Abstract
Although alteration of commensal microbiota is associated with chronic gastrointestinal (GI) diseases such as inflammatory bowel disease (IBD) in dogs, the microbiota composition in intestinal lymphoma, an important differential diagnosis of canine IBD, has not been investigated. The objective of this study was to compare the fecal microbiota in dogs with IBD, dogs with intestinal lymphoma, and healthy dogs. Eight dogs with IBD, eight dogs with intestinal lymphoma, and fifteen healthy dogs were included in the study. Fecal samples were analyzed by 16S rRNA gene next-generation sequencing. Rarefaction analysis failed to reveal any difference in bacterial diversity among healthy dogs and diseased dogs. Based on PCoA plots of unweighted UniFrac distances, the bacterial composition in dogs with intestinal lymphoma was different from those observed in dogs with IBD and healthy dogs. When compared with healthy dogs, intestinal lymphoma subjects showed significant increases in organisms belonging to the Eubacteriaceae family. The proportion of the family Paraprevotellaceae and the genus Porphyromonas was significantly higher in dogs with IBD compared to healthy dogs. These observations suggest that dysbiosis is associated with intestinal lymphoma as well as IBD in dogs.
Keywords: canine, chronic enteropathy, enteropathy-associated T-cell lymphoma, microbiota
All mammals harbor a vast and complex community of bacteria in their intestine. Recent studies have revealed that microbiota plays an important role in gastrointestinal (GI) health. Their main functions include protection against pathogens, fermentation of non-digestible dietary residues, and development of immune systems [18].
Germ-free mice are very susceptible to infection, indicating that the resident microbiota plays an important role in conferring resistance against pathogen colonization [11]. This protection is enabled by preventing the entry of pathogens into the epithelial cells [34], competing for available nutrients and space in the GI tract, and producing antimicrobial substances [43].
Microbiota metabolizes sloughed epithelial cells, endogenous mucus, and nondigested substrates that have passed through the small intestine. The fermentation of these substrates results in the production of short-chain fatty acids (SCFAs) that provide energy for bacterial metabolism and epithelial cell growth. For example, germ-free rats need 18% more caloric intake than conventional rats to sustain their energy and growth levels [40].
Microbiota has also been implicated in immune development. Dogs, as well as mice, in germ-free conditions exhibit an underdeveloped lymphoid system and decreased immunoglobulin concentrations [10, 24, 28]. Moreover, germ-free mice show a Th2 predominant immune response. These abnormalities can be corrected by restoring the conventional microbiota [18]. Therefore, GI microbiota plays a crucial role in the homeostasis of both mucosal and systemic immunity.
Inappropriate activation of immune responses to GI microbiota is thought to contribute to the pathogenesis of inflammatory bowel disease (IBD). The evidence supporting this hypothesis was obtained from experiments demonstrating the inhibition of intestinal inflammation in IBD model mice under germ-free conditions [8, 30]. Recent studies have shown distinct shifts of GI microbiota composition in human IBD, defined as “dysbiosis” [12, 29, 33]. Dysbiosis has also been reported in canine IBD cases. Most commonly, a decrease in the phyla, Firmicutes and Bacteroidetes, combined with increases in Proteobacteria and Actinobacteria are observed [19]. These observations suggest that dysbiosis is associated with IBD in both humans and dogs. However, the dysbiosis network underlying IBD in dogs differs from that in humans, with some bacteria such as Fusobacterium switching roles between the two species [38]. Similarly, there are differences in the dysbiosis network with IBD between humans and mice [14]. Therefore, further studies are needed to investigate host-specific dysbiosis networks and point the way towards a generalized understanding of IBD across different mammalian models.
Intestinal lymphoma is an important differential diagnosis of IBD in dogs [39]. Since the clinical presentation of both diseases is similar, their diagnoses are often difficult to perform on the basis of laboratory tests. Histopathologic examination, immunohistochemistry (IHC), and polymerase chain reaction for antigen receptor gene rearrangement (PARR) are helpful toward confirming the diagnosis [9]. While several studies revealed bacterial-induced lymphoma in humans [42], there has been no report on the investigation of fecal microbiomes in dogs with intestinal lymphoma. Characterizing fecal microbiota of dogs with IBD and intestinal lymphoma may provide insights into the pathogenesis of these chronic GI diseases. The aim of this study was to compare the fecal microbiota in dogs with either IBD or intestinal lymphoma, and healthy dogs.
MATERIALS AND METHODS
Animals and sample collection
This study included 16 dogs with clinical signs of chronic GI disease that underwent endoscopic examination in the Veterinary Medical Center of the University of Tokyo (VMC-UT) between 2009 and 2012. The study protocol was approved by the Animal Care Committee of VMC-UT, and informed consent was obtained from the owners of each dog. Diagnosis of IBD was based on histopathologic evidence of lymphocytic-plasmacytic enteritis, the absence of clonal gene rearrangement of IgH or TCRγ in PARR, and exclusion of food- and antibiotic-responsive enteropathies [1, 26, 39]. Diagnosis of intestinal lymphoma was based on histopathologic examination, the presence of clonal IgH or TCRγ gene rearrangement, and IHC for CD3 and CD20 [15, 17, 25]. All cases were scored for severity according to the canine IBD activity index (CIBDAI) and canine chronic enteropathy clinical activity index (CCECAI) [1, 23].
As healthy controls, 15 dogs with no clinical signs of GI disease were included. They showed no abnormalities in urinalysis and blood examinations, including complete blood count and measurements of blood urea nitrogen and creatinine concentrations as well as alanine aminotransferase and alkaline phosphatase enzyme activities. Moreover, there were no abnormalities in the parasitic and bacterial analyses of their fecal samples.
Dogs that had been treated with corticosteroids, antibiotics, or prebiotics/probiotics within 2 weeks prior to the study were excluded. The antibiotic-free interval of each case is shown in Table S1. All dogs enrolled in this study were privately owned and came from diverse environments.
