ABSTRACT
Two scaffolding proteins orchestrate ϕX174 morphogenesis. The internal scaffolding protein B mediates the formation of pentameric assembly intermediates, whereas the external scaffolding protein D organizes 12 of these intermediates into procapsids. Aromatic amino acid side chains mediate most coat-internal scaffolding protein interactions. One residue in the internal scaffolding protein and three in the coat protein constitute the core of the B protein binding cleft. The three coat gene codons were randomized separately to ascertain the chemical requirements of the encoded amino acids and the morphogenetic consequences of mutation. The resulting mutants exhibited a wide range of recessive phenotypes, which could generally be explained within a structural context. Mutants with phenylalanine, tyrosine, and methionine substitutions were phenotypically indistinguishable from the wild type. However, tryptophan substitutions were detrimental at two sites. Charged residues were poorly tolerated, conferring extreme temperature-sensitive and lethal phenotypes. Eighteen lethal and conditional lethal mutants were genetically and biochemically characterized. The primary defect associated with the missense substitutions ranged from inefficient internal scaffolding protein B binding to faulty procapsid elongation reactions mediated by external scaffolding protein D. Elevating B protein concentrations above wild-type levels via exogenous, cloned-gene expression compensated for inefficient B protein binding, as did suppressing mutations within gene B. Similarly, elevating D protein concentrations above wild-type levels or compensatory mutations within gene D suppressed faulty elongation. Some of the parental mutations were pleiotropic, affecting multiple morphogenetic reactions. This progressively reduced the flux of intermediates through the pathway. Accordingly, multiple mechanisms, which may be unrelated, could restore viability.
IMPORTANCE Genetic analyses have been instrumental in deciphering the temporal events of many biochemical pathways. However, pleiotropic effects can complicate analyses. Vis-à-vis virion morphogenesis, an improper protein-protein interaction within an early assembly intermediate can influence the efficiency of all subsequent reactions. Consequently, the flux of assembly intermediates cumulatively decreases as the pathway progresses. During morphogenesis, ϕX174 coat protein participates in at least four well-defined reactions, each one characterized by an interaction with a scaffolding or structural protein. In this study, genetic analyses, biochemical characterizations, and physiological assays, i.e., elevating the protein levels with which the coat protein interacts, were used to elucidate pleiotropic effects that may alter the flux of intermediates through a morphogenetic pathway.
KEYWORDS: bacteriophage phiX174, coat protein, scaffolding protein, virus assembly
INTRODUCTION
Scaffolding protein-induced conformational switches mediate capsid morphogenesis in many viral systems. These switches typically occur in the coat protein, lowering the thermodynamic barriers needed for productive assembly relative to those that promote off-pathway reactions (1–3). In systems requiring a single internal scaffolding protein capsid, morphogenesis can be broadly divided into three stages: coat-scaffolding protein binding followed by procapsid nucleation and elongation (1–3). However, proper morphogenesis can also include the incorporation of minor structural proteins, such as those composing portals (4, 5).
Genetic analyses have been instrumental in defining the temporal events of morphogenetic pathways (6–8). When combined with biochemical and structural studies, they can also define functional domains within coat and scaffolding proteins (4, 5, 9–23). However, the fluid nature of assembly and/or the products of defective morphogenesis can still complicate analyses. For example, a mutation that eliminates ϕX174 coat-internal scaffolding interactions moves coat protein pentamers into the insoluble fraction. As seen in other systems (24), but not rigorously documented with ϕX174, improperly folded coat proteins are often insoluble. Moreover, improper elongation reactions could theoretically produce insoluble products.
Unlike most assembly systems, ϕX174 morphogenesis is mediated by two scaffolding proteins, an internal and an external species. External scaffolding proteins are unique to the ϕX174-like viruses. During early morphogenesis, pentameric intermediates are built. Five copies of the internal scaffolding protein B bind to the underside of the 9S coat protein F pentamer forming the 9S* particle (20). This induces the conformational changes that (i) inhibit 9S particle aggregation, (ii) facilitate DNA pilot protein H incorporation, and (iii) stimulate coat-spike protein interactions (20, 25–28). The last result in the formation of the 12S* particle, which is the early assembly endpoint. Late morphogenesis resembles capsid assembly in single internal scaffolding protein systems: coat-scaffolding protein binding followed by procapsid nucleation and elongation (19, 29). Accordingly, this reaction, which contains coat proteins in pentameric subassemblies and an external scaffolding protein, displays in vitro similar kinetics to those observed in single scaffolding protein systems (30).
