Abstract
We aimed to determine the resistance mechanisms of 27 T. pyogenes isolates from swine in the Jilin province of China. Drug sensitivity analysis indicated that most of the isolated strains were resistant to aminoglycosides. We investigated the genes involved in target alteration, drug inactivation, and increased efflux as potential resistance mechanisms. Two known aminoglycoside resistance genes (aphA1 and strB) were not found in the genomic DNA of any isolate. A 3-bp (CCC) deletion in one 16S rRNA operon was detected in all isolates, and efflux pumps were not active in the resistant group. Ultimately, genes encoding aminoglycoside-modifying enzymes carried by class 1 integrons were identified as the main cause of resistance to aminoglycosides in T. pyogenes.
Keywords: aminoglycosides, pig, resistance mechanism, Trueperella pyogenes
Trueperella pyogenes is a worldwide commensal pathogen of agricultural animals that can cause subcutaneous abscesses, pneumonia, suppurative arthritis and a variety of other infections. For example, 28 T. pyogenes isolates were collected from sick and dead musk deer with suppurative infections of the neck, liver, lungs, musk pods and face in Sichuan Province of China during 2009–2010 [23]. T. pyogenes has been widely detected from cases of bovine endometritis, and 32 T. pyogenes isolates were collected from cows with endometritis from 15 dairy farms in the city of Hohhot, China [13]. T. pyogenes can also infect humans and has been reported to cause infective endocarditis in individuals that have had contact with animals in China [21]. Therefore, it is important to pay closer attention to T. pyogenes in China, not only in musk deer or cows but also in pigs. T. pyogenes is of particular relevance for the hog industry due to the overuse of antibiotics, which has resulted in bacterial resistance and consequent economic losses. T. pyogenes is rarely reported to be resistant to aminoglycosides. In the United States, 29 T. pyogenes isolates were collected from the lung lesions of white-tailed deer with pneumonia, and all isolates were found to be show full or intermediate susceptibility to gentamicin (25 susceptible, 4 intermediate) and high susceptibility to spectinomycin [19]. Antimicrobial susceptibility testing of a T. pyogenes isolate obtained from a 65-year-old man with an oozing ulcer in India revealed that the isolated strain was susceptible to gentamicin [10]. One study determined the minimum inhibitory concentrations (MICs) of 18 antimicrobial agents for 49 T. pyogenes isolates in Japan and found that only 3% were resistant to gentamicin [20]. However, since the drug resistance status of T. pyogenes to aminoglycosides is different in China, we investigated the potential resistance mechanisms of T. pyogenes to aminoglycosides in China.
In this study, a total of 27 T. pyogenes isolates were collected from eight hoggeries located in the Jilin province of China. All samples were collected from the lungs of pigs with pneumonia since 2015. The isolated strains were grown on trypticase soy agar (TSA) plates containing 5% fetal bovine serum and cultured at 37°C under 5% CO2. The 27 T. pyogenes isolates were identified using 16S rRNA gene sequencing and biochemical methods with specific primers from a previous study: 27F (5′-AGAGTTTGATCCTGGC TCAG-3′), 1492R (5′-GGTTACCTTGTTACGACTT-3′) [16]. The MIC values of aminoglycoside antibiotics for the 27 strains were determined via an agar dilution method [4]. For antimicrobial susceptibility testing, the aminoglycoside antibiotics were used at a concentration of 0.03–512 µg/ml. Because there was no Clinical and Laboratory Standards Institute (CLSI) method available for the determination of MICs for T. pyogenes, the susceptibility of the 27 T. pyogenes was determined according to the breakpoints that have been reported previously: 2 µg/ml for gentamicin and 8 µg/ml for amikacin [13]. However, for spectinomycin, kanamycin and streptomycin, there were no published MIC breakpoints in the CLSI criteria or the literature. The reference strains Escherichia coli ATCC 25922 and T. pyogenes ATCC 19411 were used as controls [5].
