ABSTRACT
Many herpesviruses express small noncoding RNAs (sncRNAs), including microRNAs (miRNAs), that may play roles in regulating lytic and latent infections. None have yet been reported in varicella-zoster virus (VZV; also known as human herpesvirus 3 [HHV-3]). Here we analyzed next-generation sequencing (NGS) data for small RNAs in VZV-infected fibroblasts and human embryonic stem cell-derived (hESC) neurons. Two independent bioinformatics analyses identified more than 20 VZV-encoded 20- to 24-nucleotide RNAs, some of which are predicted to have stem-loop precursors potentially representing miRNAs. These sequences are perfectly conserved between viruses from three clades of VZV. One NGS-identified sequence common to both bioinformatics analyses mapped to the repeat regions of the VZV genome, upstream of the predicted promoter of the immediate early gene open reading frame 63 (ORF63). This miRNA candidate was detected in each of 3 independent biological repetitions of NGS of RNA from fibroblasts and neurons productively infected with VZV using TaqMan quantitative PCR (qPCR). Importantly, transfected synthetic RNA oligonucleotides antagonistic to the miRNA candidate significantly enhanced VZV plaque growth rates. The presence of 6 additional small noncoding RNAs was also verified by TaqMan qPCR in productively infected fibroblasts and ARPE19 cells. Our results show VZV, like other human herpesviruses, encodes several sncRNAs and miRNAs, and some may regulate infection of host cells.
IMPORTANCE Varicella-zoster virus is an important human pathogen, with herpes zoster being a major health issue in the aging and immunocompromised populations. Small noncoding RNAs (sncRNAs) are recognized as important actors in modulating gene expression, and this study demonstrates the first reported VZV-encoded sncRNAs. Many are clustered to a small genomic region, as seen in other human herpesviruses. At least one VZV sncRNA was expressed in productive infection of neurons and fibroblasts that is likely to reduce viral replication. Since sncRNAs have been suggested to be potential targets for antiviral therapies, identification of these molecules in VZV may provide a new direction for development of treatments for painful herpes zoster.
KEYWORDS: microRNA, noncoding RNA, varicella-zoster virus
INTRODUCTION
Varicella-zoster virus (VZV; also known as human herpesvirus-3 [HHV-3]) infects most of the nonvaccinated population during childhood in Western societies. Primary infection infects lymphocytes, resulting in a viremia that mediates spread to the skin and the ultimate generation of varicella vesicles across the body. VZV also enters peripheral neurons, where viral genomes are retained for the life of the individual. Reactivation of VZV results in viral replication and the disease herpes zoster (reviewed in reference 1). Herpes zoster is almost always associated with pain, which can subsequently become chronic in postherpetic neuralgia (PHN). Zoster can also induce multiple neurological symptoms, with or without skin disease, and zoster on the face can result in potentially blinding disease. Zoster has also been recently epidemiologically associated with cardiovascular disease, including both heart attack and stroke (2). Unlike its close cousin, herpes simplex virus (HSV), VZV shows a high degree of human specificity, which has largely prevented in vivo modeling of latent infections and reactivation. Treatment options and prevention strategies for zoster and PHN remain an important health issue in the Western world.
Small noncoding RNAs (sncRNAs), including microRNAs (miRNAs) 17 to 24 nucleotides (nt) in length, have been studied intensively for the past 15 years because of their ability to extensively modulate gene expression and translation via multiple targets. miRNAs can downregulate target protein expression in cells by interfering with translation or by mediating mRNA degradation (reviewed recently in references 3 and 4). Importantly, many operate through loose requirements for base-matching fidelity, and the short “seed” recognition sequence of miRNAs allows them to modify the expression of many proteins simultaneously. This provides an additional level of control of gene expression that often complements that governed by transcription factors. sncRNAs and miRNAs are encoded by genomes ranging from humans to small DNA viruses. In addition to miRNAs, some sncRNAs in mammalian cells can regulate gene expression at the transcriptional level (reviewed in reference 5).
There are nine human herpesviruses, which are characterized by lytic and latent or persistent infections in most of the human population. To date, miRNAs have been reported to be expressed by 7 of the 9 of the human herpesviruses. HSV-1 encodes at least 29 miRNAs (6), while Epstein-Barr virus (EBV) may encode at least 44 (reviewed in references 7 and 8). The miRNAs encoded by HSV-1, for example, are being studied for their potential roles in neuronal infection and the establishment of the latent state (i.e., reference 9). Recent studies indicate that several are made in productive infection and may act to limit viral spread (6). To the best of our knowledge, no miRNAs have yet been described for HHV-7 and VZV. An early in silico search for predicted miRNAs in herpesvirus genomes (10) revealed no VZV miRNA candidate sequences. However, it is now recognized that bioinformatics approaches may not reveal all miRNA candidates. To wit, the same bioinformatics search did not predict miRNAs in HHV-6A and -6B, but several have since been discovered in these herpesviruses by next-generation sequencing (NGS) (11, 12). An NGS study of small RNAs in human peripheral ganglia obtained from autopsy material that was latently infected with VZV did not identify any small RNA species encoded by VZV with the appropriate molecular characteristics of miRNA, even under conditions in which many HSV miRNAs were detected (13). However, the frequency and genomic load of latent VZV-infected tissues, such as the trigeminal ganglia, are known to be considerably lower than those of HSV (i.e., references 14 and 15), and it is quite possible that the sensitivity of detection was insufficient.
Both NGS technologies and the bioinformatics tools for analyzing the data generated by NGS experiments for small RNAs have been greatly improved since the first searches. Therefore, we initiated a new search for sncRNAs in NGS data of RNAs of <200 bp in length in VZV-infected cells. Two independent analyses of NGS data obtained from VZV-infected human embryonic stem cell-derived (hESC) neurons infected productively and quiescently (16) and from productively infected cultured human foreskin fibroblasts (HFFs) revealed over 20 sncRNAs mapping to the VZV genome, with several having predicted stem-loop precursors characteristic of miRNAs. Intriguingly, most of the candidate sncRNAs clustered in or near the reiterated regions of the viral genome. The presence of 7 of the identified sequences was confirmed by TaqMan stem-loop reverse transcription-PCR (RT-PCR) of small RNA in primary human fibroblasts and ARPE19 cells. We then focused on one of these sequences predicted to have a stem-loop precursor and confirmed its presence in VZV-infected fibroblasts and in productively infected neurons. Transfection of a synthetic antagonist to this sncRNA enhanced growth of VZV-infected foci. The sequence is upstream of the VZV immediate early gene open reading frame 63 (ORF63), whose transcripts have been reported to be expressed during VZV latency (i.e., reference 17 and reviewed in reference 18). We speculate that the sncRNA described herein that limits lytic VZV replication may do so via transcriptional rather than translational regulation (5). Our data establish that VZV-infected cells do encode and express sncRNAs and miRNAs, and these are likely to participate in the regulation of the life cycle of this ubiquitous pathogen.
RESULTS
Bioinformatics searches for VZV sncRNA following NGS of small RNA.
Small RNAs (<200 bp) from four experimental groups, consisting of uninfected neurons, neurons infected productively or latently with VZV, and fibroblasts infected with VZV, were made into libraries and sequenced using the Illumina HighSeq platform. Sequencing of three independent experiments in each group yielded a total of 12 data sets. Two distinct peaks of RNA length were detected in almost all the samples centered at 24 nt (close to the canonical length of miRNAs) and 35 nt. Reads mapping to the human genome were excluded from further analysis, and the remaining reads were aligned with the VZV genome (Table 1). All reads of approximately 35 nt in length mapped to the human but not the VZV genome, were suspected to be Y RNA (reviewed in reference 19), and were not studied further.