In the P22, P2, ϕ29, and herpes simplex virus (HSV) assembly systems (31–35), which utilize only an internal scaffolding protein, the scaffolding protein's C terminus mediates most coat protein contacts (36–38). This is also observed in the two-scaffolding protein ϕX174 system. In the procapsid crystal structure (37, 38), six aromatic amino acids mediate approximately 85% of the coat-internal scaffolding B contacts (Fig. 1B). At the core of the B-protein binding cleft, phenylalanine residue 120 (F120) participates in ring-ring contacts, π-stacking, with three strongly conserved coat protein F aromatic side chains: F67, Y134, and F135. In a previous study (16), we demonstrated that nonaromatic substitutions for F120 in B protein abolish coat protein binding, whereas substitutions for the other scaffolding protein residues kinetically trap early assembly intermediates. In the current study, we extended this analysis to single mutations in the coat protein at residues F67, Y134, and F135. Unlike in the previous analysis conducted with F120 of B protein, a wide variety of molecular defects were observed, ranging from inefficient internal scaffolding protein B binding to faulty procapsid elongation reactions with the external scaffolding protein D. Some mutations were pleiotropic, affecting multiple morphogenetic reactions, which may progressively reduce the flux of intermediates through the pathway.
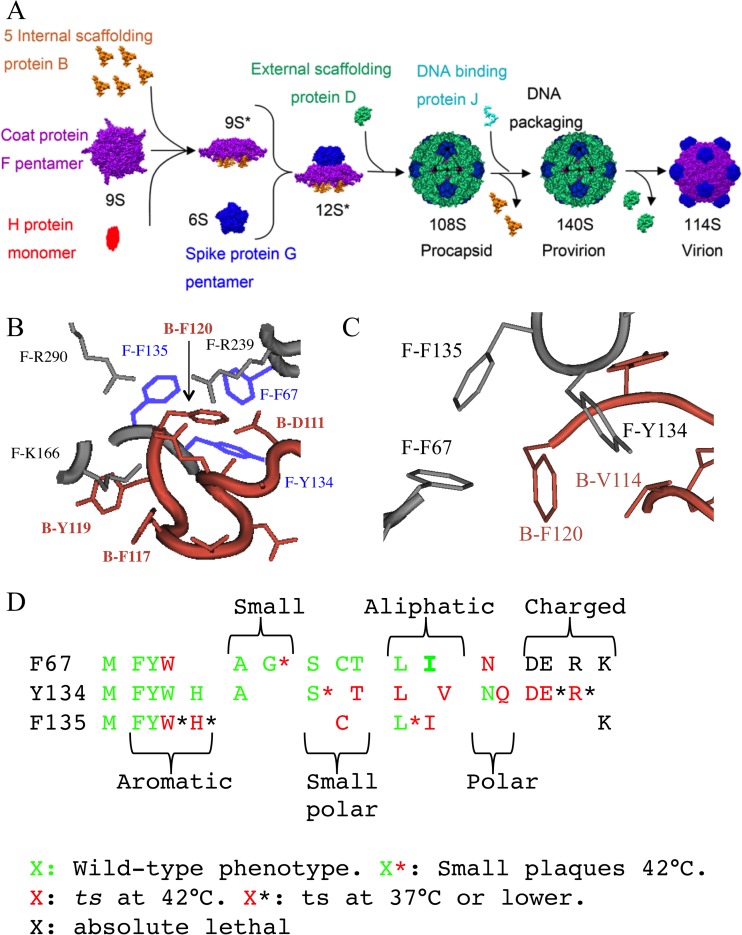
FIG 1.
The ϕX174 procapsid assembly pathway and coat-internal scaffolding protein interactions. (A) The morphogenetic pathway is dependent on two scaffolding proteins. The internal scaffolding protein B mediates early morphogenesis and is found in the 9S* and 12S* assembly intermediates. Late assembly is orchestrated by the external scaffolding protein D. Two hundred forty D proteins organize 12 12S* particles into procapsids. (B) Structure of the coat-internal scaffolding protein binding cleft in the ϕX174 procapsid crystal structure (PDB code 1CD3). Labels depict the protein (F, coat; B, internal scaffolding), amino acid (letter), and position (number) in the primary structure. The three F protein aromatic amino acid residues—F67, Y134, and F135—that participate in ring-ring contacts with internal scaffolding protein residue F120 are highlighted in lavender. (C) Structure of the coat-internal scaffolding protein binding cleft in the ϕX174 procapsid crystal structure to emphasize the position of residue B-V114F. (D) Phenotypes conferred by amino acid substitutions for coat protein residues F67, Y134, and F135. The color of the letter conveys the general phenotype; green X, no severe growth restrictions between 24°C and 42°C; red X, temperature sensitive; and black X, lethal at all temperatures. Asterisks are used to convey further phenotypic details as defined within the figure.
RESULTS
Isolation and characterization of F67, Y134, and F135 missense mutants.
Two strategies were employed to generate conditional lethal and silent missense mutations at the F67, Y134, or F135 site: (i) amber mutation reversion and (ii) codon randomization with amber codon-containing DNA templates. To recover the broadest spectrum of possible phenotypes, mutagenized progeny were plated at 24°C, 33°C, and 42°C on a sup0 host, which selected against the parental amber mutant. To isolate absolute lethal mutants, the target codon within wild-type DNA was mutagenized using randomizing primers or primers designed to introduce specific mutations. Mutagenized progeny were recovered on cells expressing a cloned, wild-type coat F gene.