The transfer of integrons plays an important role in the occurrence of drug resistance [24], and the class 1 integron is an important driver contributing to drug resistance in veterinary clinics. Class 1 integrons are the most common type of integron, and they can capture and accumulate many antibiotic resistance genes from other bacteria [7]. Gram-negative bacteria primarily contain class 1 integrons, but they can also be found in a few gram-positive bacteria [3]. There are less than six gene cassettes in class 1 integrons [6]. In this study, the template DNA for polymerase chain reaction (PCR) was prepared and class 1 integrons were detected using PCR with specific primers from a previous study, as shown in Table 1. Positive amplicons were purified and cloned for sequencing [18]. PCRs were carried out in a 25-µl volume containing 16.8 µl of PCR water, 1 µl of template DNA, 2.5 µl of 0.5 µM dNTP, 10× PCR buffer, 1 µM of each primer, and 0.2 µl of Taq polymerase (Takara, Otsu, Japan); the integrons were confirmed using sequencing.
Table 1. Primers and PCR amplification protocols used in this study.
| Amplification target | Primer | Primers sequence (5′–3′) | Product (bp) | Annealing temperature (°C) | Reference |
|---|---|---|---|---|---|
| 16S | 16S-F | AGAGTTTGATCCTGGCTCAG | ~1,500 | 50 | [16] |
| 16S-R | GGTTACCTTGTTACGACTT | ||||
| IntI1 | Int1-F | CCTCCCGCACGATGATC | 280 | 57 | [18] |
| Int1-R | TCCACGCATCGTCAGGC | ||||
| 3′ CS region | qacEΔ | CGCAATAGTTGGCGAAGTAATC | 774 | 58 | [18] |
| sul1 | GAAGAACCGCACAATCTCGTC | ||||
| Cassette | In5′CS | GGCATCCAAGCAGCAAG | unpredictable | 58.5 | [18] |
| In3′CS | AAGCAGACTTGACCTGA | ||||
| aphA1 | aphA1-F | AAAGCCGTTTCTGTAATGAAGGAG | 642 | 55 | self-design |
| aphA1-R | GGCAATCAGGTGCGACAATCT | ||||
| strB | strB-F | GCGTTGCTCCTCTTCTCCAT | 723 | 54 | self-design |
| strB-R | ACCTTTTCCAGCCTCGTTTG | ||||
| aac(6’)-Ib | aac(6′)-Ib-F | TTAGGCATCACTGCGTGTTCGCTCG | 482 | 57.5 | self-design |
| aac(6′)-Ib-R | GCATCGCAAGGTCATGCTCAGTCAT | ||||
| 16S rRNA1 | 16S rRNA1-F | GTTGGTTGTTTGTTGTGG | 1,758 | 54 | self-design |
| 16S rRNA1-R | AGCATGCCAACTCCAA | ||||
| 16S rRNA2 | 16S rRNA2-F | ACAAAAACCAAAAGATGCTCGCATC | 1,730 | 56.5 | self-design |
| 16S rRNA2-R | GTTTGATCCTGGCTCAGGACGAAC |
Aminoglycosides-modifying enzyme genes may also play a key role in antimicrobial resistance. In this study, we evaluated the presence of three relevant aminoglycoside-resistant genes in the 27 T. pyogenes isolates, including aminoglycoside 3′-phosphotransferaseIII (aphA1), aminoglycoside 6′-phosphotransferase (strB), and acetyltransferase (aac(6’)-Ib), using specifically designed primers, as shown in Table 1.
One German study of Pasteurella multocida showed that 16S rRNA mutations can lead to high-level spectinomycin resistance [11]. However, there have only been a few studies conducted on spectinomycin resistance in T. pyogenes in China. Therefore, we designed primers to identify mutations in two 16S rRNA regions of T. pyogenes that confer resistance to aminoglycosides. The two pairs of specific primers for the 16S rRNA genes of T. pyogenes and PCR settings are shown in Table 1. The PCR amplification system was the same as that used for the PCR of class 1 integron. The sequences were compared with the sequence of 2012CQ-ZSH [22]. Sequence analysis was performed using Molecular Evolutionary Genetics Analysis Version 6.0 (MEGA 6.0) software.
