Abstract
Objective
We hypothesized that DNA methylation of development-related genes may occur in endometrial cancer (EC)/ovarian cancer (OC) and may be detected in cervical scrapings.
Methods
We tested methylation status by quantitative methylation-specific polymerase chain reaction for 14 genes in DNA pools of endometrial and OC tissues. Tissues of EC/normal endometrium, OC/normal ovary, were verified in training set using cervical scrapings of 10 EC/10 OC patients and 10 controls, and further validated in the testing set using independent cervical scrapings in 30 EC/30 OC patients and 30 controls. We generated cutoff values of methylation index (M-index) from cervical scrapings to distinguish between cancer patients and control. Sensitivity/specificity of DNA methylation biomarkers in detecting EC and OC was calculated.
Results
Of 14 genes, 4 (PTGDR, HS3ST2, POU4F3, MAGI2) showed hypermethylation in EC and OC tissues, and were verified in training set. POU4F3 and MAGI2 exhibited hypermethylation in training set were validated in independent cases. The mean M-index of POU4F3 is 78.28 in EC and 20.36 in OC, which are higher than that in controls (6.59; p<0.001 and p=0.100, respectively), and that of MAGI2 is 246.0 in EC and 12.2 in OC, which is significantly higher that than in controls (2.85; p<0.001 and p=0.480, respectively). Sensitivity and specificity of POU4F3/MAGI2 were 83%–90% and 69%–75% for detection of EC, and 61% and 62%–69% for the detection of OC.
Conclusion
The findings demonstrate the potential of EC/OC detection through testing for DNA methylation in cervical scrapings.
Keywords: Endometrial Neoplasms, Ovarian Neoplasms, DNA Methylation
INTRODUCTION
Cervical cancer, endometrial cancer (EC), and ovarian cancer (OC) are the 3 major gynecologic cancers. Since Georgios Papanicolaou's introduction of cytology screening, the incidence and mortality rates of invasive cervical cancer has significantly decreased in the past decades [1]. EC is the most common, and OC is the most severe gynecologic cancer in developed countries [2,3]. Compared with cervical cancer, screening for EC and OC remains controversial and challenging.
Evaluation of endometrial thickness by transvaginal ultrasonography (TVS) is considered the first step in the detection of EC in women. However, TVS is neither a sensitive nor a specific tool [4]. The presence of abnormal endometrial cells in cervical scrapings is considered an indication of endometrial pathology, although it is not sensitive enough to consistently detect EC. The overall efficacy of detection of EC by cervical cytology is 38% [5]. Tissue proof is the gold standard in the diagnosis of EC. Dilatation and curettage and hysteroscopy are commonly used to detect EC with sensitivities of 91% and 99.6% in pre- and post-menopausal women, respectively [6]. However, endometrial biopsy is associated with significant discomfort to patients [7], and most patients undergoing these invasive procedures do not have cancer, rendering low specificity. Moreover, the yield of tissue for pathological assessment using these methods is limited, ranging from 25% to 36% [8]. Therefore, a screening policy for women with EC has not been established owing to the lack of a satisfactory method for massive endometrial evaluation.
The well-known biomarker of OC is cancer antigen 125 (CA 125). However, the fact that CA 125 is not suitable for OC screening is also well known, owing to its low specificity and to the fact that increases in CA 125 are also observed in benign gynecologic disorders, such as pelvic inflammatory disease, endometriosis, and uterine fibroids [9]. Human epididymis protein 4 (HE 4) was reported to be an alternative to CA 125. Using Risk of Ovarian Malignancy Algorithm, which utilizes the combination of HE 4 and CA 125 values to assess the risk of epithelial ovarian cancer (EOC) in women with a pelvic mass, performance in the detection of EOC in pre-menopausal women, and as a primary screening tool is still not satisfactory [10]. The value and method of OC screening remains to be defined.
Epigenetic changes play an imperative role in cancer development. Cancer formation is associated with frequent changes in DNA methylation [11]. There were several promising reports employing DNA methylation biomarkers for cervical cancer screening [12,13]. Indeed, in the clinical experience of management of atypical glandular cell (AGC), 3.9%–5.2% of AGCs in Pap smears are invasive cancers, of which 23.6%, 57.6%, and 6.4% originate from the cervix, endometrium, and ovary, respectively [14,15], suggesting the presence of cancer cells from internal genital organs in the cervix. In addition, there are occult precancerous lesions, such as endometrial hyperplasias, which significantly increase the risk of developing EC [15,16]. In 1 study, DNA methylation analysis of cervical scrapings was attempted to detect endometrial carcinomas with limited success [17]. A recent publication provides the proof-of-concept of EC and OC detection using DNA mutational analysis in cervical scrapings [18]. So far, there have been no reports of the detection of OC using DNA methylation biomarkers in cervical scrapings.
Our previous research on the epigenomics of cervical cancer using a genome-wide approach identified 14 candidate genes that were hypermethylated in cervical cancer tissues [13]. Most of these genes are development-related genes that are common in the development of various types of cancer [16,19,20,21]. We hypothesized that DNA methylation of these development-related genes may occur in EC/OC, and could be detected in cervical scrapings.
MATERIALS AND METHODS
1. Study population
The Institutional Review Board of the Tri-Service General Hospital, National Defense Medical Center and Shuang Ho Hospital, Taipei Medical University approved the study. Informed consent was acquired from all patients and control subjects.
