ABSTRACT
HIV-1-infected macrophages participate in virus dissemination and establishment of virus reservoirs in host tissues, but the mechanisms for virus cell-to-cell transfer to macrophages remain unknown. Here, we reveal the mechanisms for cell-to-cell transfer from infected T cells to macrophages and virus spreading between macrophages. We show that contacts between infected T lymphocytes and macrophages lead to cell fusion for the fast and massive transfer of CCR5-tropic viruses to macrophages. Through the merge of viral material between T cells and macrophages, these newly formed lymphocyte-macrophage fused cells acquire the ability to fuse with neighboring noninfected macrophages. Together, these two-step envelope-dependent cell fusion processes lead to the formation of highly virus-productive multinucleated giant cells reminiscent of the infected multinucleated giant macrophages detected in HIV-1-infected patients and simian immunodeficiency virus-infected macaques. These mechanisms represent an original mode of virus transmission for viral spreading and a new model for the formation of macrophage virus reservoirs during infection.
IMPORTANCE We reveal a very efficient mechanism involved in cell-to-cell transfer from infected T cells to macrophages and subsequent virus spreading between macrophages by a two-step cell fusion process. Infected T cells first establish contacts and fuse with macrophage targets. The newly formed lymphocyte-macrophage fused cells then acquire the ability to fuse with surrounding uninfected macrophages, leading to the formation of infected multinucleated giant cells that can survive for a long time, as evidenced in vivo in lymphoid organs and the central nervous system. This route of infection may be a major determinant for virus dissemination and the formation of macrophage virus reservoirs in host tissues during HIV-1 infection.
KEYWORDS: HIV-1, T lymphocytes, macrophages, cell-to-cell transfer, cell fusion
INTRODUCTION
Besides CD4+ T lymphocytes and dendritic cells, macrophages are cellular targets of human immunodeficiency virus type 1 (HIV-1) and play crucial roles in the physiopathology of infection (1–5). The presence of infected macrophages has been evidenced in vivo in HIV-1-infected patients and simian immunodeficiency virus (SIV)-infected macaques, as well as in humanized mice, where macrophages can sustain HIV-1 productive infection and HIV-1 can persist in tissue macrophages even in mice treated with antiretroviral therapy (6–8). Several specialized functions of macrophages, such as cytokine production, phagocytosis, and migration, are affected by HIV-1 infection (9–14). In addition to latently infected CD4+ T cells, infected macrophages also participate in virus dissemination and establishment of persistent virus reservoirs in numerous host tissues, including lymph nodes, spleen, lungs, genital and digestive tracts, and the central nervous system (CNS) (4, 5, 15). Virus access to the CNS is indeed mainly related to the migration of infected perivascular monocytes/macrophages through the blood-brain barrier and can result in a massive infiltration of macrophages and microglial cells, often detected as multinucleated giant cells (MGCs) (1, 16–19).
In vitro, macrophages derived from blood monocytes, which express both CD4 and the CCR5 and CXCR4 coreceptors required for virus entry, can be productively infected by HIV-1 (2, 20). However, almost all the data on macrophage infection and from analyses of the different steps of the virus life cycle in this cell type reported so far were obtained by assays performed using cell-free virus particles. Only one study evaluating the possibility of productive infection of macrophages via the selective capture of healthy or rather dying HIV-1-infected CD4+ T lymphocytes was recently reported (21).
In contrast, efficient virus dissemination in T lymphocytes by cell-to-cell transfer of virus particles between T cells or from infected macrophages or dendritic cells to T cells, mainly through the formation of the so-called virological synapse (22–26) but also through other membrane structures, such as filopodia or nanotubes (27–31), has been documented. This ability of HIV-1 to spread to T cells by cell-to-cell contacts is the major determinant for virus dissemination, as evidenced in vivo in animal models (32, 33). Moreover, these intercellular routes of infection are, at least in vitro, several orders of magnitude more efficient than T cell infection with cell-free virus particles (24, 25). Importantly, this mode of virus dissemination may enable virus to escape elimination by the immune system and antiretroviral drugs (15, 20, 22, 34–37).
Whereas infection of T lymphocytes via cell-to-cell transfer was largely investigated, there is a paucity of knowledge of the mechanisms that control infection and dissemination to macrophages by cell-to-cell transfer (21). However, it is obvious that macrophages can establish tight intercellular contacts with infected T lymphocytes, as observed in some tissues, such as lymphoid organs, colon, and brain, in HIV-infected patients and in SIV-infected monkeys (2, 19, 38). The aim of the present study was to investigate how HIV-1 is transferred from infected T cells to macrophages through cell-to-cell contacts for virus dissemination and productive infection of macrophage targets. We show here, for the first time, that the establishment of close contacts between infected T lymphocytes and macrophage targets leads to heterotypic cell fusion for the fast and efficient transfer of viral material, which subsequently triggers viral envelope-dependent homotypic fusion of macrophages able to generate new infectious particles for the intercellular dissemination of HIV-1. Altogether, our data reveal a new mechanism employed by HIV-1 for spreading between its different cell targets and an original model for the formation of macrophage virus reservoirs during HIV-1 infection.
RESULTS
Productive infection of macrophages through HIV-1 cell-to-cell transfer from infected T cells.
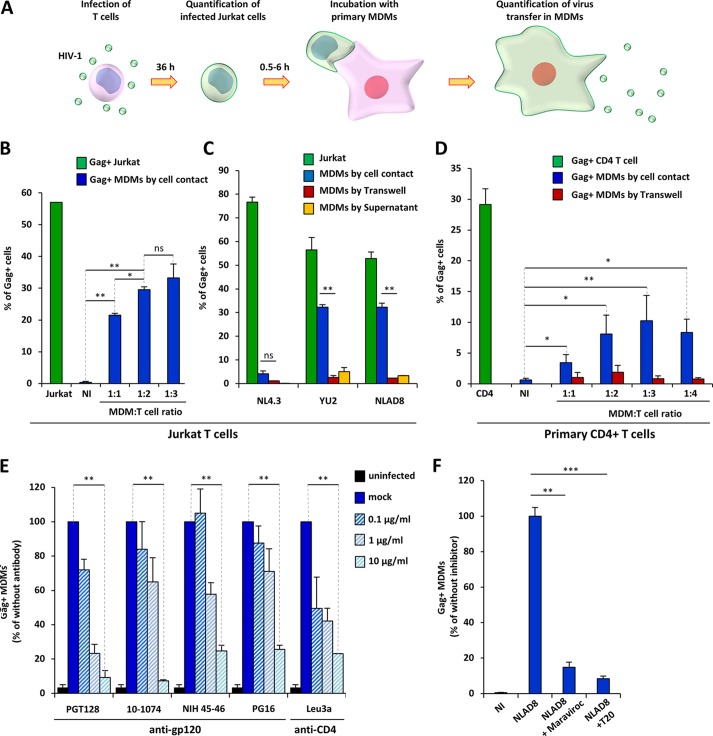
To analyze whether infected T lymphocytes could mediate cell-to-cell virus transfer to macrophages, we used Jurkat cells or purified primary CD4+ T cells as donor infected T cells and macrophages derived from blood monocytes (MDMs) as target cells (Fig. 1A). Jurkat or primary T cells were infected with CCR5-tropic (YU2 and NLAD8 strains) or CXCR4-tropic (NL4.3 strain) viruses (Fig. 1B to D, green bars). T cells that had been infected with HIV-1 for 36 h were cocultured for 6 h with MDMs using different MDM/T cell ratios. After elimination of the T cells by extensive washing (98.5% T cell removal; data not shown), MDMs were fixed and stained for intracellular Gag and cell surface CD11b and CD3, and the percentage of Gag-positive (Gag+) cells among the CD11b+ cells was quantified by flow cytometry to assess the transfer of viral material in MDMs. After 6 h of coculture with YU2- and NLAD8-infected Jurkat or primary T cells, the transfer of viral material was detected in a significant percentage (10 to 40%) of MDMs (Fig. 1B to D, blue bars). By comparison, a very low percentage of transfer of the YU2 and NLAD8 viruses was detected when infected T cells were separated from MDMs by a virus-permeable Transwell membrane (Fig. 1C and D, red bars) or when MDMs were infected with cell-free viruses produced by T cells during the 6 h of coculture (Fig. 1C, yellow bars). Since virus transfer in CD11b+ MDMs was optimal at the 1:2 MDM/T-cell ratio (Fig. 1B and D), this ratio was used in subsequent experiments.
FIG 1.