TABLE 1.
NGS of small RNAs (<200 nt) from VZV-infected cells
| Sample type and no. | No. of NGS reads: |
No. of unique reads 17–200 nt mapping to VZV genome | No. of reads 17–24 nt mapping to VZV genome | |
|---|---|---|---|---|
| 17–200 nt | 17–24 nt aligned to human genome | |||
| Neurons | ||||
| Uninfected | ||||
| 1 | 15,409,516 | 7,234,809 | 0 | 0 |
| 2 | 18,535,803 | 15,941,006 | 2 | 0 |
| 3 | 15,688,047 | 7,917,409 | 3 | 0 |
| Infected | ||||
| 1 | 14,167,260 | 5,323,812 | 6,693 | 482 |
| 2 | 16,747,763 | 13,481,851 | 12,282 | 1,645 |
| 3 | 15,012,737 | 8,171,776 | 23,339 | 858 |
| Latently infected | ||||
| 1 | 13,620,130 | 4,129,576 | 28 | 10 |
| 2 | 15,885,742 | 11,004,344 | 94 | 41 |
| 3 | 13,987,171 | 6,663,047 | 65 | 29 |
| Infected fibroblasts | ||||
| 1 | 14,092,008 | 4,830,665 | 11,458 | 3,160 |
| 2 | 11,473,036 | 8,660,687 | 8,019 | 2,015 |
| 3 | 11,633,714 | 6,879,540 | 9,693 | 2,437 |
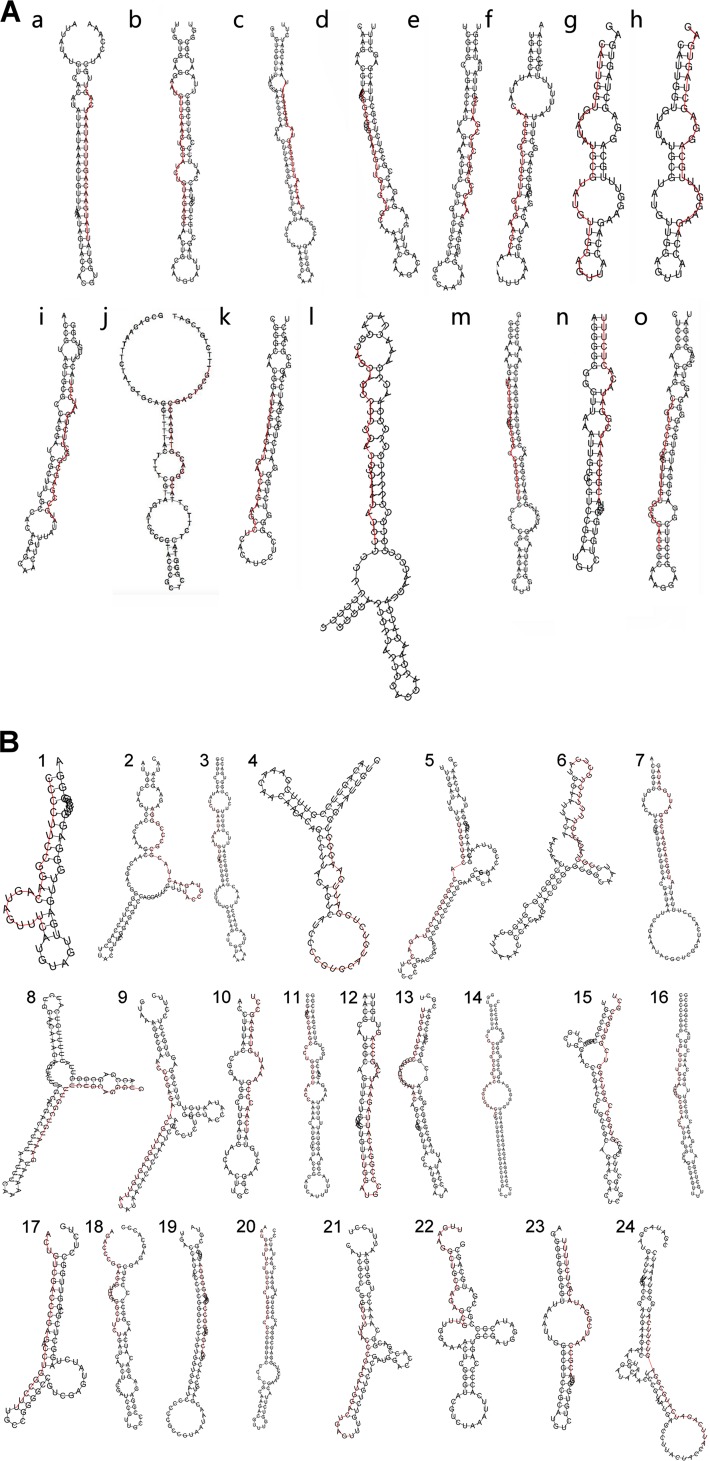
Thousands of reads of 17 to 24 nt mapping to the VZV genome were detected in neurons and fibroblasts productively infected with VZV (Table 1). RNA from latently infected neurons contained small numbers of reads mapping to the VZV genome in all three independent biological samples, but it is important to note only approximately 5% of neurons harbor VZV genomes according to our method of establishing VZV latency in vitro. The reads mapping to the VZV genome were analyzed using three widely used software packages designed to predict novel miRNAs: MirDeep2 (20), miReap (http://sourceforge.net/projects/mireap), and MiReNa (21). Each of these programs uses a different algorithm and approaches to predict miRNAs from the NGS data. Putative miRNAs (Table 2) with stem-loop precursors (Fig. 1A) were predicted by two of the programs for the infected fibroblast data (MirDeep2 and MiReap), but there was no overlap between them. In addition, there were only a few reads detected that were defined as predicted miRNAs (<10 in each sample, with the exception of sequence n in Table 2).
TABLE 2.
Potential VZV sncRNAs predicted using conventional bioinformatics tools
| Sequence letter | Sequence | Strand | nt positiona |
ORF | No. of NGS reads | Program | |
|---|---|---|---|---|---|---|---|
| Start | End | ||||||
| a | UUAUGAACAGUUUAUAAUCAGU | + | 12056 | 12077 | 4 | Mireap | |
| b | AAUGUUGAACUGGAUCUGUAGAGC | + | 15226 | 15249 | 5 | Mireap | |
| c | GAACAAUUUGGGGUAUGGCGUUU | + | 33020 | 33042 | ORF21 | 3 | Mireap |
| d | AAACGGCGUUGGAUGUUGUGUUGC | + | 39189 | 39212 | 5 | Mireap | |
| e | AAACUCGACGUCUCCGAUGCG | + | 60550 | 60570 | ORF33 antisense | 4 | Mireap |
| f | CAAGGGGCUGCUUGUGAAGCA | + | 67401 | 67421 | ORF37 | 4 | Mireap |
| g | CAUUGGUGUAUAUGCGUAUGUUGGAGUU | − | 69625 | 69652 | ORF38 | 3 | MiRDeep2 |
| h | AGAAGGUUUGCAGGAGCUAGUGAG | − | 69598 | 69621 | ORF38 | 5 | MiRDeep2 |
| i | AUGCCGAGCGUGCUUCUGAAUCGU | + | 74651 | 74674 | ORF40 | 8 | Mireap |
| j | ACGGCACCGUAGAACCGACUGCGU | + | 86606 | 86629 | ORF50 antisense | 4 | Mireap |
| k | AUCGUAGAUGAUCAGAAGCCU | + | 106549 | 106569 | ORF62 antisense | 4 | Mireap |
| l | CAGCAGGUCCGGACGCGAACACGC | + | 107927 | 107951 | ORF62 | 5 | Mireap |
| m | UUUCUGUCUCUGGCGCCCCGGUU | + | 107773 | 107795 | ORF62 antisense | 9 | Mireap |
| n | ACGCCAAUCGGAUACACUCUUU | + | 110191 | 110212 | ORF63 promoter | 90 | MiRDeep2 |
| o | CCGUGCGCGAGUUUGUGGGGGAGG | + | 111854 | 111877 | ORF64 | 4 | Mireap |
The nucleotide positions are those of the pOka genome (GenBank accession no. AB097933.1), the VZV origin of the recombinant virus used in this study.
