ABSTRACT
Coxsackievirus B (CVB) is a common enterovirus that can cause various systemic inflammatory diseases. Because CVB lacks an envelope, it has been thought to be inherently cytolytic, wherein CVB can escape from the infected host cell only by causing it to rupture. In recent years, however, we and others have observed that various naked viruses, such as CVB, can trigger the release of infectious extracellular microvesicles (EMVs) that contain viral material. This mode of cellular escape has been suggested to allow the virus to be masked from the adaptive immune system. Additionally, we have previously reported that these viral EMVs have LC3, suggesting that they originated from autophagosomes. We now report that CVB-infected cells trigger DRP1-mediated fragmentation of mitochondria, which is a precursor to autophagic mitochondrial elimination (mitophagy). However, rather than being degraded by lysosomes, mitochondrion-containing autophagosomes are released from the cell. We believe that CVB localizes to mitochondria, induces mitophagy, and subsequently disseminates from the cell in an autophagosome-bound mitochondrion-virus complex. Suppressing the mitophagy pathway in HL-1 cardiomyocytes with either small interfering RNA (siRNA) or Mdivi-1 caused marked reduction in virus production. The findings in this study suggest that CVB subverts mitophagy machinery to support viral dissemination in released EMVs.
IMPORTANCE Coxsackievirus B (CVB) can cause a number of life-threatening inflammatory diseases. Though CVB is well known to disseminate via cytolysis, recent reports have revealed a second pathway in which CVB can become encapsulated in host membrane components to escape the cell in an exosome-like particle. Here we report that these membrane-bound structures derive from mitophagosomes. Blocking various steps in the mitophagy pathway reduced levels of intracellular and extracellular virus. Not only does this study reveal a novel mechanism of picornaviral dissemination, but also it sheds light on new therapeutic targets to treat CVB and potentially other picornaviral infections.
KEYWORDS: autophagy, coxsackievirus, dissemination, mitochondria, mitophagy
INTRODUCTION
Coxsackievirus B (CVB) is a significant human pathogen that exhibits tropism toward the central nervous system, the pancreas, and the heart. CVB can cause severe acute inflammatory diseases, such as meningitis, pancreatitis, and myocarditis (1–8), and infants are particularly susceptible to generalized CVB infections (9–11). CVB is a member of the Picornaviridae family and Enterovirus genus. These viruses are nonenveloped and possess a positive-sense single-stranded RNA genome which encodes a single viral polypeptide that autocleaves to form the constituents necessary to assemble new virions. Due to its lack of an envelope, CVB has classically been thought to escape the infected cell by inducing lytic cell death once the viral copy number reaches a certain threshold and the cell succumbs to the viral burden (12–14). Though this mechanism allows for rapid release of viral particles, the death of the host cell both limits replication time and leaves the naked virus susceptible to neutralizing antibodies.
Like most viruses, CVB subverts many host cellular processes to support viral replication. Noteworthy among these hijacked pathways is macroautophagy (herein referred to as autophagy) (15, 16). Autophagy is a degradative process by which the cell targets damaged or unnecessary proteins and organelles to be destroyed by the lysosome. This begins with the initiation of a double-membrane phagophore which is thought to derive from the endoplasmic reticulum. This structure is decorated with the protein LC3-II, which binds to the adaptor protein p62/SQSTM1, allowing ubiquitinated cargos to be trafficked to the phagophore. After the phagophore fully elongates and engulfs the cargo, it is referred to as an autophagosome. The autophagosome then fuses with the lysosome, and the activation of acidic hydrolases causes the degradation of the contents within.
Not only is autophagy important in recycling cellular components, but also it can also function to eliminate foreign pathogens (xenophagy). Several bacterial strains, including Streptococcus pyogenes and Mycobacterium tuberculosis, were identified early on as pathogens that activate autophagy and become degraded (17, 18). Xenophagic clearance also extends to the clearance of viruses, such as Sindbis virus and herpes simplex virus 1 (19). However, many viruses have evolved to subvert autophagy and utilize the process to further propagate infection. The Picornaviridae family is known to activate autophagy via viral proteins 2BC and 3A (20). Studies examining CVB in the pancreata of infected mice revealed that CVB not only activates autophagy but also blocks autophagic flux, which results in an accumulation of autophagosomes that merge into large structures called megaphagosomes (16). Interestingly, the megaphagosomes were revealed to contain viral protein. A number of viruses have been reported to impair autophagosome-lysosome fusion. For example, the HIV protein Nef interacts with Beclin 1 to inhibit autophagosomal maturation and ultimately impair fusion with the lysosome (21). Similarly, the M2 protein of influenza A virus has been proposed to interfere with Beclin 1 and UVRAG-containing phosphatidylinositol 3-kinase (PI3K) complex to block autolysosome formation (22). Several groups have described the importance of autophagy during the course of CVB infection. For example, treating cells with the autophagy inhibitor 3-methyladenine results in a significant reduction in the accumulation of viral protein following infection (23). Another study in which the authors delivered CVB to mice with a pancreas-specific Atg5 deletion revealed that pancreatic viral titers were reduced and overall organ damage was limited (24). We have previously shown that CVB induces the release of extracellular microvesicles (EMVs) from the cell and that these structures not only contain virus but also are enriched for autophagosome marker LC3-II (25). Consistent with these data, other groups have observed similar structures released by other enteroviruses. Reports have shown that poliovirus induces the formation of LC3+ vesicles which contain viral particles. These vesicles are then released from the cell as a mode of viral egress. Electron micrographs revealed the presence of multiple virions contained within these extracellular vesicles, which the authors demonstrated were more efficient at infecting cells than free virions (26). As a whole, these findings reveal a link to CVB-triggered autophagy and a cytolysis-independent mechanism for subsequent viral dissemination.
Though virally mediated autophagy has been described for some time now, more recent work suggests that certain viruses manipulate mitochondrial dynamics and can induce mitochondrion-specific autophagy (mitophagy) as well. For example, a study investigating dengue virus, another RNA virus, revealed that the viral protease NS2B3 specifically cleaves mitochondrial fusion proteins Mfn1 and Mfn2 to suppress cellular antiviral immunity (27). A study investigating hepatitis C virus (HCV) revealed that the virus induces DRP1-mediated mitochondrial fission, a prerequisite for mitophagy (28). Indeed, mitochondrial networks in HCV-infected Huh7 human hepatoma cells became fragmented and were subsequently targeted to autophagosomes. These studies, coupled with our evidence that CVB-induced EMVs are autophagosomal in origin, implicate a mechanism by which CVB enters autophagosomes and subsequently disseminates from the host via membrane-bound vesicles. Viruses localize to the mitochondria for numerous reasons. The mitochondrial outer membrane is the assembly site of the mitochondrial antiviral signaling (MAVS) signalosome (29). This supramolecular assembly upregulates NF-κB and interferon regulatory factor 3 (IRF-3) pathways following the detection of double-stranded RNA. The 3Cpro viral protease encoded by CVB is responsible for destroying the MAVS protein and prolonging replication by suppressing apoptosis (30). African swine fever virus (ASFV) causes mitochondria to cluster around sites of viral replication in order to better utilize the energy produced (31). Additionally, HCV localizes to mitochondria to alter the permeability of mitochondrial membranes to cations, enhancing cell survival and prolonging viral replication (32–34).
In this study, we investigated the link between mitophagy and EMV-based viral dissemination. We found that CVB associates with mitochondria and triggers mitochondrial fragmentation. EMVs isolated from infected cells contained not only viral protein but also autophagosomal and mitochondrial protein. We hypothesize that CVB triggers mitophagy and becomes engulfed within mitophagosomes. Instead of undergoing lysosomal degradation, these virus-laden mitophagosomes are then expelled from the cell as EMVs. We propose that in addition to traditional cytolytic viral egress, mitophagy-derived EMVs serve as an alternative and possibly more prominent mode of CVB dissemination.
RESULTS
Impaired autophagy results in reduced viral dissemination.