Bacterial resistance to antibiotics is a serious problem in healthcare. The existence of various types of efflux pumps is one of the important mechanisms of bacterial resistance. Efflux pumps can expel various antibiotics and promote mutation accumulation for acquired drug resistance by lowering the intracellular antibiotic concentration [17]. Efflux pump inhibitors can reduce biofilm formation in both Klebsiella strains and E. coli [14]. Efflux pump overexpression can be found in gram-positive bacteria such as methicillin-resistant Staphylococcus aureus [12]. In this study, drug-resistant strains were selected from isolates with high MIC values of gentamicin (≥16 µg/ml) and amikacin (≥64 µg/ml), and the MIC values were compared after mixing with the potent efflux pump inhibitors verapamil and carbonyl cyanide m-chlorophenyl hydrazones (CCCP). The appropriate concentrations (MIC/2) were added to each TSA plate containing CCCP and verapamil using the broth microdilution method and cultured at 37°C under 5% CO2 for 24 hr.
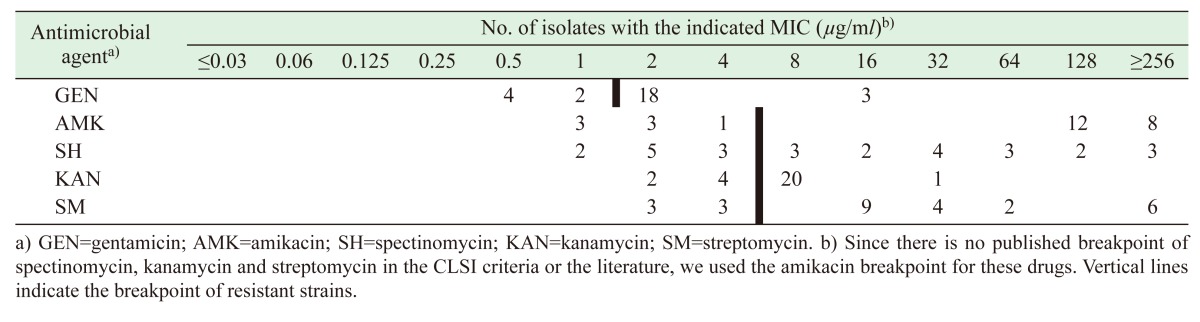
As shown in Table 2 , the MIC values of five aminoglycosides antibiotics against all T. pyogenes isolates were determined. The MIC values of gentamicin, amikacin, spectinomycin, kanamycin and streptomycin for ATCC 19411 were 0.25, 0.5, 0.5, 0.5 and 1 µg/ml, respectively. Most isolates (74.1%) were resistant to amikacin, with MIC values in the range of 1–256 µg/ml, and 21 isolates (77.8%) were resistant to gentamicin. Spectinomycin and streptomycin showed the broadest range of activity (MIC: 1–256 µg/ml). Twenty isolates (74.1%) showed a kanamycin MIC of 8 µg/ml. These values are higher than those previously reported for T. pyogenes in China, which were collected from cows in Hohhtot, Inner Mongolia [13]. However, aminoglycosides have been traditionally used to treat animal diseases, and the overuse and misuse of aminoglycosides render T. pyogenes isolates highly resistant to this class of antibiotics. Sequencing analysis of gene cassettes conducted using Basic Local Alignment Search Tool (BLAST) showed that 59.3% (n=16) of the samples contained gene cassettes, including 2–5 genes for five kinds of arrays. The combinations of gene cassettes contributing to aminoglycoside resistance are shown in Table 3; all of the gene cassettes only exist in the resistant bacteria. The most prevalent gene cassette array detected was aadA1-catB3-dfrB4 (n=6) located in the variable region of class 1 integrons. We here report a novel arrangement in the integron structure of aac(6)’-IIc-blaOXA-10; this is also the first time that the blaOXA-10 gene has been described in T. pyogenes. Class 1 integrons were highly prevalent among the T. pyogenes isolates. With respect to strains that are resistant to aminoglycosides reported in China, one study detected gene cassettes of intI I in T. pyogenes isolates obtained from cows with endometritis, and found that most of the gene cassettes encode resistant determinants of aminoglycosides (aadA1, aadA5, aadA24, and aadB) [13]. However, some amikacin-resistant strains with high MIC values did not carry any aminoglycoside resistance genes in their gene cassettes. Aminoglycoside-modifying enzymes can be chromosomally encoded or plasmid-borne, and are more often encoded on transposable elements. Integrons, as mobile genetic elements, exist in both chromosomal and extra-chromosomal DNA and are able to capture resistance gene cassettes by a site-specific recombination system [2]. We frequently detected a variety of gene cassettes encoding aminoglycoside-modifying enzymes in the isolates. Multiple aminoglycoside resistance genes (aadA1, aadA2, aacA4, aadA5, aac(6’)-IIc and aac(6’)-Ib) were carried by class 1 gene cassettes, which play important roles in the dissemination of aminoglycoside resistance. Furthermore, two relevant aminoglycoside resistance genes (aphA1 and strB) were not found in any of the T. pyogenes isolates, and only aac(6’)-Ib was detected in two isolates; this result is in accordance with the gene cassettes of intI I detected in 27 T. pyogenes isolates from swine in the Jilin province of China.