The F genes from all temperature-sensitive (ts) and lethal mutants were sequenced. No cold-sensitive mutants were recovered. To ascertain which substitutions were well tolerated at each site, genomes from at least 30 phage with wild-type or near-wild-type phenotypes were also analyzed. The substitutions and phenotypes are summarized in Fig. 1D and Table 1. Table S1 in the supplemental material provides a more detailed description of the frequency of codon isolation, recovery conditions, and phenotypes. Codon usage had no obvious effect on phenotype. Different codons for the same amino acid were observed 18 times. All such mutants at any given location exhibited the same phenotype (see Table S1). Similarly, 16 mutants were independently isolated by two or more methods. Again, no phenotypic disparities were observed.
TABLE 1.
Plating efficiency of temperature-sensitive and lethal mutants
| Mutanta | Efficiency at indicated type of temp (°C)b |
Efficiency under restrictive conditionc with exogenous gene expression (temp, °C) |
||
|---|---|---|---|---|
| Permissive | Restrictive | F gene | B gene | |
| ts(F)F67N | 1.0 (33) | 10−6 (42) | 1.1 (42) | 0.3 (42) |
| ts(F)F67W | 1.0 (33) | <10−4 (42) | 1.0 (42) | <10−4 (42) |
| l(F)F67D | 10−4 (37) | 1.0 (37) | 1.0 (37) | |
| l(F)F67E | 10−4 (37) | 1.0 (37) | 10−4 (37) | |
| l(F)F67K | 10−4 (37) | 1.0 (37) | 10−4 (37) | |
| l(F)F67R | 10−4 (37) | 1.0 (37) | 10−4 (37) | |
| ts(F)Y134D | 1.0 (33) | 10−3 (42) | 0.9 (42) | 0.1 (42) |
| ts(F)Y134E | 1.0 (28) | <10−6 (37) | 0.5 (37) | <10−6 (37) |
| ts(F)Y134L | 1.0 (33) | 10− (42) | 1.0 (42) | 0.6 (42) |
| ts(F)Y134Q | 1.0 (33) | 10−5 (42) | 0.9 (42) | 0.9 (42) |
| ts(F)Y134T | 1.0 (33) | 10−5 (42) | 0.7 (42) | 0.7 (42) |
| ts(F)Y134V | 1.0 (33) | 10−5 (42) | 1.1 (42) | 1.0 (42) |
| ts(F)Y134R | 1.0 (33) | 10−3 (37) | 1.0 (37) | 0.9 (37) |
| ts(F)F135C | 1.0 (33) | <10−4 (42) | 1.1 (42) | 0.6 (42) |
| ts(F)F135H | 1.0 (33) | <10−5 (37) | 1.0 (37) | 10−5 (37) |
| ts(F)F135I | 1.0 (33) | <10−6 (42) | 1.0 (42) | 0.7 (42) |
| ts(F)F135W | 1.0 (28) | 10−4 (33) | 1.0 (33) | <10−3 (33) |
| l(F)F135K | 10−4 (37) | 1.0 (37) | 10−4 (37) | |
Mutant names reflect the phenotype (ts, temperature sensitive; l, lethal) and conferred substitution. The letters in parentheses indicate the genes. Thus, the designation ts(F)F67N means that an F→N at position 67 within gene F confers a temperature-sensitive phenotype.
For ts mutants, the restrictive and permissive temperatures are given in parentheses. For lethal mutants, all experiments were conducted at 37°C. The permissive condition includes the exogenous expression of a cloned F gene.
For ts mutants, experiments involving the exogenous expression of either the cloned B or F gene were conducted at the restrictive temperature.
The randomization technique did exhibit isolation bias: many codons were disproportionally represented (see Table S1). Moreover, several mutants that were not recovered by randomization were readily generated when the primers specifically introduced the requisite codons (see Materials and Methods). Thus, in this study and in other studies utilizing randomizing protocols, no conclusions should be drawn by the absence of a specific mutation within a mutagenized pool.
The missense and lethal substitutions are both necessary and sufficient to confer the defective phenotype.
The exogenous expression of a cloned F gene rescued all lethal and ts mutants at their restrictive temperatures (Table 1), indicating that the identified mutations were both necessary and sufficient to confer phenotype. Moreover, in subsequent reversion analyses, temperature sensitivity was lost via same-site reversion events. Nonetheless, the entire genomes of six mutants—ts(F)F67N, ts(F)F67W, ts(F)Y134Q, ts(F)F135C, ts(F)F135I, and ts(F)F135W—were sequenced. No additional changes were detected.
The phenotypes can be interpreted within a structural context. In general, smaller polar and aliphatic amino acids conferred less severe ts phenotypes than larger polar and aliphatic substitutions. The introduction of charged amino acids (D, E, K, and R) appeared to have more detrimental effects, producing more dramatic ts or lethal phenotypes. The geometry of the binding pocket may explain phenotypic differences conferred by the same amino acid, such as tryptophan, at the three different positions (see Discussion).