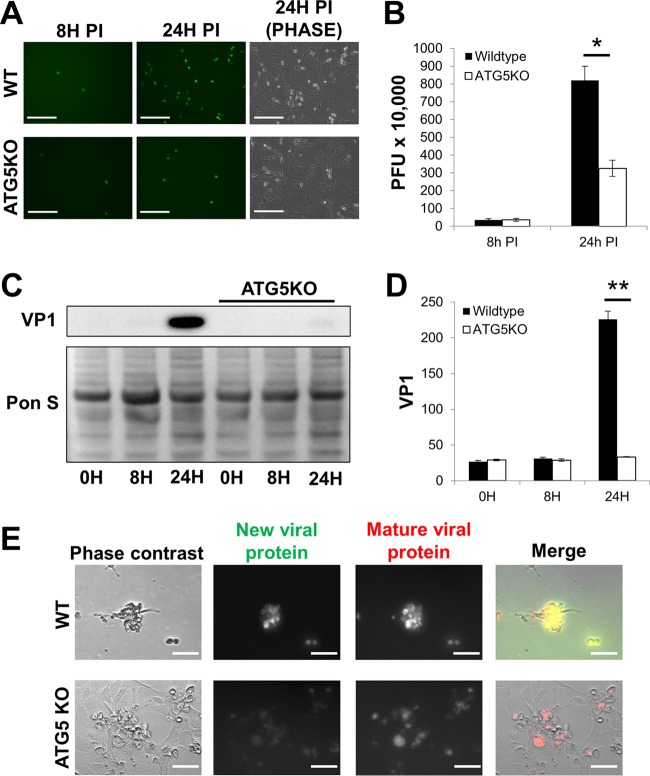
As mentioned above, many viruses, including CVB, upregulate host autophagy (15, 16); however, it is unclear what role autophagy plays during the course of CVB infection. To explore this, we utilized mouse embryonic fibroblasts with ATG5 knocked out (ATG5KO MEFs). ATG5 is an essential autophagy protein required for autophagosome biogenesis. We infected ATG5KO and wild-type (WT) MEFs with an enhanced green fluorescent protein (eGFP)-expressing CVB (eGFP-CVB) at a multiplicity of infection of 10 (MOI10). We found that at 24 h postinfection (p.i.), WT MEFs exhibited brisk infection which was marked by numerous detached, eGFP+ cells (Fig. 1A). ATG5KO MEFs showed much less infection than WT MEFs at 24 h p.i. Though plaque assays on the media from these infected cells showed no statistically significant difference in extracellular infectious virus at 8 h p.i., ATG5KO MEFs showed a substantial reduction in viral release by 24 h p.i. compared to that of WT cells (Fig. 1B). Western blots revealed reduced cellular levels of viral capsid protein VP1 by 24 h p.i. (Fig. 1C and D). These data are consistent with other reports regarding the importance of autophagy in the propagation of CVB infection (23, 24, 35). We next infected WT and ATG5KO MEFs with CVB expressing timer protein (timer-CVB) to determine if new rounds of infection occur normally at a later time point (25). Timer protein is a mutated DsRed which transitions from green to red fluorescence within 24 h after initial synthesis (36). Therefore, green fluorescence represents newly synthesized viral protein, whereas red represents viral protein that was synthesized >24 h earlier. We observed a heterogeneous distribution of green and red fluorescence in infected WT MEFs at 48 h p.i., suggesting new infection was ongoing (Fig. 1E). Infected ATG5KO MEFs, on the other hand, exclusively exhibited red fluorescence, indicating that new rounds of infection did not occur. In all, these data illustrate that throughout the duration of CVB infection, autophagy plays an integral role in viral egress.
FIG 1.
Impaired autophagy results in reduced viral dissemination. Wild-type (WT) and ATG5 knockout (ATG5KO) mouse embryonic fibroblasts (MEFs) were infected with eGFP-expressing coxsackievirus B (eGFP-CVB) at a multiplicity of infection of 10 (MOI10). (A) Fluorescence microscopy of infected MEFs at 8 h and 24 h postinfection (p.i.). Phase-contrast images show similar cell numbers at 24 h p.i. Scale bars represent 200 μm. (B) Extracellular viral titers of infected cells at 8 h and 24 h p.i. as measured by plaque assay. *, P < 0.05, Student t test; n = 3. (C) Western blots of infected cells at 0 h, 8 h, and 24 h p.i. (D) Densitometric quantification of Western blots in panel C. A representative Western blot is shown. **, P < 0.01, Student t test; n = 3. (E) Phase-contrast and fluorescence microscopy images of MEFs infected with timer protein-expressing CVB. Scale bars represent 30 μm.
Autophagy is essential for the release of virus-induced extracellular microvesicles.
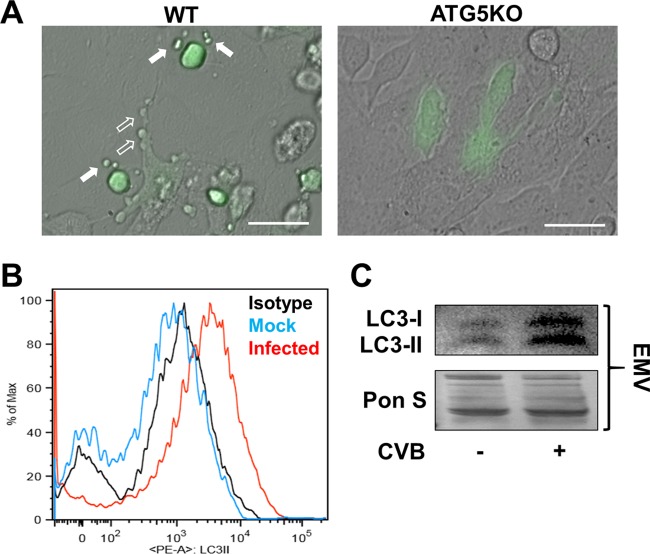
Having established the importance of autophagy for CVB dissemination, we next examined specifically how autophagy promotes viral spread. Inspection of infected WT cells at high magnification revealed EMVs budding off the surface of infected WT cells 24 h p.i., which was consistent with previous findings for CVB and other viruses (Fig. 2A) (25, 26, 37). Interestingly, infected ATG5KO MEFs did not produce eGFP+ EMVs at this time point. Next, we isolated EMVs from mock- and eGFP-CVB-infected C2C12 skeletal myoblasts and stained them with a phycoerythrin (PE)-conjugated antibody specific to the autophagosome marker LC3-II. We chose C2C12 myoblasts because we have observed that these cells can be infected with high concentrations of eGFP-CVB and survive infection for several days, permitting longer shedding of viral EMVs. We performed flow cytometry on these stained EMVs and found that those shed from infected C2C12 myoblasts were enriched for LC3-II, whereas the EMVs from mock-infected cells were not (Fig. 2B). Because CVB exhibits cardiac tropism, causing myocarditis, we next examined viral EMV release in HL-1 mouse atrial cardiomyocytes. Western blots of EMVs from infected HL-1 cells revealed a pronounced increase in both LC3-I and -II compared to the levels in mock-infected cells, which was consistent with our findings in C2C12 myoblasts (Fig. 2C). These results indicate that CVB induces release of viral EMVs whose membranes likely originate from autophagosomes.
FIG 2.
Autophagy is essential for the release of virus-induced extracellular microvesicles. WT and ATG5KO MEFs were infected with eGFP-CVB at MOI10. (A) Merged fluorescence and phase-contrast images of infected MEFs 24 h p.i. Scale bars represent 30 μm. Open arrows indicate blebs stemming from the surfaces of infected cells, whereas solid arrows indicated fully detached eGFP+ EMVs. (B) EMVs were isolated from C2C12 skeletal myoblasts that were either infected with eGFP-CVB at MOI10 or mock infected. EMVs were stained with a phycoerythrin-conjugated LC3-II antibody and analyzed via flow cytometry. EMVs from mock-infected cells were used for isotype control staining. (C) Western blots of EMVs isolated from HL-1 cells infected with eGFP-CVB at MOI1.
CVB induces fragmentation of mitochondrial networks and subsequent EMV release.
HCV was recently reported to induce mitochondrial fission and subsequent mitophagy (28). Knowing that CVB-induced EMVs displayed LC3, we next investigated whether mitophagy might be specifically involved in EMV biogenesis. We used eGFP-CVB to infect HL-1 cells expressing DsRed targeted to the mitochondrial matrix (mito-DsRed). Whereas mitochondria in uninfected (eGFP−) cells were arranged in a typical filamentous network, infected (eGFP+) cells showed fragmented mitochondrial networks (Fig. 3A). These results were reflected in area/perimeter ratio measurements of mitochondria showing that CVB infection significantly reduced mitochondrial network interconnectivity (Fig. 3B). We next examined whether EMVs shed from infected cells contained both mitochondrial fragments and virus. Though eGFP is synthesized concurrently with viral replication, individual virions are not actually associated with eGFP molecules. Therefore, the presence of green fluorescence does not guarantee the presence of actual virions. To address this, we immunostained eGFP-CVB-infected mito-DsRed cells to detect VP1, which is a virion structural protein (stained in blue). We observed distinct EMVs containing not only viral eGFP but also VP1 and mito-DsRed, confirming simultaneous release of virions and mitochondria in EMVs (Fig. 4A and B). Consistent with this, Western blots revealed that isolated EMVs from infected cells contained VP1 as well as the outer mitochondrial membrane marker TOM70 (Fig. 4C). Furthermore, viral EMVs also contained mitochondrial fission protein DRP1. None of these proteins were detectable in the EMVs from mock-infected controls, which likely represent conventional exosomes. However, much like exosomes, EMVs from infected cells contained the tetraspanin CD63 and lacked intracellular proteins, such as endoplasmic reticulum protein ERp72 and histone H3 (Fig. 4D). Previous reports have shown that not only does CVB become engulfed in autophagosomes (16), but also the viral protein 3Cpro localizes with mitochondria (30). However, because 3Cpro is a nonstructural protein of the virion, we investigated whether VP1 could be detected within mitochondria isolated from infected cells. Subcellular fractionation of infected cells revealed the presence of VP1 in the mitochondrial fraction (Fig. 4E). Washing the mitochondrial pellet did not reduce the amount of VP1 protein, which indicated that the virus was specifically associated with the mitochondria. To further support the idea that CVB is associates with mitochondria, we performed submitochondrial fractionation to separate mitoplasts (inner membrane and matrix) from the outer membrane and intermembrane space. Interestingly, we found more VP1 in the mitoplast fraction than in the outer membrane and intermembrane space fraction, suggesting that rather than anchoring to the surface of mitochondria, CVB embeds itself within the organelle (Fig. 4F). These data indicate that CVB associates with mitochondria following infection, triggers mitochondrial fragmentation (a precursor to mitophagy), and disseminates from the cell in a mitophagosome-virus complex.