FIG 1.
(A) Predicted folding of precursors to potential sncRNAs identified in the first (A) and second (B) bioinformatics analyses. Several of the folding predictions reveal a stem-loop structure consistent with the mature sequences being miRNAs. The red lines indicate the NGS bioinformatically predicted sncRNA sequences.
The low numbers of reads and inconsistency between the prediction tools led us to seek potential VZV sncRNAs using another strategy. In this second strategy, similar sequences of small RNAs that aligned to the VZV genome starting at approximately the same genomic position were displayed in an Excel spreadsheet and aligned with one another manually. An example is shown in Table 3, where 14 different sequence reads detected in a total of 179 reads were aligned. Using this method, 24 unique sequences were detected a minimum of 25 times in pooled transcriptome sequencing (RNA-Seq) data. The sequences and the numbers of reads from 2 independent experiments for each cell type are shown in Table 4. Most of the sequences corresponded to predicted mRNAs of known coding ORFs, but several were antisense to ORFs (mapping opposite the coding strand), and some were located in between documented ORFs.
TABLE 3.
Manual alignment of similar sequences used in the second bioinformatics analysis
| Start nt | Sequencea | Length (nt) | No. of readsb |
|---|---|---|---|
| 110191 | ACGCCAAUCGGAUACACUCUUU | 22 | 41 |
| 110191 | ACGCCAAUCGGAUACACUCUUUU | 23 | 40 |
| 110192 | CGCCAAUCGGAUACACUCUUUU | 22 | 21 |
| 110192 | CGCCAAUCGGAUACACUCUUU | 21 | 15 |
| 110193 | GCCAAUCGGAUACACUCUUUU | 21 | 14 |
| 110193 | GCCAAUCGGAUACACUCUUU | 20 | 12 |
| 110190 | UACGCCAAUCGGAUACACUCUUUU | 24 | 9 |
| 110190 | UACGCCAAUCGGAUACACUCUUU | 23 | 6 |
| 110189 | GUACGCCAAUCGGAUACACUCUUU | 24 | 5 |
| 110191c | ACGCCAAUCGGAUACACUCUUX | 21 | 4 |
| 110195c | XXCAAUCGGAUACACUCUUUU | 19 | 4 |
| 110192c | CGCCAAUCGGAUgCACUCUUX | 20 | 3 |
| 110194c | XCCAAUCGGAUACACUCUUUU | 20 | 3 |
| 110190c | UACGCCAAUCGGAUgCACUCUUU | 23 | 2 |
X represents missing nucleotides from the read, and lowercase letters represent deviations from the consensus.
The total count is 179 reads.
Sequences with mismatches from the consensus.
TABLE 4.
Potential VZV sncRNAs predicted using the second bioinformatics analysis
| sncRNA no. | Sequence | Strand | nt positiona |
ORF | No. of NGS reads |
||||
|---|---|---|---|---|---|---|---|---|---|
| Start | End | Fibroblasts | Latent neurons | Productive neurons | Total | ||||
| 1 | CCCCUUCCGGACAGUAGUUUC | − | 560 | 580 | ORF0 | 18 | 3 | 6 | 27 |
| 2b | UACCGUAGAACUACGCGCCCGGGA | + | 12641 | 12664 | ORF10 | 349 | 0 | 26 | 375 |
| 3c | UCUGAAUAAAAUGUUUAGC | − | 30836 | 30854 | ORF21 | 2 | 0 | 28 | 30 |
| 4c | CCGUGCAUGUCUGGAUUGAAGGG | − | 68227 | 68249 | ORF37 antisense | 0 | 0 | 25 | 25 |
| 5 | UUGUUGCACCGGGGGACGUAGACC | + | 73097 | 73120 | ORF40 | 14 | 0 | 12 | 26 |
| 6 | CGCUAGACUGUUUUUCUGCUCA | + | 80199 | 80220 | ORF42 antisense | 22 | 0 | 5 | 27 |
| 7 | AUGGCACGAUCGGGAUUGGAUAG | + | 84527 | 84549 | ORF48 | 24 | 0 | 7 | 31 |
| 8c | GAAAUGUCGGUUUCUGCAGGGUG | − | 85352 | 85374 | ORF48 antisense | 17 | 0 | 46 | 63 |
| 9b | ACCCGAGAUCGAUUGGAGUGUUAU | − | 100153 | 100176 | ORF59 | 612 | 0 | 126 | 738 |
| 10c | UCAACCCGAAAUUGUAGAGCCU | − | 103626 | 103647 | ORF61 | 12 | 0 | 30 | 42 |
| 11c | GAGACUCGAGCCCUGGUGUAACC | − | 103938 | 103960 | ORF61 | 8 | 0 | 18 | 26 |
| 12 | GCCCGGACAUUAGAAUACAGCCAG | − | 104456 | 104479 | ORF61 promoter | 15 | 0 | 13 | 28 |
| 13 | UUGGAUGCCCGGACAUUAGAAUAC | − | 104462 | 104485 | ORF61 promoter | 49 | 0 | 24 | 73 |
| 14b | CGCCCGGCGGUCAAGGUGGAGCU | − | 105251 | 105273 | ORF62 | 29 | 0 | 4 | 33 |
| 15c | CGUGGCCGGUCGGUCGGUGGCGCU | − | 105626 | 105649 | ORF62 | 14 | 0 | 37 | 51 |
| 16c | GUGUUUGCAUCGUCGCGCACCU | − | 106089 | 106110 | ORF62 | 6 | 0 | 19 | 25 |
| 17b | ACUGUCGACCCGAGACCUGGCCUU | − | 106319 | 106342 | ORF62 | 115 | 0 | 61 | 176 |
| 18c | AGACCCGGAGGAUGUGAGGCUUCU | − | 106562 | 106585 | ORF62 | 9 | 0 | 32 | 41 |
| 19c | GACGGACCGGCCCCAAACGGGGGA | − | 106846 | 106869 | ORF62 | 6 | 0 | 24 | 30 |
| 20b | AAUGAUUUCUGUCUCUGGCGCC | + | 107768 | 107789 | ORF62 antisense | 30 | 0 | 12 | 42 |
| 21c | CUUUUUACCCGAGAUGGACUGAGU | + | 108451 | 108474 | ORF62 antisense | 8 | 0 | 50 | 58 |
| 22b,c | UUGAAGGCUGCGAGAGCGUU | − | 109136 | 109155 | ORF62 promoter | 505 | 0 | 1,047 | 1,552 |
| 23b | UACGCCAAUCGGAUACACUCUUUU | + | 110190 | 110213 | ORF63 promoter | 108 | 0 | 15 | 123 |
| 24c | AUUCAGAUCAUGCGGAGUCUUCA | + | 116079 | 116101 | ORF68 | 14 | 0 | 21 | 35 |
The nucleotide positions are those of the pOka genome (GenBank accession no. AB097933.1), the VZV origin of the recombinant virus used in this study. VZVsncRNA14 to -23 lie in the terminal repeat region of the VZV genome; the positions given are for the first copy in the genome.
sncRNAs whose presence was confirmed in VZV-infected fibroblasts by stem-loop RT-qPCR.
sncRNAs present at higher levels in VZV-infected neurons than in VZV-infected fibroblasts.