The lethal and ts mutations have recessive phenotypes.
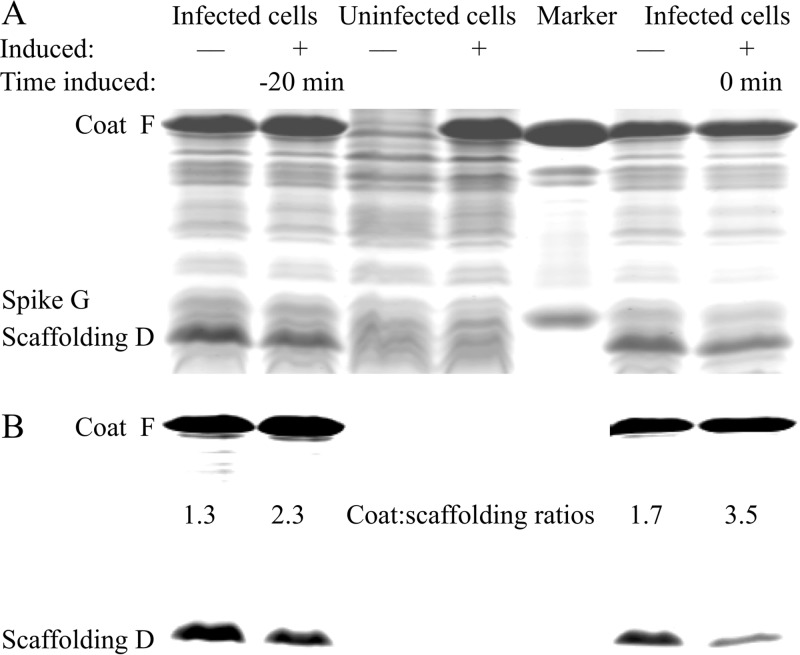
The exogenous expression of the cloned F gene efficiently complemented the ts(F) and lethal(F) mutants. However, the molecular basis of rescue is dependent on the relative contribution of the plasmid- and genome-borne F genes to the intracellular coat protein level. For example, if cloned-gene expression resulted in a wild-type protein level that greatly exceeded the mutant protein level, rescue was driven by concentration disparities between the two coat protein species. All mutations would appear to have a recessive phenotype; dominant phenotypes would be obscured. Consequently, mechanistic insights underlying a recessive phenotype would be limited. In contrast, if the plasmid- and genome-borne genes contributed equally to the coat protein level, rescue would indicate that the mutations are indeed recessive. Therefore, the relative contribution of mutant and wild-type protein levels was determined. Lysis-resistant cells harboring the inducible F gene were infected with wild-type ϕX174 with and without induction (Fig. 2A). Whole-cell lysates were analyzed by SDS-PAGE and the coat/external scaffolding protein ratio was determined (Fig. 2B). Cloned-gene induction approximately doubled coat/external scaffolding protein ratios. Thus, the plasmid- and genome-borne genes equally contributed to the coat protein pool. Induction time, 20 min preinfection or concurrent with infection, did not significantly alter the results. By this assay, the mutations appeared to be recessive. To further test this hypothesis, the lethal mutations were placed into the clone gene and assayed for the ability to affect wild-type plaque formation and morphology; no inhibition was observed (data not shown). At least three mechanisms may underlie the recessive phenotype. (i) Unfolded mutant proteins were not incorporated into pentamers. (ii) Cotranslational subunit assembly produced only homogeneous pentamers; consequently, only wild-type pentamers entered the assembly pathway. (iii) Heterogeneous pentamers were assembly competent.
FIG 2.
The relative contribution of the cloned and genome encoded coat F genes to coat protein levels during infections. Cells harboring the cloned F gene were infected with (+) and without (−) cloned gene induction at an MOI of 3. The cloned gene was induced at the time of infection (0 min) or 20 min before infection (−20 min). (A) The digitally unadjusted, Coomassie blue-stained, SDS-PAGE gel of whole-cell lysates. The marker lane contained purified virions. (B) The digitally modified gel used for more accurate densitometry measurements using the ImageJ (NIH) program. Coat/external scaffolding protein ratios were determined and are shown.
Extragenic rescue mechanisms can distinguish between folding and assembly defects.
The wild-type coat protein residues at the mutated sites directly contact the internal scaffolding protein (37, 38). If the mutations weaken coat-internal scaffolding protein interactions, increasing intracellular B protein concentrations, via exogenous expression of cloned B gene, may drive inefficient reactions forward and rescue mutants. In contrast, coat protein folding defects are less likely to be suppressed by this extragenic mechanism. Exogenous B gene expression efficiently rescued 10 out of the 18 mutants (Table 1). These data indicate that the primary defect associated with those mutants rescued via this mechanism is inefficient B protein binding, not coat protein folding. However, the converse may not be true. Other defects besides protein folding could prevent rescue. Indeed, the results of the second-site genetic analyses (see below) indicate that most mutants used in these studies could be suppressed by at least one extragenic mechanism.