FIG 3.
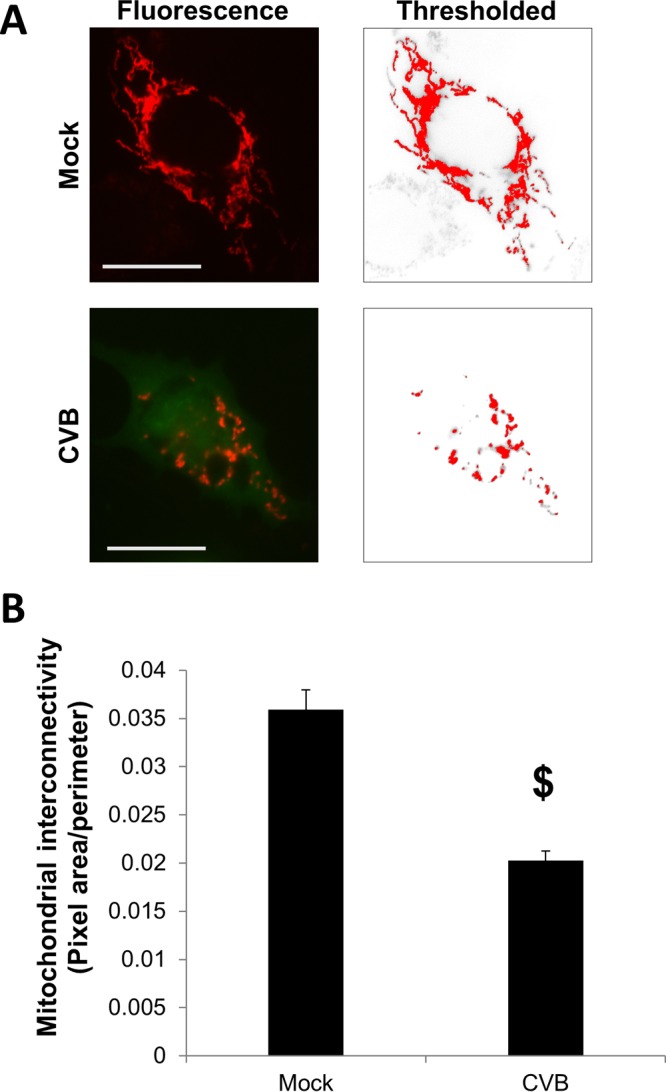
CVB induces fragmentation of mitochondrial networks. HL-1 cells expressing mitochondrion-targeted DsRed (mito-DsRed) were either infected with eGFP-CVB at MOI1 or mock infected. (A) On the left are representative fluorescence microscopy images of a mock-infected cell or an eGFP-CVB-infected cell 24 h p.i. Red, mitochondria; green, viral protein. On the right are thresholded images used for quantifications in panel B. Scale bars represent 20 μm. (B) Quantification of mitochondrial interconnectivity as measured by area/perimeter ratio calculations based on thresholded images of the red channel. $, P < 4.843 × 10−8, Student t test; n = 25 cells.
FIG 4.
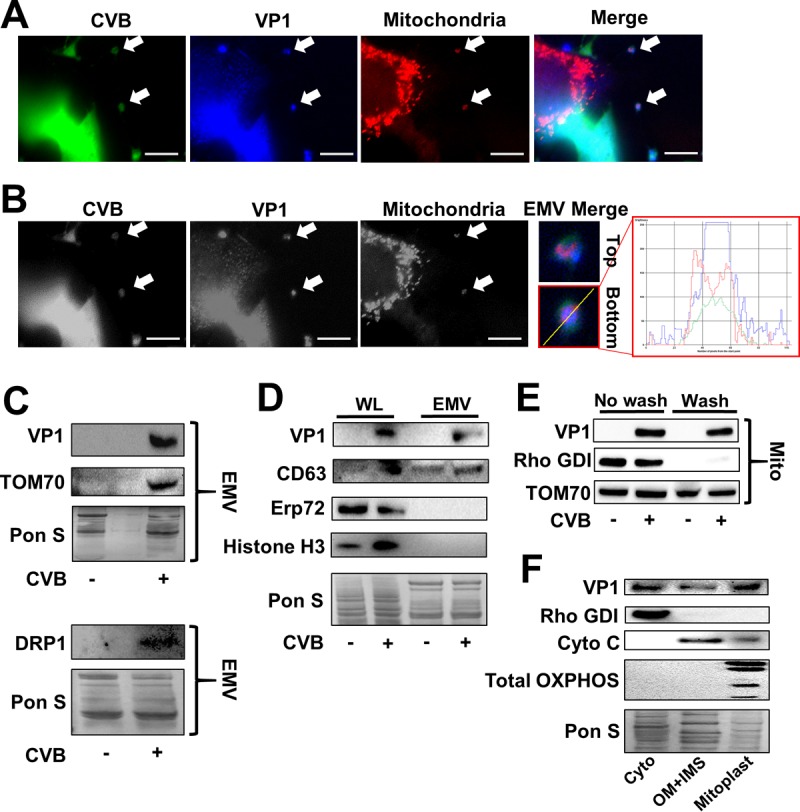
CVB-induced viral EMVs contain mitochondrial fragments. Mito-DsRed-expressing HL-1 cells were infected with eGFP-CVB at MOI1. (A) Fluorescence microscopy images of infected cells immunostained for VP1 24 h p.i. Scale bars represent 10 μm. (B) Grayscale images from panel A. Arrows indicate EMVs containing mito-DsRed, viral eGFP, and VP1. Images are presented in grayscale to enhance contrast of all channels. Scale bars represent 10 μm. Further-magnified colored images of EMVs indicated by top and bottom arrows are presented to the right, accompanied by representative line profiling depicting color composition of bottom EMV. (C) Western blots on EMVs isolated from HL-1 cells infected with eGFP-CVB at MOI1 24 h p.i. Two separate membranes are shown. (D) Western blots of cell lysates and EMVs isolated from infected HL-1 cells 24 h p.i. (E) Western blots of washed and nonwashed mitochondria isolated from infected HL-1 cells 24 h p.i. (F) Western blots of cytoplasm (Cyto), outer membrane plus intermembrane space (OM+IMS), and mitoplast fractions from infected HL-1 cells 24 h p.i. Blots were probed for fraction-specific markers to ensure efficient fractionation. These were Rho GDP-dissociation inhibitor 1 (Rho GDI) (cyto), cytochrome c (OM+IMS), and total OXPHOS complexes (mitoplast). From largest to smallest, the bands labeled by the total OXPHOS complex antibody cocktail are as follows: complex 5 (CV), ATP5A; CIII, UQRC2; CIV, MTCO1; CII, SDHB; and CI, NDUFB8. Equal amounts of protein were loaded into each lane.
Suppressing DRP1 activity limits CVB infection.
Our findings thus far revealed that CVB infection causes fragmentation of the mitochondrial network. It is unclear if this virally induced fission is simply a by-product of infection-mediated stress or if mitochondrial fragmentation plays a part in viral propagation; therefore, we tested whether blocking DRP1 could alter CVB infection. To accomplish this, we first treated HL-1 cells with small interfering RNA (siRNA) targeting DRP1 (siDRP1) or scrambled RNA (siSCRAMBLE) as a control. Following treatment, we infected the cells with eGFP-CVB. As was the case when we infected ATG5KO MEFs (Fig. 1A), at 24 h p.i., fewer eGFP+ cells were observed in cell cultures treated with siDRP1 than in siSCRAMBLE controls (Fig. 5A). In addition, extracellular viral titers were reduced by ∼80% when cells were treated with siDRP1 (Fig. 5B). Western blots confirmed efficient DRP1 silencing; strikingly, VP1 was barely detectable at 24 h p.i. in cells treated with siDRP1 (Fig. 5C and D).
FIG 5.
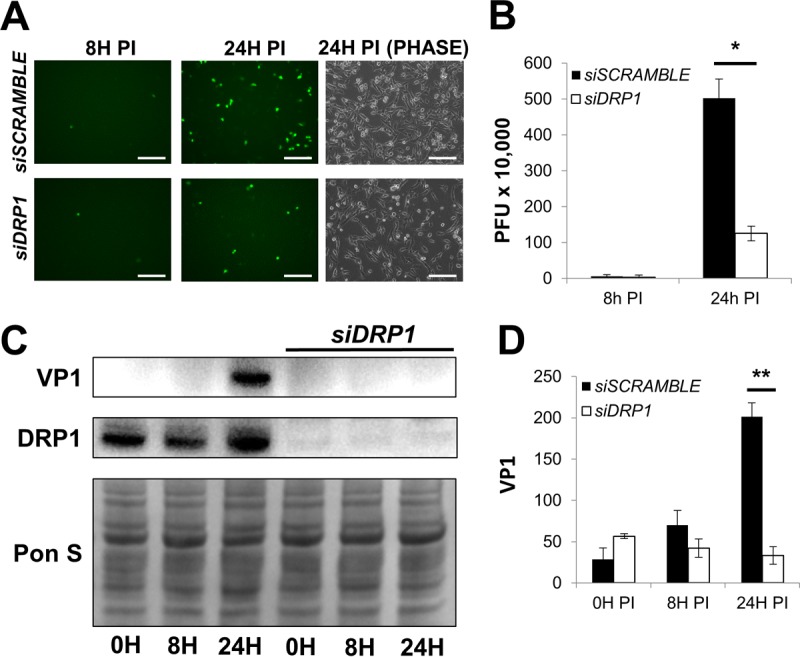
Silencing DRP1 limits CVB infection. HL-1 cells were treated with siRNA targeting DRP1 (siDRP1) or scrambled RNA (siSCRAMBLE) and subsequently infected with eGFP-CVB at MOI1. (A) Fluorescence microscopy of infected HL-1 cells at 8 h and 24 h p.i. Phase-contrast images show similar cell numbers at 24 h p.i. Scale bars represent 200 μm. (B) Extracellular viral titers of infected cells at 8 h and 24 h p.i. as measured by plaque assay. *, P < 0.05, Student t test; n = 3. (C) Western blots of infected cells at 0 h, 8 h, and 24 h p.i. (D) Densitometric quantification of Western blots in panel C. A representative Western blot is shown. **, P < 0.01, Student t test; n = 3.