Fresh rectal feces were collected from all dogs, and immediately frozen at −80°C until DNA extraction.
DNA extraction
Fecal samples (20 mg) were resuspended in 450 µl extraction buffer (100 mM Tris/HCl, 40 mM EDTA, pH 9.0), and 50 µl 10% SDS. Glass beads (300 mg, 0.1 mm diameter) and 500 µl buffer-saturated phenol were added to the suspension, and the mixture was vortexed vigorously for 30 sec using a FastPrep FP 100A (MP Biomedicals, Santa Ana, CA, U.S.A.) at a power level of 5. After centrifugation at 14,000 × g for 5 min, 400 µl of the supernatant was extracted with phenol/chloroform, and 250 µl of supernatant was precipitated with propan-2-ol. Purified DNA was rinsed with 300 µl 70% ethanol and then resuspended in 200 µl Tris/EDTA buffer (pH 8.0).
16S rRNA gene sequencing
Amplification and sequencing of the V3-V4 region of the bacterial 16S rRNA gene was performed according to previously described methods optimized for the Illumina MiSeq (Illumina, San Diego, CA, U.S.A.), with primers forward 357F: CGCTCTTCCGATCTCTGTACGGRAGGCAGCAG and reverse 806R: CGCTCTTCCGATCTGACGGACTACHVGGGTWTCTAAT, as described previously [21]. Dual barcoded amplicons were generated using TaKaRa Ex Taq HS (Takara Bio, Otsu, Japan). The amplification conditions were as follows: 94°C for 3 min, 25 cycles of PCR (94°C for 30 sec, 50°C for 1 min, and 72°C for 1 min), and a final elongation step at 72°C for 10 min. The amplicons were pooled in equimolar concentrations and sequenced with an Illumina MiSeq platform using MiSeq Reagent Kit v2 (Illumina).
Raw 150 bp paired-end sequence reads were combined using the script fastq-join (ea-utils-1.1.2-301.x86_64.rtp: https://expressionanalysis.github.io/ea-utils/) with the default settings. Further data processing included filtering and denoising by clustering similar sequences with less than 3% dissimilarity using USEARCH v5.2.32 (http://drive5.com/usearch/), as well as de-novo chimera detection and removal in UCHIME (http://drive5.com/usearch/manual/uchime_algo.html). The 16S rRNA operational taxonomic units (OTUs) were selected from the combined reads using a de-novo OTU picking protocol clustered at 97% identity through the Quantitative Insights Into Microbial Ecology (QIIME) pipeline software version 1.6.0 (http://qiime.org), with USEARCH against the Greengenes database (http://greengenes.secondgenome.com/downloads/database/12_10; Oct. 2012 release). The representative sequences for each OTU were compared with those present in the Greengenes database for taxonomy assignment. To account for unequal sequencing depth across samples, subsequent analyses were performed on a randomly selected subset of 53,580 sequences per sample.
Statistics
Continuous variables were compared between groups by the Kruskal-Wallis test, and categorical variables were determined by the chi-square test or Fisher’s exact test using Prism v5.0 (GraphPad Software, San Diego, CA, U.S.A.).
To estimate the bacterial diversity of each sample, three indices (number of OTUs, Shannon index, and Chao1) were calculated and rarefaction curves were depicted using QIIME. Differences in microbial communities among samples were investigated using phylogeny-based unweighted or weighted UniFrac distance matrices, which were calculated using the Greengenes reference tree. Principal coordinate analysis (PCoA) and hierarchical dendrogram construction were performed using QIIME. Differences in microbiota compositions between samples from three groups were tested using the one-way analysis of similarity (ANOSIM) function in the statistical software package PRIMER 6 (PRIMER-E, Luton, U.K.). To determine differences in bacterial diversity indices and proportions of bacterial taxa between three groups, Kruskal-Wallis test was performed and adjusted for multiple comparison using Bonferroni correction. Post hoc Dunn’s multiple comparison test was used. Only bacterial taxa that were present in at least half of the dogs (either IBD, intestinal lymphoma, or healthy) were included in the analysis. The chi-square test was used to determine the proportion of dogs that harbored specific bacterial taxa. A value of P<0.05 was considered to be statistically significant for all analyses.
RESULTS
Animals
A summary of the characteristics of the dogs enrolled in this study is shown in Table 1. Eight dogs were diagnosed with IBD. All dogs with IBD had evidence of inflammation within the intestinal mucosa and a histopathologic diagnosis of lymphocytic-plasmacytic enteritis. The breeds were Miniature Dachshund (n=2), Beagle (n=1), French Bulldog (n=1), Siberian husky (n=1), Miniature Schnauzer (n=1), Pembroke Welsh Corgi (n=1), and Yorkshire Terrier (n=1).
Table 1. Comparison of characteristics and bacterial diversity indices.
| Healthy | IBD | Intestinal lymphoma | P value | |
|---|---|---|---|---|
| Gender (female/male) | 8/7 | 4/4 | 3/5 | 0.895 |
| Age (months) | 108 (18–198) | 105 (56–140) | 91.5 (52–143) | 0.789 |
| Body weight (kg) | 7.3 (1.8–31.2) | 7.6 (2.0–18.8) | 5.6 (2.7–28.9) | 0.946 |
| CIBDAI | 0a (0–0) | 4.5b (2–12) | 6.5b (6–14) | <0.0001 |
| CCECAI | 0a (0–0) | 7b (4–12) | 7b (6–18) | <0.0001 |
| OTU | 1,908 (929–2,603) | 1,971 (1,350–2,296) | 1,897 (1,247–2,721) | 0.935 |
| Shannon index | 6.13 (3.84–8.07) | 6.10 (5.12–6.87) | 6.20 (3.52–7.47) | 0.830 |
| Chao1 | 2,929 (1,822–3,701) | 3,027 (2,163–3,568) | 2,890 (2,077–3,836) | 0.815 |
IBD, inflammatory bowel disease; CIBDAI, canine IBD activity index; CCECAI, canine chronic enteropathy clinical activity index; OUT, operational taxonomic unit. Age, body weight, CIBDAI, and CCECAI scores are represented by the median value (range), while bacterial diversity indices (OTU, Shannon index, and Chao1) are represented by the mean (range). *Medians not sharing a common superscript are significantly different (P<0.01 based on a Dunn’s multiple comparison test).