The coat protein interacts with three other proteins after protein B: in temporal order, the spike G, the external scaffolding D and the DNA binding J proteins. If the coat mutations result in structurally altered assembly intermediates, interactions with the proteins G, D, and J may be compromised. Increasing the intracellular concentration of these proteins may drive assembly forward. No mutants were rescued by the exogenous gene G or J expression. However, exogenous, external scaffolding D gene expression rescued ts(F)Y134Q. Plating efficiency increased 4 orders of magnitude, from 10−5 to 0.1 at 42°C, but plaques displayed a pinprick morphology. Rescue by exogenous D gene expression was weaker than rescue by cloned B gene expression. These data indicate that the Y134Q mutation confers pleiotropic effects that reduce the flux through the assembly pathway at least two steps (see Discussion).
In vivo characterization of mutant assembly pathways.
The assembly pathway was characterized for all 18 mutants listed in Table 1. For the temperature-sensitive mutants, phage were preattached to lysis-resistant cells by incubation at 16°C for 30 min. The infections were then split between two medium-containing flasks prewarmed to the permissive and restrictive assay temperatures. For the lethal mutants, lysis-resistant cells carrying a complementing clone of gene F were infected at 37°C; immediately after infection, the cultures were split and the cloned F gene was induced in only one culture. Cell extracts were analyzed by rate zonal sedimentation. After centrifugation, gradients were separated into approximately 40 fractions by removing 125-μl aliquots from the bottom of the tube. Thus, lower fraction numbers would contain particles with larger S values. The presence of assembled particles—virions (114S), procapsids (108S), and degraded procapsids (70S)—was determined by UV spectroscopy.
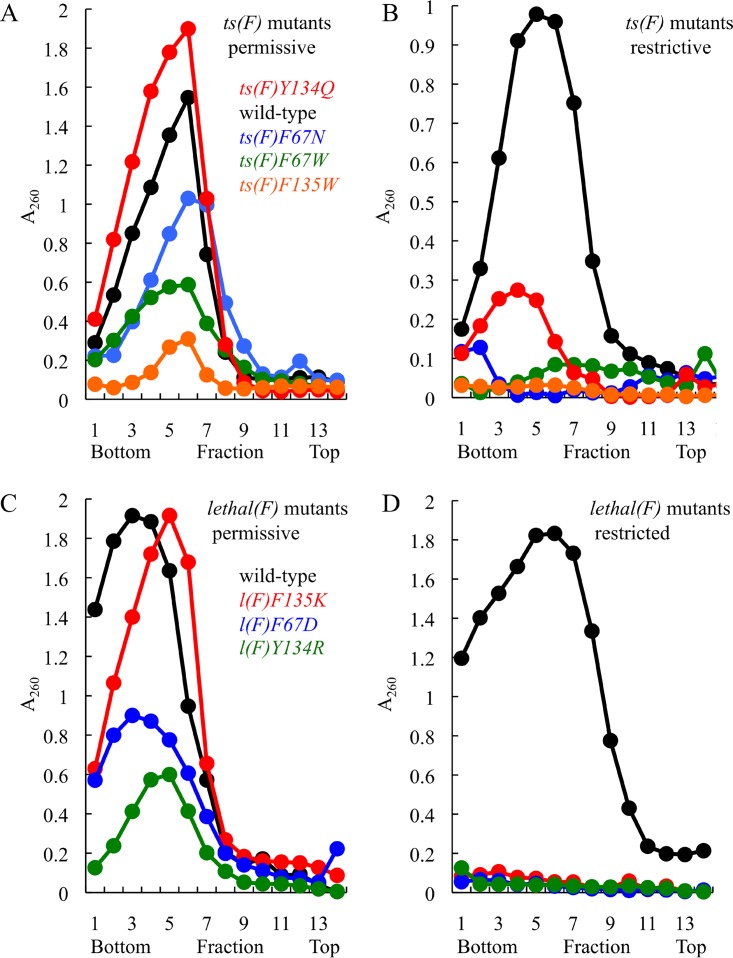
Figure 3 presents the sedimentation profiles generated with four ts (Fig. 3A and B) and three lethal mutants (Fig. 3C and D). In extracts produced from permissive infections (Fig. 3A and C), particles sedimenting in the region from 108S to 114S were detected in all samples. No significant differences in specific infectivity (PFU/optical density at 260 nm [OD260]) were observed between the mutant and wild-type particles, indicating that the peaks primarily contained virions, as opposed to procapsids (data not shown). Rigorous conclusions cannot by drawn by comparing the magnitudes of the peaks, which could result from variations between infections, such as multiplicities of infection (MOI). In contrast, permissive and restrictive infections for individual mutants were generated in pairs: a common cell culture was infected and subsequently split between assays conditions. Therefore, each permissive mutant infection serves as an internal control for the restrictive assay. Under restrictive conditions, virions were detected only in wild-type and ts(F)Y134Q extracts. However, ts(F)Y134Q particle yield was greatly reduced compared to that in the permissive infection. Nonetheless, the specific infectivity was comparable to that of the wild type (data not shown). These data indicate that the coat protein mutations block particle assembly before procapsid formation. Similar results were obtained for the remaining 10 mutants not depicted in Fig. 3.