Since we do not yet know if these sequences are authentic miRNAs, we provisionally named them as VZV small noncoding RNAs 1 to 24 (VZVsncRNA1 to -24). Flanking sequences (available upon request) were added to the NGS-detected sequences (Table 4) to predict precursor stem-loop structures (Fig. 1B).
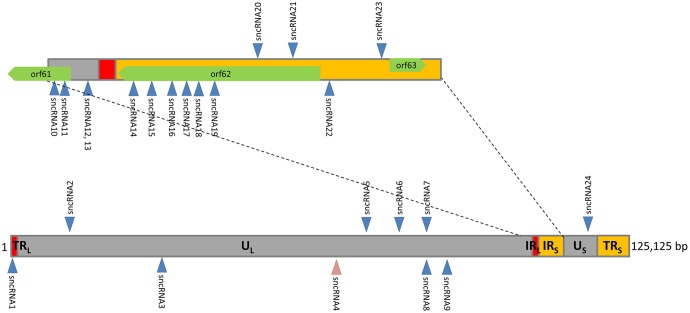
Mapping of the genomic positions of the 24 sncRNAs using the annotated VZV genome Dumas (see Materials and Methods) revealed that most are clustered in the genomic region between ORF56 and ORF63 (Fig. 2). miRNA clustering in other herpesviruses has been well documented (7, 8). Indeed, 9 of the 24 unique sncRNA sequences mapped to the ORF or promoter of the transcriptional activator ORF62. Of the nine mapping to ORF62, seven are predicted to be expressed as part of the mRNA for the coding sequence and immediately upstream of ORF62, while two are antisense to the ORF. An additional four sequences are coded within ORF61 and immediately upstream of it. The sequences upstream of ORF61, VZVsncRNA12 and VZVsncRNA13, partially overlap, but the overlap is not in their presumptive seed regions. Importantly, many of the sequences were detected in more than 10 copies in the NGS analysis of samples of both neurons and fibroblasts that were infected productively. This suggests that they may have physiological roles (11).
FIG 2.
Locations in the genome of 20- to 24-nt VZV sncRNAs obtained from VZV-infected cells. Sequences were identified by manual alignment (2nd analysis in Table 3). A large portion of these sequences are present in the last 1/3 of the VZV genome, with a concentration in the coding and promoter and downstream regions of VZV transcriptional activator ORF62. The upward arrows indicate the sncRNA coded for by the positive strand and the downward arrows indicate the negative strand of the VZV genome. sncRNA4 was only found in the NGS analysis of productively infected neurons; all of the other sequences were found in VZV-infected neurons and fibroblasts.
The numbers of reads of several of these sequences differed between the cell types. While most of the sncRNA sequences were present at highest levels in infected fibroblasts, many of these were detected preferentially in productively infected neurons. While only one sequence was detected in latently infected neurons at low copy number, we suspect this may be an underrepresentation, since our previous studies have indicated that only about 5% of the neurons in our latently infected hESC-derived neuron cultures harbor VZV genomes (16). Therefore, reads from the latently infected neurons are likely to be significantly diluted by the “noise” of the uninfected neurons in the sample (see quantitative PCR [qPCR] results below). The lowest free energy folding of potential precursors to these sncRNAs as predicted by RNAfold are shown in Fig. 1B and indicate some have stem-loop-like structural predictions.
Of particular note is that 1 of the 24 sequences detected using this analysis was the most abundant short RNA identified in the previously described “conventional” bioinformatics analysis (sequence n in Table 2). This sequence was predicted by the miRNA prediction software miRDeep2 to be a potential miRNA. Using our “manual alignment of similar sequences” strategy, we found many more copies of the consensus sequence in the data than were revealed by the initial bioinformatics analysis, including sequences in samples of productively infected neurons. Intriguingly, this sncRNA lies in a genomic position that has been proposed to be a “bidirectional promoter for ORF62 and ORF63” (22). We chose to further study this candidate miRNA, designated VZVsncRNA23.
Confirmation of expression of VZV sncRNAs by qRT-PCR.
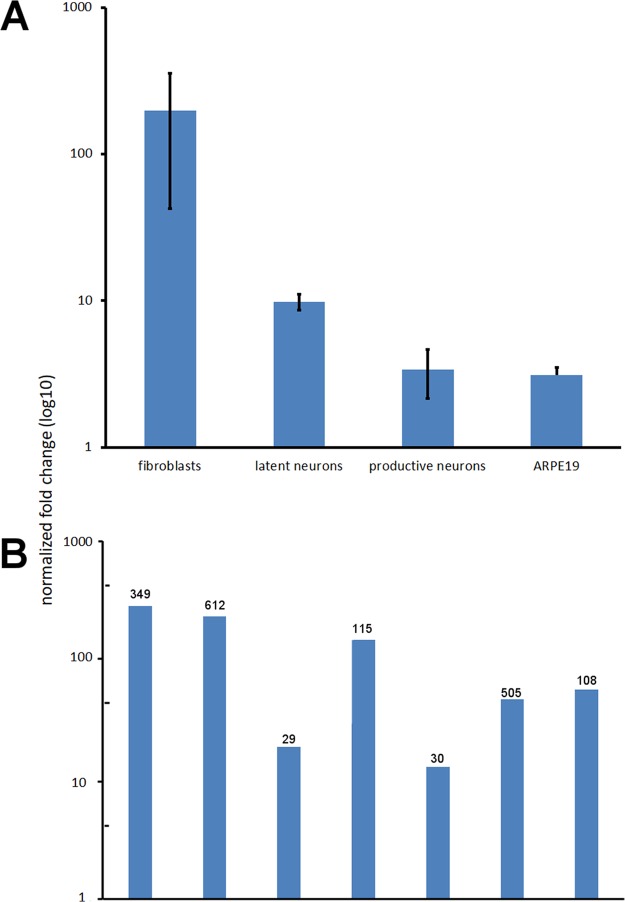
The presence of VZVsncRNA23 was next confirmed by an independent method, using a stem-loop primer qPCR approach. The methods, primers, and probe sequences are listed in Table 5. Primers and a TaqMan probe were designed for detection of VZVsncRNA23, and qPCR was performed on RNA from the same samples used in the NGS analysis. As shown in Fig. 3A, specific reactions for VZVsncRNA23 were found in all samples of cells infected by VZV, but not in uninfected controls. Importantly, small amounts of VZVsncRNA23 sequence were detected by qPCR in latently infected neurons, although analysis of the NGS data failed to detect reads with this sequence. Primer-TaqMan probe sets to six additional small RNA sequences present in NGS at more than 30 reads also successfully confirmed the presence of these small RNAs as well as VZVsncRNA23 in VZV-infected fibroblasts in two independent biological repeats distinct from the RNA used for the NGS (Fig. 3B).
TABLE 5.
