dehydrogenase. Eur J Biochem. 1977;72:425–442. doi: 10.1111/j.1432-1033.1977.tb11267.x. [DOI] [PubMed] [Google Scholar]
- 55.Li H, Wongkongkathep P, Van Orden SL, Ogorzalek Loo RR, Loo JA. Revealing ligand binding sites and quantifying subunit variants of noncovalent protein complexes in a single native top-down FTICR MS experiment. J Am Soc Mass Spectrom. 2014;25:2060–2068. doi: 10.1007/s13361-014-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saito R, Sato T, Ikai A, Tanaka N. Structure of bovine carbonic anhydrase II at 1.95 A resolution. Acta Crystallogr D Biol Crystallogr. 2004;60:792–795. doi: 10.1107/S0907444904003166. [DOI] [PubMed] [Google Scholar]
- 57.Lauer HH, Mcmanigill D. Capillary Zone Electrophoresis of Proteins in Untreated Fused-Silica Tubing. Analytical Chemistry. 1986;58:166–170. [Google Scholar]
- 58.Chen FTA. Rapid Protein-Analysis by Capillary Electrophoresis. Journal of Chromatography. 1991;559:445–453. [Google Scholar]
- 59.Schwartz H, Pritchett T. Beckman. 1994 doi: 10.1038/nbt0494-408. [DOI] [PubMed] [Google Scholar]
- 60.Alexandru I, Kells DIC, Dorrington KJ, Klein M. Non-Covalent Association of Heavy and Light-Chains of Human Immunoglobulin-G - Studies Using Light Chain Labeled with a Fluorescent-Probe. Molecular Immunology. 1980;17:1351–1363. doi: 10.1016/0161-5890(80)90004-8. [DOI] [PubMed] [Google Scholar]
- 61.Redman EA, Mellors JS, Starkey JA, Ramsey JM. Characterization of Intact Antibody Drug Conjugate Variants Using Microfluidic Capillary Electrophoresis-Mass Spectrometry. Analytical Chemistry. 2016;88:2220–2226. doi: 10.1021/acs.analchem.5b03866. [DOI] [PubMed] [Google Scholar]
- 62.Beck A, Sanglier-Cianferani S, Van Dorsselaer A. Biosimilar, biobetter, and next generation antibody characterization by mass spectrometry. Analytical Chemistry. 2012;84:4637–4646. doi: 10.1021/ac3002885. [DOI] [PubMed] [Google Scholar]
- 63.Harris RJ, Kabakoff B, Macchi FD, Shen FJ, Kwong M, Andya JD, et al. Identification of multiple sources of charge heterogeneity in a recombinant antibody. J Chromatogr B Biomed Sci Appl. 2001;752:233–245. doi: 10.1016/s0378-4347(00)00548-x. [DOI] [PubMed] [Google Scholar]
- 64.Xie H, Chakraborty A, Ahn J, Yu YQ, Dakshinamoorthy DP, Gilar M, et al. Rapid comparison of a candidate biosimilar to an innovator monoclonal antibody with advanced liquid chromatography and mass spectrometry technologies. mAbs. 2010;2:379–394. doi: 10.4161/mabs.2.4.11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ha S, Ou Y, Vlasak J, Li Y, Wang S, Vo K, et al. Isolation and characterization of IgG1 with asymmetrical Fc glycosylation. Glycobiology. 2011;21:1087–1096. doi: 10.1093/glycob/cwr047. [DOI] [PubMed] [Google Scholar]
- 66.Sahin E, Roberts CJ. Size-exclusion chromatography with multi-angle light scattering for elucidating protein aggregation mechanisms. Methods Mol Biol. 2012;899:403–423. doi: 10.1007/978-1-61779-921-1_25. [DOI] [PubMed] [Google Scholar]
- 67.Indrawati L, Stroshine RL, Narsimhan G. Low-field NMR: A tool for studying protein aggregation. J Sci Food Agr. 2007;87:2207–2216. [Google Scholar]
- 68.Brandt JP, Patapoff TW, Aragon SR. Construction, MD simulation, and hydrodynamic validation of an all-atom model of a monoclonal IgG antibody. Biophys J. 2010;99:905–913. doi: 10.1016/j.bpj.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu H, Kroe-Barrett R, Singh S, Robinson AS, Roberts CJ. Competing aggregation pathways for monoclonal antibodies. FEBS Lett. 2014;588:936–941. doi: 10.1016/j.febslet.