The retrospective study included 60 archived EC (tissues, n=20 and scrapings, n=40), 59 OCs (tissues, n=19 and scrapings, n=40) and 74 normal controls (tissues, n=34 and cervical scrapings, n=40). All tissues samples and scrapings were not paired from the same patients. In the cases of EC and OC, cervical scrapings were collected before surgery from women undergoing surgery as their treatment guideline, and normal control tissues specimens were collected from women who had previously undergone hysterectomy due to benign gynecologic disease (uterine fibroids or uterine prolapse). Patients attending our gynecologic outpatient department whose cervical scrapings exhibited normal cytology and pelvic sonography did not reveal significant abnormalities served as control subjects of cervical scrapings. In all women, a cytobrush (CooperSurgical, Inc., Trumbull, CT, USA) was used to collect samples through physician cervical sampling.
The patients were diagnosed and treated and had their tissues placed in a bank at the Tri-Service General Hospital and Shuang Ho Hospital, Taipei, Taiwan [22]. All invasive cancers were confirmed by histopathology.
2. DNA extraction, bisulfite conversion, quantitative methylation-specific polymerase chain reaction (QMSP) and generation of cutoff value of methylation index (M-index)
In tissue, we cut a 5 μm-thick section from each tissue block and stained it with hematoxylin and eosin to confirm the histologic diagnosis and to define the purity of endometrial/ovarian normal and cancer cells. Genomic DNA was extracted from tissues and cervical scraping cells using the QIAmp DNA Mini Kit (QIAGEN, Hilden, Germany). DNA pool was comprised of a mixture of an equal amount of DNA from 3 randomly collected specimens of archive EC and OC tissues of individual patients. We used 1 μg genomic DNA to do bisulfite modified using the EZ DNA Methylation Kit (Zymo Research Corp., Irvine, CA, USA), according to the manufacturer's recommendations, and dissolved in 70 μL of nuclease-free water. Polymerase chain reaction (PCR) products were amplified with the LightCycler 480 SYBR Green I Master (Roche, Indianapolis, IN, USA) and LightCycler 480. The 20 μL reaction contained 2 μL bisulfite-converted DNA, 250 nM of each primer, and 10 μL of Master Mix. The reactions were conducted on a LightCycler 480 under the following thermal profiles: 95°C for 5 minutes, 50 cycles consisting of 95°C for 10 seconds, 60°C for 30 seconds, and 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. Duplicate testing for each gene was conducted for all specimens. To normalize the input DNA, we designed primers to measure the amount of a non-CpG region of COL2A1 in each methylation-independent assay. The DNA methylation level was estimated from the M-index using the formula [24]:
| 10,000×2[(Cp ofCOL2A)–(Cp of gene)] |
Test results with Cp-values of COL2A >36 were defined as detection failures. The primers were designed by Oligo 7.0 Primer Analysis software (Molecular Biology Insights, Inc., Colorado Springs, CO, USA).
We used fluorescence-based real-time PCR for QMSP. The type II collagen gene (COL2A1) was used as an internal reference gene by amplifying non-CpG sequences. Gene symbols, primers, and probes for QMSP were listed in our previous study [13] and summarized in Supplementary Table 1. QMSP was performed in a TaqMan probe system using the LightCycler 480 (Roche). The 5′ and 3′ ends of the probes were separately labeled with 6-carboxy-fluorescein and with a quencher dye. The 20 μL reaction contained 2 μL of bisulfite template DNA, 250 nM of each primer, 225 nM of TaqMan probe, and 10 μL of FastStart PCR Master (ROX; Roche). For the TaqMan-based QMSP, each sample was analyzed in duplicate. The reactions were performed by using an initial incubation at 95°C for 4 minutes, followed by 45 cycles of 95°C for 30 seconds, 60°C for 30 seconds, 72°C for 30 seconds and a final extension of 72°C for 7 minutes. The level of DNA methylation was labeled as the M-index [23]:
| 10,000×2[(Cp ofCOL2A)–(Cp of gene)] |
If the Cp-value of COL2A was >36, the reaction was defined as a detection failure.
QMSP was done to assess the methylation status of ADRA1D, AJAP1, COL6A2, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, SOX17, ST6GAL2, SYT9, and ZNF614 in study samples. The QMSP primers and amplified region of each gene was previously reported by Chen et al. [13]. The primers used for QMSP reside from −1.2 kb to +0.8 kb, spanning the transcription start site of each gene. We generated cutoff values of M-index from the tissues and cervical scrapings to distinguish between normal and cancer subjects. The optimal cutoff values were determined by the most significant difference of area under the receiver operating characteristic curve (AUC).
3. Statistical analysis
We matched inpatient year and age in 1:1 ratio. Age differences between the normal and the cancer subjects were compared using Student's t-tests (2-tailed). Mann-Whitney U tests were used to test differences in gene methylation levels between different disease statuses. Receiver operating characteristic curves were generated to determine the optimal cutoff value of gene methylation included average and median for distinguishing cancers from normal controls [24]. The sensitivity and specificity of each gene were calculated with 95% confidence intervals (CIs). All differences were considered to be statistically significant at p<0.05. All analyses were performed using IBM SPSS Statistics software, version 20.0 (IBM Corp., Armonk, NY, USA) and the Prism software, version 4.03 (Graphpad Software Inc., La Jolla, CA, USA).