Eight dogs were diagnosed with intestinal lymphoma. Among these, seven neoplasms were classified as T-cell lymphoma, and one was classified as B-cell lymphoma according to IHC and PARR results (Table S1). Six dogs were diagnosed with small cell type and two dogs with large cell type. The breeds were Shiba inu (n=2), Chihuahua (n=1), Jack Russell Terrier (n=1), Labrador Retriever (n=1), Shih-tzu (n=1), Miniature Dachshund (n=1), and Toy Manchester Terrier (n=1).
The control group included 15 healthy dogs: Miniature Dachshund (n=2), Chihuahua (n=2), Beagle (n=1), French Bulldog (n=1), Jack Russell Terrier (n=1), Labrador Retriever (n=1), Maltese (n=1), Miniature Schnauzer (n=1), Papillon (n=1), Siberian husky (n=1), Shiba inu (n=1), Yorkshire Terrier (n=1) and Pembroke Welsh corgi (n=1).
Group compositions in terms of sex, age, and body weight showed no difference between the three groups (Table 1). CIBDAI and CCECAI scores differed significantly between the three groups; however, there was no significant difference between the IBD and intestinal lymphoma groups.
Bacterial diversity
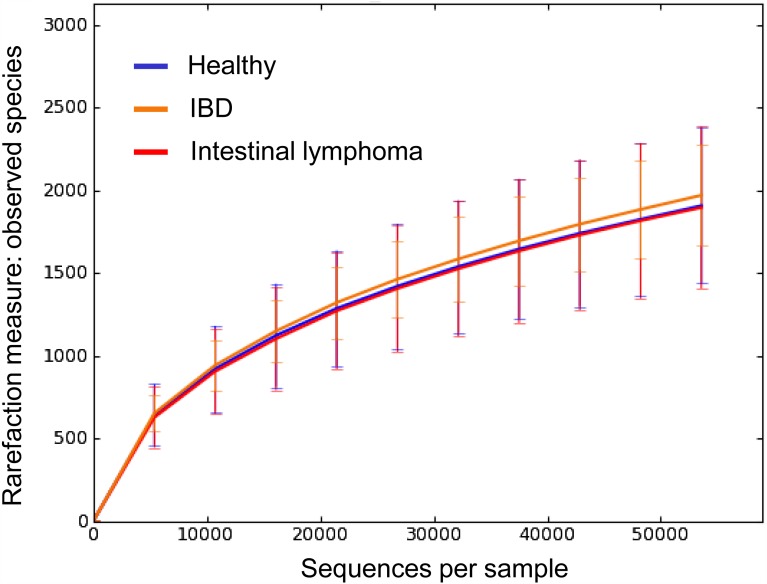
Rarefaction curves are shown in Fig. 1. No significant differences in bacterial diversity indices were observed between the three groups (Table 1).
Fig. 1.
Rarefaction analysis of 16S rRNA gene sequences obtained from canine fecal samples. Lines represent the average for each group (blue: healthy, yellow: inflammatory bowel disease, red: intestinal lymphoma). The error bars represent the standard deviations. The analysis was performed on a randomly selected subset of 53,580 sequences per sample.
Bacterial composition
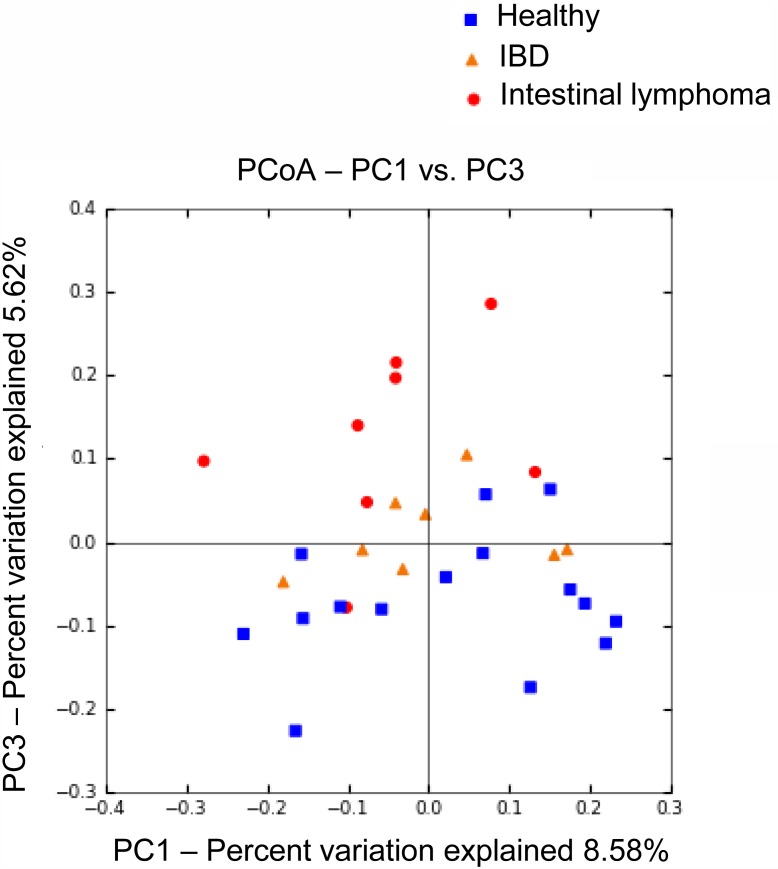
Bacterial composition of fecal microbiota from dogs with intestinal lymphoma was significantly different from those observed in IBD and healthy dogs. PCoA plots based on the unweighted UniFrac distance matrices (Fig. 2) showed significant separations between the samples obtained from intestinal lymphoma dogs and those retrieved from the other two groups (ANOSIM; global R=0.133, P=0.023; IBD vs intestinal lymphoma, R=0.289, P=0.006; intestinal lymphoma vs healthy, R=0.247, P=0.003). The effects of gender, age, diet, as well as CIBDAI and CCECAI scores on bacterial compositions were also evaluated by ANOSIM, and there was no clustering (Table 2). Therefore, the separation was predominantly due to disease rather than to other factors. In contrast, no significant difference was observed between IBD and healthy dogs (ANOSIM; IBD vs healthy, R=−0.012, P=0.540). The PCoA plots and hierarchical dendrogram constructed with the weighted UniFrac distance matrices showed no difference between the three groups.