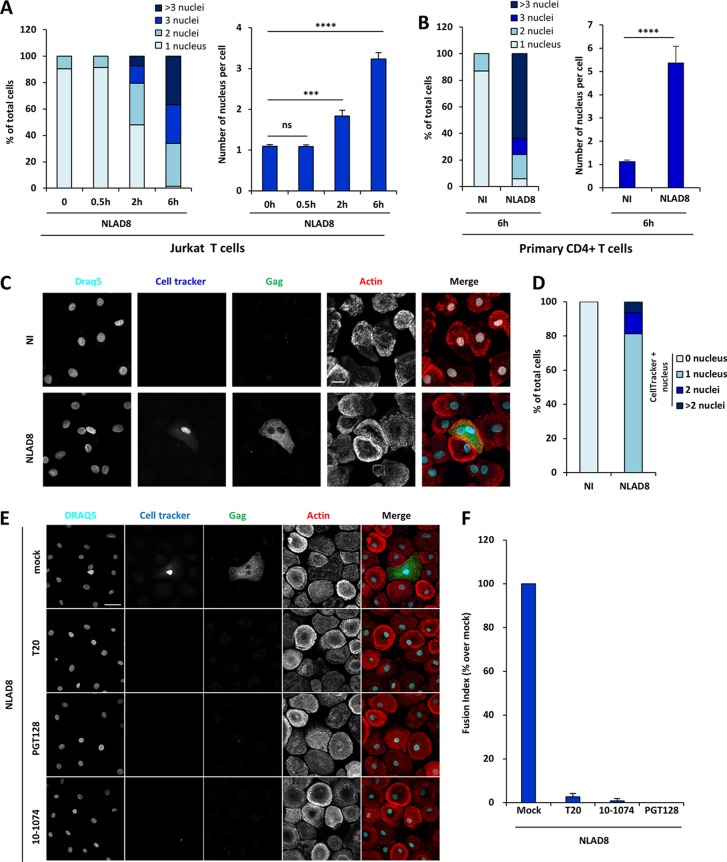
HIV-1 transfer from infected T cells to macrophages. (A) Experimental protocol. (B) Jurkat cells were infected with the NLAD8 strain, and the percentage of infected cells was evaluated 36 h later by flow cytometry after Gag staining (green bars). Infected Jurkat cells were cocultured for 6 h with MDMs at the indicated cell ratio (1:1, 1:2, or 1:3 MDM/Jurkat cell ratio). After elimination of the Jurkat cells, the percentage of CD11b+/Gag+ MDMs was quantified by flow cytometry (blue bars). As a negative control, noninfected (NI) Jurkat cells were cocultured with MDMs. (C) Jurkat cells were infected with the NL4.3, YU2, or NLAD8 strain, and the percentage of infected cells was evaluated 36 h later by flow cytometry (green bars). The infected Jurkat cells were then cocultured with MDMs directly (blue bars) or through a Transwell membrane (red bars) for 6 h. In parallel, culture supernatants from Jurkat cells collected during the 6-h coculture with MDMs were used to infect autologous MDMs (yellow bars). The percentage of CD11b+/Gag+ MDMs was evaluated by flow cytometry. (D) Purified primary CD4+ T cells were infected with the YU2 strain, and the percentage of infected cells was evaluated 36 h later by flow cytometry (green bar). Infected T cells were then cocultured either directly (blue bars) or through a Transwell membrane (red bars) with autologous MDMs using different cell ratios (1:1, 1:2, 1:3, or 1:4 MDM/T cell ratios) for 6 h. After elimination of T cells, the percentage of CD11b+/Gag+ MDMs was quantified by flow cytometry. As a negative control, noninfected CD4+ T cells were cocultured with MDMs. (E and F) NLAD8-infected Jurkat cells were pretreated with anti-gp120 antibodies or T20, while MDMs were pretreated with anti-CD4 or maraviroc. Infected T cells were cocultured with MDMs for 6 h, and virus transfer to MDMs was quantified as described above. The results are expressed as the percentage of Gag+ MDMs relative to the number of Gag+ MDMs determined without antibodies or inhibitors. The results are the means from 5 independent experiments performed with MDMs from 5 donors. Error bars represent 1 SEM. Statistical significance was determined using a paired Student's t test (ns, not significant [P > 0.05]; *, P < 0.05; **, P < 0.01; ***, P < 0.001).
Interestingly, the NLAD8 CCR5-tropic viral strain, which differs from the NL4.3 CXCR4-tropic strain only by substitution of a fragment of the gp120 envelope glycoprotein from the ADA CCR5-tropic strain (39, 40), was efficiently transferred to MDMs, whereas the NL4.3 viral strain was not (Fig. 1C, blue bars). This indicates that the process for virus transfer to MDMs is related to a mechanism dependent on the tropism and coreceptor usage of the viral envelope (Env). To confirm that HIV-1 Env contributed to virus transfer, we tested various viral entry inhibitors, such as anti-gp120 neutralizing antibodies, an anti-CD4 Leu3 antibody, the fusion inhibitor T20, and the CCR5 antagonist maraviroc. As shown in Fig. 1E and F, all the entry inhibitors blocked virus transfer to MDMs, demonstrating that virus cell-to-cell transfer to MDMs is Env dependent.
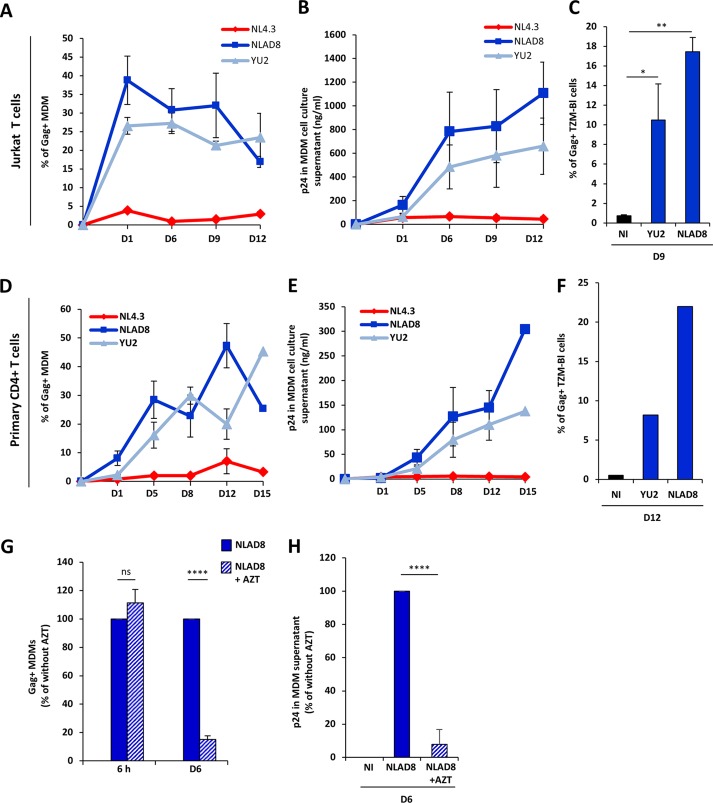
We next investigated whether the transfer of viral material leads to the productive infection of MDMs. Infected Jurkat or primary CD4+ T cells were cocultured for 6 h with MDMs as described above and eliminated by washing, and the percentage of Gag+ MDMs was evaluated 1, 6, 9, 12, and 15 days later (Fig. 2A and D). Again, a very low level of virus transfer was observed with NL4.3-infected Jurkat cells, whereas both the YU2 and NLAD8 macrophage-tropic strains propagated efficiently in MDMs, as evidenced by the level of Gag+ MDMs detected during the 15-day observation period after the initial transfer from infected T cells. As expected, high levels of viral p24 production were detected in the supernatants from MDMs cocultured with YU2- and NLAD8-infected T cells (Fig. 2B and E). We checked that the virions produced by MDMs were fully infectious by infecting the TZM-bl reporter cell line (Fig. 2C and F). Additionally, virus dissemination and production in MDMs were inhibited by the reverse transcriptase inhibitor zidovudine (AZT) (Fig. 2G, day 6, and H) without affecting the initial virus transfer to MDMs (Fig. 2G, 6 h).
FIG 2.
Productive infection of macrophages by virus cell-to-cell transfer from infected T cells. (A to F) NLAD8-infected Jurkat cells (A to C) or primary CD4+ T cells (D to F) were cocultured with MDMs for 6 h and eliminated, and the percentage of CD11b+/Gag+ MDMs was then evaluated 1, 6, 9, 12, or 15 days later by flow cytometry (A and D). In parallel, cell culture supernatants from MDMs were collected and p24 was quantified (B and E). Culture supernatants (100 ng of p24) of MDMs collected 9 days (C) or 12 days (F) after the coculture with YU2- or NLAD8-infected Jurkat (C) or CD4+ T (F) cells were used to infect TZM-bl cells, and the percentage of Gag+ TZM-bl cells was evaluated 48 h later by flow cytometry. (G and H) NLAD8-infected Jurkat cells were cocultured for 6 h with MDMs pretreated or not pretreated with AZT. (G) The percentage of CD11b+/Gag+ MDMs was then evaluated just after coculture (6 h) and 6 days later. (H) In parallel, culture supernatants of MDMs were collected 6 days after coculture and p24 was quantified. The results shown in panels A, B, D, E, G, and H are the means from 5 independent experiments performed with MDMs from 5 donors, while the results shown in panels C and F are representative of those from 3 independent experiments. Error bars represent 1 SEM. Statistical significance was determined using one-way analysis of variance (ns, not significant [P > 0.05]; *, P < 0.05; **, P < 0.01; ****, P < 0.0001). D, day.
Visualization of cell contacts and virus transfer between infected T cells and macrophages.
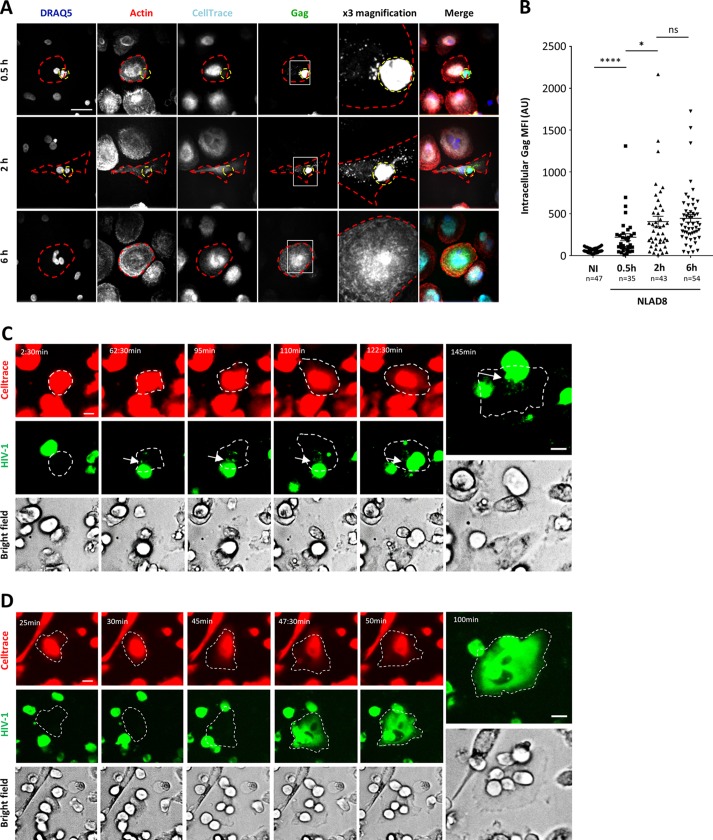
To visualize virus transfer, fluorescence microscopy was performed first using infected Jurkat cells cocultured for 30 min, 2 h, or 6 h with MDMs preloaded with the CellTrace Far Red dye before fixation and intracellular Gag staining. As shown in Fig. 3A and B, infected T cells established contacts with MDMs, leading to the visualization of Gag+ dots in the cytoplasm of the MDMs after 30 min of coculture (Fig. 3A, top row, 0.5 h). Virus transfer subsequently resulted in the accumulation of larger Gag+ puncta (Fig. 3A, middle row, 2 h). Intriguingly, a diffuse cytoplasmic staining was observed in almost all Gag+ MDMs after 6 h of coculture (Fig. 3A, bottom row, 6 h); at this time point, all the Gag+ MDMs contained at least 2 nuclei. In agreement with this finding, fluorescence quantification from images showed a significant increase in the level of intracellular Gag staining of MDMs from 0.5 h to 6 h of coculture (Fig. 3B). Similarly, infected primary CD4+ T cells also established contacts with autologous MDMs (leading to the accumulation of intracytoplasmic Gag+ dots in MDMs after 2 h of coculture) (data not shown), exhibited a diffuse cytoplasmic Gag staining, and contained several nuclei after 6 h of coculture with infected T cells (see Fig. 6G).