RESULTS
1. Demographics of study subjects in testing set of cervical scraping groups of EC and OC
The ages of the control groups were matched within ±3 years old from the archived specimen. The ages of the study subjects in cancer and control groups from whom the cervical scrapings were collected were not significantly different (53.2±11.7 vs. 50.2±14.3, p=0.38 in the endometrial group, 52.3±13.7 vs. 50.2±14.3, p=0.57 in the ovarian group, respectively) (Supplementary Table 2). We confined the histology of study groups in endometrioid adenocarcinoma of endometrium (n=30) and EOC (n=30; 18 papillary serous adenocarcinomas, 5 clear cell carcinomas, and 7 mucinous adenocarcinomas) and regardless of clinical stage and pathological grading (Supplementary Table 2).
The distributions of pathological stages were 83.3%, 6.7%, 10.0% for EC and 30.0%, 13.3%, 56.7% for OC in stages I, II, III, respectively. And the distribution of tumor grades were 56.7%, 26.7%, 16.6% for EC and 16.7%, 13.3%, 70.0% for OC in grades I, II, III, respectively (Supplementary Table 2).
2. Candidate gene selection in EC and OC tissues
The flowchart for the study is shown in Fig. 1. We tested the methylation levels of 14 candidate genes (ADRA1D, AJAP1, COL6A2, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, SOX17, ST6GAL2, SYT9, and ZNF614) in DNA pools of EC and OC tissues. There were 8 hypermethylated genes (AJAP1, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, ST6GAL2, and SYT9) in EC tissues (Supplementary Fig. 1), and 11 hypermethylated genes (ADRA1D, AJAP1, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX17, SYT9, and ZNF614) in OC tissues (Supplementary Fig. 2). These genes were further tested in individual tissues of 20 EC/20 normal endometrial tissues, and in 19 OC/14 normal ovarian tissues. Four genes (MAGI2, POU4F3, AJAP1, and HS3ST2) showed significant hypermethylation in both EC and OC tissues (Fig. 2).
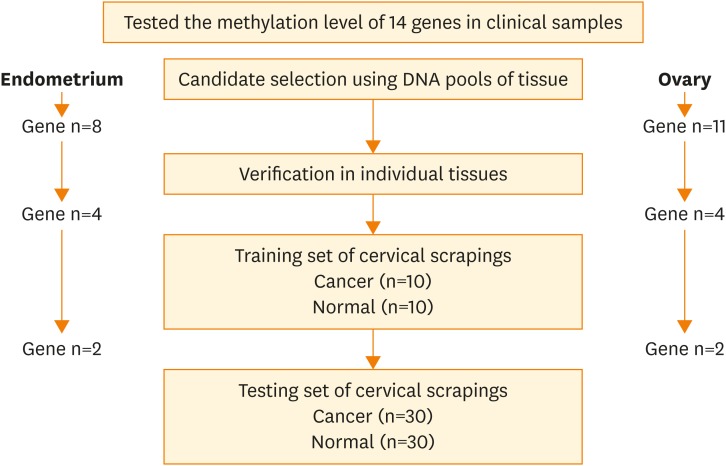
Fig. 1.
The flow chart of the study. The methylation status of 14 candidate genes (ADRA1D, AJAP1, COL6A2, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, SOX17, ST6GAL2, SYT9, and ZNF614) was tested in this study. Genes methylated in DNA pools from cancer tissues were further tested in individual tissues and cervical scrapings.
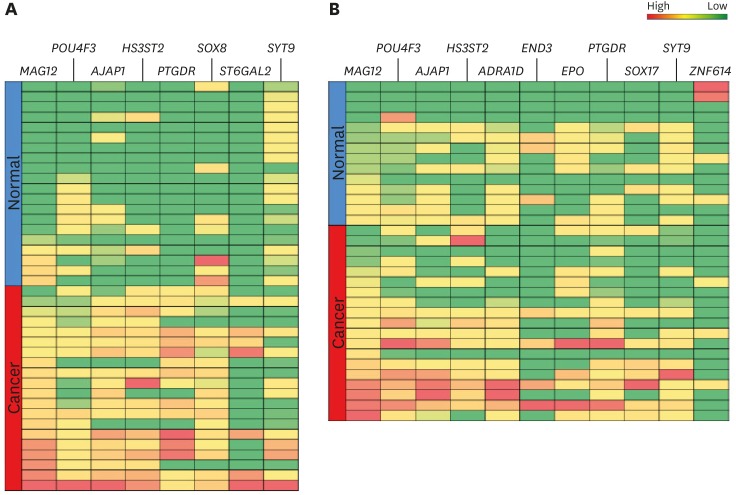
Fig. 2.
The heat map of hypermethylated genes (MAGI2, POU4F3 AJAP1, and HS3ST2) in endometrial tissues (A) and ovarian tissues (B). The blue and dark red colors represent normal and cancerous tissue, respectively. The green and red colors represent low and high methylation, respectively.