Fig. 2.
Principal Coordinates Analysis (PCoA) of unweighted UniFrac distances of 16S rRNA genes. The samples clustered along principal coordinates (PC) 1 and 3. Samples from dogs with intestinal lymphoma were separated from those obtained from healthy and IBD subjects (ANOSIM; intestinal lymphoma vs healthy, P=0.003; intestinal lymphoma vs IBD, P=0.006), while no separation was observed between healthy and IBD dogs (ANOSIM; P=0.54).
Table 2. One-way analysis of similarities (ANOSIM) in bacterial compositions considering several variables.
| Grouping | R value | P value | ||
|---|---|---|---|---|
| Clinical disease | Healthy, IBD, Intestinal lymphoma | 0.133 | 0.023 | |
| Healthy vs Intestinal lymphoma | 0.247 | 0.003 | ||
| IBD vs Intestinal lymphoma | 0.289 | 0.006 | ||
| CIBDAI | clinicaly insignificant, mild, moderate, severe, very severe | 0.079 | 0.216 | |
| CCECAI | clinicaly insignificant, mild, moderate, severe, very severe | −0.001 | 0.516 | |
| Age | <7, 7–10, >10 (years) | −0.028 | 0.723 | |
| Sex | male, female | 0.057 | 0.077 | |
| Food composition | ||||
| Fat | <3.0, 3.0–3.9, >3.9 (g/100 kcal) | −0.047 | 0.872 | |
| Protein | <6.2, 6.2–6.5, >6.5 (g/100 kcal) | −0.039 | 0.824 | |
| Carbohydrate | <10.0, 10.0–13.5, >13.5 (g/100 kcal) | 0.054 | 0.123 | |
| Coarse fiber | <1.1, 1.1–1.5, >1.5 (g/100 kcal) | −0.059 | 0.893 | |
IBD, inflammatory bowel disease; CIBDAI, canine IBD activity index; CCECAI, canine chronic enteropathy clinical activity index.
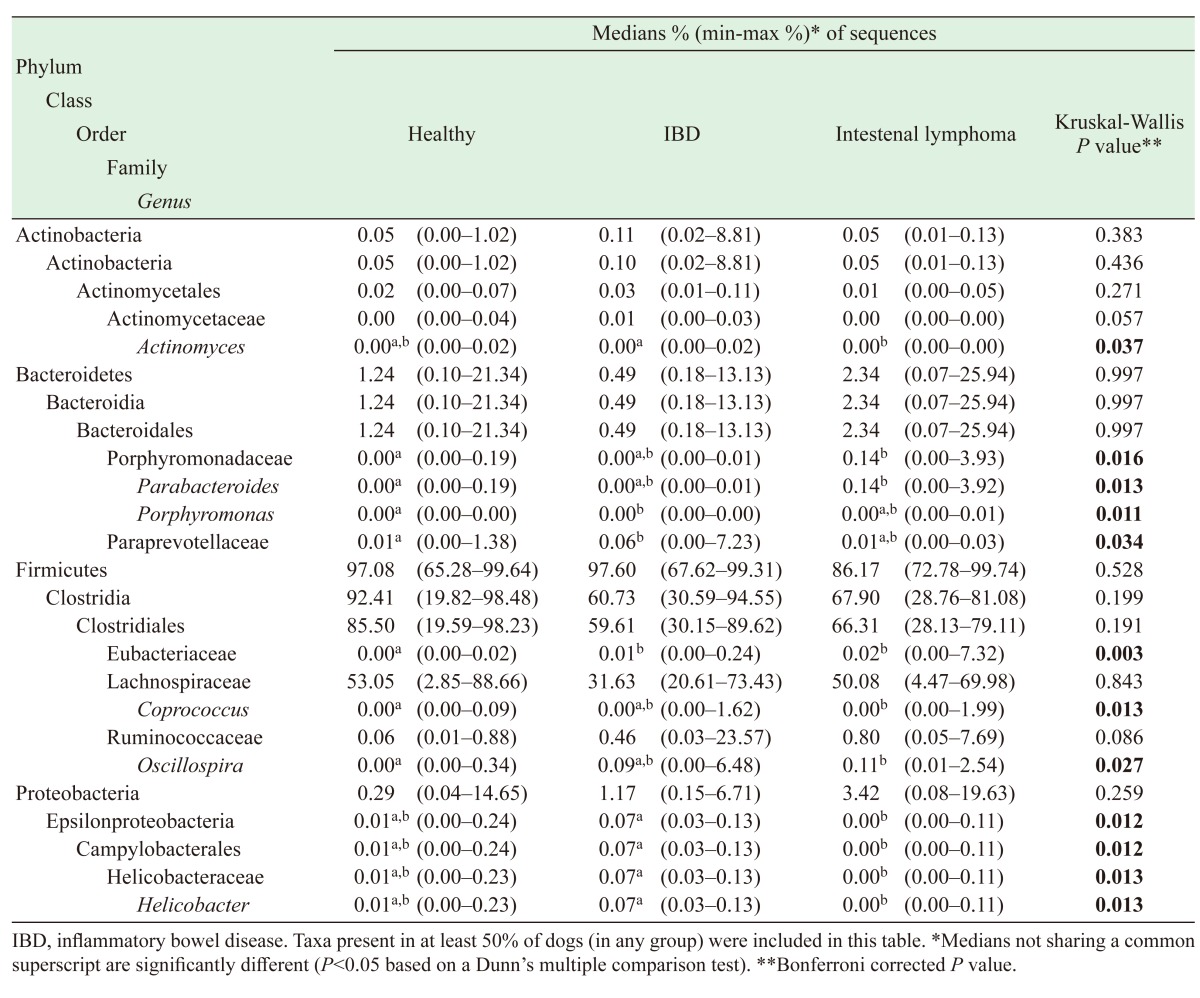
Table 3 summarizes the relative proportions of bacterial taxa differing between three groups. In addition, the proportions of all bacterial taxa analyzed in this study are represented in Table S2. At phylum level, there was no difference between diseased and healthy dogs (Table S2); however, significant differences were identified at lower phylogenetic levels.