FIG 3.
Fluorescence microscopy analysis of intercellular contacts and virus transfer between infected T cells and macrophages. (A and B) NLAD8-infected Jurkat cells were cocultured for 0.5, 2, or 6 h with MDMs prestained with CellTrace. Cells were then fixed, stained with anti-Gag, phalloidin, and DRAQ5, and analyzed by confocal microscopy. (A) Images of a 5-μm-thick medial stack are shown. Infected donor T cells and MDM targets are indicated by dashed yellow and red lines, respectively. The areas in the white squares in the Gag column were magnified three times, and the images are shown in the ×3 magnification column. Bar, 25 μm. (B) The intracellular Gag mean fluorescence intensity (MFI) was quantified as indicated in the Materials and Methods section. Each dot corresponds to 1 cell, and the number of cells analyzed (n) is indicated. Horizontal bars represent the mean ± 1 SEM. Statistical significance was determined by an unpaired t test (ns, not significant [P > 0.05]; *, P < 0.05; ****, P < 0.0001). AU, absorbance units. (C) Jurkat cells infected with HIV-1R5-GFP (green) were cocultured with MDMs that had previously been plated onto an Ibidi dish and labeled with CellTrace (red). Fluorescence images were acquired using a 20× air objective on a spinning-disk microscope every 2.5 min for 145 min. A 5-μm-thick medial stack of representative images is shown, and the time lapse is indicated. Bars, 25 μm. The discharge of viral material (arrows) into MDMs (dashed lines) from infected T cells is shown. (D) Jurkat cells infected with HIV-1R5-GFP (green) were cocultured with MDMs that had previously been labeled with CellTrace (red). Fluorescence images were then acquired using a 20× air objective on a spinning-disk microscope every 2.5 min for 100 min. A 5-μm-thick medial stack of representative images is shown, and the time lapse is indicated. Bars, 25 μm. The labeled MDM target is indicated with a dashed line.
FIG 6.
Macrophages express T cell-specific markers after fusion with infected T cells. NLAD8-infected Jurkat cells (A to F) or primary CD4+ T cells (G, H) were cocultured for 6 h with MDMs. After elimination of T cells, MDMs were stained with anti-CD2 (A and B) and anti-CD3 (G and H) before permeabilization. The cells were then permeabilized and stained with anti-Gag, anti-CD3 (C and D), anti-Lck (E and F), and DAPI. Representative images of cell surface CD2 (A), CD3 (C and G), and Lck (E) staining are shown. Bars, 25 μm. Cell surface CD2 (B) or CD3 (H) and intracellular CD3 (D) or Lck (F) mean fluorescence intensities were quantified as indicated in the Materials and Methods section. Each dot corresponds to 1 cell, and the number of cells analyzed (n) is indicated. Horizontal bars represent the mean ± 1 SEM. Statistical significance was determined by the Mann-Whitney U-test (****, P < 0.0001).
We also performed live-cell imaging using Jurkat cells infected with fluorescent HIV-1–green fluorescent protein (GFP) and cocultured with MDMs preloaded with CellTrace. As shown in Fig. 3C (see also Movie S1 in the supplemental material), we could visualize contacts between infected T cells and MDMs, showing a continuous discharge of fluorescent viral material (white arrows) into MDMs during the first hour of coculture. Interestingly, longer live-cell experiments showed a rapid and massive diffusion of the virus-associated fluorescence from infected T cells to the cytoplasm of MDMs, strongly suggesting that these cell contacts lead to the fusion of infected T cells with the MDM targets (Fig. 3D and Movie S2). Since our experiments were carried out 36 h after T cell infection, when less than 2% of infected T cells were apoptotic (data not shown), we did not observe phagocytosis of infected T cells by MDMs, as recently reported (21). In addition, virus transfer from infected T cells to macrophages was not significantly affected by high concentrations of latrunculin A, an actin-binding drug that prevents F-actin polymerization and that is known to totally inhibit phagocytosis at a low concentration, in our experimental system (data not shown). Of note, we did not observe the formation of Gag+ T cell syncytia in our experimental system.
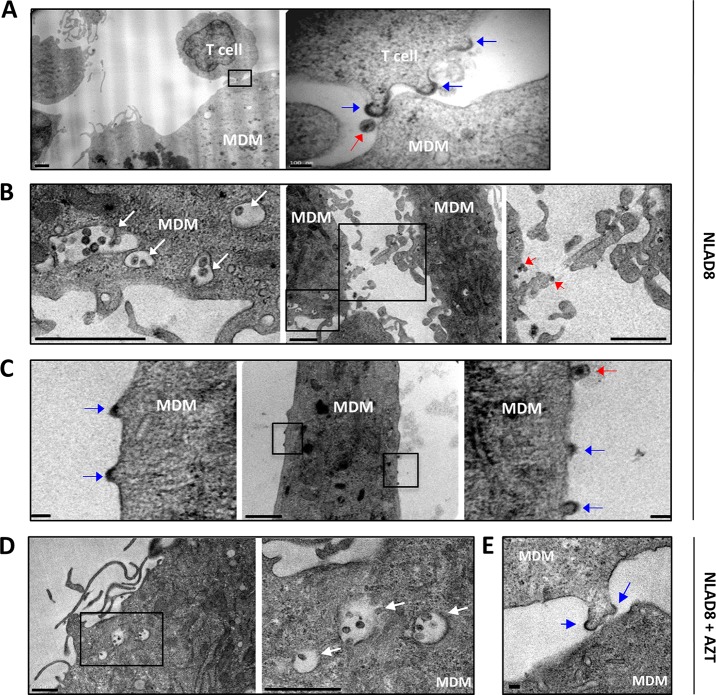
Transmission electron microscopy was also used to visualize the contacts between infected T cells and MDMs after 6 h of coculture. As shown in Fig. 4A, electron-dense material corresponding to viral buds protruding from the plasma membrane of the donor T cell (blue arrows), as well as mature virions (red arrow), was observed at the site of contact with MDMs. Interestingly, we also observed the accumulation of mature viruses in cytoplasmic membrane compartments of MDMs (Fig. 4B, white arrows). More surprisingly, electron-dense material reminiscent of Gag assembly and virus budding (blue arrows) and mature virions (red arrows) were observed at the plasma membrane of MDMs as well as between MDMs which established tight contacts (Fig. 4B and C), suggesting that virus assembly and budding took place at the cell surface of MDMs only 6 h after coculture with infected T cells, before de novo virus production. In agreement with this finding, virus-containing cytoplasmic compartments as well as plasma membrane viral buds were still observed when MDMs were treated with AZT during coculture with infected T cells (Fig. 4D and E). In contrast, we could not observe such virus-containing compartments or viral buds 6 h after infection of MDMs with cell-free viruses (data not shown). These observations could result from the fusion of infected T cells with MDMs revealed by live-cell imaging, suggesting plasma membrane exchanges and the merge of Gag and Env material between infected T cells and MDM targets.
FIG 4.
Transmission electron microscopy analysis of virus transfer to macrophages. NLAD8-infected Jurkat cells were cocultured for 6 h with MDMs with (D and E) or without (A to C) AZT (5 μM). Cells were then fixed and dehydrated. Ultrathin sections were cut, stained, and observed with a transmission electron microscope. Blue arrows, assembling and budding viruses; red arrows, mature virions; white arrows, MDM cytoplasmic membrane compartments containing mature viruses (B and D). In panel A, the right image (bar, 100 nm) corresponds to a higher magnification of the boxed area in the left image (bar, 1 μm). In panel B, the left and right images (bars, 1 μm) correspond to higher magnifications of the small and larger boxed areas in the middle image (bar, 1 μm), respectively. In panel C, the left and right images (bars, 100 nm) correspond to higher magnifications of the left and right boxed areas in the middle image (bar, 1 μm), respectively. In panel D, the right image (bar, 1 μm) corresponds to a higher magnification of the boxed area in the left image (bar, 1 μm). The bar in panel E is 100 nm. The images shown are representative of those obtained from analysis of MDMs from 3 independent donors.
Virus transfer to macrophages through heterotypic cell fusion with infected T cells.
Because we observed from immunofluorescence images that almost all Gag+ MDMs contained at least 2 nuclei after virus transfer (Fig. 3A, bottom row, 6 h), we first quantified the number of nuclei in MDMs after coculture with infected Jurkat cells (Fig. 5A). Before coculture, more than 90% of the MDMs contained 1 nucleus, whereas about 50% of the Gag+ MDMs contained at least 2 nuclei (mean nucleus number = 1.83) after 2 h of coculture, and this percentage increased to 99% (mean nucleus number = 3.24) after 6 h (Fig. 5A). Similarly, MDMs cocultured for 6 h with autologous infected primary CD4+ T cells contained several nuclei (mean nucleus number = 5.36; Fig. 5B), suggesting that these Gag+ multinucleated MDMs are generated through cell fusion events with infected T cells, as documented by live-cell imaging analysis (Fig. 3D and Movie S2).
FIG 5.