Table 2. MIC distribution of T. pyogenes isolates.
Table 3. Distribution of gene cassette arrays in 16 T. pyogenes isolates.
| Gene cassette array in Class 1 integrons | Resistance pattern | Number |
|---|---|---|
| aadA1-catB3-dfrB4 | GEN-AMK-SM-KAN-SHa) | 6 |
| aac(6’)-IIc-blaOXA-10 | GEN-AMK-SM-KAN-SH | 2 |
| aac(6’)-Ib-aacA4-arr-2-arr-3 | GEN-AMK-SM-KAN-SH | 2 |
| ORF1-aadA5 | GEN-AMK-SM-KAN-SH | 2 |
| dfrA12-orfF-aadA2 | GEN-AMK-SM-KAN-SH | 4 |
a) Due to the lack of the breakpoints of spectinomycin, kanamycin and streptomycin, we considered the breakpoints for amikacin for these drugs.
The 16S rRNA gene sequences of T. pyogenes isolates, including both the resistant and susceptible strains, were analyzed using MEGA 6.0. The results showed that both resistant and susceptible isolates harbored a 3-bp (CCCC) deletion (from nucleotide 98 to 100 in the locus compared to the reference strain 2012CQ-ZSH) in one rRNA operon of the 16S rRNA gene. However, there were no mutations detected in another rRNA operon. In addition, our results indicated that the MIC values of gentamicin and amikacin were not reduced in the presence of CCCP or verapamil. Therefore, the mechanism of the high-MIC group (MIC for gentamicin ≥16 µg/ml, amikacin ≥64 µg/ml) is not related to the efflux pump but instead involves the presence of aminoglycoside-modifying enzyme resistance genes carried by class 1 integrons.
The resistance issue of many bacteria has become a major threat to public health in humans [17] and animals. A case of spondylodiscitis caused by T. pyogenes in a veterinary surgeon was the first report of a zoonotic disease for this bacterium [9]. Therefore, we must attach importance to the resistance mechanism of T. pyogenes. The antimicrobial resistance of T. pyogenes has been poorly studied to date, especially for aminoglycosides. By contrast, most of the antimicrobial resistance research has focused on Enterobacteriaceae isolated from livestock animals [1]. In this study, we investigated three different resistance mechanisms (target alteration, drug inactivation, and increased efflux); thus, this is the first comprehensive report about resistance mechanisms to aminoglycosides of T. pyogenes in China. In conclusion, this study showed that the main drug resistance mechanisms of T. pyogenes involve the presence of aminoglycoside resistance genes carried by class 1 integrons.
Bacteria are frequently exposed to various antibiotics, which increases the potential for the acquisition and dissemination of antibiotic resistance genes [8]. The analysis of antimicrobial resistance gene cassettes in this study revealed that aminoglycoside resistance determinants (aadA1, aadA2, aacA4, aadA5, aac(6’)-IIc and aac(6’)-Ib) were prevalent among T. pyogenes strains isolated from the lungs of pigs. However, isolates showing high resistance to amikacin did not harbor aminoglycoside resistance genes in the gene cassettes. This could be attributed to the presence of other mobile genetic elements or unknown aminoglycoside resistance genes. OXA-type carbapenemases are frequently detected among Acinetobacter isolates [15]. This issue is worthy of further research because the blaOXA-10 gene belongs to the group of extended-spectrum beta-lactamases. Two T. pyogenes isolates with the blaOXA-10 gene showed higher MIC values of cefotaxime and imipenem compared to the other isolates. Therefore, the blaOXA-10 gene in gene cassettes was found to play a role in the drug resistance of cefotaxime and imipenem in this study. Overall, the present findings send a strong warning about the usage of antimicrobial drugs in livestock production.