FIG 3.
Assembled particles produced in mutant infected cells under permissive and restrictive conditions. The curves represent 280-nm absorbance profiles of infected cell extracts analyzed by rate zonal sedimentation. Gradient parameters were designed to detect assembled particles sedimenting between 132S and 70S, which would contain infectious provirions (132S), virions (114S), procapsids (108S), and degraded procapsids (70S). Fraction 1 represents the gradient bottom and the profiles. (A and B) sedimentation profiles from extracts of ts(F) mutant-infected cells at permissive (A) and restrictive (B) temperatures. (C and D) Sedimentation profiles from extracts of lethal(F) mutant-infected cells with (C) and without (D) the induction of cloned F gene.
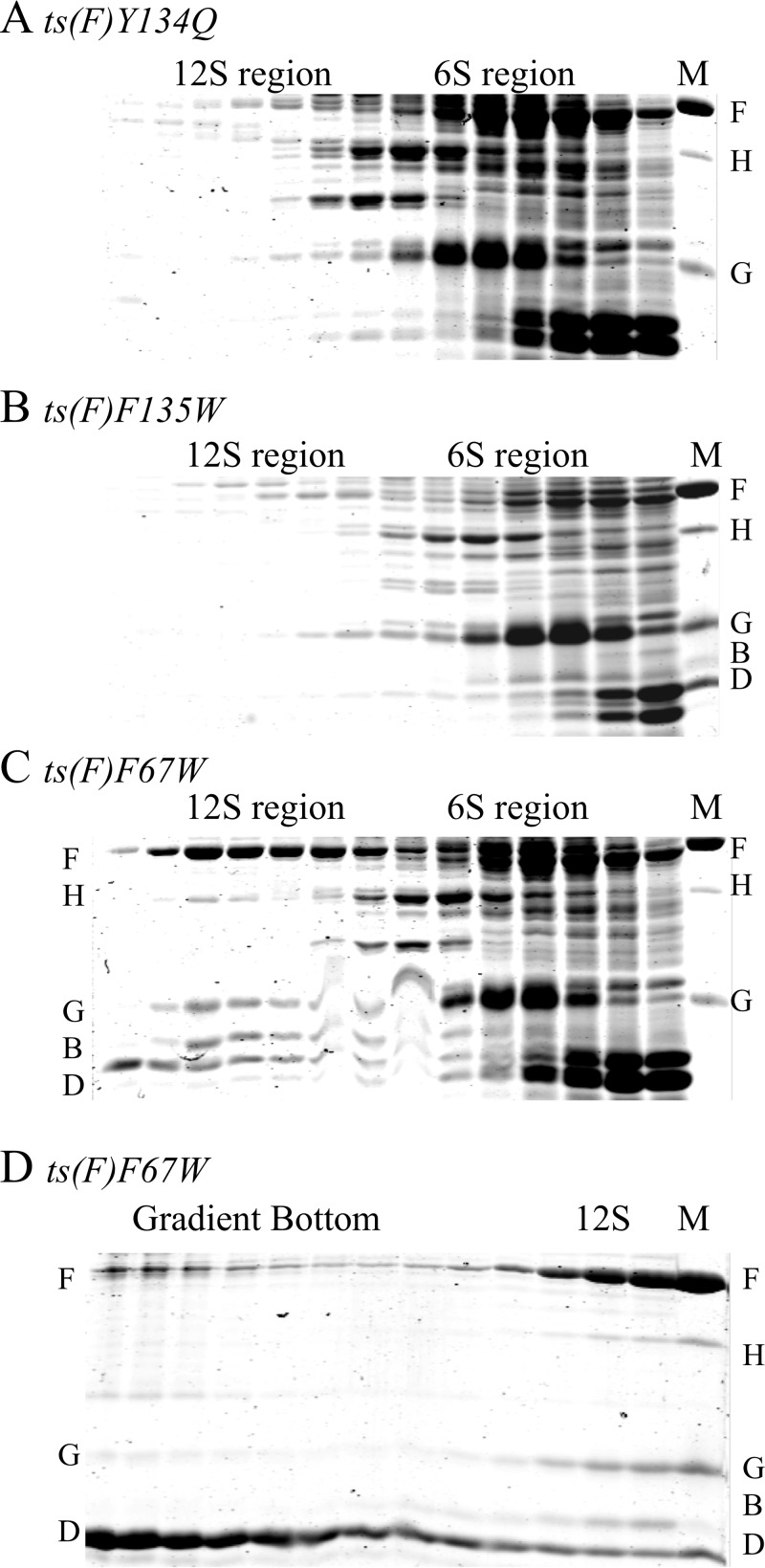
Using SDS-PAGE, gradient fractions were examined for the presence of soluble, early assembly intermediates: 6S, 9S*, and 12S* particles (Fig. 4). Fourteen mutants yielded results similar to those presented in Fig. 4A and B (data not shown). Although the coat protein was detected in whole-cell lysates (data not shown) and soluble 6S G protein spikes are readily apparent with the 6S region of the gel, soluble coat protein was not detected in gradient fractions. Thus, the mutant coat proteins were most likely insoluble, which can arise by at least two documented mechanisms: blocking coat interactions with either protein B (16) or protein G (39). As seen in other systems, but not rigorously documented with ϕX174, improperly folded coat protein monomers may also result in aggregation (40, 41).