Primers used for RT-qPCR detection of sncRNA in VZV-infected cells
| sncRNA no. or universal primer or probe | Primer orientationa | Sequenceb |
|---|---|---|
| sncRNA | ||
| 2 | Reverse | GGTCGTATGCAAGAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCTCCCGGGC |
| Forward | GACTGATACCGTAGAACTACGC | |
| 9 | Reverse | GGTCGTATGCAAGAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCCACCCTGC |
| Forward | CGCGGAAATGTCGGTTTCT | |
| 14 | Reverse | GGTCGTATGCAAGAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCGGTTACAC |
| Forward | TAAAGTATGAGACTCGAGCCCTG | |
| 17 | Reverse | GGTCGTATGCAAGAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCAAGGCCAG |
| Forward | TATGATACTGTCGACCCGAGAC | |
| 20 | Reverse | GGTCGTATGCAAGAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCGGCGCCAG |
| Forward | GCGGGCAATGATTTCTGTCT | |
| 22 | Reverse | GGTCGTATGCAAGAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCTCCCCCGT |
| Forward | TATATATAGACGGACCGGCCCCAA | |
| 23 | Reverse | GGTCGTATGCAAGAGCAGGGTCCGAGGTATCCATCGCACGCATCGCACTGCATACGACCCACTGG |
| Forward | AGACGTACGCCAATCGGATA | |
| Universal reverse primer | GAGCAGGGTCCGAGGT | |
| Universal probe | FAM–5′-TCCATCGCACGCATCGCACT-3′–BHQ-1 |
The orientations of the specific reverse and forward primers are shown.
FAM, 6-carboxyfluorescein; BHQ-1, black hole quencher 1.
FIG 3.
(A) TaqMan RT-qPCR detection of the presence of VZVsncRNA23. RT-qPCR confirmed the NGS data for fibroblasts and in neurons productively infected with VZV. In addition, the sequence was detected in RNA from neurons latently infected with VZV, although the NGS data did not contain reads for this sequence. ARPE19 cells infected with VZV also contained detectable VZVsncRNA23. (B) TaqMan qPCR detection of the presence of an additional six sncRNAs in VZV-infected fibroblasts. The numbers above the columns indicate the number of reads in the NGS analysis for comparison. All seven sequences (including VZVsncRNA23) were detected in two separate RNA extractions from infected fibroblasts independent of the NGS experiments.
Inhibition of VZVsncRNA23 increases spread of VZV.
In order to evaluate the potential physiological effect of VZVsncRNA23 expression on infection, we acquired a specific and stabilized RNA inhibitor designed to block potential miRNA activities. This inhibitor or a control fluorescein-labeled 20-nt RNA was transfected into ARPE19 cells that had been inoculated 24 h earlier with small numbers (<100) of VZV66RFP-infected ARPE19 cells. This approach was used so that formation and growth of individual infectious foci could be identified and monitored for quantitation.
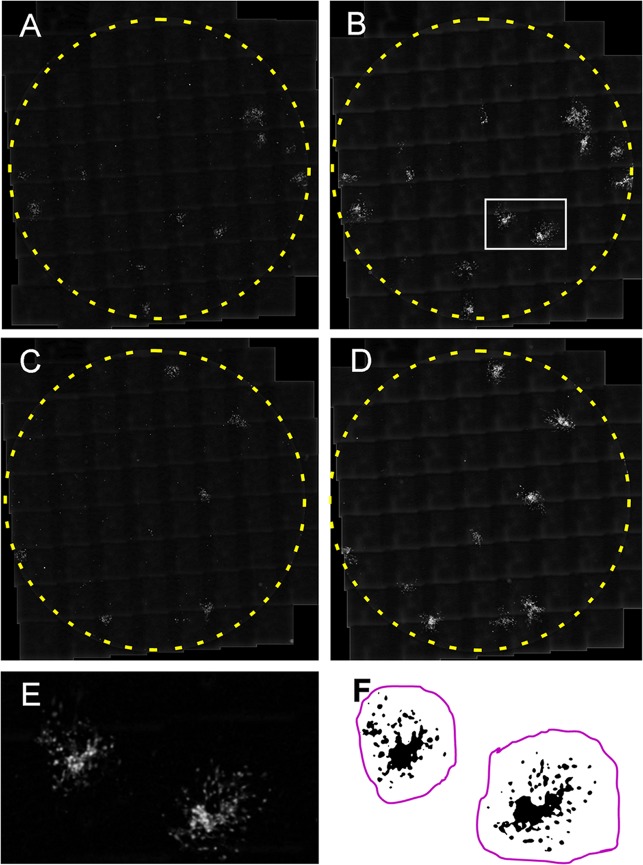
At 2 and 7 days postinfection (dpi [i.e., 1 and 6 days after transfection with the inhibitor and control RNA]), more than 90% of the surface area of infected wells was photographed with an automated microscope. The images were then “stitched” together in order to visualize the infected cells in the wells (Fig. 4A to D). Copies of the images were prepared with the background subtracted (median filter and “rolling ball”), and the contrast was improved with gamma adjustment. A Gaussian blur was then performed in order to remove pixel artifacts and make the ORF66 RFP fluorescence more homogeneous (Fig. 4E). All of the processed images were prepared automatically as a batch without user intervention, and all of the procedures were performed across the entire photographed portion of the well.
FIG 4.
Analysis method for growth of individual foci of VZV infection in ARPE19 cells after transfection of a control RNA (A and B) or a locked antagonist RNA to VZVsncRNA23 (C and D). Eighty-one micrographs covering more than 95% of wells in a 96-well culture plate were taken with an automated microscope 1 day after transfection (2 dpi in panels A and C) and 6 days after transfection (7 dpi in panels B and D) and assembled (“stitched” together) using the program Fiji. The area enclosed by the white rectangle in panel B is shown at higher magnification in panel E. The infectious focus is blurred because it has been treated with a Gaussian filter to eliminate photographic artifacts and smooth the outlines of the infected cells. Panel F shows a thresholded image used for measurements. Black indicates RFP fluorescence indicating the presence of virus, and the magenta line indicates the area to be measured as one infectious focus.
The image files were then renamed alphabetically in pairs according to date (2 and 7 days postinfection [dpi] were used in the analyses) without indicating whether they were transfected with the inhibitor or controls, and the quantitative analysis was performed blind by three other individuals in the following manner: Using the unmodified, stitched image of the well as a reference, the fluorescence was separated from the background in a thresholding step (Fig. 4F) so that only infected cells were selected. The area (in pixel units) occupied by fluorescence of individual foci of infection (FOI) was then measured by delineating an area encompassing the FOI. The analysis was performed first on the 7-dpi image of the pair, and then same FOI in the 2-dpi image were identified (still blind in terms of which were experimental and which were control) and quantified. FOI that touched the edges of the well were excluded from the analysis, since they were growth constrained. Finally, the areas of the 2-dpi FOI were subtracted from the 7-dpi images, giving a value correlating directly to spread of each FOI.
These measurements were performed on a total of 150 foci (80 for control RNA and 70 for inhibitor RNA) from 18 wells in three experiments. In each of the three experiments, transfection with the inhibitor resulted in a statistically significant increase in the average growth of FOI in each experiment (73, 39, and 35%; all P < 0.05), indicating increased and/or more rapid infection by VZV. When the growth of FOI data were pooled from all three experiments, the average enhancement of FOI growth was 46% (P < 0.001) (Fig. 5). These results strongly suggest that VZVsncRNA23 inhibits the spread of VZV and therefore is a virally encoded, physiologically relevant small noncoding RNA.
FIG 5.