Having established that silencing DRP1 could suppress viral propagation, we next tested the antiviral potential of the specific DRP1 inhibitor Mdivi-1. This cell-permeative drug blocks assembly of DRP1 on the mitochondria and effectively inhibits DRP1 fission activity without impacting mitochondrial fusion (38). Consistent with our siDRP1 findings, at 24 h p.i., we saw a marked reduction in eGFP+ cells following Mdivi-1 treatment and plaque assays showed a more modest, but statistically significant, reduction in extracellular viral titers in Mdivi-1-treated cells (Fig. 6A and B). We also observed a reduction in cellular VP1 in infected Mdivi-1-treated cells (Fig. 6C and D). Similarly, Western blots of isolated EMVs from infected siDRP1-treated and Mdivi-1-treated HL-1 cells showed dramatic reductions in both VP1 and TOM70 (Fig. 7A and B) compared to their respective controls, suggesting that blocking DRP1 with siRNA or Mdivi-1 disrupted EMV-mediated viral dissemination. These findings reveal a novel role for DRP1 in the biogenesis of CVB-EMVs and also highlight the antiviral capacity of Mdivi-1.
FIG 6.
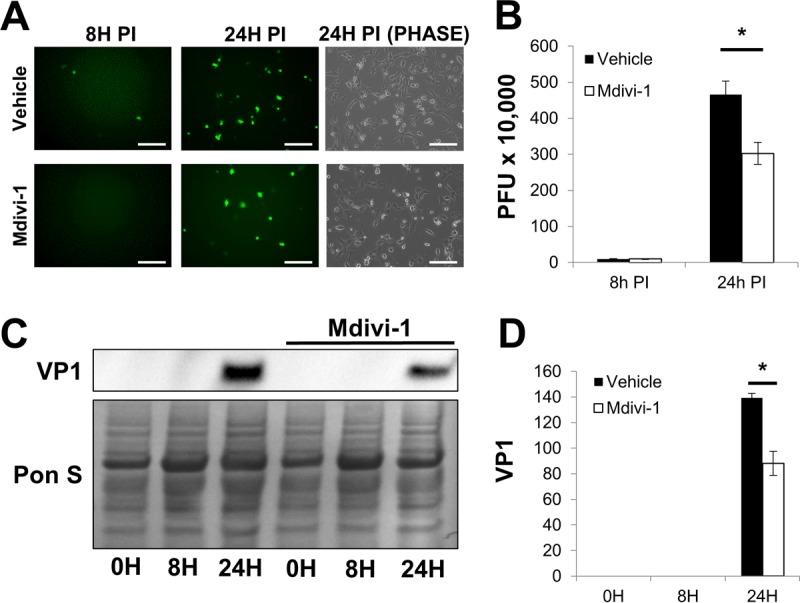
Mdivi-1 treatment reduces CVB infection. A 50 μM concentration of Mdivi-1 or an equal volume of DMSO (vehicle control) was added to HL-1 cells 40 min before infection with eGFP-CVB at MOI1 (51). Mdivi-1 was maintained in medium throughout the infection time course. (A) Fluorescence microscopy of infected HL-1 cells at 8 h and 24 h p.i. Phase-contrast images show similar cell numbers at 24 h p.i. Scale bars represent 200 μm. (B) Extracellular viral titers of infected cells at 8 h and 24 h p.i. as measured by plaque assay. *, P < 0.05, Student t test; n = 3. (C) Western blots of infected cells at 0 h, 8 h, and 24 h p.i. (D) Densitometric quantification of Western blots in panel C. A representative Western blot is shown. *, P < 0.01, Student t test; n = 3.
FIG 7.
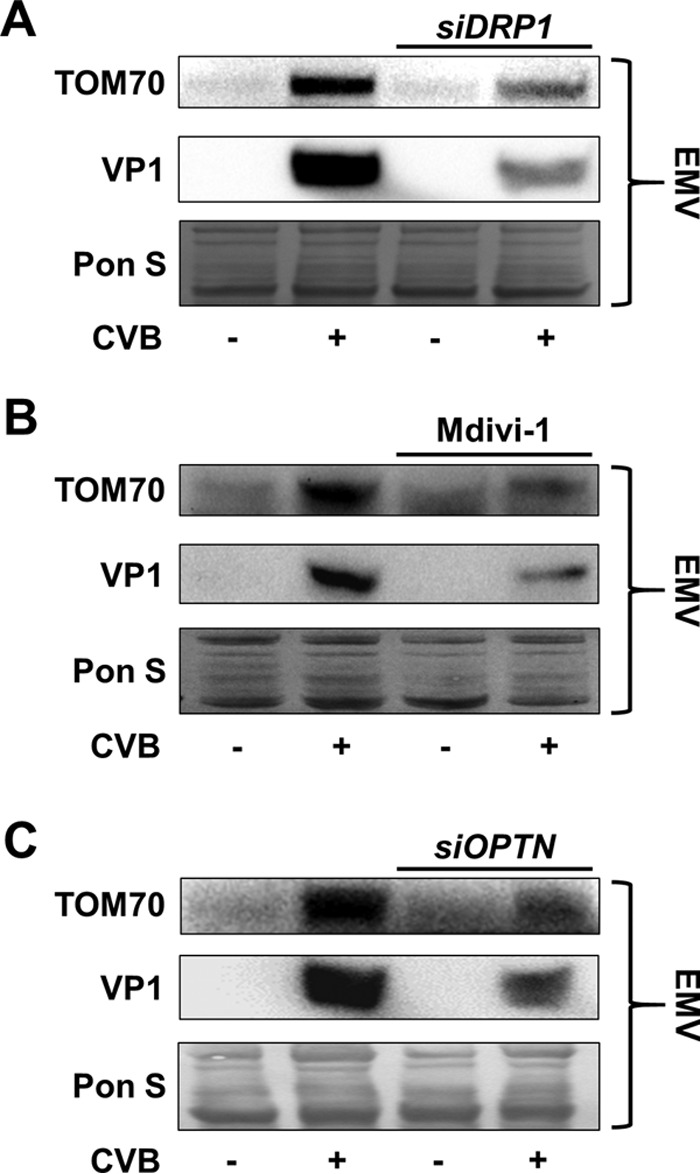
Blocking mitophagy suppresses mitochondrial and viral secretion into EMVs. Shown are Western blots of EMVs isolated from siDRP1-treated (A), Mdivi-1-treated (B), or siOPTN-treated (C) HL-1 cells infected with eGFP-CVB at MOI1 24 h p.i. Untreated cells were given respective controls.
Silencing optineurin limits CVB infection.
We have shown that blocking DRP1 disrupts CVB release through EMVs, and we hypothesize that this is due to impaired induction of mitophagy. To focus more specifically on the involvement of mitophagy in EMV-mediated viral egress, we treated cells with siRNA targeting optineurin (siOPTN) prior to infection. Similar to p62/SQSTM, optineurin is an autophagy adaptor which recruits LC3 to ubiquitinated substrates. Recently optineurin has been shown to be specifically recruited to damaged mitochondria targeted for autophagic degradation (39–41). At 24 h p.i. we observed fewer eGFP+ cells in infected siOPTN-treated cultures than in siSCRAMBLE controls (Fig. 8A), and plaque assays showed blunted release of extracellular virus (Fig. 8B). VP1 expression was reduced in siOPTN-treated cells as measured by Western blotting (Fig. 8C and D). Additionally, Western blots of EMVs isolated from infected siOPTN-treated cells showed a reduction in both VP1 and TOM70 compared to the levels in EMVs from infected siSCRAMBLE-treated cells (Fig. 7C). This is consistent with results from DRP1 inhibition experiments showing a reduction in viral propagation and VP1 levels. We hypothesize that following virally mediated DRP1-dependent mitochondrial fragmentation, optineurin coordinates viral escape from the host cell by targeting CVB-associated mitochondria for mitophagy. When coupled with previously described autophagic flux impairment (16, 26), the resultant virion-containing mitophagosomes, in turn, get released from the cell as viral EMVs.
FIG 8.