Table 3. Relative proportions of predominant bacterial taxa.
Within the phylum Actinobacteria, an increased proportion of Actinomyces was observed in IBD compared to intestinal lymphoma (P<0.05).
Within the phylum Bacteroidetes, the families Porphyromonadaceae and Paraprevotellaceae showed significant changes. In comparison with healthy dogs, Porphyromonadaceae Parabacteroides increased in dogs with intestinal lymphoma (P<0.01), while Porphyromonas increased in dogs with IBD (P=0.004). The family Paraprevotellaceae increased in dogs with IBD (P=0.01).
Firmicutes was the most abundant phylum among all dogs, however, there was no significance difference between the three groups in this study. At the lower levels, some families and genera in the order Clostridiales increased in dogs with intestinal lymphoma compared to healthy ones: Lachnospiraceae Coprococcus (P<0.01) and Ruminococcaceae Oscillospira (P<0.05). Eubacteriaceae increased in both IBD and intestinal lymphoma dogs compared to healthy ones (IBD vs healthy, P<0.01; intestinal lymphoma vs healthy, P<0.05).
Within the phylum Proteobacteria, Helicobacteraceae Helicobacter increased in dogs with IBD compared to those suffering from intestinal lymphoma (P<0.01). In addition, Escherichia increased in intestinal lymphoma dogs, even though this trend was not statistically significant (P=0.06).
DISCUSSION
This study evaluated the fecal microbiota in healthy dogs and dogs with IBD and intestinal lymphoma. Although the rarefaction analysis failed to show any differences in bacterial diversities, we revealed significant alterations in microbiota compositions in dogs with IBD and intestinal lymphoma. In terms of individual bacterial taxa, unique compositions were observed in dogs with IBD and intestinal lymphoma, respectively. Therefore, there is a possibility that dogs with intestinal lymphoma or IBD have their own distinctive characteristics in terms of dysbiosis in GI microbiota.
The proportion of the family Eubacteriaceae was significantly higher in dogs with intestinal lymphoma. Eubacteriaceae is a Gram-positive bacterial family from the order Clostridiales. This family includes members of clusters IV and XIVa of the genus Clostridium, producing important SCFAs butyrate. Atarashi et al. revealed that 46 and 17 strains of Clostridium, including clusters IV and XIVa, had the potential to induce regulatory T cells (Tregs) in mice and humans, respectively [2, 3]. Furthermore, Furusawa et al. have reported that commensal bacteria-derived butyrate induces the differentiation of colonic Tregs via enhancement of histone H3 acetylation in the promoter regions of the Foxp3 gene locus [13]. The results of Treg number in our previous study [25] showed that half of the dogs with intestinal lymphoma used in this study exhibited a high density of Tregs in the intestinal lesions (Table S1). Collectively, these observations suggest that the increase in Eubacteriaceae might be involved in the increased number of Tregs in canine intestinal lymphoma through overproduction of butyrate. Further analyses are needed to clarify the association between the family Eubacteriaceae and Treg infiltration in dogs with intestinal lymphoma.
In humans, mucosal-associated lymphoid tissue (MALT) lymphomas are strongly associated with the presence of Helicobacter spp [22, 32]. Wotherspoon et al. reported that Helicobacter infection was detected in 92% of patients with MALT lymphomas [41]. Eradication of Helicobacter leads to complete remission in 70% of MALT lymphoma cases [5], suggesting that the development of lymphoma is caused by Helicobacter infection. In this study, the proportion of Helicobacter was not increased in dogs with intestinal lymphoma. Histologically, the lesions associated with canine intestinal lymphoma are mainly localized in the mucosal lamina propria [9]. Additionally, MALT lymphomas have histological features allowing the neoplastic cells to proliferate around reactive B-cell follicles in the region corresponding to the Peyer’s patch marginal zone, spreading diffusely into the surrounding tissues [22]. Therefore, the mechanisms governing tumorigenesis in canine intestinal lymphoma may be different from those driving MALT lymphomas in humans.
Parabacteroides spp. significantly increased in dogs with intestinal lymphoma. Parabacteroides is a genus of Gram-negative obligate anaerobic bacteria belonging to the phylum Bacteroidetes and the order Bacteroidales. In colorectal cancer model mice and rats, the members of the Bacteroidales, including Bacteroides and Parabacteroides, have been associated with tumorigenesis [4, 44]; therefore, there is a possibility that the presence of Parabacteroides play a role in the development of canine intestinal lymphoma. However, the observed microbiome change might just reflect higher disease severity rather than being causal. Further investigations will be necessary to confirm this microbiota-induced lymphomagenesis hypothesis in dogs.