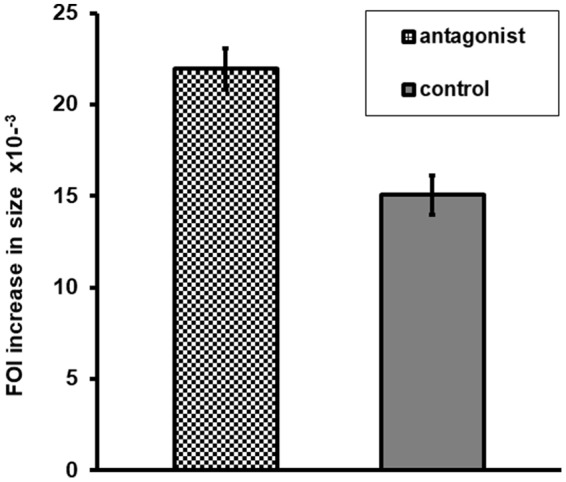
An sncRNA encoded by VZV interferes with VZV replication. Shown is quantification of the effect of transfection of an antagonist to VZVsncRNA23 1 day after infection of ARPE19 cells on the growth of individual infectious foci. The columns represent the increase in numbers of pixels representing fluorescence in antagonist-treated versus control RNA-treated wells. Seventy antagonist-treated and 80 control RNA-treated foci were analyzed, and the difference between the groups was significant (P < 0.001). Error bars represent the standard error of the mean.
DISCUSSION
Virus-encoded miRNAs have been the topic of intensive study for more than a decade, and herpesviruses encode the largest number of reported virus-encoded miRNAs. Of the nine human herpesviruses, miRNAs have been reported for seven, the exceptions being HHV-3 (VZV) and HHV-7. Our NGS of small RNA in VZV-infected fibroblasts and neurons strongly suggests that that VZV expresses many sncRNAs, at least one of which may fold like a classical miRNA and affect virus growth. Strikingly, many of these sncRNAs map as a cluster near the end of the genome to the sequences of ORFs 60 to 62. Indeed, no fewer than nine map to ORF62 and its presumed promoter.
Our experiments were performed with a fluorescent derivative of the pOka strain of VZV, which belongs to VZV clade 2. All 24 sequences discovered in our second bioinformatics analysis were found to be identical in strains Dumas (clade 1) and Ellen (clade 3). This perfect conservation of sequences between three different clades is consistent with their having importance to the life cycle of the virus. These 24 sequences are also identical in the vaccine strain vOka, which has multiple genomic changes from its parent strain, pOka, as a result of extensive passaging in guinea pig embryo fibroblasts to reduce its virulence. This conservation is again consistent with the sncRNAs having a functional role in VZV infection. The presence of seven of the eight sncRNAs detected in 30 or more reads in fibroblasts by analysis of NGS data was confirmed in VZV-infected fibroblasts by stem-loop PCR. Six of the seven (excluding VZVsncRNA22) were also detected using stem-loop RT-qPCR in ARPE19 cells (not shown).
VZVsncRNA23, the sncRNA predicted to have a stem-loop precursor, was detected in both of our bioinformatics analyses of NGS of neurons and fibroblasts during productive infection and also in neurons latently infected with VZV. Furthermore, inhibition of its activity leads to a consistent and statistically significant increase in growth of VZV infectious foci in ARPE19 cells. We therefore think it likely that VZVsncRNA23 acts by reducing replication of the virus by a yet undetermined mechanism. This is similar to a recent report that two newly discovered HSV miRNAs, 28 and 29, reduce viral replication in nonneuronal cells (6). We cannot exclude the possibility that VZVsnc23 acts in other aspects of infection and spread of infection.
Several HSV miRNAs have been shown to inhibit productive infection in neurons and have been implicated in the lytic-latent decisions governing infection using rodent models. We are currently evaluating the possible roles of the predicted VZV sncRNAs in our model for VZV latency and reactivation using the stem-cell-derived human neuron system.
It has been suggested that sncRNAs are attractive targets for antiviral therapy (23–25). Indeed, since the antagonist of VZVsncRNA23 increases viral spread, it is possible that topical administration of a locked RNA with sequence based on VZVsncRNA23 could reduce virus growth. Therefore, our description of multiple sncRNAs encoded by VZV may open the door to development of new therapies for herpes zoster.
An important issue to be addressed in future studies is the precursor RNAs from which these sncRNAs are derived and their genomic location. In particular, the strongest candidate miRNA, VZVsncRNA23, lies in the intergenic region between ORF62 and ORF63, lying very close to the origin of replication of VZV in the terminal repeats of short region of the VZV genome. This area has been studied and was shown by mutational analysis to be part of a bidirectional promoter containing sequences that regulate expression of both of these proteins (22). Multiple transcripts encoding ORF63 protein are transcribed from this region from several start sites (17), and it is possible that one or more of these transcripts are precursors to both the ORF63 protein and VZVsncRNA23. It is even possible that that there are some transcripts that may cross the region and the origin of replication. Indeed, HSV has been suggested to carry transcripts that span its origin of replication only under certain conditions, named L/ST transcripts (26).
It is also intriguing that many of the VZV sncRNAs lie in the ORF62 region, since HSV ICP4 is the homolog of VZV ORF62, and several HSV miRNAs are proposed to target ICP4 (27). In the case of the VZV sncRNAs described here, most are encoded on the same strand as ORF62, but two of them (VZVsncRNA20 and VZVsncRNA21) are encoded by the opposite strand and could therefore potentially target the RNAs containing the ORFs. Six VZVsncRNAs are located close to one another within the ORF62 coding sequence; therefore it is possible that the ORF62 transcript, in addition to encoding the viral transactivator, also gives rise to sncRNA with independent activities. It would be of great interest to compare the sncRNAs of all three human alphaherpesviruses to one another in terms of their function and position once more is learned about the VZV sncRNA.
miRNAs usually modulate protein expression by preventing translation or destabilizing transcripts in the cytoplasm. Both VZVsncRNA22 and VZVsncRNA23 map within the presumed bidirectional promoter region of immediate early genes ORF62 and ORF63 containing sequences that regulate their expression (22). These sncRNAs could therefore act in the nucleus by a mechanism known as transcriptional gene silencing (reviewed in reference 5). The presence of both of these sncRNAs was confirmed by stem-loop RT-qPCR. If this mechanism is validated for VZV, it will be the first example of the use by a virus of transcriptional gene silencing, which has only been shown so far in mammalian cells. Therefore, VZV, in addition to joining the majority of human herpesviruses by encoding miRNAs in their genome, may yet reveal another mechanism for these ubiquitous and extremely evolutionarily successful viruses to control their replication.
MATERIALS AND METHODS
Cells, viruses, and infection.
Human perinatal foreskin fibroblasts (HFFs) and neurons generated from the hESC line H9 were prepared and grown as detailed in reference 28. Infections for NGS analysis and for stem-loop RT-PCR were performed using a recombinant VZV expressing green fluorescent protein (GFP) as an N-terminal fusion to ORF66 (VZV66GFP) as detailed previously (29). This study also used a new recombinant VZV derived from the parent of Oka bacterial artificial chromosome (pOka BAC), as detailed in reference 30. Briefly, the VZV strain pOka was obtained at passage 7 in human lung fibroblasts by Kinchington directly from Takahashi in 1986 and grown for three to five further passages in MeWo cells into stocks. The amplified stock was then supplied to Ann Arvin (Stanford University, CA), and her group derived from it an overlapping 4-cosmid-based genetic system (31). The cosmid system was then used to generate the VZV BAC single-plasmid systems through a rescued infectious virus intermediate (30). In the present study, a VZV similar to the VZVGFP66 recombinant described previously (29) was developed by recombineering of the pOka BAC to place the gene for monomeric red fluorescent protein (mRFP) at the N terminus of and in frame with ORF66, using lambda Red recombination, as previously described (15). Virus was generated by transfecting MeWo cells with the BAC DNA, and cell-free virus preparations were prepared in ARPE19 cells as described previously (32).