3. Verification of hypermethylated genes from cervical scrapings in EC and OC in training set
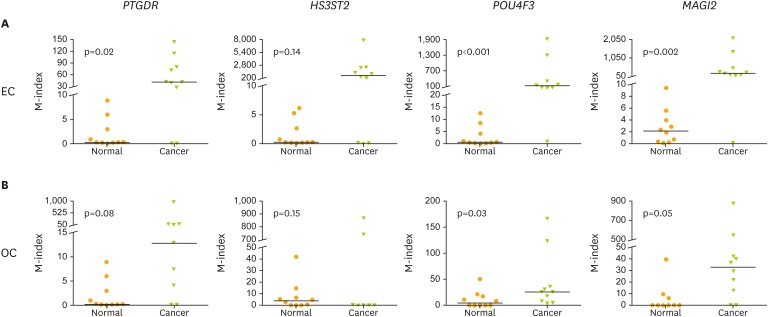
For use in clinical applications, 4 candidate hypermethylated genes (PTGDR, HS3ST2, POU4F3, and MAGI2) in cancer tissues were verified using cervical scrapings of EC/OC patients and normal controls (n=10 in each group, Fig. 3). The M-indexes observed in tests of POU4F3 and MAGI2 recovered from cervical scrapings of EC and OC were significantly higher than in normal controls. The mean M-index of POU4F3 was 155.2 in EC and 25.87 in OC, which were significantly higher than those in controls (0.5 and 4.75; p<0.001 and p=0.03, respectively). The mean M-index of MAGI2 was 209.2 in EC and 32.93 in OC, which was higher than that in controls (2.09 and 0.22; p=0.002 and p=0.050, respectively) (Fig. 3). An M-index of POU4F3/MAGI2 greater than 155.2/209.2 in the endometrial group was considered to be positive for cancer, and an M-index of POU4F3/MAGI2 greater than 25.87/21.75 in the ovarian group was considered to be positive (Fig. 3).
Fig. 3.
The M-index of 4 genes (PTGDR, HS3ST2, POU4F3, and MAGI2) in cervical scrapings of EC/OC patient and normal control in training set. (A) Endometrium and (B) ovary. Ten samples of each group were tested.
EC, endometrial cancer; M-index, methylation index; OC, ovarian cancer.
4. Validation of clinical performance for the detection of EC and OC from cervical scrapings in testing set
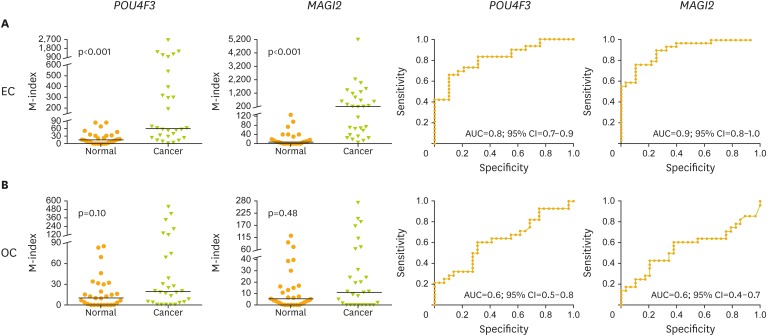
The results of training set were further validated in an independent testing set of patients that included ECs (n=30), OCs (n=30), and normal controls (n=30). The mean M-index of POU4F3 was 78.28 in EC and 20.36 in OC, which were higher than those in controls (6.59; p<0.001 and p=0.100, respectively). The mean M-index of MAGI2 was 246.0 in EC and 12.2 in OC, which were both higher than that in controls (2.85; p<0.001 and p=0.480, respectively) (Fig. 4).
Fig. 4.
The M-index of POU4F3 and MAGI2 in cervical scrapings from EC/OC patients and normal controls in testing set. (A) Endometrium and (B) ovary. Receiver operating characteristic curve analysis of POU4F3 and MAGI2. The AUC of the receiver operating characteristic curve of each gene was calculated for the diagnosis of EC and OC.
AUC, area under the receiver operating characteristic curve; CI, confidence interval; EC, endometrial cancer; M-index, methylation index; OC, ovarian cancer.
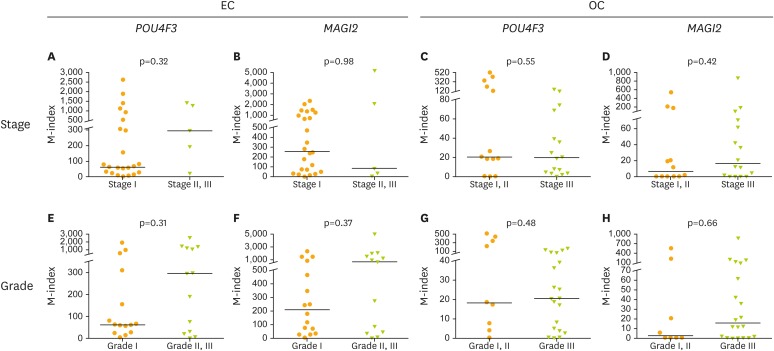
The clinical performance of POU4F3 and MAGI2 genes was calculated in the detection of EC and OC from cervical scrapings. POU4F3 resulted in a comparable performance with MAGI2, with 83% sensitivity and 69% specificity in the detection of EC (AUC=0.8; 95% CI=0.7–0.9) and 61% sensitivity and 69% specificity in the detection of OC (AUC=0.6; 95% CI=0.5–0.8), MAGI2 with 90% sensitivity and 75% specificity in the detection of EC (AUC=0.9; 95% CI=0.8–1.0) and 61% sensitivity and 62% specificity in the detection of OC (AUC=0.6; 95% CI=0.4–0.7) (Table 1). The methylation level of POU4F3 and MAGI2 were not significantly different between early and late stages of EC/OC (Fig. 5).