In this study, we found increased proportions of the family Paraprevotellaceae and the genus Porphyromonas in dogs with IBD compared to healthy dogs. Both of them are Gram-negative bacteria belonging to the phylum Bacteroidetes and the order Bacteroidales. To our knowledge, no study has showed significant change of the family Paraprevotellaceae and the genus Porphyromonas in canine IBD. In humans, the one study, using biopsy samples from IBD patients, has revealed an increase in the genus Porphyromonas [7]; however, the meaning is unknown. Therefore, we would like to examine the function of the family Paraprevotellaceae and the genus Porphyromonas in canine IBD.
Previous studies showed that the phyla Firmicutes and Fusobacteria decreased in dogs with IBD, whereas Proteobacteria and Actinobacteria increased [35, 37]. However, in this study, we could not detect these shifts in canine IBD. Regarding the trend for higher proportions of Proteobacteria and Actinobacteria in dogs with IBD, the discrepancy among the present and previous studies may be due to the large inter-animal variation and small sample size. The number of dogs with IBD in this study (n=8) was less than that in the previous studies (n=14−19) [35, 37]. Another possible cause for the lack of reproducibility is the differences between the types of samples used for the respective microbiome analyses. Over half of the previous studies used mucosal samples including duodenum and colon [19]. More extensive studies are necessary to investigate the phyla alterations in dogs with IBD.
Medications, such as antibiotics, are capable of changing bacterial compositions dramatically. Therefore, a withdrawal period is required before performing any microbiome analysis. A previous study included IBD dogs without antibiotic treatment for 2 months prior to sampling [37]. Another investigation included dogs without antibiotics and immunomodulating drugs administrations for a month prior to sampling [31]. In this study, we adopted a 2-weeks washout of antibiotics and corticosteroids. In dogs, changes in GI microbiota induced by macrolide antibiotic tylosin were not reversed within 4 weeks [36]. Our previous study showed that metronidazole induced an alteration of the intestinal microbiota, and that the altered bacterial composition could return to the pre-treatment state after a 2-weeks cessation period [20]. Therefore, the inconsistencies in microbiota compositions among dogs with IBD between studies could be explained by the discrepancies in the washout periods applied. Food is another potential factor influencing changes in GI microbiota. To date, several studies have shown the effect of dietary interventions, such as fiber, animal-derived protein, and carbohydrates supplementations on GI microbiota in dogs [6, 16, 27]. However, no significant effects of dietary components on GI microbiota were observed in this study, and therefore, it is unlikely that the differences in diets affected the present results.
In conclusion, we observed that the compositions of fecal microbiota in dogs with IBD and intestinal lymphoma were significantly different from that detected in healthy dogs. These results form a roadmap for additional studies focused on the mechanisms governing microbiota-induced lymphomagenesis in dogs. Since a limited number of diseased dogs was included in this study, a further follow-up study using a larger number of dogs with IBD and intestinal lymphoma should be performed in the future.
Supplementary
REFERENCES
- 1.Allenspach K., Wieland B., Gröne A., Gaschen F.2007. Chronic enteropathies in dogs: evaluation of risk factors for negative outcome. J. Vet. Intern. Med. 21: 700–708. doi: 10.1111/j.1939-1676.2007.tb03011.x [DOI] [PubMed] [Google Scholar]
- 2.Atarashi K., Tanoue T., Shima T., Imaoka A., Kuwahara T., Momose Y., Cheng G., Yamasaki S., Saito T., Ohba Y., Taniguchi T., Takeda K., Hori S., Ivanov I. I., Umesaki Y., Itoh K., Honda K.2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331: 337–341. doi: 10.1126/science.1198469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atarashi K., Tanoue T., Oshima K., Suda W., Nagano Y., Nishikawa H., Fukuda S., Saito T., Narushima S., Hase K., Kim S., Fritz J. V., Wilmes P., Ueha S., Matsushima K., Ohno H., Olle B., Sakaguchi S., Taniguchi T., Morita H., Hattori M., Honda K.2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500: 232–236. doi: 10.1038/nature12331 [DOI] [PubMed] [Google Scholar]
- 4.Baxter N. T., Zackular J. P., Chen G. Y., Schloss P. D.2014. Structure of the gut microbiome following colonization with human feces determines colonic tumor burden. Microbiome 2: 20. doi: 10.1186/2049-2618-2-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayerdörffer E., Neubauer A., Rudolph B., Thiede C., Lehn N., Eidt S., Stolte M., MALT Lymphoma Study Group. 1995. Regression of primary gastric lymphoma of mucosa-associated lymphoid tissue type after cure of Helicobacter pylori infection. Lancet 345: 1591–1594. doi: 10.1016/S0140-6736(95)90113-2 [DOI] [PubMed] [Google Scholar]
- 6.Beloshapka A. N., Dowd S. E., Suchodolski J. S., Steiner J. M., Duclos L., Swanson K. S.2013. Fecal microbial communities of healthy adult dogs fed raw meat-based diets with or without inulin or yeast cell wall extracts as assessed by 454 pyrosequencing. FEMS Microbiol. Ecol. 84: 532–541. doi: 10.1111/1574-6941.12081 [DOI] [PubMed] [Google Scholar]