Generation of small RNA libraries and NGS sequencing.
Neurons (with a minimum of 2 weeks of terminal differentiation following plating) and fibroblasts at 95% confluence were infected with cell-free VZV preparations containing approximately 4 × 105 PFU/ml. (The exact multiplicity of infection [MOI] could not be determined for neurons because neuron density and numbers in these cultures are difficult to assess due to their three-dimensional nature.) RNA was prepared from infected cultures showing 80% to 100% GFP or mRFP expression, before plaque formation in fibroblasts and development of obvious cytopathology in neurons (33). Cultures containing approximately 5% latently infected human neurons were prepared as we detailed previously (16). Total RNA was extracted with TRIzol reagent (Sigma-Aldrich) as described previously (16), and libraries were prepared with an Illumina TruSeq small RNA library preparation kit. Twelve samples (3 from infected fibroblasts, 3 from lytically infected neurons, 3 from mock-infected neurons, and 3 from latently infected neurons established as previously detailed [16]) were sequenced using an Illumina 2500 Hiseq instrument at the Israel National Center for Personalized Medicine.
Conventional bioinformatics analysis.
NGS reads obtained after trimming of the Illumina sequences were aligned to the human genome (GenBank accession no. hg19) at all possible positions using Bowtie2 (-n 0) with perfect matches (0 mismatches). The remaining reads that did not map perfectly to the human genome were passed on to the following analysis steps: reads of a length of <17 nt or >35 nt were removed, and unique sequences were summarized (i.e., when multiple reads had exactly the same sequence, they were replaced by a single “pseudo-read” with an ID containing read number information). Small RNA-Seq reads were then collected that aligned perfectly to the widely studied and well-annotated VZV genome Dumas (NCBI accession no. NC_001348.1).
After alignment to the Dumas genome, the reads were fed into the web-based miRNA prediction programs MirDeep2 (https://www.mdc-berlin.de/8551903/en/), miReap (https://sourceforge.net/projects/mireap/), and MiReNa (21). miRDeep2 identifies potential miRNA precursors in a genome using mapped reads as guidelines. It identifies reads that could correspond to mature miRNAs and excises the genomic sequences and flanking sequences. miRDeep2 then invokes Bowtie to map the sequencing reads to potential precursors and invokes RNAfold to predict the RNA secondary structures of precursors that resemble a typical miRNA hairpin structure. Then, the miRDeep2 core algorithm determines whether to call a candidate a potential miRNA.
MIReNA was run with the following parameters: (i) a minimum free energy (MFE) allowed for a miRNA precursor of 18 kcal/mol and (ii) a minimum number of reads matching a pre-miRNA of 3. Like miRDeep2, MIReNA computes secondary structures of all extended sequences containing the potential miRNA and scans them to filter out those that do not satisfy suitable combinatorial and physicochemical conditions. MIReNA also evaluates the most stable potential folding structures or, in case of equal stability of the matching (i.e., an equal MFE value), the longest sequence. Predicted stem-loop precursors were then generated by the programs.
Search for sncRNAs in VZV by manual compilation of similar sequences.
Illumina sequences were trimmed and reads mapping perfectly to the human genome were removed as just described. The remaining reads with similar sequences starting in approximately the same position in the VZV genome were combined from all the samples and aligned visually in an Excel spreadsheet. Consensus sequences from the alignment were subjected to BLAST searches against three additional VZV genomes: pOka (GenBank accession no. AB097933.1 [the virus from which the recombinant VZV used in this study was derived]), vOka (GenBank accession no. AB097932.1), and Ellen (GenBank accession no. JQ972913.1). The obtained sequences with additional 5′ and 3′ VZV genomic sequences from Dumas (sequences available on request) were fed into RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) in order to determine if stem-loop precursors could be predicted to exist for each putative sncRNA.
Stem-loop TaqMan PCR for sncRNA.
Infection of HFFs or ARPE19 cells was performed using cell-associated or cell-free VZV66GFP until approximately 90% of cells were GFP positive. RNA was extracted using miRNeasy (Qiagen) or the Hybrid-R kit for mRNA/small RNA (Geneall, South Korea). All primers and probes were designed as described in reference 34. Primers were generated by IDT (Israel), and the probe was DLO-RFB-5 from LGC Biosearch Technologies (Petaluma, CA).
The cDNA for each sncRNA was prepared in a separate reaction. cDNA was made using Moloney murine leukemia virus (MMLV) reverse transcriptase (M1701; Promega) at increasing temperatures: 25°C for 15 min, 37°C for 15 min, 42°C for 40 min, and finally 95°C for enzyme inactivation. All qPCRs were done in triplicates and averaged to compensate for pipetting errors.
The RNA used for qPCR of latently and productively infected neurons was from samples used for the NGS just described. Evaluation of the NGS results for human sncRNAs showed that the expression of the miRNA hsa-mir26 was at a high level and showed little variation from sample to sample in those used for NGS (not shown); therefore qRT-PCR was performed for this sncRNA in each sample and used to normalize the expression of the VZV sncRNAs (35).
Effect of an inhibitor of VZVsncRNA23 on VZV replication.
A locked RNA antagonist to the VZV sncRNA identified in both bioinformatics analyses was designed by and purchased from Exiqon. Approximately 100 infected ARPE19 cells from a culture with approximately 70% infection with VZVRFP66 were added to confluent ARPE19 cells (a ratio of approximately 1:100) in wells of a 96-well plate in order to obtain a relatively low-density infection, where infected foci could be followed individually. At 24 h postinfection, the antagonist or a fluorescein-labeled scrambled control RNA (SIC007; Sigma-Aldrich) was transfected using Dharmafect 2 (Dharmacon) according to the manufacturer's protocol. Growth of individual foci of infection (FOI) was measured from images of living cultures using a quantitative analysis method that is detailed in Results.
ACKNOWLEDGMENTS
We are very grateful to Noam Shomron (Tel Aviv University) for many helpful discussions that allowed us to pursue this project successfully, as well as reading of an earlier version of the manuscript. We also thank Dena Leshkowitz, Shlomit Gilad, and Sima Benjamin of The Crown Genomics institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science, for expert help and advice. Aryeh Weiss (Bar-Ilan University Faculty of Engineering) provided invaluable advice in the stitching of multiple micrographs.
This research was supported by NIH grants R01 AI122640 (P.R.K. and R.G.), R21 NS082662 (P.R.K. and R.G.), US-Israel Bi-National Science Foundation 2013072 (R.G. and P.R.K.). R.G. was also supported by Israel Science Foundation grant 254/16. P.R.K. also acknowledges an NEI CORE grant for Vison Research (EY08098) and unrestricted funds from Research to Prevent Blindness, Inc., and the Eye & Ear Foundation of Pittsburgh.