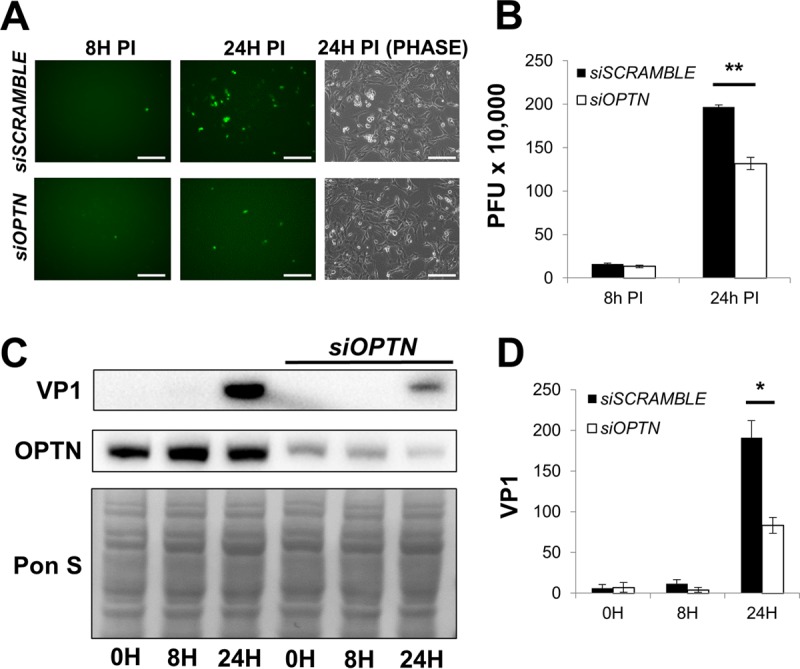
Silencing optineurin reduces CVB infection. HL-1 cells were treated with siRNA targeting optineurin (siOPTN) or siSCRAMBLE and subsequently infected with eGFP-CVB at MOI1. (A) Fluorescence microscopy of infected HL-1 cells at 8 h and 24 h p.i. Phase-contrast images show similar cell numbers at 24 h p.i. Scale bars represent 200 μm. (B) Extracellular viral titers of infected cells at 8 h and 24 h p.i. as measured by plaque assay. **, P < 0.01, Student t test; n = 3. (C) Western blots of infected cells at 0 h, 8 h, and 24 h p.i. (D) Densitometric quantification of Western blots in panel C. A representative Western blot is shown. *, P < 0.05, Student t test; n = 3.
Blocking mitophagy suppresses viral egress predominantly by limiting release of viral EMVs.
Knowing that the disruption of mitophagy reduces EMV-based viral dissemination and CVB infection as a whole, we next investigated how reliant CVB is on EMV release. We compared viral titers in EMVs isolated from HL-1 cells transfected with either siDRP1, siOPTN, or siSCRAMBLE. We then performed plaque assays to compare infectious virus in EMV-depleted media (free virus) versus EMV pellets resuspended in the same volume of media (EMV bound). Consistent with our earlier findings, total cumulative extracellular titers were lower following siDRP1 and siOPTN treatment (Table 1). Surprisingly though, in all samples, the bulk of infectious virus was found in the EMV fraction. Inhibiting mitochondrial fragmentation or mitophagy itself reduced EMV-based viral release but did not appear to affect free-virus release. We also investigated EMV release in ATG5KO MEFs. Whereas wild-type MEFs had more virus bound in EMVs, ATG5KO MEFs had a greater proportion of free-virus, suggesting that autophagy impairment may cause a bias toward cytolytic viral release. Interestingly, even wild-type MEFs exhibited a substantial amount of free virus release compared to HL-1 cells, which may shed light on differences in infection dynamics from cell type to cell type.
TABLE 1.
EMV-bound versus free virus in media from infected mitophagy-impaired cells
| Cell type | PFU of: |
% EMV bound | |
|---|---|---|---|
| Free virus | EMV-bound virus | ||
| siSCRAMBLE-transfected HL-1 cells | 125,000 | 2,525,000 | 95.3 |
| siDRP1-transfected HL-1 cells | 117,500 | 1,350,000 | 92.0 |
| siOPTN-transfected HL-1 cells | 167,500 | 1,450,000 | 89.6 |
| Wild-type MEFs | 2,000,000 | 2,550,000 | 56.0 |
| ATG5KO MEFs | 1,600,000 | 750,000 | 31.9 |
Blocking mitophagy does not impair viral replication.
Our findings thus far demonstrate how mitochondrial fragmentation and subsequent mitophagy are crucial for viral egress via EMVs; however, it is not entirely clear if disruption of the mitophagy pathway may directly impair viral replication itself. To our knowledge, proteins such as DRP1 and optineurin have not been reported to influence viral replication machinery, but these proteins could have alternate functions that could interface with various aspects of cellular machinery related to viral synthesis. To determine if silencing DRP1 or optineurin could impair viral replication, we silenced HeLa cells and infected them with a low MOI, 0.01. We then examined the early hours postinfection to assess when viral protein first appeared and if this differed among the groups. We performed these experiments with HeLa cells because they are infected very rapidly even with a low concentration of virus; thus, they are ideal for teasing apart the early stages of infection. VP1 was first detected 7 h p.i. regardless of siRNA treatment (siSCRAMBLE, siDRP1, or siOPTN), indicating similar susceptibilities to viral infection and similar rates of viral replication (Fig. 9A and B). However, by 24 h p.i., VP1 levels were reduced in siOPTN-treated cells but minimally altered in siDRP1-treated cells (both compared to siSCRAMBLE-treated cells) (Fig. 9C). The silencing effect of siDRP1 appeared to have declined by this time point. As for extracellular viral titers, infectious virus in the media was first detectable at the same time point (7 h p.i.) for siSCRAMBLE, siDRP1, and siOPTN groups (1,500, 2,000, and 1,000 PFU/ml, respectively). Interestingly, media collected 24 h p.i. from siDRP1- and siOPTN-treated cells contained ∼80% and ∼90% less infectious virus, respectively, than media from siSCRAMBLE-treated cells (Fig. 9D). It is worth noting that levels of both DRP1 and optineurin were simultaneously reduced whether cells were transfected with siDRP1 or siOPTN. Though it is logical for optineurin recruitment to be blunted when upstream DRP1-mediated fission is inhibited, it is interesting that DRP1 levels are diminished when optineurin is silenced. This may reflect a feedback effect that these proteins (and possibly other mitophagy components) have on each other. Together these data demonstrate that cell infectibility and viral synthesis are preserved whether DRP1 or optineurin activity is intact; however, viral dissemination is impaired when mitophagy is blocked, and this could cause accumulation of virus within the cell and limit subsequent rounds of infection.
FIG 9.
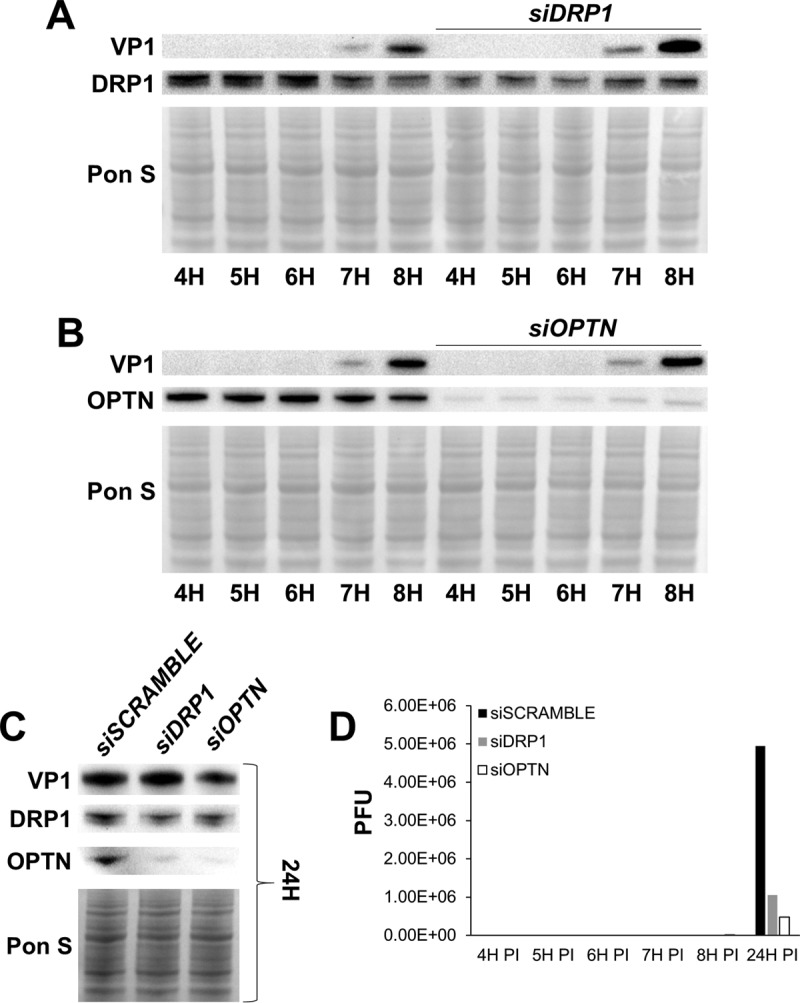
Blocking mitophagy does not impair viral replication. HeLa cervical cancer cells were transfected with either siDRP1, siOPTN, or siSCRAMBLE. Cells were then infected with eGFP-CVB at MOI0.01. Culture media and cell lysates were isolated at various time points. (A) Western blots of infected cells at 4 h, 5 h, 6 h, 7 h, and 8 h following treatment with either siSCRAMBLE or siDRP1. (B) Western blots of infected cells at 4 h, 5 h, 6 h, 7 h, and 8 h following treatment with either siSCRAMBLE or siOPTN. (C) Western blots of infected cells at 24 h p.i. following treatment with either siSCRAMBLE, siDRP1, or siOPTN. (D) Extracellular viral titers of infected cells at 4 h, 5 h, 6 h, 7 h, 8 h, and 24 h p.i. as measured by plaque assay.