Table 1. Utility of POU4F3 and MAGI2 in the detection of EC and OC.
| Utility | EC (n=30) vs. Normal (n=30) | OC (n=30) vs. Normal (n=30) | |||
|---|---|---|---|---|---|
| POU4F3 | MAG12 | POU4F3 | MAG12 | ||
| M-Index* cutoff value | 21.7 | 19.6 | 16.5 | 7.7 | |
| Sensitivity (%) | 83 | 90 | 61 | 61 | |
| 95% CI | 64–94 | 73–98 | 41–79 | 41–79 | |
| Specificity (%) | 69 | 75 | 69 | 62 | |
| 95% CI | 49–85 | 55–89 | 49–85 | 42–79 | |
| AUC† | 0.8 | 0.9 | 0.6 | 0.6 | |
| 95% CI | 0.7–0.9 | 0.8–1.0 | 0.5–0.8 | 0.4–0.7 | |
AUC, area under the receiver operating characteristic curve; CI, confidence interval; EC, endometrial cancer; M-Index, methylation index; OC, ovarian cancer.
*M-Index: the value cutoff for cancer cases or normal specimens; †AUC with 95% CI of each gene was calculated for the diagnosis of cancer compare with normal.
Fig. 5.
The M-index of POU4F3 and MAGI2 genes divided by clinical stage or tumor grade in cervical scrapings from EC and OC patients.
EC, endometrial cancer; M-index, methylation index; OC, ovarian cancer.
DISCUSSION
Our results support the hypothesis that EC/OC can be detected by cervical DNA methylation, which also broaden the scope of the Pap smear in the detection of EC and OC with DNA methylation molecular biomarkers.
Recent progress in EC and OC detection using genetic mutations in cervical scrapings or uterine lavage are encouraging [17,18,25]. It is most likely the upper gynecological tract DNA that is shed from primary EC or OC passing through and on the cervix [26]. Kinde et al. [18] used a liquid Pap, which showed that cells originating from gynecologic Müllerian duct cancers can be present in the uterine cervix. Massively parallel sequencing for tumor-specific mutations was performed on DNA from liquid Pap smear specimens. This technique was successfully applied to 24 (100%) of 24 patients with EC. In patients with OC, the sensitivity was lower, with mutations identified in 9 (41%) of 22 patients. Maritschnegg et al. [25] used uterine lavage as material for genetic testing. All of the 5 cases of ECs can be detected using this approach. Furthermore, in 24 (80%) of 30 patients with OC, specific mutations could also be identified [25]. The role for this relatively invasive and inconvenient method may be reserved for specific situations such as EC/OC detection in high-risk populations.
DNA methylation biomarkers for cervical cancer screening are promising [12,13]. DNA methylation testing on self-sampled brush material could be a better collection method, if markers are sensitive and specific enough [27]. In addition to cervical cancer, DNA methylation testing of candidate genes CADM1/MAL/miR124-2 developed from a cervical cancer project using cervical scrapings was reported to have 76.2% sensitivity for EC [17]. Genome-wide attempts to discover novel DNA methylation markers in EC have been conducted using a limited array of 807 genes known to be relevant in cancer [28]. Wentzensen et al. [28] reported on 8 genes that are significantly hypermethylated in EC tissues and tested their performance in identifying EC using endometrial brush materials. They estimated a sensitivity of >90% and a specificity of >50% in endometrial brush specimens. However, they did not test these genes in cervical scrapings. Bakkum-Gamez et al. [26] reported on the feasibility of a minimally invasive biospecimen sampling technique using vaginal tampons coupled with methylation markers for EC detection. They evaluated DNA methylation in vagina tampons using bisulfite pyrosequencing in 38 ECs and in 28 benign endometrial tumor samples. The best AUC is 0.82 for distinguishing between benign and malignant endometrial tumors [26]. The broader scope of the Pap smear detection capability, ranging from cervical cancer to EC, is appealing, but its sensitivity and specificity are not satisfactory [17]. Recently, we analyzed 2 methylomic data sets from EC of endometrioid type, and validated using cervical scrapings from hospital-based samples [29]. Three genes, BHLHE22, CDO1, and CELF4, exhibited the best performance with a sensitivity and specificity of individual genes in the range of 83.7%–96.0% and 78.7%–96.0%, respectively. A panel testing hypermethylations in any 2 of these 3 genes can reach a sensitivity of 91.8%, a specificity of 95.5%, and an odds ratio (OR) of 236.3 (95% CI of OR=56.4–989.6) in EC detection. We demonstrate that methylation biomarkers from cervical scrapings hold great potential for EC detection. Combined with previous reports, this study provides proof-of-principle that tumor DNA from the contiguous female reproductive tract may be detected in a minimally invasive manner from the lower tract [30]. So far, we know of no reports using DNA methylation biomarkers from cervical scrapings to detect OC. To our knowledge, this is the first report demonstrating the clinical potential of a methylation biomarker for the simultaneous detection of EC and OC.
Why are cells taken from cervical scrapings susceptible to the detection of EC and OC? Indeed, psammoma bodies of OC can be detected in cytology in 0%–27% of OC patients, depending on the series [31], indicating that OC tissue can migrate through the endometrial cavity and cervix where it can be collected by Pap smear [32]. A high incidence of abnormal cervical cytology was also observed in women with high-grade EC, a lesson learned about “cancerous floater” [33,34]. In addition to the mechanical aspect, frequent changes in DNA methylation showed evidence of an extensive field effect in breast cancer and methylation was also frequently found in histologically ‘normal’ cervical tissues adjacent to cancer lesions [35,36]. One possible explanation is that there are DNA methylation changes in the cervix, reflecting disease in the endometrium or ovary, although further evidence is needed to prove this speculation.