REFERENCES
- 1.Abendroth A, Arvin AM, Moffat JF. 2010. Varicella-zoster virus. Springer, New York, NY. [Google Scholar]
- 2.Kim M-C, Yun S-C, Lee H-B, Lee PH, Lee S-W, Choi S-H, Kim YS, Woo JH, Kim S-H, Kwon SU. 2017. Herpes zoster increases the risk of stroke and myocardial infarction. J Am Coll Cardiol 70:295–296. doi: 10.1016/j.jacc.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Jonas S, Izaurralde E. 2015. Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 4.Iwakawa H, Tomari Y. 2015. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 25:651–665. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg MS, Morris KV. 2016. Transcriptional gene silencing in humans. Nucleic Acids Res 44:6505–6517. doi: 10.1093/nar/gkw139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han Z, Liu X, Chen X, Zhou X, Du T, Roizman B, Zhou G. 2016. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci U S A 113:E894–E901. doi: 10.1073/pnas.1525674113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cullen BR. 2011. Herpesvirus microRNAs: phenotypes and functions. Curr Opin Virol 1:211–215. doi: 10.1016/j.coviro.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piedade D, Azevedo-Pereira JM. 2016. The role of microRNAs in the pathogenesis of herpesvirus infection. Viruses 8:E156. doi: 10.3390/v8060156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan D, Flores O, Umbach JL, Pesola JM, Bentley P, Rosato PC, Leib DA, Cullen BR, Coen DM. 2014. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe 15:446–456. doi: 10.1016/j.chom.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods 2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 11.Nukui M, Mori Y, Murphy EA. 2015. A human herpesvirus 6A-encoded microRNA: role in viral lytic replication. J Virol 89:2615–2627. doi: 10.1128/JVI.02007-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuddenham L, Jung JS, Chane-Woon-Ming B, Dölken L, Pfeffer S. 2012. Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B. J Virol 86:1638–1649. doi: 10.1128/JVI.05911-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR. 2009. Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol 83:10677–10683. doi: 10.1128/JVI.01185-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohrs RJ, Laguardia JJ, Gilden D. 2005. Distribution of latent herpes simplex virus type-1 and varicella zoster virus DNA in human trigeminal ganglia. Virus Genes 31:223–227. doi: 10.1007/s11262-005-1799-5. [DOI] [PubMed] [Google Scholar]
- 15.Wang K, Lau TY, Morales M, Mont EK, Straus SE. 2005. Laser-capture microdissection: refining estimates of the quantity and distribution of latent herpes simplex virus 1 and varicella-zoster virus DNA in human trigeminal ganglia at the single-cell level. J Virol 79:14079–14087. doi: 10.1128/JVI.79.22.14079-14087.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Markus A, Lebenthal-Loinger I, Yang IH, Kinchington PR, Goldstein RS. 2015. An in vitro model of latency and reactivation of varicella zoster virus in human stem cell-derived neurons. PLoS Pathog 11:e1004885. doi: 10.1371/journal.ppat.1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinchington PR, Vergnes JP, Defechereux P, Piette J, Turse SE. 1994. Transcriptional mapping of the varicella-zoster virus regulatory genes encoding open reading frames 4 and 63. J Virol 68:3570–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinchington PR, Leger AJS, Guedon J-MG, Hendricks RL. 2012. Herpes simplex virus and varicella zoster virus, the house guests who never leave. Herpesviridae 3:5. doi: 10.1186/2042-4280-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall AE, Turnbull C, Dalmay T. 2013. Y RNAs: recent developments. Biomol Concepts 4:103–110. doi: 10.1515/bmc-2012-0050. [DOI] [PubMed] [Google Scholar]
- 20.Friedländer MR, Mackowiak SD, Li N, Chen W, Rajewsky N. 2012. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52. doi: 10.1093/nar/gkr688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathelier A, Carbone A. 2010. MIReNA: finding microRNAs with high accuracy and no learning at genome scale and from deep sequencing data. Bioinformatics 26:2226–2234. doi: 10.1093/bioinformatics/btq329. [DOI] [PubMed] [Google Scholar]
- 22.Jones JO, Sommer M, Stamatis S, Arvin AM. 2006. Mutational analysis of the varicella-zoster virus ORF62/63 intergenic region. J Virol 80:3116–3121. doi: 10.1128/JVI.80.6.3116-3121.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Louten J, Beach M, Palermino K, Weeks M, Holenstein G. 2016. MicroRNAs expressed during viral infection: biomarker potential and therapeutic considerations. Biomark Insights 10:25–52. doi: 10.4137/BMI.S29512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurzyńska-Kokorniak A, Jackowiak P, Figlerowicz M, Figlerowicz M. 2009. Human- and virus-encoded microRNAs as potential targets of antiviral therapy. Mini Rev Med Chem 9:927–937. doi: 10.2174/138955709788681573. [DOI] [PubMed] [Google Scholar]
- 25.Moens U. 2009. Silencing viral microRNA as a novel antiviral therapy? J Biomed Biotechnol 2009:419539. doi: 10.1155/2009/419539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee LY, Schaffer PA. 1998. A virus with a mutation in the ICP4-binding site in the L/ST promoter of herpes simplex virus type 1, but not a virus with a mutation in open reading frame P, exhibits cell-type-specific expression of γ134.5 transcripts and latency-associated transcripts. J Virol 72:4250–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang S, Bosch-Marce M, Patel A, Margolis TP, Krause PR. 2015. Characterization of herpes simplex virus 2 primary microRNA transcript regulation. J Virol 89:4837–4848. doi: 10.1128/JVI.03135-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pomp O, Brokhman I, Ziegler L, Almog M, Korngreen A, Tavian M, Goldstein RS. 2008. PA6-induced human embryonic stem cell-derived neurospheres: a new source of human peripheral sensory neurons and neural crest cells. Brain Res 1230:50–60. doi: 10.1016/j.brainres.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 29.Erazo A, Yee MB, Osterrieder N, Kinchington PR. 2008. Varicella-zoster virus open reading frame 66 protein kinase is required for efficient viral growth in primary human corneal stromal fibroblast cells. J Virol 82:7653–7665. doi: 10.1128/JVI.00311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tischer BK, Kaufer BB, Sommer M, Wussow F, Arvin AM, Osterrieder N. 2007. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J Virol 81:13200–13208. doi: 10.1128/JVI.01148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zerboni L, Hinchliffe S, Sommer MH, Ito H, Besser J, Stamatis S, Cheng J, Distefano D, Kraiouchkine N, Shaw A, Arvin AM. 2005. Analysis of varicella zoster virus attenuation by evaluation of chimeric parent Oka/vaccine Oka recombinant viruses in skin xenografts in the SCIDhu mouse model. Virology 332:337–346. doi: 10.1016/j.virol.2004.10.047. [DOI] [PubMed] [Google Scholar]
- 32.Sloutskin A, Goldstein RS. 2014. Laboratory preparation of varicella-zoster virus: concentration of virus-containing supernatant, use of a debris fraction and magnetofection for consistent cell-free VZV infections. J Virol Methods 206:128–132. doi: 10.1016/j.jviromet.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 33.Markus A, Waldman Ben-Asher H, Kinchington PR, Goldstein RS. 2014. Cellular transcriptome analysis reveals differential expression of pro- and anti-apoptosis genes by varicella zoster virus-infected neurons and fibroblasts. J Virol 88:7674–7677. doi: 10.1128/JVI.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mohammadi-Yeganeh S, Paryan M, Mirab Samiee S, Soleimani M, Arefian E, Azadmanesh K, Mostafavi E, Mahdian R, Karimipoor M. 2013. Development of a robust, low cost stem-loop real-time quantification PCR technique for miRNA expression analysis. Mol Biol Rep 40:3665–3674. doi: 10.1007/s11033-012-2442-x. [DOI] [PubMed] [Google Scholar]
- 35.Watson S, Mercier S, Bye C, Wilkinson J, Cunningham AL, Harman AN. 2007. Determination of suitable housekeeping genes for normalisation of quantitative real time PCR analysis of cells infected with human immunodeficiency virus and herpes viruses. Virol J 4:130. doi: 10.1186/1743-422X-4-130. [DOI] [PMC free article] [PubMed] [Google Scholar]