- 7.Bibiloni R., Mangold M., Madsen K. L., Fedorak R. N., Tannock G. W.2006. The bacteriology of biopsies differs between newly diagnosed, untreated, Crohn’s disease and ulcerative colitis patients. J. Med. Microbiol. 55: 1141–1149. doi: 10.1099/jmm.0.46498-0 [DOI] [PubMed] [Google Scholar]
- 8.Blumberg R. S., Saubermann L. J., Strober W.1999. Animal models of mucosal inflammation and their relation to human inflammatory bowel disease. Curr. Opin. Immunol. 11: 648–656. doi: 10.1016/S0952-7915(99)00032-1 [DOI] [PubMed] [Google Scholar]
- 9.Carrasco V., Rodríguez-Bertos A., Rodríguez-Franco F., Wise A. G., Maes R., Mullaney T., Kiupel M.2015. Distinguishing Intestinal Lymphoma From Inflammatory Bowel Disease in Canine Duodenal Endoscopic Biopsy Samples. Vet. Pathol. 52: 668–675. doi: 10.1177/0300985814559398 [DOI] [PubMed] [Google Scholar]
- 10.Cohn I., Jr., Heneghan J. B.1991. Germfree animals and technics in surgical research. Am. J. Surg. 161: 279–283. doi: 10.1016/0002-9610(91)91145-9 [DOI] [PubMed] [Google Scholar]
- 11.Collins F. M., Carter P. B.1978. Growth of salmonellae in orally infected germfree mice. Infect. Immun. 21: 41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frank D. N., St Amand A. L., Feldman R. A., Boedeker E. C., Harpaz N., Pace N. R.2007. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104: 13780–13785. doi: 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furusawa Y., Obata Y., Fukuda S., Endo T. A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N. N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J. M., Topping D. L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H.2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504: 446–450. doi: 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 14.Gevers D., Kugathasan S., Denson L. A., Vázquez-Baeza Y., Van Treuren W., Ren B., Schwager E., Knights D., Song S. J., Yassour M., Morgan X. C., Kostic A. D., Luo C., González A., McDonald D., Haberman Y., Walters T., Baker S., Rosh J., Stephens M., Heyman M., Markowitz J., Baldassano R., Griffiths A., Sylvester F., Mack D., Kim S., Crandall W., Hyams J., Huttenhower C., Knight R., Xavier R. J.2014. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15: 382–392. doi: 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto-Koshino Y., Mochizuki H., Sato M., Nakashima K., Hiyoshi S., Fujiwara-Igarashi A., Maeda S., Nakamura K., Uchida K., Fujino Y., Ohno K., Tsujimoto H.2015. Construction of a multicolor GeneScan analytical system to detect clonal rearrangements of immunoglobulin and T cell receptor genes in canine lymphoid tumors. Vet. Immunol. Immunopathol. 165: 81–87. doi: 10.1016/j.vetimm.2015.03.005 [DOI] [PubMed] [Google Scholar]
- 16.Hang I., Rinttila T., Zentek J., Kettunen A., Alaja S., Apajalahti J., Harmoinen J., de Vos W. M., Spillmann T.2012. Effect of high contents of dietary animal-derived protein or carbohydrates on canine faecal microbiota. BMC Vet. Res. 8: 90. doi: 10.1186/1746-6148-8-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiyoshi S., Ohno K., Uchida K., Goto-Koshino Y., Nakashima K., Fukushima K., Kanemoto H., Maeda S., Tsujimoto H.2015. Association between lymphocyte antigen receptor gene rearrangements and histopathological evaluation in canine chronic enteropathy. Vet. Immunol. Immunopathol. 165: 138–144. doi: 10.1016/j.vetimm.2015.03.009 [DOI] [PubMed] [Google Scholar]
- 18.Honda K., Littman D. R.2012. The microbiome in infectious disease and inflammation. Annu. Rev. Immunol. 30: 759–795. doi: 10.1146/annurev-immunol-020711-074937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Honneffer J. B., Minamoto Y., Suchodolski J. S.2014. Microbiota alterations in acute and chronic gastrointestinal inflammation of cats and dogs. World J. Gastroenterol. 20: 16489–16497. doi: 10.3748/wjg.v20.i44.16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igarashi H., Maeda S., Ohno K., Horigome A., Odamaki T., Tsujimoto H.2014. Effect of oral administration of metronidazole or prednisolone on fecal microbiota in dogs. PLOS ONE 9: e107909. doi: 10.1371/journal.pone.0107909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igarashi H., Ohno K., Horigome A., Fujiwara-Igarashi A., Kanemoto H., Fukushima K., Odamaki T., Tsujimoto H.2016. Fecal dysbiosis in miniature dachshunds with inflammatory colorectal polyps. Res. Vet. Sci. 105: 41–46. doi: 10.1016/j.rvsc.2016.01.005 [DOI] [PubMed] [Google Scholar]
- 22.Isaacson P. G., Du M. Q.2004. MALT lymphoma: from morphology to molecules. Nat. Rev. Cancer 4: 644–653. doi: 10.1038/nrc1409 [DOI] [PubMed] [Google Scholar]
- 23.Jergens A. E., Schreiner C. A., Frank D. E., Niyo Y., Ahrens F. E., Eckersall P. D., Benson T. J., Evans R.2003. A scoring index for disease activity in canine inflammatory bowel disease. J. Vet. Intern. Med. 17: 291–297. doi: 10.1111/j.1939-1676.2003.tb02450.x [DOI] [PubMed] [Google Scholar]
- 24.Lamm C. G., Ferguson A. C., Lehenbauer T. W., Love B. C.2010. Streptococcal infection in dogs: a retrospective study of 393 cases. Vet. Pathol. 47: 387–395. doi: 10.1177/0300985809359601 [DOI] [PubMed] [Google Scholar]
- 25.Maeda S., Ohno K., Fujiwara-Igarashi A., Uchida K., Tsujimoto H.2016. Changes in Foxp3-Positive Regulatory T Cell Number in the Intestine of Dogs With Idiopathic Inflammatory Bowel Disease and Intestinal Lymphoma. Vet. Pathol. 53: 102–112. doi: 10.1177/0300985815591081 [DOI] [PubMed] [Google Scholar]