POU4F3 is located on chromosome 5q32 and has various biological functions, such as regulation of transcription, cellular and metabolic processes, organ development, cellular differentiation, nervous system development, neurogenesis, and generation of neurons [37]. The biological function of POU4F3 in cancer remains largely unclear. Hypermethylation of POU4F3 in cervical cancer and in glioma suggests that it has a role as a suppressor in cancer [13,37]. Methylation of POU4F3 is a promising molecular marker for triage in detecting CIN3 or worse lesions in high-risk human papillomavirus-positive women [38,39], which also can detect endometrial complex hyperplasia in AGC of Pap smear [16]. MAGI2 is located on chromosome 7q21 and serves as a scaffold for the assembly of synaptic protein complexes and is crucial for development and maintenance of synapses. MAGI2 gene can be reflected as a biomarker for the detection of prostate cancer [40]. This study illustrates the potential of POU4F3 and MAGI2 methylation for the possible triple screening of gynecological cancers.
The present study provides the proof-of-principle that methylation testing of cervical scrapings is potential for EC/OC detection. However, the limitation of this study is its small sample size, and the specificity observed in a broader spectrum of patients remains to be determined. The results from using some genes may be confounded by the presence of other common gynecologic diseases, such as ovarian cyst and uterine fibroids [29]. The performance of POU4F3 and MAGI2 methylation testing in detecting EC and OC from cervical scraping needs further validation in a larger sample size that includes various benign diseases.
Footnotes
Funding: This work was supported in part by the following grants from the Tri-Service General Hospital (TSGH-C104-006-006-008-S01&S02;TSGH-C105-010), Ministry of Science and Technology (NSC 105-2628-B-038-011-MY3 and NSC 103-2325-B-195-002), Taipei Medical University (104TMU-SHH-07), Ministry of Health and Welfare (MOHW105-TDU-PB-212-000007), National Health Research Institutes (NHRI-EX106-10406BI), and the Teh-Tzer Study Group for Human Medical Research Foundation.
Conflict of Interest: Dr. Hung-Cheng Lai declared that the application of the genes in the manuscript in cancer detection has been patented. National Defense Medical Center, Taipei, Taiwan holds the IP. Dr. Hung-Cheng Lai is the inventor.
The other authors declared that they have no competing interests.
- Conceptualization: C.C.C., L.H.C.
- Data curation: C.C.C., Y.M.H., L.H.C.
- Formal analysis: L.Y.P.
- Investigation: W.H.C., W.Y.C.
- Resources: C.Y.C.
- Writing - original draft: C.C.C., L.H.C.
- Writing - review & editing: C.C.C., L.H.C.
Supplementary Materials
QMSP primers and probes in this study
Demographics of study subjects in testing set of cervical scraping groups of EC and OC
The M-index of 14 candidate genes (ADRA1D, AJAP1, COL6A2, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, SOX17, ST6GAL2, SYT9, and ZNF614) in DNA pools of endometrial tissues. Each dot represent one pool and each pool comprised of 3 patients tissues of ECs.
The M-index of 14 candidate genes (ADRA1D, AJAP1, COL6A2, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, SOX17, ST6GAL2, SYT9, and ZNF614) in DNA pools of ovarian tissues. Each dot represent one pool and each pool comprised of 3 patients tissues of OCs.
References
- 1.Traut HF, Papanicolaou GN. Cancer of the uterus: the vaginal smear in its diagnosis. Cal West Med. 1943;59:121–122. [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Timmermans A, Opmeer BC, Khan KS, Bachmann LM, Epstein E, Clark TJ, et al. Endometrial thickness measurement for detecting endometrial cancer in women with postmenopausal bleeding: a systematic review and meta-analysis. Obstet Gynecol. 2010;116:160–167. doi: 10.1097/AOG.0b013e3181e3e7e8. [DOI] [PubMed] [Google Scholar]
- 5.Lai CR, Hsu CY, Hang JF, Li AF. The diagnostic value of routine papanicolaou smears for detecting endometrial cancers: an update. Acta Cytol. 2015;59:315–318. doi: 10.1159/000438975. [DOI] [PubMed] [Google Scholar]
- 6.Dijkhuizen FP, Mol BW, Brölmann HA, Heintz AP. The accuracy of endometrial sampling in the diagnosis of patients with endometrial carcinoma and hyperplasia: a meta-analysis. Cancer. 2000;89:1765–1772. [PubMed] [Google Scholar]
- 7.Stovall TG, Ling FW, Morgan PL. A prospective, randomized comparison of the Pipelle endometrial sampling device with the Novak curette. Am J Obstet Gynecol. 1991;165:1287–1290. doi: 10.1016/0002-9378(91)90351-q. [DOI] [PubMed] [Google Scholar]