Virus transfer to macrophages by cell fusion with infected T cells. (A and B) NLAD8-infected Jurkat (A) or primary CD4+ T (B) cells were cocultured with MDMs for 0.5, 2, or 6 h. After elimination of T cells, MDMs were stained with anti-Gag, phalloidin, and DAPI. Cells were analyzed by confocal microscopy, and the number of nuclei per cell was analyzed from images of at least 50 cells. The results are expressed as the percentage of cells with 1, 2, 3, or more than 3 nuclei (left) and as the mean nucleus number per cell (right). Error bars represent 1 SEM. Statistical significance was determined by the Mann-Whitney U test (ns, not significant [P > 0.05]; ***, P < 0.001; ****, P < 0.0001). (C and D) NLAD8-infected Jurkat cells that had been prelabeled with CellTracker were cocultured for 6 h with MDMs. (C) After elimination of T cells, MDMs were fixed, stained with anti-Gag, phalloidin, and DRAQ5, and analyzed by confocal microscopy. Bar, 25 μm. (D) The number of CellTracker-positive (CellTracker+) nuclei per cell was analyzed from images of at least 50 cells. The results are expressed as the percentage of cells with 1, 2, or more than 2 CellTracker-positive nuclei. (E and F) NLAD8-infected Jurkat cells that had been prelabeled with CellTracker were pretreated with anti-gp120 antibodies (PGT128 or 10-1074) or T20 for 1 h and cocultured with MDMs for 6 h in the presence of the inhibitors. Cells were then fixed, permeabilized, stained with anti-Gag, phalloidin, and DRAQ5, and analyzed by confocal microscopy. (E) Images were acquired and processed as described in the text. Bar, 25 μm. (F) The number of CellTracker-positive nuclei per DRAQ5-positive MDM was analyzed from images of at least 1,200 cells for each condition. The results are expressed as the fusion index, corresponding to the percentage of cells containing at least 1 CellTracker-positive nucleus relative to the number of NLAD8-infected Jurkat cells cocultured with MDMs without drugs with at least 1 CellTracker-positive nucleus. The results shown are representative of those from 4 independent experiments performed with MDMs from 4 donors. NI, noninfected Jurkat or primary CD4+ T cells cocultured with MDMs.
To test this hypothesis, infected Jurkat cells were preloaded with the CellTracker 7-amino-4-chloromethylcoumarin (CMAC) dye and cocultured with MDMs as described above. After 6 h of coculture, all Gag+ MDMs contained several nuclei, including at least 1 CellTracker dye-stained nucleus (Fig. 5C and D), demonstrating cell fusion between infected T cells and MDMs. This cell fusion between infected T cells and MDMs was mediated by viral envelope-receptor interactions, since it was totally blocked by anti-gp120 neutralizing antibodies and the fusion inhibitor T20 (Fig. 5E and F). Finally, immunofluorescence staining confirmed that all Gag+ MDMs contained membrane- and cytoplasm-specific T cell markers, such as CD3, CD2, and Lck, after 6 h of coculture with infected Jurkat cells (Fig. 6A to F) or primary CD4 T cells (Fig. 6G to H).
Together, these results demonstrate that HIV-1 is mainly transferred from infected T cells to macrophages through a heterotypic envelope-dependent cell fusion process leading to the formation of lymphocyte-macrophage fused cells (LMFCs).
Virus dissemination through homotypic cell fusion between macrophages.
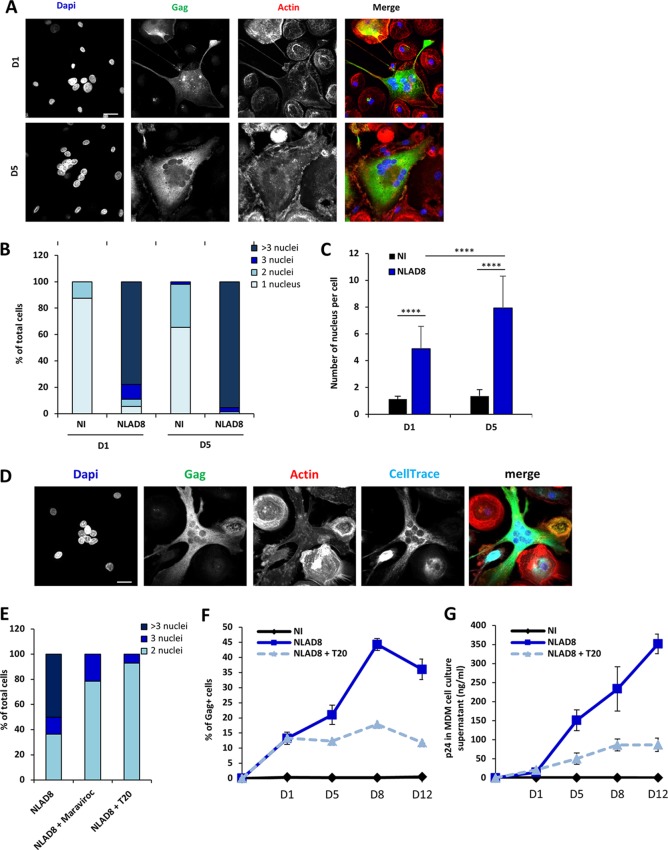
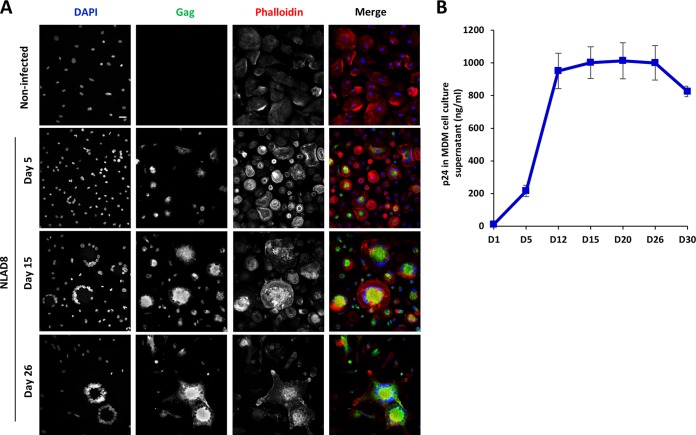
Interestingly, since we observed that a large majority of the Gag+ newly formed LMFCs contained more than 2 nuclei after 6 h of coculture (Fig. 5A and B), we explored whether LMFCs could fuse with surrounding MDMs, leading to the formation of Gag+ multinucleated giant cells. After coculture for 6 h with infected Jurkat cells to allow initial cell fusion and virus transfer, LMFCs cultured for 1 or 5 days after elimination of infected T cells indeed contained more than 2 nuclei, with the average nucleus number being 4.8 and 7.9, respectively (Fig. 7A to C). To confirm LMFC-MDM fusion, infected T cells were cocultured for 6 h with MDMs and eliminated, and autologous noninfected MDMs preloaded with the CellTrace dye were added. Twenty-four hours later, Gag+ MDMs with several nuclei were also labeled with CellTrace (Fig. 7D), resulting from the fusion of Gag+ LMFCs with neighboring uninfected MDMs to form Gag+ multinucleated giant cells (MGCs). Fusion of Gag+ MDMs with surrounding MDMs was also observed when prelabeled noninfected MDMs were added 5 days after coculture and elimination of infected T cells (data not shown). Since the initial fusion with infected T cells led to the presence of T cell-specific markers at the cell surface of LMFCs (Fig. 6), we hypothesized that the latter cells could also express the viral envelope and explored whether the fusion of Gag+ LMFCs with MDMs was mediated by viral envelope-receptor interactions. Following the 6-h coculture to allow initial cell fusion and virus transfer, infected T cells were eliminated and MDMs were cultured in the presence of the fusion inhibitor T20 and the CCR5 antagonist maraviroc. As expected, about 60% of Gag+ LMFCs cultured without inhibitors contained at least 3 nuclei 1 day later, but this percentage decreased to 20 and 10% when LMFCs were cultured with maraviroc and T20, respectively (Fig. 7E). Importantly, inhibition of LMFC fusion with neighboring MDMs by T20 after initial virus transfer led to a net decrease in virus dissemination (Fig. 7F) and virus production by LMFCs (Fig. 7G). In agreement with this finding, infected MGCs with many nuclei could be still detected after 26 days of culture (Fig. 8A) and were indeed still able to produce high levels of p24 30 days after elimination of infected T cells (Fig. 8B). These results show that the LMFC-MDM cell fusion is required for optimal virus spreading and production by long-lived multinucleated giant cells.
FIG 7.
Viral dissemination between macrophages by homotypic cell fusion. (A to C) NLAD8-infected Jurkat cells were cocultured with MDMs for 6 h. After elimination of T cells, MDMs were cultured for 1 or 5 more days and then stained with anti-Gag, phalloidin, and DAPI. (A) Cells were analyzed by confocal microscopy. Bar, 25 μm. (B and C) The number of nuclei per MDM was quantified from images of at least 50 cells. The results are expressed as the percentage of cells with 1, 2, 3, or more than 3 nuclei (B) and as the mean nucleus number per cell (C). Error bars represent 1 SEM. Statistical significance was determined by the Mann-Whitney U-test (****, P < 0.0001). (D) Infected Jurkat cells were cocultured with MDMs for 6 h. After elimination of T cells, autologous MDMs prelabeled with CellTrace were added and cultured for 1 day. MDMs were then stained with anti-Gag, phalloidin, and DAPI. Cells were analyzed by confocal microscopy. Bar, 25 μm. (E) Infected Jurkat cells were cocultured with MDMs for 6 h. After elimination of T cells, MDMs were cultured for 1 day with or without T20 (10 μg/ml) or maraviroc (10 μM) before staining with anti-Gag, phalloidin, and DAPI. Cells were analyzed by confocal microscopy. The number of nuclei was analyzed from images of at least 50 cells. The results are expressed as the percentages of cells with 2, 3, or more than 3 nuclei. (F and G) Infected Jurkat cells were cocultured with MDMs for 6 h. After elimination of T cells, MDMs were cultured for 1, 5, 8, or 12 days with or without T20 (10 μg/ml). (F) The percentage of Gag+ MDMs was then evaluated by flow cytometry. (G) In parallel, culture supernatants from MDMs were collected and p24 was quantified. The results shown in panels A to E are representative of those from 4 independent experiments performed with MDMs from 4 donors, while the results shown in panels F and G correspond to the means from 3 independent experiments performed with MDMs from 3 donors. NI, noninfected Jurkat cells cocultured with MDMs.
FIG 8.
Formation and survival of infected multinucleated giant cells. NLAD8-infected Jurkat cells were cocultured with MDMs for 6 h and eliminated, and MDMs were then cultured for the indicated periods of time. (A) Cells were fixed, permeabilized, and stained with anti-Gag, phalloidin, and DAPI. Cells were analyzed by confocal microscopy, and images were acquired and processed as described in the text. Bar, 25 μm. (B) Culture supernatants of MDMs were collected at the indicated days after coculture, and p24 was quantified. The results correspond to the means from 3 independent experiments. Error bars represent 1 SEM. Noninfected, noninfected Jurkat cells cocultured with MDMs.