FIG 4.
SDS-PAGE gels of early assembly intermediates (S value < 15) isolated from ts(F) mutant-infected cells at restrictive temperatures. Gels depict gradient fractions. Faster-sedimenting fractions are on the left. Approximate S values are given atop the gels. The marker (M) lane contained either purified virions (A, C, and D) or a mix of virions and procapsids (B). The positions of the viral coat F, major spike G, minor spike H, internal scaffolding B and external scaffolding D proteins are indicated. (A, B, and C) Small assembly intermediates respectively isolated from ts(F)Y134Q mutant-, ts(F)F135W mutant-, and ts(F)F67W mutant-infected cells at restrictive temperatures. (D) SDS-PAGE of fractions containing faster-sedimenting particles (S value > 15) isolated from ts(F)F67W mutant-infected cells. The gels in panels C and D are from the same gradient.
The results obtained for the ts(F)F67W mutant were unique (Fig. 4C and D). A soluble coat protein-containing intermediate was detected within the 12S region of the gradient. However, the stoichiometry of this particle differs from that of the well-characterized 12S* assembly intermediate. In addition to containing the coat F, internal scaffolding B, spike G, and DNA pilot H proteins, this particle also contained the external scaffolding D protein (Fig. 4C). The relative intensities of the coat and external scaffolding protein bands do not remain constant. As can be seen in Fig. 4D, the external scaffolding D/coat F protein ratios increase as S values increase. This suggests that after capsid morphogenesis was nucleated, 12S* particles were not incorporated during elongation.
Reversion analyses: both intragenic and extragenic second-site suppressors can restore viability.
To further elucidate the defects conferred by the parental mutations, a reversion analysis was conducted. Most likely due to recombination with the cloned F gene required for absolute lethal mutant propagation, it was extremely difficult to grow stocks of these strains with suitably low reversion frequencies. Thus, they were excluded from the analysis. Although ts+ revertants were isolated for all 14 starting parental mutants, same-site reversion events predominated. Seven of the 14 parental mutants did not yield second-site suppressors. However, the selections were far from exhaustive. In many instances, only one stock was used. Therefore, one revertant genotype could be highly overrepresented within a single ts+ revertant pool (42). To circumvent this problem, known second-site suppressors were crossed into several backgrounds for which suppressors were not isolated by direct selection (see below).
The suppressors and the parental background in which they were isolated are listed in Table 2. The entire genome of each strain was sequenced. No other mutations were detected, indicating that the su substitutions were both necessary and sufficient to confer the ts+ phenotype. The suppressors are named to reflect the amino acid changed within the protein in which they reside. For example, su-F L236H indicates that the suppressor mutation is in gene F (su-F) and changes the leucine residue at position 236 to histidine (L236H). Two suppressors were extragenic, one conferring an amino acid change in the external scaffolding protein D, su-D E148K, and the other conferring an amino acid substitution in the internal scaffolding protein B, su-B V114F. As with rescue by exogenous cloned B gene expression (Table 1), extragenic suppressors indicate that the primary defect conferred by the parental mutations, F67W and F135W, was not coat protein folding.
TABLE 2.
Isolation of second-site suppressors
| Starting mutant | Suppressor frequency(ies)a (no. of suppressors/total) | Suppressorb isolated | No. of independent events | Restrictivec plating efficiency |
|---|---|---|---|---|
| ts(F)F67N | 2/12 | su-F L236H | 1 | 0.9 |
| ts(F)F67W | 6/6, 5/12 | su-F Y199F | 1 | 0.8 |
| su-F H203R | 1 | 1.0 | ||
| su-F V318A | 2 | 0.8 | ||
| su-F Y353C | 1 | 0.9 | ||
| su-D E148K | 1 | 1.0 | ||
| su-FRBSA991G/-F S1Fd | 1 | 0.5 | ||
| su-F S1F | 0.2 | |||
| su-FRBSA991G | 10−4 | |||
| ts(F)Y134Q | 6/6, 5/6, 2/8 | su-F L49F | 3 | 0.7 |
| ts(F)Y134T | 1/12 | su-F L49F | 1 | 1.0 |
| ts(F)F135C | 0/5, 5/18 | su-F S62A | 1 | 0.9 |
| ts(F)F135I | 12/12, 0/4 | su-F L49F | 1 | 0.4 |
| ts(F)F135W | 1/3, 3/10. 1/2 | su-B V114F | 3 | 1.0 |
| l(F)F67D | 0/8 | NAe | NA | NA |
| ts(F)Y134D | 0/12 | NA | NA | NA |
| ts(F)Y134E | 0/12 | NA | NA | NA |
| ts(F)Y134L | 0/12 | NA | NA | NA |
| ts(F)Y134V | 0/12 | NA | NA | NA |
| ts(F)Y134R | 0/10 | NA | NA | NA |
| ts(F)F135H | 0/12 | NA | NA | NA |
Total number of suppressors/total number of revertants sequenced. Multiple ratios represent the results from independent selections.