- 8.Duffy S, Jackson TL, Lansdown M, Philips K, Wells M, Pollard S, et al. The ATAC adjuvant breast cancer trial in postmenopausal women: baseline endometrial subprotocol data. BJOG. 2003;110:1099–1106. [PubMed] [Google Scholar]
- 9.Dorigo O, Berek JS. Personalizing CA125 levels for ovarian cancer screening. Cancer Prev Res (Phila) 2011;4:1356–1359. doi: 10.1158/1940-6207.CAPR-11-0378. [DOI] [PubMed] [Google Scholar]
- 10.Montagnana M, Danese E, Ruzzenente O, Bresciani V, Nuzzo T, Gelati M, et al. The ROMA (Risk of Ovarian Malignancy Algorithm) for estimating the risk of epithelial ovarian cancer in women presenting with pelvic mass: is it really useful? Clin Chem Lab Med. 2011;49:521–525. doi: 10.1515/CCLM.2011.075. [DOI] [PubMed] [Google Scholar]
- 11.Heichman KA, Warren JD. DNA methylation biomarkers and their utility for solid cancer diagnostics. Clin Chem Lab Med. 2012;50:1707–1721. doi: 10.1515/cclm-2011-0935. [DOI] [PubMed] [Google Scholar]
- 12.Hesselink AT, Heideman DA, Steenbergen RD, Coupé VM, Overmeer RM, Rijkaart D, et al. Combined promoter methylation analysis of CADM1 and MAL: an objective triage tool for high-risk human papillomavirus DNA-positive women. Clin Cancer Res. 2011;17:2459–2465. doi: 10.1158/1078-0432.CCR-10-2548. [DOI] [PubMed] [Google Scholar]
- 13.Chen YC, Huang RL, Huang YK, Liao YP, Su PH, Wang HC, et al. Methylomics analysis identifies epigenetically silenced genes and implies an activation of β-catenin signaling in cervical cancer. Int J Cancer. 2014;135:117–127. doi: 10.1002/ijc.28658. [DOI] [PubMed] [Google Scholar]
- 14.Schnatz PF, Guile M, O'Sullivan DM, Sorosky JI. Clinical significance of atypical glandular cells on cervical cytology. Obstet Gynecol. 2006;107:701–708. doi: 10.1097/01.AOG.0000202401.29145.68. [DOI] [PubMed] [Google Scholar]
- 15.Cheng WF, Chen YL, You SL, Chen CJ, Chen YC, Hsieh CY, et al. Risk of gynaecological malignancies in cytologically atypical glandular cells: follow-up study of a nationwide screening population. BJOG. 2011;118:34–41. doi: 10.1111/j.1471-0528.2010.02769.x. [DOI] [PubMed] [Google Scholar]
- 16.Chang CC, Ou YC, Wang KL, Chang TC, Cheng YM, Chen CH, et al. Triage of atypical glandular cell by SOX1 and POU4F3 methylation: a Taiwanese Gynecologic Oncology Group (TGOG) Study. PLoS One. 2015;10:e0128705. doi: 10.1371/journal.pone.0128705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Strooper LM, van Zummeren M, Steenbergen RD, Bleeker MC, Hesselink AT, Wisman GB, et al. CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J Clin Pathol. 2014;67:1067–1071. doi: 10.1136/jclinpath-2014-202616. [DOI] [PubMed] [Google Scholar]
- 18.Kinde I, Bettegowda C, Wang Y, Wu J, Agrawal N, Shih IM, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med. 2013;5:167ra4. doi: 10.1126/scitranslmed.3004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu Y, Li Z, Guo L, Wang L, Zhang L, Cai X, et al. MAGI-2 Inhibits cell migration and proliferation via PTEN in human hepatocarcinoma cells. Arch Biochem Biophys. 2007;467:1–9. doi: 10.1016/j.abb.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 20.Lin N, Di C, Bortoff K, Fu J, Truszkowski P, Killela P, et al. Deletion or epigenetic silencing of AJAP1 on 1p36 in glioblastoma. Mol Cancer Res. 2012;10:208–217. doi: 10.1158/1541-7786.MCR-10-0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Glöckner SC, Guo M, Machida EO, Wang DH, Easwaran H, et al. Epigenetic inactivation of the canonical Wnt antagonist SRY-box containing gene 17 in colorectal cancer. Cancer Res. 2008;68:2764–2772. doi: 10.1158/0008-5472.CAN-07-6349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chu TY, Hwang KS, Yu MH, Lee HS, Lai HC, Liu JY. A research-based tumor tissue bank of gynecologic oncology: characteristics of nucleic acids extracted from normal and tumor tissues from different sites. Int J Gynecol Cancer. 2002;12:171–176. doi: 10.1046/j.1525-1438.2002.01085.x. [DOI] [PubMed] [Google Scholar]
- 23.Huang RL, Chang CC, Su PH, Chen YC, Liao YP, Wang HC, et al. Methylomic analysis identifies frequent DNA methylation of zinc finger protein 582 (ZNF582) in cervical neoplasms. PLoS One. 2012;7:e41060. doi: 10.1371/journal.pone.0041060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–35. doi: 10.1002/1097-0142(1950)3:1<32::aid-cncr2820030106>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Maritschnegg E, Wang Y, Pecha N, Horvat R, Van Nieuwenhuysen E, Vergote I, et al. Lavage of the uterine cavity for molecular detection of müllerian duct carcinomas: a proof-of-concept study. J Clin Oncol. 2015;33:4293–4300. doi: 10.1200/JCO.2015.61.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakkum-Gamez JN, Wentzensen N, Maurer MJ, Hawthorne KM, Voss JS, Kroneman TN, et al. Detection of endometrial cancer via molecular analysis of DNA collected with vaginal tampons. Gynecol Oncol. 