- 26.Maeda S., Ohno K., Uchida K., Nakashima K., Fukushima K., Tsukamoto A., Nakajima M., Fujino Y., Tsujimoto H.2013. Decreased immunoglobulin A concentrations in feces, duodenum, and peripheral blood mononuclear cells of dogs with inflammatory bowel disease. J. Vet. Intern. Med. 27: 47–55. doi: 10.1111/jvim.12023 [DOI] [PubMed] [Google Scholar]
- 27.Middelbos I. S., Vester Boler B. M., Qu A., White B. A., Swanson K. S., Fahey G. C., Jr.2010. Phylogenetic characterization of fecal microbial communities of dogs fed diets with or without supplemental dietary fiber using 454 pyrosequencing. PLoS ONE 5: e9768. doi: 10.1371/journal.pone.0009768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishio J., Honda K.2012. Immunoregulation by the gut microbiota. Cell. Mol. Life Sci. 69: 3635–3650. doi: 10.1007/s00018-012-0993-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Packey C. D., Sartor R. B.2009. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr. Opin. Infect. Dis. 22: 292–301. doi: 10.1097/QCO.0b013e32832a8a5d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peloquin J. M., Nguyen D. D.2013. The microbiota and inflammatory bowel disease: insights from animal models. Anaerobe 24: 102–106. doi: 10.1016/j.anaerobe.2013.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi G., Pengo G., Caldin M., Palumbo Piccionello A., Steiner J. M., Cohen N. D., Jergens A. E., Suchodolski J. S.2014. Comparison of microbiological, histological, and immunomodulatory parameters in response to treatment with either combination therapy with prednisone and metronidazole or probiotic VSL#3 strains in dogs with idiopathic inflammatory bowel disease. PLOS ONE 9: e94699. doi: 10.1371/journal.pone.0094699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito Y., Suzuki H., Tsugawa H., Imaeda H., Matsuzaki J., Hirata K., Hosoe N., Nakamura M., Mukai M., Saito H., Hibi T.2012. Overexpression of miR-142-5p and miR-155 in gastric mucosa-associated lymphoid tissue (MALT) lymphoma resistant to Helicobacter pylori eradication. PLOS ONE 7: e47396. doi: 10.1371/journal.pone.0047396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L. G., Gratadoux J. J., Blugeon S., Bridonneau C., Furet J. P., Corthier G., Grangette C., Vasquez N., Pochart P., Trugnan G., Thomas G., Blottière H. M., Doré J., Marteau P., Seksik P., Langella P.2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105: 16731–16736. doi: 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Srikanth C. V., McCormick B. A.2008. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: a three-way crosstalk. Interdiscip. Perspect. Infect. Dis. 2008: 626827. doi: 10.1155/2008/626827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suchodolski J. S., Dowd S. E., Wilke V., Steiner J. M., Jergens A. E.2012. 16S rRNA gene pyrosequencing reveals bacterial dysbiosis in the duodenum of dogs with idiopathic inflammatory bowel disease. PLoS ONE 7: e39333. doi: 10.1371/journal.pone.0039333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suchodolski J. S., Dowd S. E., Westermarck E., Steiner J. M., Wolcott R. D., Spillmann T., Harmoinen J. A.2009. The effect of the macrolide antibiotic tylosin on microbial diversity in the canine small intestine as demonstrated by massive parallel 16S rRNA gene sequencing. BMC Microbiol. 9: 210. doi: 10.1186/1471-2180-9-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suchodolski J. S., Markel M. E., Garcia-Mazcorro J. F., Unterer S., Heilmann R. M., Dowd S. E., Kachroo P., Ivanov I., Minamoto Y., Dillman E. M., Steiner J. M., Cook A. K., Toresson L.2012. The fecal microbiome in dogs with acute diarrhea and idiopathic inflammatory bowel disease. PLOS ONE 7: e51907. doi: 10.1371/journal.pone.0051907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vázquez-Baeza Y., Hyde E. R., Suchodolski J. S., Knight R.2016. Dog and human inflammatory bowel disease rely on overlapping yet distinct dysbiosis networks. New Microbiol. 1: 16177. [DOI] [PubMed] [Google Scholar]
- 39.Washabau R. J., Day M. J., Willard M. D., Hall E. J., Jergens A. E., Mansell J., Minami T., Bilzer T. W., Group W. I. G. S. WSAVA. International Gastrointestinal Standardization Group. 2010. Endoscopic, biopsy, and histopathologic guidelines for the evaluation of gastrointestinal inflammation in companion animals. J. Vet. Intern. Med. 24: 10–26. doi: 10.1111/j.1939-1676.2009.0443.x [DOI] [PubMed] [Google Scholar]
- 40.Wostmann B. S., Larkin C., Moriarty A., Bruckner-Kardoss E.1983. Dietary intake, energy metabolism, and excretory losses of adult male germfree Wistar rats. Lab. Anim. Sci. 33: 46–50. [PubMed] [Google Scholar]
- 41.Wotherspoon A. C., Ortiz-Hidalgo C., Falzon M. R., Isaacson P. G.1991. Helicobacter pylori-associated gastritis and primary B-cell gastric lymphoma. Lancet 338: 1175–1176. doi: 10.1016/0140-6736(91)92035-Z [DOI] [PubMed] [Google Scholar]
- 42.Yamamoto M. L., Schiestl R. H.2014. Lymphoma caused by intestinal microbiota. Int. J. Environ. Res. Public Health 11: 9038–9049. doi: 10.3390/ijerph110909038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang S. C., Lin C. H., Sung C. T., Fang J. Y.2014. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front. Microbiol. 5: 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Q., Jin Z., Wu W., Gao R., Guo B., Gao Z., Yang Y., Qin H.2014. Analysis of the intestinal lumen microbiota in an animal model of colorectal cancer. PLOS ONE 9: e90849. doi: 10.1371/journal.pone.0090849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.