DISCUSSION
Until recently, the nonenveloped family of viruses Picornaviridae has been thought to escape from the host cell exclusively via cytolysis. This mode of dissemination releases free “naked” virions and renders the virus susceptible to neutralizing antibodies. We were among the first groups to discover that certain nonenveloped viruses can hijack cell membranes and disseminate within a released vesicle (25, 26, 37). In the case of CVB, the viral EMVs are enriched for the autophagosome protein LC3-II tethered to the external face of the EMVs, compared to basally released vesicles. A previous study investigating CVB infection in pancreatic acinar cells showed that the virus initiates autophagy but inhibits autophagic flux (16). This results in an accumulation of autophagosomes which eventually fuse into larger megaphagosomes. A common observation regarding the intracellular buildup of virus-induced vesicles is that these structures somehow evade lysosomal degradation (16, 26, 42). Though it is not entirely clear how vesicular clearance is subverted, Chen et al. observed that several different enteroviruses, including CVB, escape the cell in phosphatidylserine vesicles which may have bypassed lysosomal degradation because they lack the autophagosomal SNARE protein syntaxin 17 required for lysosomal fusion (26). In the study presented here, we have found that these vesicles contain mitochondrial fragments in addition to infectious virus. Subcellular fractionation of infected cells showed that mitochondrial fractions contain viral protein. Thus, we hypothesize that CVB associates with mitochondria and induces mitophagy to gain entry into autophagosomes which the virus hijacks as a mechanism to escape the cell. Many viruses localize to and interact with mitochondrial networks. For example, adenovirus piggybacks onto the mitochondrion-associated protein p32 to translocate its genome to the nucleus (43). The enveloped hepatitis C virus activates mitophagy, which prevents apoptosis, prolonging viral replication (28). Similarly, the 2B protein of coxsackievirus can depolarize mitochondria, a trigger for mitophagy (44, 45). Since mitochondria are scaffolds for the assembly of the antiviral MAVS signalosome, the coxsackieviral protease 3Cpro is adept at destroying MAVS to prevent downstream antiviral cytokine production. However, while antiviral proteins are present on the mitochondrial outer membrane, mitochondria also provide the ATP needed for viral replication (46–48); therefore, many viruses rely on mitochondrial function to establish infection. In this study, we observed that CVB causes profound remodeling of mitochondrial networks. This is consistent with a recent report suggesting that hepatitis C virus (HCV) also induces mitochondrial fission following infection (28). Kim et al. (28) showed not only that HCV triggers a DRP1-mediated fragmentation and subsequent autophagic engulfment of mitochondria but also that this process facilitates viral dissemination; however, it is unclear how this induction of fission results in viral spread. A study by Feng et al. described how hepatitis A virus (another picornavirus) induces the release of infectious exosome-like vesicles (37). This is similar to our recent observations that CVB escapes infected cells via vesicles decorated with the autophagosome marker LC3-II (25). We investigated further and found that CVB localizes to mitochondria and triggers mitochondrial fission; the fragments are packaged into autophagosomes which are then hijacked, enabling virus to escape the cell in mitophagosomes. Interestingly, the degree of reduction in PFU from the media of autophagy/mitophagy-impaired cells was not as pronounced as the reduction of VP1 in respective cell lysates. This is likely attributable to the ability for EMVs to harbor multiple virions (26, 49). Therefore, even though an EMV could contain more than one virion, it would still produce a single plaque.
The discovery that CVB relies on mitophagy to promote infection reveals another novel therapeutic strategy to block CVB infection. We found that silencing the mitochondrial fission protein DRP1 dramatically suppressed CVB infection, which was marked by statistically significant reductions in both intracellular and extracellular virus (Fig. 8). With this finding, we proceeded to test the antiviral potential of the DRP1 inhibitor Mdivi-1. Consistent with our DRP1-silencing study, treatment with Mdivi-1 resulted in reductions in cellular viral content and viral dissemination. Silencing of the mitophagy adaptor optineurin also resulted in a blunting of both intracellular and extracellular virus. When we examined EMVs from infected cells pretreated with mitochondrial fission and mitophagy inhibitors, we found that all these treatments inhibited viral secretion into EMVs, and we hypothesize that the mitophagy pathway facilitates production of viral EMVs. The relative importance of cytolytic viral dissemination compared to EMV-mediated dissemination may vary according to cell type. In the context of CVB infection in vivo, cytolysis versus EMV release may vary by tissue type. Blocking mitophagy may cause the virus to rely more heavily on cytolysis, but this may be beneficial in vivo, as the immune system can better recognize and dispatch naked virions (Fig. 10). It should be noted, however, that we did not see an elevation in free virus release upon silencing DRP1 or optineurin in vitro; therefore, blocking mitophagy may only disrupt EMV release. In addition, blocking mitophagy did not seem to have any bearing on initial infection and viral synthesis; therefore, a mitophagy-impaired cell may be equally susceptible to infection, but viral secretion and subsequent rounds of infection would be hampered.
FIG 10.
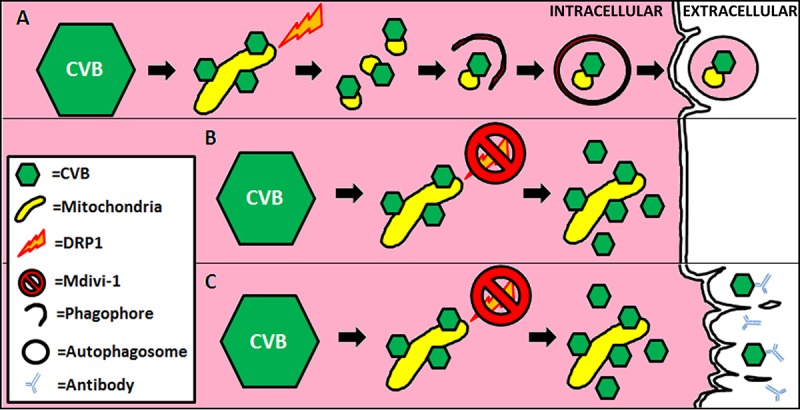
Schematic of mitophagy subversion by CVB. (A) Upon infection, CVB virions localize to mitochondria and trigger DRP1-mediated mitochondrial fission. The resultant mitochondrial fragments undergo autophagic engulfment. Rather than undergoing lysosomal degradation, these mitophagosomes containing both mitochondrial components and CVB virions are instead ejected from the cell as viral EMVs. Accumulation of mitophagosomes may also cause them to fuse together as megaphagosomes prior to becoming released. (B) Blocking DRP1 with Mdivi-1 (or siDRP1) inhibits mitochondrial fragmentation and subsequent initiation of mitophagy. This results in impaired EMV biogenesis, causing CVB virions to accumulate in the cell. (C) Due to a lack in dissemination, the virus may rely more on cytolytic cell death resulting in the release of free virions. In vivo, this mode of viral release would leave the virus more susceptible to neutralizing antibodies.
The effects of Mdivi-1 are already being investigated in the context of tumor suppression and cardioprotection; thus, its efficacy as a therapeutic has already been validated in vivo. In fact, a recent study described how Mdivi-1 could suppress CVB-induced myocarditis in mice (50). The authors hypothesized that this was due to preservation of mitochondrial integrity, reduced reactive oxygen species production, and increased cell survival. However, the authors did not examine viral replication and egress. Their findings are compatible with our hypothesis that Mdivi-1 could serve as an antiviral drug and therefore limit viral myocarditis. We believe that our observations regarding the protective nature of Mdivi-1 could extend beyond coxsackieviral suppression; therefore, it is important to test the efficacy of the drug as well as other mitophagy inhibitors in limiting infection caused by other RNA viruses.
MATERIALS AND METHODS
Production of recombinant coxsackievirus.
The production of recombinant coxsackievirus B3 (pMKS1) expressing protein inserts has been previously described. Briefly, an infectious CVB3 clone (pH 3) was engineered with a unique SfiI restriction site allowing for the insertion of foreign DNA fragments. Enhanced green fluorescent protein (eGFP) and timer protein were amplified from expression plasmids using sequence-specific primers with flanking SfiI sequences. PCR products were cloned into pMKS1 to generate eGFP-CVB and timer-CVB. Constructs were transfected into HeLa RW cells maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco; 11995-073), and infectious virus was produced. Cells were then scraped, freeze-thawed three times, and centrifuged at 2,000 rpm to remove cellular debris. Concentrations of viral stocks were determined by plaque assay.
Cell culture and treatments.
Wild-type mouse embryonic fibroblasts, ATG5KO mouse embryonic fibroblasts, C2C12 mouse skeletal myoblasts, and HeLa cervical cancer cells were maintained in DMEM growth medium consisting of DMEM supplemented with 10% fetal bovine serum (FBS; Life Technologies; 16010-159) and antibiotic/antimycotic (Life Technologies; 15240-062). HL-1 mouse atrial cardiomyocytes were maintained in Claycomb medium (Sigma-Aldrich; 51800C) supplemented with 10% FBS, 0.1 nM norepinephrine (Sigma-Aldrich; A0937), 2 mM l-glutamine (Thermo Fisher Scientific; 25030-081), and antibiotic/antimycotic.