2015;137:14–22. doi: 10.1016/j.ygyno.2015.01.552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boers A, Bosgraaf RP, van Leeuwen RW, Schuuring E, Heideman DA, Massuger LF, et al. DNA methylation analysis in self-sampled brush material as a triage test in hrHPV-positive women. Br J Cancer. 2014;111:1095–1101. doi: 10.1038/bjc.2014.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wentzensen N, Bakkum-Gamez JN, Killian JK, Sampson J, Guido R, Glass A, et al. Discovery and validation of methylation markers for endometrial cancer. Int J Cancer. 2014;135:1860–1868. doi: 10.1002/ijc.28843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang RL, Su PH, Liao YP, Wu TI, Hsu YT, Lin WY, et al. Integrated epigenomics analysis reveals a DNA methylation panel for endometrial cancer detection using cervical scrapings. Clin Cancer Res. 2017;23:263–272. doi: 10.1158/1078-0432.CCR-16-0863. [DOI] [PubMed] [Google Scholar]
- 30.Fiegl H, Gattringer C, Widschwendter A, Schneitter A, Ramoni A, Sarlay D, et al. Methylated DNA collected by tampons--a new tool to detect endometrial cancer. Cancer Epidemiol Biomarkers Prev. 2004;13:882–888. [PubMed] [Google Scholar]
- 31.Fadare O, Chacho MS, Parkash V. Psammoma bodies in cervicovaginal smears: significance and practical implications for diagnostic cytopathology. Adv Anat Pathol. 2004;11:250–261. doi: 10.1097/01.pap.0000131823.69694.62. [DOI] [PubMed] [Google Scholar]
- 32.Smith JH. Psammoma bodies in cervical smears: sifting the grains of sand. Cytopathology. 2007;18:140–142. doi: 10.1111/j.1365-2303.2007.00456.x. [DOI] [PubMed] [Google Scholar]
- 33.Skaznik-Wikiel ME, Ueda SM, Frasure HE, Rose PG, Fleury A, Grumbine FC, et al. Abnormal cervical cytology in the diagnosis of uterine papillary serous carcinoma: earlier detection of a poor prognostic cancer subtype? Acta Cytol. 2011;55:255–260. doi: 10.1159/000324052. [DOI] [PubMed] [Google Scholar]
- 34.Bossuyt V, Buza N, Ngo NT, Much MA, Asis MC, Schwartz PE, et al. Cancerous ‘floater’: a lesson learned about tissue identity testing, endometrial cancer and microsatellite instability. Mod Pathol. 2013;26:1264–1269. doi: 10.1038/modpathol.2013.63. [DOI] [PubMed] [Google Scholar]
- 35.Rauscher GH, Kresovich JK, Poulin M, Yan L, Macias V, Mahmoud AM, et al. Exploring DNA methylation changes in promoter, intragenic, and intergenic regions as early and late events in breast cancer formation. BMC Cancer. 2015;15:816. doi: 10.1186/s12885-015-1777-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang KH, Liu HW, Lin SR, Ding DC, Chu TY. Field methylation silencing of the protocadherin 10 gene in cervical carcinogenesis as a potential specific diagnostic test from cervical scrapings. Cancer Sci. 2009;100:2175–2180. doi: 10.1111/j.1349-7006.2009.01285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu X, Rauch TA, Zhong X, Bennett WP, Latif F, Krex D, et al. CpG island hypermethylation in human astrocytomas. Cancer Res. 2010;70:2718–2727. doi: 10.1158/0008-5472.CAN-09-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pun PB, Liao YP, Su PH, Wang HC, Chen YC, Hsu YW, et al. Triage of high-risk human papillomavirus-positive women by methylated POU4F3. Clin Epigenetics. 2015;7:85. doi: 10.1186/s13148-015-0122-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kocsis A, Takacs T, Jeney C, Schaff Z, Koiss R, Jaray B, et al. Performance of a new HPV and biomarker assay in the management of hrhpv positive women: subanalysis of the ongoing multicenter TRACE clinical trial (n>6,000) to evaluate POU4F3 methylation as a potential biomarker of cervical precancer and cancer. Int J Cancer. 2017;140:1119–1133. doi: 10.1002/ijc.30534. [DOI] [PubMed] [Google Scholar]
- 40.Mahdian R, Nodouzi V, Asgari M, Rezaie M, Alizadeh J, Yousefi B, et al. Expression profile of MAGI2 gene as a novel biomarker in combination with major deregulated genes in prostate cancer. Mol Biol Rep. 2014;41:6125–6131. doi: 10.1007/s11033-014-3491-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
QMSP primers and probes in this study
Demographics of study subjects in testing set of cervical scraping groups of EC and OC
The M-index of 14 candidate genes (ADRA1D, AJAP1, COL6A2, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, SOX17, ST6GAL2, SYT9, and ZNF614) in DNA pools of endometrial tissues. Each dot represent one pool and each pool comprised of 3 patients tissues of ECs.
The M-index of 14 candidate genes (ADRA1D, AJAP1, COL6A2, EDN3, EPO, HS3ST2, MAGI2, POU4F3, PTGDR, SOX8, SOX17, ST6GAL2, SYT9, and ZNF614) in DNA pools of ovarian tissues. Each dot represent one pool and each pool comprised of 3 patients tissues of OCs.