DISCUSSION
In the present study, we reveal the mechanisms involved in the rapid and massive cell-to-cell transfer of HIV-1 from infected T cells to MDMs and the subsequent virus spreading between MDMs by a two-step cell fusion process leading to the productive infection of MDM targets. Both cell fusion steps are mediated by viral envelope-receptor interactions at the cell surface of T cells and MDMs and are completed in less than 2 h. This route of infection may be a major determinant in vivo for virus dissemination to macrophages.
As evidenced by fluorescence microscopy analyses, including live-cell imaging, the first step is related to the establishment of contacts with infected T cells, resulting in the fusion of infected T cells with MDM targets. This cell fusion process is evidenced by the massive and rapid transfer of Gag+ material as well as membrane and cytosolic T cell-specific markers, such as CD2, CD3, and Lck, which thus ensures efficient virus dissemination between these two important target cells of HIV-1. We did not observe in our experimental system the formation of HIV-1-induced T cell syncytia using either Jurkat cells or CD4+ primary T cells as virus donor cells, suggesting that HIV-1-induced cell-to-cell fusion could be restricted to myeloid cell targets, such as macrophages, and inhibited between T cells (41–45). In agreement with this first step of T cell-to-MDM fusion for virus transfer, it was shown that myeloid cells from lymphoid tissues of SIV-infected macaques, such as spleen and lymph nodes, contain T cell markers and viral RNA and DNA originating from infected T cells (46, 47).
Electron microscopy analysis confirmed that infected T cells establish contacts with macrophages, with evidence of virus assembly at the site of cell-to-cell contacts, leading to virus transfer and the accumulation of mature virus particles in cytoplasmic membrane compartments in macrophages after only 6 h of coculture with infected T cells, before de novo virus production by the macrophage targets. We hypothesize that the mature and immature viruses found in these early-formed virus-containing compartments (VCCs) could result from the initial discharge of viral material before cell fusion, observed by fluorescence microscopy on both fixed and lived cells. Such early-formed VCCs have been observed in target T cells following virus cell-to-cell transfer in enclosed endocytic compartments from infected donor T cells (48–51). The formation of VCCs has also been largely documented when macrophages were infected with cell-free viruses, but they appeared later, at least 5 to 6 days after infection, and contained newly synthesized virus particles (52–57). While the exact mechanisms for the formation of these early and late VCCs need to be further investigated, such VCCs could participate in the long-term storage of HIV-1 in tissue macrophages and lead to the establishment of viral reservoirs for virus maintenance and spreading (1–5). Interestingly, some events of assembly and budding of virus particles were detected at the plasma membrane of the macrophage targets before de novo virus production after only 6 h of coculture with infected T cells, suggesting that the newly formed Gag+ lymphocyte-macrophage fused cells also expose the viral envelope at their cell surface. These cells then acquire the ability to fuse with neighboring uninfected MDMs, leading to the formation of multinucleated giant cells that can survive and produce large amounts of fully infectious viruses even after 30 days of cell culture. Similarly, the formation of HIV-1-infected giant cells was reported when macrophages were infected with cell-free viruses (14, 58), but they appeared several days after infection. Despite their large size, these infected multinucleated macrophages migrate faster than their infected mononucleated counterpart and may participate in virus dissemination (13). More importantly, these Gag+ multinucleated giant cells observed in vitro are reminiscent of the infected multinucleated giant macrophages detected in vivo in lymphoid organs and the CNS of HIV-1-infected patients and SIV-infected macaques (3, 17, 59–64).
We found that virus transfer to macrophages through initial T cell fusion and subsequent virus dissemination in multinucleated giant macrophages were restricted to macrophage-tropic CCR5-using viral strains. In contrast, Baxter et al. (21) reported that internalization of healthy or dying infected T cells by MDMs was related to a nonconventional mechanism independent of the viral envelope and showed that the NL4.3 CXCR4-tropic strain could be efficiently transferred to macrophages using GFP-tagged NL4.3-infected T lymphocytes as donor cells. This discrepancy could be related to the use of nondying T cells infected with the untagged wild-type replication-competent NL4.3 strain to analyze transfer in our experimental system. While initial studies suggested that virus transfer between CD4+ T cells at the virological synapse was independent of coreceptor usage (65, 66), subsequent reports showed that this transfer required coreceptor expression and was inhibited by coreceptor antagonists (24, 49, 67, 68). Here, we found that the initial virus transfer to macrophages was inhibited by neutralizing monoclonal antibodies targeting the HIV-1 gp120 envelope glycoprotein as well as by the Leu3a antibody targeting the CD4 receptor, indicating that early interactions between T cells and recipient macrophages involve recognition of CD4 by gp120. Virus transfer to macrophages leading to the formation of infected lymphocyte-macrophage fused cells was also blocked by the fusion inhibitor T20, targeting the viral transmembrane gp41 envelope glycoprotein, and the CCR5 antagonist maraviroc. These results indicate that virus transfer and T cell fusion with macrophages are mediated by initial viral envelope-receptor and -coreceptor interactions. In addition, we show that the second cell fusion step between Gag+ lymphocyte-macrophage fused cells and neighboring MDMs, required for virus dissemination and virus production by multinucleated giant cells, is also dependent on the viral envelope, since it is inhibited by T20 and maraviroc. While our data clearly show that both cell fusion processes for virus transfer and dissemination in macrophages are dependent on the viral envelope, we cannot exclude the possibility that other mechanisms related to the propensity of macrophages to mediate homotypic cell fusion could also participate in the cell fusion processes for virus transfer and dissemination. It would be interesting to investigate the potential role in HIV-1-mediated T cell-MDM and MDM-MDM fusions of some effector proteins known to be involved in programming of macrophages into a fusion-competent state and then in cell fusion for the formation of multinucleated giant macrophages (for a review, see reference 69).
Our analyses indicate that this cell-to-cell transfer of virus from infected T cells to macrophages is largely more efficient than virus infection with cell-free viruses and is certainly the most potent experimental system to infect macrophages in vitro described so far (1–5). Indeed, we failed to detect a significant transfer of viral material when macrophages were directly infected with the cell-free virus particles released by infected donor T cells during the time of coculture with macrophages. In sharp contrast, virus transfer from the infected CD4+ T cells evaluated here resulted in a robust productive infection of macrophages, as evidenced by the high levels of virus production in the supernatant of the multinuclear giant macrophages detected by monitoring of these macrophages for 30 days.
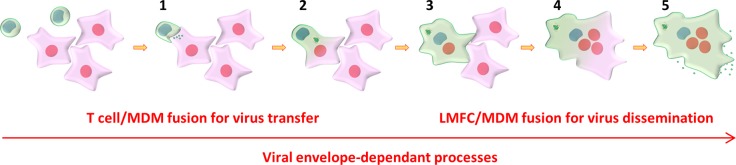
In summary, our results show that HIV-1 can be efficiently transferred to macrophages from infected donor T cells, without implying internalization of infected CD4+ T cells by the macrophage targets (21). While the two mechanisms are not mutually exclusive, we reveal that a novel fast and very efficient mechanism is involved in cell-to-cell transfer from nondying infected T cells to macrophages and subsequent virus spreading between macrophages by a two-step cell fusion process (the model is shown in Fig. 9). In the first step, infected T cells establish tight contacts and initially discharge viral material to MDMs, resulting in the fusion of infected T cells with MDM targets. The newly formed Gag+ lymphocyte-macrophage fused cells then acquire the ability to fuse with surrounding noninfected MDMs, leading to the formation of infected multinucleated giant cells that can survive for a long time in host tissues to produce infectious virus particles, as has been shown in vivo in lymphoid organs and the CNS of HIV-1-infected patients and SIV-infected macaques (3, 17, 59–64). Similarly, the first step related to the initial T cell-to-MDM fusion agrees with results showing that myeloid cells from lymphoid tissues of SIV-infected macaques contain T cell markers and viral DNA originating from infected T cells (46, 47). These in vivo observations support the importance of the molecular mechanisms revealed here, which contribute to a better understanding of virus dissemination from infected T cells to macrophages and the formation of long-lived macrophage viral reservoirs in host tissues.
FIG 9.
Model for virus cell-to-cell transfer from infected T cells to MDMs and virus spreading between MDMs. Initial virus transfer and subsequent virus spreading are mediated by a two-step cell fusion process. In the first step, infected T cells establish contacts, initially discharge viral material to MDMs (step 1), and then fuse with MDM targets (step 2), with accumulation of viruses in intracytoplasmic compartments and virus assembly and budding at the cell surface. Gag+ newly formed LMFCs (step 3) then acquire the ability to fuse with surrounding uninfected MDMs, leading to the formation of Gag+ multinucleated giant cells (step 4) that could survive for a long time to produce infectious viruses (step 5).
MATERIALS AND METHODS
Plasmids and reagents.
The proviral plasmids pNL4-3 and pNLAD8 were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID. The proviral plasmid HIV-1R5-GFP was a gift of Michael Schindler (Munich, Germany) (70), while pYU-2 and the plasmid encoding the vesicular stomatitis virus (VSV) envelope G glycoprotein (pVSVg) have been described previously (71). The following antibodies were used: phycoerythrin (PE)- or fluorescein isothiocyanate (FITC)-conjugated anti-CD11b (clone ICRF44; BD Biosciences), PE-Cy7-conjugated anti-CD11b (clone ICRF44; BioLegend), RD1- or FITC-conjugated anti-Gag (clone KC57; Beckman Coulter), anti-CD4 (clone Leu3a; BioLegend), anti-CD3 (clone UCHT1; BioLegend), and anti-CD2 (TS2/18 clone; a gift from Andres Alcover, Paris, France [72]). Alexa Fluor 647-conjugated phalloidin (Life Technologies) was also used. The following reagents were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID: anti-gp120 antibodies (PG16 and NIH45-46), HIV-1 CAp24 hybridoma (183-H12-5C), HIV-Ig, maraviroc, T20, and AZT. Anti-gp120 antibodies (10-1074 and PGT128) were gifts from Paul Zhou (Shanghai, China).
Cell culture.