Mdivi-1 (Sigma-Aldrich; M0199) was dissolved in dimethyl sulfoxide (DMSO) at a concentration of 50 mM. HL-1 cells were treated with 50 μM Mdivi-1 (diluted in Claycomb medium) or equivalent amounts of DMSO for 40 min prior to infection.
DRP1 siRNA (mouse [Santa Cruz Biotechnology; SC-45953] and human [Santa Cruz Biotechnology; SC-43732]) was reconstituted by following the manufacturer-provided datasheet. HL-1 cells were transfected using Effectene transfection reagent (Qiagen; B00118) by following the manufacturer's guidelines for reagent volumes. Forty-eight hours following transfection, the medium was refreshed and cells were subsequently infected.
Optineurin siRNA (mouse [Santa Cruz Biotechnology; SC-39055] and human [Santa Cruz Biotechnology; SC-39054]) was reconstituted by following the manufacturer-provided datasheet. HL-1 cells were transfected using Effectene transfection reagent by following the manufacturer's guidelines for reagent volumes. Forty-eight hours following transfection, the medium was refreshed and cells were subsequently infected.
EMV isolation.
EMVs were isolated with ExoQuick-TC (Systems Biosciences; EXOTCxxA-1) as described previously (25). Briefly, medium was removed from infected cells and centrifuged at 3,000 × g for 15 min to pellet cells and cell debris. Supernatant was transferred to new tube and ExoQuick-TC was added at a 1:6 dilution. After refrigeration overnight, the mixture was centrifuged at 1,500 × g for 30 min to pellet EMVs. The pellet was then washed in phosphate-buffered saline (PBS) and then lysed in radioimmunoprecipitation assay (RIPA) buffer containing Tris (pH 8.0) (50 mM; Sigma-Aldrich; T1503), NaCl (150 mM; Sigma-Aldrich; S7653), EGTA (1 mM; Sigma-Aldrich; E4884), NP-40 (1%; Sigma-Aldrich; I3021), sodium deoxycholate (0.5%; Sigma-Aldrich; D6750), SDS (0.1%; Bio-Rad Laboratories Inc.; 161-0302), and protease inhibitors (Sigma-Aldrich; 05056489001) pH adjusted to 7.4.
Western blots.
Whole-cell lysates were obtained by applying RIPA buffer directly to adherent cells and scraping. Detached cells were pelleted from culture media and combined with the rest of the corresponding lysate. For isolation of mitochondria, cells were scraped in mitochondrial isolation buffer containing sucrose (250 mM; Sigma-Aldrich; 179949), EDTA (1 mM; Sigma-Aldrich; E4884), HEPES (10 mM; Sigma-Aldrich; H3375), and protease inhibitors pH adjusted to 7.4. Cells were then mechanically disrupted by passage through a 27 1/2-gauge needle five times. Cell slurries were centrifuged at 600 × g for 5 min at 4°C. Supernatants (whole lysate) were collected and centrifuged again at 8,000 × g for 15 min at 4°C to pellet the mitochondrial fraction from the cytosol fraction. The mitochondrial pellet was washed twice by resuspending it in mitochondrial isolation buffer, spinning it again at 8,000 × g for 15 min at 4°C, and resuspending it in mitochondrial isolation buffer. To separate mitoplasts from the outer membrane and intermembrane space, isolated mitochondria were resuspended for 10 min on ice in 4 parts hypoosmotic buffer containing sucrose (5 mM), HEPES (5 mM), and EDTA (1 mM) pH adjusted to 7.4. One part hyperosmotic solution consisting of KCl (750 mM; Sigma-Aldrich; P5405), HEPES (100 mM), and EDTA (1 mM) pH adjusted to 7.4 was then added. Mitochondria were then centrifuged at 2,350 × g for 5 min at 4°C to pellet mitoplasts from the supernatant containing both outer membrane and intermembrane space.
Proteins were quantified using bicinchoninic acid solution (Sigma-Aldrich; B9643). Equal amounts of protein were run in 4 to 20% Tris-glycine SDS-PAGE gels (Life Technologies; EC6025) and transferred to nitrocellulose membranes. Membranes were blocked in 5% nonfat dry milk in Tris-buffered saline with Tween 20 (TBS-T) for 1 h at room temperature and then incubated in primary antibody diluted in 5% nonfat dry milk overnight at 4°C. Primary antibodies used were as follows: VP1 (1:1,000; Vector Laboratories; VP-E603), LC3 (1:1,000; Cell Signaling; 4108), TOM70 (1:1,000; Proteintech; 14528-1-AP), DRP1 (1:1,000; Millipore; ABT 155), optineurin (1:1,000; Santa Cruz Biotechnology; SC-60013), CD63 (1:1,000; System Biosciences; EXOAB-CD63A-1), Erp72 (1:1,000; Cell Signaling; 5033), histone H3 (1:1,000; Cell Signaling; 9715), Rho GDI (Santa Cruz Biotechnology; SC-373724), cytochrome c (Santa Cruz Biotechnology; SC-13560), and total OXPHOS complex cocktail (Abcam; ab110413).
Flow cytometry.
EMVs were fixed in 4% formalin for 10 min and then stained with an LC3-II antibody (Cell Signaling Technology; 2775) conjugated to phycoerythrin (PE). Flow cytometry was performed on a BD FACSAria (BD Biosciences) located in the San Diego State University Flow Cytometry Core Facility. Data were analyzed using BD FACSDIVA software.
Measurement of mitochondrial interconnectivity.
HL-1 cells expressing mitochondrion-targeted DsRed were imaged using a Keyence BZ-9000 microscope. Images were then analyzed using ImageJ software with “Mito-Morphology Macro” (http://imagejdocu.tudor.lu/doku.php?id=plugin:morphology:mitochondrial_morphology_macro_plug-in:start). Pixel area/perimeter ratios were used as a measure of mitochondrial interconnectivity.
Statistical analysis.
Statistical significance was determined using a two-tailed Student t test. Differences were measured relative to growth medium controls. Groups were considered significantly different if P values were less than 0.05. Error bars in figures indicate standard errors.
ACKNOWLEDGMENTS
This work was funded in part by NIH grant P01 HL112730 (R.A.G.), NIH grant T32 HL116273 (J.S.), a Myocarditis Foundation fellowship research grant (J.S.), and NIH grant KL2 TR001882 (J.S.).
Fluorescence-activated cell sorting (FACS) experiments were generously performed by the Flow Cytometry Core Facility at San Diego State University.
The CVB3 clone (pH 3) was generously provided by Kirk Knowlton (University of California, San Diego). Wild-type and ATG5 knockout mouse embryonic fibroblasts were generously provided by Noboru Mizushima (University of Tokyo).
R.A.G. is a consultant for Takeda Pharmaceuticals and is a cofounder of TissueNetix, Inc. The other authors have no potential conflicts of interest to disclose.