Acknowledgments
This research was funded by the National Key Research and Development Plan (Grant number: 2016YFD0501301), National Natural Science Foundation of China (Grant number: 31702293), and Science and Technology Development Plan of Jilin Province (Grant number: 20170520074JH).
REFERENCES
- 1.Arcangioli M. A., Leroy-Setrin S., Martel J. L., Chaslus-Dancla E.2000. Evolution of chloramphenicol resistance, with emergence of cross-resistance to florfenicol, in bovine Salmonella Typhimurium strains implicates definitive phage type (DT) 104. J. Med. Microbiol. 49: 103–110. doi: 10.1099/0022-1317-49-1-103 [DOI] [PubMed] [Google Scholar]
- 2.Bennett P. M.1999. Integrons and gene cassettes: a genetic construction kit for bacteria. J. Antimicrob. Chemother. 43: 1–4. doi: 10.1093/jac/43.1.1 [DOI] [PubMed] [Google Scholar]
- 3.Cambray G., Guerout A. M., Mazel D.2010. Integrons. Annu. Rev. Genet. 44: 141–166. doi: 10.1146/annurev-genet-102209-163504 [DOI] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2013. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. approved standard, 4th ed. In: ClSI Document VET01-A4, CLSI, Wayne. [Google Scholar]
- 5.de Boer M., Heuer C., Hussein H., McDougall S.2015. Minimum inhibitory concentrations of selected antimicrobials against Escherichia coli and Trueperella pyogenes of bovine uterine origin. J. Dairy Sci. 98: 4427–4438. doi: 10.3168/jds.2014-8890 [DOI] [PubMed] [Google Scholar]
- 6.Gillings M., Boucher Y., Labbate M., Holmes A., Krishnan S., Holley M., Stokes H. W.2008. The evolution of class 1 integrons and the rise of antibiotic resistance. J. Bacteriol. 190: 5095–5100. doi: 10.1128/JB.00152-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grape M., Farra A., Kronvall G., Sundström L.2005. Integrons and gene cassettes in clinical isolates of co-trimoxazole-resistant Gram-negative bacteria. Clin. Microbiol. Infect. 11: 185–192. doi: 10.1111/j.1469-0691.2004.01059.x [DOI] [PubMed] [Google Scholar]
- 8.Gu B., Pan S., Wang T., Zhao W., Mei Y., Huang P., Tong M.2008. Novel cassette arrays of integrons in clinical strains of Enterobacteriaceae in China. Int. J. Antimicrob. Agents 32: 529–533. doi: 10.1016/j.ijantimicag.2008.06.019 [DOI] [PubMed] [Google Scholar]
- 9.Ide L., Decostere A., Stuer A., Stuer P., Laere E. D., Verlinde A., Spiritus T., Surmont I.2006. Arcanobacterium pyogenes, spondylodiscitis in a veterinary surgeon: a plea for cooperation between medical and veterinary microbiologists in identification of causal agents of zoonotic infections. Clin. Microbiol. Newsl. 28: 163–167. doi: 10.1016/j.clinmicnews.2006.10.004 [DOI] [Google Scholar]
- 10.Kavitha K., Latha R., Udayashankar C., Jayanthi K., Oudeacoumar P.2010. Three cases of Arcanobacterium pyogenes-associated soft tissue infection. J. Med. Microbiol. 59: 736–739. doi: 10.1099/jmm.0.016485-0 [DOI] [PubMed] [Google Scholar]
- 11.Kehrenberg C., Schwarz S.2007. Mutations in 16S rRNA and ribosomal protein S5 associated with high-level spectinomycin resistance in Pasteurella multocida. Antimicrob. Agents Chemother. 51: 2244–2246. doi: 10.1128/AAC.00229-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosmidis C., Schindler B. D., Jacinto P. L., Patel D., Bains K., Seo S. M., Kaatz G. W.2012. Expression of multidrug resistance efflux pump genes in clinical and environmental isolates of Staphylococcus aureus. Int. J. Antimicrob. Agents 40: 204–209. doi: 10.1016/j.ijantimicag.2012.04.014 [DOI] [PubMed] [Google Scholar]