Cells of the HEK293T, TZM-bl, and Jurkat cell lines were obtained from the ATCC collection. HEK293T and TZM-bl cells were maintained in Dulbecco minimal essential medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 IU of penicillin/ml, and 100 μg of streptomycin/ml (ATB; Invitrogen). Jurkat cells were maintained in RPMI 1640 complete culture medium supplemented with 10% FCS and ATB. Peripheral blood mononuclear cells (PBMCs) were isolated from the blood of healthy anonymous donors by density gradient sedimentation using Histopaque (Sigma-Aldrich), and monocytes were purified using a CD14-positive selection kit (CD14 microbeads, Miltenyi) according to the manufacturer's guidelines. Blood samples from anonymous healthy donors were purchased from the Etablissement Français du Sang Paris-Saint-Antoine-Crozatier, Paris, France. The monocytes were differentiated into macrophages for 8 days in RPMI 1640 culture medium supplemented with 20% FCS, ATB, and 25 ng/ml of granulocyte-macrophage colony-stimulating factor and granulocyte-macrophage colony-stimulating factor (Miltenyi). Human primary CD4+ T cells were isolated from PBMCs by density gradient sedimentation using Histopaque and then purified by negative selection (CD4+ T cell isolation kit; Miltenyi) following the manufacturer's recommendation. CD4+ T cells were activated for 3 days in RPMI 1640 medium containing 20% fetal bovine serum (FBS), interleukin-2 (IL-2; Miltenyi) at 10 U/ml, and phytohemagglutinin-P (PHA-P; Sigma-Aldrich) at 5 μg/ml. After activation, the CD4+ T cells were kept in RPMI 1640 medium supplemented with 20% FBS and IL-2. All cells were grown at 37°C under 5% CO2.
Viral production, titration, and infection.
Replication-competent HIV-1 strains YU2, NL4.3, and NLAD8 and HIV-1R5-GFP were produced in HEK293T cells by cotransfection of the proviral plasmid in combination with pVSVg using the calcium phosphate precipitation technique as described previously (71). The amounts of CAp24 produced were determined by enzyme-linked immunosorbent assay (ELISA; Innogenetics). The viral titer was determined by flow cytometry (Accuri C6; BD Biosciences) using Jurkat cells (J77 clone) as described previously (71).
Virus transfer and dissemination.
To study the transfer from Jurkat or primary CD4+ T cells to MDMs, T cells were infected using a multiplicity of infection (MOI) of 0.5 for 16 h. The cells were then washed and cultured for 20 h. After washing, the T cells were cocultured at a ratio of 2:1 (unless stated otherwise in the text or figure legends) with MDMs, seeded at a density of 0.5 × 106 cells/well, for 0.5 to 6 h. To remove T cells, MDMs were then washed extensively with phosphate-buffered saline (PBS) once, with PBS containing 10 mM EDTA, and then 2 more times with PBS. MDMs were harvested or cultured for several days and then collected. Cells were then surface stained using anti-CD11b and anti-CD3 antibodies and then fixed with 4% paraformaldehyde (PFA), permeabilized using permeabilization buffer (PBS, 1% BSA, 0.1% Triton X-100), and stained with an anti-Gag (KC57, 1/500). The percentage of Gag+ cells among the CD11b+ cells corresponding to the MDM population was determined by flow cytometry. To analyze viral production, MDM culture supernatants were collected, and the amount of CAp24 produced was determined by ELISA. For MDM infection with cell-free viruses, T cells were infected as described above and incubated with MDMs for 6 h through a 0.4-μm-pore-size Transwell membrane. Alternatively, the supernatant of producer Jurkat cells corresponding to the 6-h coculture was added to the MDMs, and the percentage of Gag+ cells was analyzed by flow cytometry as described above.
Infectivity assay.
A total of 2 × 105 TZM-bl cells (HeLa-CD4 cells stably expressing CD4 and CCR5) were incubated with viral supernatants (200 ng of CAp24) for 18 h. The TZM-bl cells were then cultured for 2 days, fixed with 3.7% formaldehyde (Sigma), and treated with permeabilization buffer. The cells were stained with an anti-Gag antibody, and the percentage of Gag+ cells was determined by flow cytometry.
Effect of inhibitors on virus transfer and dissemination.
To analyze the effect of inhibitors on virus transfer, infected donor T cells or MDM targets were pretreated for 1 h with anti-gp120 neutralizing antibodies (PGT128, 10-1074, NIH 45-46, and PG16) or anti-CD4 (Leu3a), respectively, using 3 concentrations (0.1, 1, or 10 μg/ml) of antibodies in the presence of 10 μg/ml of Fc-Block reagent (Sigma). Infected T cells were then cocultured for 6 h with MDMs and removed by washings, and the percentage of Gag+/CD11b+ MDMs was determined by flow cytometry. Results were expressed as the percentage of Gag+ MDMs relative to the number of Gag+ MDMs detected without antibody treatment. The effects of the CCR5 receptor antagonist maraviroc as well as the fusion inhibitor T20 on virus transfer were also tested. Infected donor T cells or MDM targets were pretreated for 1 h with T20 or maraviroc at 10 μM and 10 μg/ml, respectively. Infected T cells were then cocultured for 6 h with MDMs and removed, and the percentage of Gag+/CD11b+ MDMs was determined by flow cytometry. Results were expressed as the percentage of Gag+ MDMs relative to the number of Gag+ MDMs detected without inhibitor treatment. To show that virus transfer from infected T cells to MDMs leads to productive infection, MDM targets were pretreated with AZT (5 μM) for 2 h prior to coculture for 6 h with infected T cells. After removal of infected T cells, the percentage of Gag+/CD11b+ MDMs was determined by flow cytometry either directly after the 6 h of coculture or 6 days later, while the level of viral p24 production from MDMs was determined by ELISA after 6 days of culture. Results are expressed as the percentage of Gag+ MDMs or as the amount of p24 relative to that determined without AZT. To analyze the effect of inhibitors on virus dissemination between MDMs, infected T cells were first cocultured for 6 h with MDMs and removed by washings, and MDMs were cultured for 1, 5, 8, or 12 days in the presence of 10 μM T20. The percentage of Gag+/CD11b+ MDMs was then determined by flow cytometry, while the level of CAp24 production from MDMs was determined by ELISA. Results are expressed as the percentage of Gag+ MDMs or as the amount of CAp24 relative to that determined without T20 treatment.
Fluorescence microscopy analysis.
To visualize cell contacts and virus transfer, 106 Jurkat or primary CD4+ T cells infected with NLAD8 as described above were prelabeled with 2 μM CellTrace Far Red (Life Technologies) and cocultured for 0.5, 2, or 6 h with 0.5 × 106 MDMs that had been plated onto coverslips. MDMs were then fixed with 4% PFA, blocked for 10 min in PBS containing 1% bovine serum albumin (BSA), stained with 2 μM deep red fluorescing agent Q5 (DRAQ5; eBioscience) for 20 min in PBS, permeabilized, and stained using KC57 FITC-conjugated antibody and phalloidin-Alexa Fluor 647 (Molecular Probes) diluted in permeabilization buffer for 1 h. The coverslips were washed with PBS and mounted on slides using 10 μl of Fluoromount mounting medium (Sigma). Images were acquired on a spinning-disk (CSU-X1M1; Yokogawa) inverted microscope (DMI6000; Leica) and then processed using Fiji software (ImageJ; NIH) (73). Quantitative image analysis was performed using Fiji software, in which a region of interest was defined by using actin staining and measuring the whole fluorescence intensity of the Gag staining with respect to that for noninfected cells. To analyze fusion between infected T cells and MDMs, infected Jurkat or primary CD4+ T cells were cocultured for 6 h with MDMs. Cells were then fixed with 4% PFA and blocked in PBS containing 1% BSA. Anti-CD2 (TS2/18, 1/200 dilution) surface staining was performed on nonpermeabilized cells in PBS-BSA for 1 h. The coverslips were then rinsed in PBS-BSA and incubated for 1 h with anti-mouse immunoglobulin-Alexa Fluor 555 in PBS-BSA. Anti-CD3 (10 μg/ml), anti-Lck (2 μg/ml), anti-Gag (KC57-FITC, 1/200 dilution), and F-actin intracellular staining was done by incubating the coverslips with the primary antibody indicated above and phalloidin-Alexa Fluor 647 diluted in the permeabilization buffer. The coverslips were then rinsed with PBS-BSA and incubated for 1 h with the corresponding fluorescence-coupled secondary antibody. After three washes, the coverslips were mounted on microscope slides, using 10 μl of Fluoromount mounting medium with DAPI (4′,6-diamidino-2-phenylindole; Sigma-Aldrich). Cells were examined under an epifluorescence microscope (DMI6000; Leica), and quantitative image analysis was performed using Fiji software, in which a region of interest was defined using actin staining and measuring the whole fluorescence intensity of the indicated marker (i.e., CD3, CD2, or Lck) in Gag+ cells with respect to that in noninfected cells. For the CellTracker experiment, NLAD8-infected Jurkat cells were labeled with 5 μM CellTracker (7-amino-4-chloromethylcoumarin [CMAC]) dye (Life Technologies) for 30 min and then cocultured for 6 h with MDMs. The cells were then fixed in 4% PFA for 20 min, blocked for 10 min in PBS containing 1% BSA, stained with 2 μM DRAQ5 for 20 min in PBS, permeabilized, and stained using KC57 FITC-conjugated antibody and phalloidin-Alexa Fluor 555 (Molecular Probes) diluted in permeabilization buffer for 1 h. The coverslips were washed with PBS and mounted on slides using 10 μl of Fluoromount mounting medium. Images were acquired and then processed as described above. To analyze the effect of inhibitors on fusion between T cells and macrophages, infected donor T cells or MDM targets were pretreated for 1 h with anti-gp120 neutralizing antibodies (PGT128, 10-1074) or T20, using a concentration of 10 μg/ml in the presence of 10 μg/ml of Fc-Block reagent (Sigma). Infected T cells were then labeled with 5 μM CellTracker CMAC and cocultured for 6 h with MDMs in the presence of inhibitors. Cells were then fixed and stained as described above using KC57 FITC-conjugated antibody and phalloidin-Alexa Fluor 647. Images were acquired and then processed as described above. To analyze fusion between MDMs, infected T cells were initially cocultured with MDMs for 6 h. After elimination of T cells by extensive washing, autologous MDMs prelabeled with CellTrace Far Red or CellTracker were added to the MDMs just after the 6-h coculture or 4 days later, respectively, and cultured for 1 more day. The coverslips were then washed and mounted with Fluoromount mounting medium containing DAPI or the NucRed reagent. Images were acquired and processed as described above. To analyze the effect of T20 and maraviroc on fusion between MDMs, infected T cells were initially cocultured with MDMs for 6 h. After removal of T cells by extensive washings, MDMs were cultured for 1 more day with or without T20 (10 μg/ml) or maraviroc (10 μM) and the cells were stained for nuclei, Gag, and F actin following performance of the fixation and blocking steps described above. The number of nuclei per cell was analyzed from images of at least 50 cells. To analyze the survival of infected giant multinucleated cells in culture, infected Jurkat cells and MDMs were cocultured for 6 h, and MDMs were then cultured for up to 30 days after the removal of T cells by extensive washings as described above. Cells were then collected, fixed, and stained for Gag and F actin as described above. The coverslips were washed with PBS and mounted on slides using 10 μl of Fluoromount mounting medium containing DAPI. Images were acquired on a spinning-disk inverted microscope and then processed using Fiji software as described above.