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 70.Yin LS, Chen XY, Tiwari A, Vicini P, Hickling TP. The Role of Aggregates of Therapeutic Protein Products in Immunogenicity: An Evaluation by Mathematical Modeling. J Immunol Res. 2015 doi: 10.1155/2015/401956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xue S, Barna M. Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slavov N, Semrau S, Airoldi E, Budnik B, van Oudenaarden A. Differential Stoichiometry among Core Ribosomal Proteins. Cell reports. 2015;13:865–873. doi: 10.1016/j.celrep.2015.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilbert WV. Functional specialization of ribosomes? Trends Biochem Sci. 2011;36:127–132. doi: 10.1016/j.tibs.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Filipovska A, Rackham O. Specialization from synthesis: how ribosome diversity can customize protein function. FEBS Lett. 2013;587:1189–1197. doi: 10.1016/j.febslet.2013.02.032. [DOI] [PubMed] [Google Scholar]
- 75.Byrgazov K, Vesper O, Moll I. Ribosome heterogeneity: another level of complexity in bacterial translation regulation. Current Opinion in Microbiology. 2013;16:133–139. doi: 10.1016/j.mib.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ilag LL, Videler H, McKay AR, Sobott F, Fucini P, Nierhaus KH, et al. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc Natl Acad Sci U S A. 2005;102:8192–8197. doi: 10.1073/pnas.0502193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmitt ND, Agar JN. Parsing disease-relevant protein modifications from epiphenomena: perspective on the structural basis of SOD1-mediated ALS. J Mass Spectrom. 2017;52:480–491. doi: 10.1002/jms.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gervais D, King D. Capillary isoelectric focusing of a difficult-to-denature tetrameric enzyme using alkylurea-urea mixtures. Anal Biochem. 2014;465:90–95. doi: 10.1016/j.ab.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 79.Ngounou Wetie AG, Sokolowska I, Woods AG, Roy U, Loo JA, Darie CC. Investigation of stable and transient protein-protein interactions: Past, present, and future. Proteomics. 2013;13:538–557. doi: 10.1002/pmic.201200328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, et al. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 81.Stock A, Clarke S, Clarke C, Stock J. N-terminal methylation of proteins: structure, function and specificity. FEBS Lett. 1987;220:8–14. doi: 10.1016/0014-5793(87)80866-9. [DOI] [PubMed] [Google Scholar]
- 82.Nesterchuk MV, Sergiev PV, Dontsova OA. Posttranslational Modifications of Ribosomal Proteins in Escherichia coli. Acta Naturae. 2011;3:22–33. [PMC free article] [PubMed] [Google Scholar]
- 83.Wittmann-Liebold B, Greuer B. The primary structure of protein S5 from the small subunit of the Escherichia coli ribosome. FEBS Lett. 1978;95:91–98. doi: 10.1016/0014-5793(78)80059-3. [DOI] [PubMed] [Google Scholar]
- 84.Yaguchi M. Primary structure of protein S18 from the small Escherichia coli ribosomal subunit. FEBS Lett. 1975;59:217–220. doi: 10.1016/0014-5793(75)80378-4. [DOI] [PubMed] [Google Scholar]
- 85.Terhorst C, Moller W, Laursen R, Wittmann-Liebold B. The primary structure of an acidic protein from 50-S ribosomes of Escherichia coli which is involved in GTP hydrolysis dependent on elongation factors G and T. Eur J Biochem. 1973;34:138–152. doi: 10.1111/j.1432-1033.1973.tb02740.x. [DOI] [PubMed] [Google Scholar]
- 86.Tanaka S, Matsushita Y, Yoshikawa A, Isono K. Cloning and molecular characterization of the gene rimL which encodes an enzyme acetylating ribosomal protein L12 of Escherichia coli K12. Mol Gen Genet. 1989;217:289–293. doi: 10.1007/BF02464895. [DOI] [PubMed] [Google Scholar]
- 87.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, et al. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