- 13.Liu M. C., Wu C. M., Liu Y. C., Zhao J. C., Yang Y. L., Shen J. Z.2009. Identification, susceptibility, and detection of integron-gene cassettes of Arcanobacterium pyogenes in bovine endometritis. J. Dairy Sci. 92: 3659–3666. doi: 10.3168/jds.2008-1756 [DOI] [PubMed] [Google Scholar]
- 14.Matsumura K., Furukawa S., Ogihara H., Morinaga Y.2011. Roles of multidrug efflux pumps on the biofilm formation of Escherichia coli K-12. Biocontrol Sci. 16: 69–72. doi: 10.4265/bio.16.69 [DOI] [PubMed] [Google Scholar]
- 15.Mendes R. E., Bell J. M., Turnidge J. D., Castanheira M., Jones R. N.2009. Emergence and widespread dissemination of OXA-23, -24/40 and -58 carbapenemases among Acinetobacter spp. in Asia-Pacific nations: report from the SENTRY Surveillance Program. J. Antimicrob. Chemother. 63: 55–59. doi: 10.1093/jac/dkn434 [DOI] [PubMed] [Google Scholar]
- 16.Moreno C., Romero J., Espejo R. T.2002. Polymorphism in repeated 16S rRNA genes is a common property of type strains and environmental isolates of the genus Vibrio. Microbiology 148: 1233–1239. doi: 10.1099/00221287-148-4-1233 [DOI] [PubMed] [Google Scholar]
- 17.Sun J., Deng Z., Yan A.2014. Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 453: 254–267. doi: 10.1016/j.bbrc.2014.05.090 [DOI] [PubMed] [Google Scholar]
- 18.Sun N., Liu J. H., Yang F., Lin D. C., Li G. H., Chen Z. L., Zeng Z. L.2012. Molecular characterization of the antimicrobial resistance of Riemerella anatipestifer isolated from ducks. Vet. Microbiol. 158: 376–383. doi: 10.1016/j.vetmic.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 19.Tell L. A., Brooks J. W., Lintner V., Matthews T., Kariyawasam S.2011. Antimicrobial susceptibility of Arcanobacterium pyogenes isolated from the lungs of white-tailed deer (Odocoileus virginianus) with pneumonia. J. Vet. Diagn. Invest. 23: 1009–1013. doi: 10.1177/1040638711416618 [DOI] [PubMed] [Google Scholar]
- 20.Yoshimura H., Kojima A., Ishimaru M.2000. Antimicrobial susceptibility of Arcanobacterium pyogenes isolated from cattle and pigs. J. Vet. Med. B Infect. Dis. Vet. Public Health 47: 139–143. doi: 10.1046/j.1439-0450.2000.00315.x [DOI] [PubMed] [Google Scholar]
- 21.Zhang H., Shi Z., Yang Q., Chen Y., Xu Y.2014. Endocarditis caused by Arcanobacterium pyogenes. Chin. Med. J. (Engl.) 127: 3510–3511. [PubMed] [Google Scholar]
- 22.Zhang S. H., Qiu J. J., Yang R., Shen K. F., Xu G. Y., Fu L. Z.2016. Complete genome sequence of Trueperella pyogenes, isolated from infected farmland goats. Genome Announc. 4: 6. doi: 10.1128/genomeA.01421-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao K. L., Liu Y., Zhang X. Y., Palahati P., Wang H. N., Yue B. S.2011. Detection and characterization of antibiotic-resistance genes in Arcanobacterium pyogenes strains from abscesses of forest musk deer. J. Med. Microbiol. 60: 1820–1826. doi: 10.1099/jmm.0.033332-0 [DOI] [PubMed] [Google Scholar]
- 24.Zhu Y., Yi Y., Liu F., Lv N., Yang X., Li J., Hu Y., Zhu B.2014. Distribution and molecular profiling of class 1 integrons in MDR Acinetobacter baumannii isolates and whole genome-based analysis of antibiotic resistance mechanisms in a representative strain. Microbiol. Res. 169: 811–816. doi: 10.1016/j.micres.2014.04.002 [DOI] [PubMed] [Google Scholar]