Live-cell imaging of HIV-1 transfer.
Jurkat cells (2 × 106) infected with HIV-1R5-GFP were cocultured for 1 h with 106 MDMs prelabeled with CellTrace Far Red and plated onto a 35-mm Ibidi dish in RPMI 1640 medium not containing phenol red but containing 10% FBS and ATB. Images were recorded using a 20× air objective every 1 or 2.5 min for 1, 2, or 3 h on a spinning-disk (CSU-X1M1; Yokogawa) confocal inverted microscope (DMI6000; Leica) equipped with a heated chamber. z-stack optical sections were acquired at 0.5-μm depth increments, and movies were analyzed using Fiji software.
Transmission electron microscopy analysis.
Jurkat cells (2 × 106) infected with NLAD8 as described above were cocultured for 6 h with 106 MDMs that had been plated onto coverslips with or without AZT (5 μM). In parallel, MDMs were infected by cell-free NLAD8 for 6 h. The coverslips were then fixed using 2.8% glutaraldehyde and 2% PFA for 20 min. After 2 washes in PBS, the cells were dehydrated and embedded into epoxy (Electron Microscopy Sciences). Ultrathin sections of 90 nm were cut with an ultramicrotome, stained with uranyl acetate and Reynold's lead, and observed with a transmission electron microscope (JEOL 1011). Acquisition was performed with a Gatan ES1000W charge-coupled-device camera.
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael Schindler and Andres Alcover for the gift of reagents. We thank the members of the Flow Cytometry (Cybio) and the Cellular Imaging core facilities of the Cochin Institute for their technical help. We thank Clarisse Vigne for technical help. We thank Andres Alcover, Georges Bismuth, Clarisse Berlioz-Torrent, and Mark Scott for discussions and critical readings of the manuscript.
This work was supported in part by INSERM, CNRS, and the University Paris-Descartes. It was also funded by grants from the Agence Nationale de Recherche sur le SIDA et les Hépatites Virales (ANRS). L.B. is supported by grants from the Institut Pasteur International Network and the Chinese Academy of Sciences. M.L. is supported by a grant from the European HIVERA Program. M.X. is supported by a grant from the China Scholarship Council. J.B. is supported by a grant from Sidaction. The research was conducted within the context of the Associated International Laboratory (LIA) VIRHOST, a collaboration among CNRS, Institut Pasteur, Université Paris-Descartes, and the Institut Pasteur Shanghai-Chinese Academy of Sciences.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01237-17.
REFERENCES
- 1.Burdo TH, Lackner A, Williams KC. 2013. Monocyte/macrophages and their role in HIV neuropathogenesis. Immunol Rev 254:102–113. doi: 10.1111/imr.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carter CA, Ehrlich LS. 2008. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol 62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- 3.Harbison C, Zhuang K, Gettie A, Blanchard J, Knight H, Didier P, Cheng-Mayer C, Westmoreland S. 2014. Giant cell encephalitis and microglial infection with mucosally transmitted simian-human immunodeficiency virus SHIVSF162P3N in rhesus macaques. J Neurovirol 20:62–72. doi: 10.1007/s13365-013-0229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan J, Sattentau QJ. 2013. The HIV-1-containing macrophage compartment: a perfect cellular niche? Trends Microbiol 21:405–412. doi: 10.1016/j.tim.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Watters SA, Mlcochova P, Gupta RK. 2013. Macrophages: the neglected barrier to eradication. Curr Opin Infect Dis 26:561–566. doi: 10.1097/QCO.0000000000000014. [DOI] [PubMed] [Google Scholar]
- 6.Honeycutt JB, Wahl A, Baker C, Spagnuolo RA, Foster J, Zakharova O, Wietgrefe S, Caro-Vegas C, Madden V, Sharpe G, Haase AT, Eron JJ, Garcia JV. 2016. Macrophages sustain HIV replication in vivo independently of T cells. J Clin Invest 126:1353. doi: 10.1172/JCI84456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araínga M, Edagwa B, Mosley RL, Poluektova LY, Gorantla S, Gendelman HE. 2017. A mature macrophage is a principal HIV-1 cellular reservoir in humanized mice after treatment with long acting antiretroviral therapy. Retrovirology 14:17. doi: 10.1186/s12977-017-0344-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Honeycutt JB, Thayer WO, Baker CE, Ribeiro RM, Lada SM, Cao Y, Cleary RA, Hudgens MG, Richman DD, Garcia JV. 2017. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med 23:638–643. doi: 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumas A, Lê-Bury G, Marie-Anaïs F, Herit F, Mazzolini J, Guilbert T, Bourdoncle P, Russell DG, Benichou S, Zahraoui A, Niedergang F. 2015. The HIV-1 protein Vpr impairs phagosome maturation by controlling microtubule-dependent trafficking. J Cell Biol 211:359–372. doi: 10.1083/jcb.201503124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kedzierska K, Ellery P, Mak J, Lewin SR, Crowe SM, Jaworowski A. 2002. HIV-1 down-modulates γ signaling chain of FcγR in human macrophages: a possible mechanism for inhibition of phagocytosis. J Immunol 168:2895–2903. doi: 10.4049/jimmunol.168.6.2895. [DOI] [PubMed] [Google Scholar]
- 11.Koppensteiner H, Brack-Werner R, Schindler M. 2012. Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology 9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazzolini J, Herit F, Bouchet J, Benmerah A, Benichou S, Niedergang F. 2010. Inhibition of phagocytosis in HIV-1-infected macrophages relies on Nef-dependent alteration of focal delivery of recycling compartments. Blood 115:4226–4236. doi: 10.1182/blood-2009-12-259473. [DOI] [PubMed] [Google Scholar]
- 13.Vérollet C, Souriant S, Bonnaud E, Jolicoeur P, Raynaud-Messina B, Kinnaer C, Fourquaux I, Imle A, Benichou S, Fackler OT, Poincloux R, Maridonneau-Parini I. 2015. HIV-1 reprograms the migration of macrophages. Blood 125:1611–1622. doi: 10.1182/blood-2014-08-596775. [DOI] [PubMed] [Google Scholar]
- 14.Vérollet C, Zhang YM, Cabec VL, Mazzolini J, Charrière G, Labrousse A, Bouchet J, Medina I, Biessen E, Niedergang F, Bénichou S, Maridonneau-Parini I. 2010. HIV-1 Nef triggers macrophage fusion in a p61Hck- and protease-dependent manner. J Immunol 184:7030–7039. doi: 10.4049/jimmunol.0903345. [DOI] [PubMed] [Google Scholar]
- 15.Costiniuk CT, Jenabian M-A. 2014. The lungs as anatomical reservoirs of HIV infection. Rev Med Virol 24:35–54. doi: 10.1002/rmv.1772. [DOI] [PubMed] [Google Scholar]
- 16.Fischer-Smith T, Bell C, Croul S, Lewis M, Rappaport J. 2008. Monocyte/macrophage trafficking in acquired immunodeficiency syndrome encephalitis: lessons from human and nonhuman primate studies. J Neurovirol 14:318–326. doi: 10.1080/13550280802132857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geny C, Gherardi R, Boudes P, Lionnet F, Cesaro P, Gray F. 1991. Multifocal multinucleated giant cell myelitis in an AIDS patient. Neuropathol Appl Neurobiol 17:157–162. doi: 10.1111/j.1365-2990.1991.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 18.González-Scarano F, Martín-García J. 2005. The neuropathogenesis of AIDS. Nat Rev Immunol 5:69–81. doi: 10.1038/nri1527. [DOI] [PubMed] [Google Scholar]
- 19.Gras G, Kaul M. 2010. Molecular mechanisms of neuroinvasion by monocytes-macrophages in HIV-1 infection. Retrovirology 7:30. doi: 10.1186/1742-4690-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorry PR, Francella N, Lewin SR, Collman RG. 2014. HIV-1 envelope-receptor interactions required for macrophage infection and implications for current HIV-1 cure strategies. J Leukoc Biol 95:71–81. doi: 10.1189/jlb.0713368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter AE, Russell RA, Duncan CJA, Moore MD, Willberg CB, Pablos JL, Finzi A, Kaufmann DE, Ochsenbauer C, Kappes JC, Groot F, Sattentau QJ.. 2014. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe 16:711–721. doi: 10.1016/j.chom.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agosto LM, Uchil PD, Mothes W. 2015. HIV cell-to-cell transmission: effects on pathogenesis and antiretroviral therapy. Trends Microbiol 23:289–295. doi: 10.1016/j.tim.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez RA, Barría MI, Chen BK. 2014. Unique features of HIV-1 spread through T cell virological synapses. PLoS Pathog 10:e1004513. doi: 10.1371/journal.ppat.1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dale BM, Alvarez RA, Chen BK. 2013. Mechanisms of enhanced HIV spread through T-cell virological synapses. Immunol Rev 251:113–124. doi: 10.1111/imr.12022. [DOI] [PubMed] [Google Scholar]
- 25.Schiffner T, Sattentau QJ, Duncan CJA. 2013. Cell-to-cell spread of HIV-1 and evasion of neutralizing antibodies. Vaccine 31:5789–5797. doi: 10.1016/j.vaccine.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 26.Law KM, Komarova NL, Yewdall AW, Lee RK, Herrera OL, Wodarz D, Chen BK. 2016. In vivo HIV-1 cell-to-cell transmission promotes multicopy micro-compartmentalized infection. Cell Rep 15:2771–2783. doi: 10.1016/j.celrep.2016.05.059. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. 2005. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol 170:317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Llewellyn GN, Hogue IB, Grover JR, Ono A. 2010. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog 6:e1001167. doi: 10.1371/journal.ppat.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ménager MM, Littman DR. 2016. Actin dynamics regulates dendritic cell-mediated transfer of HIV-1 to T cells. Cell 164:695–709. doi: 10.1016/j.cell.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherer NM, Lehmann MJ, Jimenez-Soto LF, Horensavitz C, Pypaert M, Mothes W. 2007. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol 9:310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sowinski S, Jolly C, Berninghausen O, Purbhoo MA, Chauveau A, Köhler K, Oddos S, Eissmann P, Brodsky FM, Hopkins C, Onfelt B, Sattentau Q, Davis DM. 2008. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat Cell Biol 10:211–219. doi: 10.1038/ncb1682. [DOI] [PubMed] [Google Scholar]