REFERENCES
- 1.Alexander JP Jr, Chapman LE, Pallansch MA, Stephenson WT, Torok TJ, Anderson LJ. 1993. Coxsackievirus B2 infection and aseptic meningitis: a focal outbreak among members of a high school football team. J Infect Dis 167:1201–1205. doi: 10.1093/infdis/167.5.1201. [DOI] [PubMed] [Google Scholar]
- 2.Burch GE, Sun SC, Chu KC, Sohal RS, Colcolough HL. 1968. Interstitial and coxsackievirus B myocarditis in infants and children. A comparative histologic and immunofluorescent study of 50 autopsied hearts. JAMA 203:1–8. [PubMed] [Google Scholar]
- 3.Horwitz MS, Krahl T, Fine C, Lee J, Sarvetnick N. 1999. Protection from lethal coxsackievirus-induced pancreatitis by expression of gamma interferon. J Virol 73:1756–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber S, Ramsingh AI. 2004. Coxsackievirus-induced pancreatitis. Viral Immunol 17:358–369. doi: 10.1089/vim.2004.17.358. [DOI] [PubMed] [Google Scholar]
- 5.Marier R, Rodriguez W, Chloupek RJ, Brandt CD, Kim HW, Baltimore RS, Parker CL, Artenstein MS. 1975. Coxsackievirus B5 infection and aseptic meningitis in neonates and children. Am J Dis Child 129:321–325. [DOI] [PubMed] [Google Scholar]
- 6.Reyes MP, Lerner AM. 1985. Coxsackievirus myocarditis—with special reference to acute and chronic effects. Prog Cardiovasc Dis 27:373–394. doi: 10.1016/0033-0620(85)90001-5. [DOI] [PubMed] [Google Scholar]
- 7.Tracy S, Hofling K, Pirruccello S, Lane PH, Reyna SM, Gauntt CJ. 2000. Group B coxsackievirus myocarditis and pancreatitis: connection between viral virulence phenotypes in mice. J Med Virol 62:70–81. doi:. [DOI] [PubMed] [Google Scholar]
- 8.Zaragoza C, Ocampo CJ, Saura M, Bao C, Leppo M, Lafond-Walker A, Thiemann DR, Hruban R, Lowenstein CJ. 1999. Inducible nitric oxide synthase protection against coxsackievirus pancreatitis. J Immunol 163:5497–5504. [PubMed] [Google Scholar]
- 9.Kaplan MH, Klein SW, McPhee J, Harper RG. 1983. Group B coxsackievirus infections in infants younger than three months of age: a serious childhood illness. Rev Infect Dis 5:1019–1032. doi: 10.1093/clinids/5.6.1019. [DOI] [PubMed] [Google Scholar]
- 10.Chiou CC, Liu WT, Chen SJ, Soong WJ, Wu KG, Tang RB, Hwang B. 1998. Coxsackievirus B1 infection in infants less than 2 months of age. Am J Perinatol 15:155–159. doi: 10.1055/s-2007-993917. [DOI] [PubMed] [Google Scholar]
- 11.Isacsohn M, Eidelman AI, Kaplan M, Goren A, Rudensky B, Handsher R, Barak Y. 1994. Neonatal coxsackievirus group B infections: experience of a single department of neonatology. Isr J Med Sci 30:371–374. [PubMed] [Google Scholar]
- 12.Herzum M, Ruppert V, Kuytz B, Jomaa H, Nakamura I, Maisch B. 1994. Coxsackievirus B3 infection leads to cell death of cardiac myocytes. J Mol Cell Cardiol 26:907–913. doi: 10.1006/jmcc.1994.1108. [DOI] [PubMed] [Google Scholar]
- 13.Polacek C, Ekstrom JO, Lundgren A, Lindberg AM. 2005. Cytolytic replication of coxsackievirus B2 in CAR-deficient rhabdomyosarcoma cells. Virus Res 113:107–115. doi: 10.1016/j.virusres.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Feuer R, Mena I, Pagarigan R, Slifka MK, Whitton JL. 2002. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J Virol 76:4430–4440. doi: 10.1128/JVI.76.9.4430-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kweon HS, Lee JY, Kim DH. 2008. Coxsackievirus B4 uses autophagy for replication after calpain activation in rat primary neurons. J Virol 82:11976–11978. doi: 10.1128/JVI.01028-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemball CC, Alirezaei M, Flynn CT, Wood MR, Harkins S, Kiosses WB, Whitton JL. 2010. Coxsackievirus infection induces autophagy-like vesicles and megaphagosomes in pancreatic acinar cells in vivo. J Virol 84:12110–12124. doi: 10.1128/JVI.01417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knodler LA, Celli J. 2011. Eating the strangers within: host control of intracellular bacteria via xenophagy. Cell Microbiol 13:1319–1327. doi: 10.1111/j.1462-5822.2011.01632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levine B. 2005. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 19.Cheng CY, Chi PI, Liu HJ. 2014. Commentary on the regulation of viral proteins in autophagy process. Biomed Res Int 2014:962915. doi: 10.1155/2014/962915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein KA, Jackson WT. 2011. Picornavirus subversion of the autophagy pathway. Viruses 3:1549–1561. doi: 10.3390/v3091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyei GB, Dinkins C, Davis AS, Roberts E, Singh SB, Dong C, Wu L, Kominami E, Ueno T, Yamamoto A, Federico M, Panganiban A, Vergne I, Deretic V. 2009. Autophagy pathway intersects with HIV-1 biosynthesis and regulates viral yields in macrophages. J Cell Biol 186:255–268. doi: 10.1083/jcb.200903070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, Garcia-Sastre A, Munz C. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon SY, Ha YE, Choi JE, Ahn J, Lee H, Kim DH. 2009. Autophagy in coxsackievirus-infected neurons. Autophagy 5:388–389. doi: 10.4161/auto.5.3.7723. [DOI] [PubMed] [Google Scholar]
- 24.Alirezaei M, Flynn CT, Wood MR, Whitton JL. 2012. Pancreatic acinar cell-specific autophagy disruption reduces coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe 11:298–305. doi: 10.1016/j.chom.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, Williams W, Kha N, Cruz C, Hancock BM, Nguyen DP, Sayen MR, Hilton BJ, Doran KS, Segall AM, Wolkowicz R, Cornell CT, Whitton JL, Gottlieb RA, Feuer R. 2014. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog 10:e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, Jiang P, Wimmer E, Altan-Bonnet G, Altan-Bonnet N. 2015. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu CY, Liang JJ, Li JK, Lee YL, Chang BL, Su CI, Huang WJ, Lai MM, Lin YL. 2015. Dengue virus impairs mitochondrial fusion by cleaving mitofusins. PLoS Pathog 11:e1005350. doi: 10.1371/journal.ppat.1005350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SJ, Syed GH, Khan M, Chiu WW, Sohail MA, Gish RG, Siddiqui A. 2014. Hepatitis C virus triggers mitochondrial fission and attenuates apoptosis to promote viral persistence. Proc Natl Acad Sci U S A 111:6413–6418. doi: 10.1073/pnas.1321114111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koshiba T. 2013. Mitochondrial-mediated antiviral immunity. Biochim Biophys Acta 1833:225–232. doi: 10.1016/j.bbamcr.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Mukherjee A, Morosky SA, Delorme-Axford E, Dybdahl-Sissoko N, Oberste MS, Wang T, Coyne CB. 2011. The coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog 7:e1001311. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojo G, Chamorro M, Salas ML, Vinuela E, Cuezva JM, Salas J. 1998. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J Virol 72:7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pavlović D, Neville DC, Argaud O, Blumberg B, Dwek RA, Fischer WB, Zitzmann N. 2003. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc Natl Acad Sci U S A 100:6104–6108. doi: 10.1073/pnas.1031527100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez ME, Carrasco L. 2003. Viroporins. FEBS Lett 552:28–34. doi: 10.1016/S0014-5793(03)00780-4. [DOI] [PubMed] [Google Scholar]
- 34.Griffin SD, Harvey R, Clarke DS, Barclay WS, Harris M, Rowlands DJ. 2004. A conserved basic loop in hepatitis C virus p7 protein is required for amantadine-sensitive ion channel activity in mammalian cells but is dispensable for localization to mitochondria. J Gen Virol 85:451–461. doi: 10.1099/vir.0.19634-0. [DOI] [PubMed] [Google Scholar]
- 35.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. 2008. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol 82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terskikh A, Fradkov A, Ermakova G, Zaraisky A, Tan P, Kajava AV, Zhao X, Lukyanov S, Matz M, Kim S, Weissman I, Siebert P. 2000. “Fluorescent timer”: protein that changes color with time. Science 290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 37.Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496:367–371. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanaka A, Youle RJ. 2008. A chemical inhibitor of DRP1 uncouples mitochondrial fission and apoptosis. Mol Cell 29:409–410. doi: 10.1016/j.molcel.2008.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen TN, Padman BS, Lazarou M. 2016. Deciphering the molecular signals of PINK1/Parkin mitophagy. Trends Cell Biol 26:733–744. doi: 10.1016/j.tcb.2016.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, Sideris DP, Fogel AI, Youle RJ. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524:309–314. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong YC, Holzbaur EL. 2014. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc Natl Acad Sci U S A 111:E4439–E4448. doi: 10.1073/pnas.1405752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alirezaei M, Flynn CT, Wood MR, Harkins S, Whitton JL. 2015. Coxsackievirus can exploit LC3 in both autophagy-dependent and -independent manners in vivo. Autophagy 11:1389–1407. doi: 10.1080/15548627.2015.1063769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matthews DA, Russell WC. 1998. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J Gen Virol 79(Part 7):1677–1685. doi: 10.1099/0022-1317-79-7-1677. [DOI] [PubMed] [Google Scholar]
- 44.van Kuppeveld FJ, de Jong AS, Melchers WJ, Willems PH. 2005. Enterovirus protein 2B po(u)res out the calcium: a viral strategy to survive? Trends Microbiol 13:41–44. doi: 10.1016/j.tim.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 45.Nieva JL, Agirre A, Nir S, Carrasco L. 2003. Mechanisms of membrane permeabilization by picornavirus 2B viroporin. FEBS Lett 552:68–73. doi: 10.1016/S0014-5793(03)00852-4. [DOI] [PubMed] [Google Scholar]
- 46.Chang CW, Li HC, Hsu CF, Chang CY, Lo SY. 2009. Increased ATP generation in the host cell is required for efficient vaccinia virus production. J Biomed Sci 16:80. doi: 10.1186/1423-0127-16-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anand SK, Tikoo SK. 2013. Viruses as modulators of mitochondrial functions. Adv Virol 2013:738794. doi: 10.1155/2013/738794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Klumpp K, Ford MJ, Ruigrok RW. 1998. Variation in ATP requirement during influenza virus transcription. J Gen Virol 79(Part 5):1033–1045. doi: 10.1099/0022-1317-79-5-1033. [DOI] [PubMed] [Google Scholar]
- 49.Altan-Bonnet N, Chen YH. 2015. Intercellular transmission of viral populations with vesicles. J Virol 89:12242–12244. doi: 10.1128/JVI.01452-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin L, Zhang M, Yan R, Shan H, Diao J, Wei J. 2017. Inhibition of Drp1 attenuates mitochondrial damage and myocardial injury in coxsackievirus B3 induced myocarditis. Biochem Biophys Res Commun 484:550–556. doi: 10.1016/j.bbrc.2017.01.116. [DOI] [PubMed] [Google Scholar]
- 51.Ong SB, Subrayan S, Lim SY, Yellon DM, Davidson SM, Hausenloy DJ. 2010. Inhibiting mitochondrial fission protects the heart against ischemia/reperfusion injury. Circulation 121:2012–2022. doi: 10.1161/CIRCULATIONAHA.109.906610. [DOI] [PubMed] [Google Scholar]