- 32.Murooka TT, Deruaz M, Marangoni F, Vrbanac VD, Seung E, von Andrian UH, Tager AM, Luster AD, Mempel TR. 2012. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 490:283–287. doi: 10.1038/nature11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sewald X, Gonzalez DG, Haberman AM, Mothes W. 2012. In vivo imaging of virological synapses. Nat Commun 3:1320. doi: 10.1038/ncomms2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abela IA, Berlinger L, Schanz M, Reynell L, Günthard HF, Rusert P, Trkola A. 2012. Cell-cell transmission enables HIV-1 to evade inhibition by potent CD4bs directed antibodies. PLoS Pathog 8:e1002634. doi: 10.1371/journal.ppat.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agosto LM, Zhong P, Munro J, Mothes W. 2014. Highly active antiretroviral therapies are effective against HIV-1 cell-to-cell transmission. PLoS Pathog 10:e1003982. doi: 10.1371/journal.ppat.1003982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Malbec M, Sourisseau M, Guivel-Benhassine F, Porrot F, Blanchet F, Schwartz O, Casartelli N. 2013. HIV-1 Nef promotes the localization of Gag to the cell membrane and facilitates viral cell-to-cell transfer. Retrovirology 10:80. doi: 10.1186/1742-4690-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, Milo R, Baltimore D. 2011. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477:95–98. doi: 10.1038/nature10347. [DOI] [PubMed] [Google Scholar]
- 38.Orenstein JM. 2001. The macrophage in HIV infection. Immunobiology 204:598–602. doi: 10.1078/0171-2985-00098. [DOI] [PubMed] [Google Scholar]
- 39.Theodore TS, Englund G, Buckler-White A, Buckler CE, Martin MA, Peden KW. 1996. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retroviruses 12:191–194. doi: 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- 40.Freed EO, Englund G, Martin MA. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol 69:3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krementsov DN, Weng J, Lambelé M, Roy NH, Thali M. 2009. Tetraspanins regulate cell-to-cell transmission of HIV-1. Retrovirology 6:64. doi: 10.1186/1742-4690-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weng J, Krementsov DN, Khurana S, Roy NH, Thali M. 2009. Formation of syncytia is repressed by tetraspanins in human immunodeficiency virus type 1-producing cells. J Virol 83:7467–7474. doi: 10.1128/JVI.00163-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roy NH, Lambelé M, Chan J, Symeonides M, Thali M. 2014. Ezrin is a component of the HIV-1 virological presynapse and contributes to the inhibition of cell-cell fusion. J Virol 88:7645–7658. doi: 10.1128/JVI.00550-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Symeonides M, Murooka T, Bellfy L, Roy N, Mempel T, Thali M. 2015. HIV-1-induced small T cell syncytia can transfer virus particles to target cells through transient contacts. Viruses 7:6590–6603. doi: 10.3390/v7122959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Compton AA, Schwartz O. 2017. They might be giants: does syncytium formation sink or spread HIV infection? PLoS Pathog 13:e1006099. doi: 10.1371/journal.ppat.1006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calantone N, Wu F, Klase Z, Deleage C, Perkins M, Matsuda K, Thompson EA, Ortiz AM, Vinton CL, Ourmanov I, Loré K, Douek DC, Estes JD, Hirsch VM, Brenchley JM. 2014. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity 41:493–502. doi: 10.1016/j.immuni.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DiNapoli SR, Ortiz AM, Wu F, Matsuda K, Twigg HL, Hirsch VM, Knox K, Brenchley JM. 2017. Tissue-resident macrophages can contain replication-competent virus in antiretroviral-naive, SIV-infected Asian macaques. JCI Insight 2:e91214. doi: 10.1172/jci.insight.91214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosch B, Grigorov B, Senserrich J, Clotet B, Darlix J-L, Muriaux D, Este JA. 2008. A clathrin-dynamin-dependent endocytic pathway for the uptake of HIV-1 by direct T cell-T cell transmission. Antiviral Res 80:185–193. doi: 10.1016/j.antiviral.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 49.Hübner W, McNerney GP, Chen P, Dale BM, Gordon RE, Chuang FYS, Li X-D, Asmuth DM, Huser T, Chen BK. 2009. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 323:1743–1747. doi: 10.1126/science.1167525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sloan RD, Kuhl BD, Mesplède T, Münch J, Donahue DA, Wainberg MA. 2013. Productive entry of HIV-1 during cell-to-cell transmission via dynamin-dependent endocytosis. J Virol 87:8110–8123. doi: 10.1128/JVI.00815-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, Eng ET, Law K, Gordon RE, Rice WJ, Chen BK. 2017. Visualization of HIV T cell virological synapses and virus-containing compartments by three-dimensional correlative light and electron microscopy. J Virol 91:e01605–16. doi: 10.1128/JVI.01605-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pelchen-Matthews A, Kramer B, Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol 162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol 177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. 2007. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe 2:85–95. doi: 10.1016/j.chom.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 55.Bennett AE, Narayan K, Shi D, Hartnell LM, Gousset K, He H, Lowekamp BC, Yoo TS, Bliss D, Freed EO, Subramaniam S. 2009. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog 5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Kräusslich H-G. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog 3:e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chu H, Wang J-J, Qi M, Yoon J-J, Chen X, Wen X, Hammonds J, Ding L, Spearman P. 2012. Tetherin/BST-2 is essential for the formation of the intracellular virus-containing compartment in HIV-infected macrophages. Cell Host Microbe 12:360–372. doi: 10.1016/j.chom.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kadiu I, Ricardo-Dukelow M, Ciborowski P, Gendelman HE. 2007. Cytoskeletal protein transformation in HIV-1-infected macrophage giant cells. J Immunol 178:6404–6415. doi: 10.4049/jimmunol.178.10.6404. [DOI] [PubMed] [Google Scholar]
- 59.Dargent JL, Lespagnard L, Kornreich A, Hermans P, Clumeck N, Verhest A. 2000. HIV-associated multinucleated giant cells in lymphoid tissue of the Waldeyer's ring: a detailed study. Mod Pathol 13:1293–1299. [DOI] [PubMed] [Google Scholar]
- 60.Lewin-Smith M, Wahl SM, Orenstein JM. 1999. Human immunodeficiency virus-rich multinucleated giant cells in the colon: a case report with transmission electron microscopy, immunohistochemistry, and in situ hybridization. Mod Pathol 12:75–81. [PubMed] [Google Scholar]
- 61.Ryzhova EV, Crino P, Shawver L, Westmoreland SV, Lackner AA, González-Scarano F. 2002. Simian immunodeficiency virus encephalitis: analysis of envelope sequences from individual brain multinucleated giant cells and tissue samples. Virology 297:57–67. doi: 10.1006/viro.2002.1395. [DOI] [PubMed] [Google Scholar]
- 62.Soulas C, Conerly C, Kim W-K, Burdo TH, Alvarez X, Lackner AA, Williams KC. 2011. Recently infiltrating MAC387(+) monocytes/macrophages: a third macrophage population involved in SIV and HIV encephalitic lesion formation. Am J Pathol 178:2121–2135. doi: 10.1016/j.ajpath.2011.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Teo I, Veryard C, Barnes H, An SF, Jones M, Lantos PL, Luthert P, Shaunak S. 1997. Circular forms of unintegrated human immunodeficiency virus type 1 DNA and high levels of viral protein expression: association with dementia and multinucleated giant cells in the brains of patients with AIDS. J Virol 71:2928–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vicandi B, Jiménez-Heffernan JA, López-Ferrer P, Patrón M, Gamallo C, Colmenero C, Viguer JM. 1999. HIV-1 (p24)-positive multinucleated giant cells in HIV-associated lymphoepithelial lesion of the parotid gland. A report of two cases. Acta Cytol 43:247–251. [DOI] [PubMed] [Google Scholar]
- 65.Blanco J, Bosch B, Fernández-Figueras MT, Barretina J, Clotet B, Esté JA. 2004. High level of coreceptor-independent HIV transfer induced by contacts between primary CD4 T cells. J Biol Chem 279:51305–51314. doi: 10.1074/jbc.M408547200. [DOI] [PubMed] [Google Scholar]
- 66.Chen P, Hübner W, Spinelli MA, Chen BK. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol 81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Felts RL, Narayan K, Estes JD, Shi D, Trubey CM, Fu J, Hartnell LM, Ruthel GT, Schneider DK, Nagashima K, Bess JW, Bavari S, Lowekamp BC, Bliss D, Lifson JD, Subramaniam S. 2010. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc Natl Acad Sci U S A 107:13336–13341. doi: 10.1073/pnas.1003040107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Martin N, Welsch S, Jolly C, Briggs JAG, Vaux D, Sattentau QJ. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol 84:3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Helming L, Gordon S. 2009. Molecular mediators of macrophage fusion. Trends Cell Biol 19:514–522. doi: 10.1016/j.tcb.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 70.Koppensteiner H, Banning C, Schneider C, Hohenberg H, Schindler M. 2012. Macrophage internal HIV-1 is protected from neutralizing antibodies. J Virol 86:2826–2836. doi: 10.1128/JVI.05915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Herate C, Vigne C, Guenzel CA, Lambele M, Rouyez M-C, Benichou S. 2016. Uracil DNA glycosylase interacts with the p32 subunit of the replication protein A complex to modulate HIV-1 reverse transcription for optimal virus dissemination. Retrovirology 13:26. doi: 10.1186/s12977-016-0257-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bouchet J, Del Río-Iñiguez I, Lasserre R, Agüera-Gonzalez S, Cuche C, Danckaert A, McCaffrey MW, Di Bartolo V, Alcover A. 2016. Rac1-Rab11-FIP3 regulatory hub coordinates vesicle traffic with actin remodeling and T-cell activation. EMBO J 35:1160–1174. doi: 10.15252/embj.201593274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.