Suppressor nomenclature: the letter after “su” indicates the gene in which the suppressor resides. The letter and numbers indicate the wild-type amino acid, the position within the primary structure, and the amino acid change conferred by the suppressor. Thus, in the designation su-F L236H, the suppressor is in gene F and confers an L→H substitution for amino acid 236.
Titer at the parental mutant's restrictive temperature/titer at the parental mutant's permissive temperature.
The suppressor contained two mutations, one at the beginning of gene F and one directly upstream of the gene A991G. The two putative suppressors were separated. Each one was then tested for the ability to suppress the ts phenotype.
NA, not applicable.
One suppressor, su-F L49F, was independently isolated in three different genetic backgrounds. A non-allele-specific suppressor may indicate that the molecular defects conferred by the parental mutations are related. One strain, su-FRBSA991G/-F S1F/ts(F)F67W, contained two mutations. The A991G mutation confers a nucleotide change in the gene F ribosome-binding site. These two mutations were separated and individually assayed for the suppressing phenotype. Only su-F S1F displayed activity on the level of plaque formation (Table 2). Several other assays were performed with the su-FRBSA991G/-F S1F/ts(F)F67W and su-FS1F/ts(F)F67W strains to determine whether the A991G mutation contributed to suppression. Although the mutation did confer a modest reduction in F protein levels (approximately 25%), no significant differences were observed in plaque size, burst size, or the kinetics of virion production (data not shown). Thus, if the A991G mutation contributed to suppression, its effects were too subtle to detect with these assays.
The broad allele specificity of the second-site suppressors and its functional implications.
To determine whether parental defects were related, suppressors were moved into backgrounds in which they were not isolated. Three criteria were used to select the parental mutants for this study. The first criterion was rescue by exogenous B gene expression; both rescued [ts(F)F67N, ts(F)Y134, ts(F)F135I, ts(F)Y134V, and ts(F)Y134Q] and nonrescued [ts(F)F67W, ts(F)Y134E, ts(F)F135H, and ts(F)F135W] mutants were used. The second criterion was the inability to isolate second-site suppressors in direct genetic selections, e.g., ts(F)Y134E, ts(F)F135H and ts(F)Y134V. The third criterion was whether soluble coat protein-containing intermediates were detected at restrictive temperatures: ts(F)F67W versus all other mutants.
Two intragenic suppressors and the two extragenic suppressors were placed in the aforementioned genetic backgrounds. The extragenic suppressors were chosen to distinguish between assembly and protein folding defects as previously discussed. The intragenic su-F L49F was isolated in several backgrounds and might display the most relaxed allele specificity. The su-F H203R mutation, which was isolated in the F67W background, resides in a large α-helix at the 3-fold axis of symmetry (43, 44). Mutations in this helix are known to suppress defects in external scaffolding protein D function (17, 18, 20, 45, 46).
To ascertain whether the suppressor was both necessary and sufficient to restore viability, the newly constructed su/ts(F) strains were assayed for plaque formation at their restrictive temperatures (Table 3). Each suppressor exhibited activity in at least one additional background. However, there was no apparent correlation between su/ts(F) viability at restrictive temperatures and (i) parental mutant rescue by exogenous B gene expression or (ii) the solubility of coat protein-containing intermediates. For example, the su-F H203R suppressor was active in the ts(F)F67W, ts(F)F67N, and ts(F)Y134E backgrounds. Restrictive ts(F)F67W infections produced soluble intermediates, whereas ts(F)Y134E and ts(F)F67N infections did not. Exogenous expression of cloned B gene rescued ts(F)F67N but not ts(F)F67W or ts(F)Y134E. Similarly, the su-B V114F and su-F L49F mutations suppress parental defects that were both rescued and not rescued by exogenous B gene expression.
TABLE 3.
Cross suppressor analyses
| Parental mutant | B gene rescue and expression conditionb | EOP at parental restrictive temp with suppressors in the indicated genetic backgrounda |
||||
|---|---|---|---|---|---|---|
| No suppressor | su-BV114F | su-FL49F | su-FH203R | su-DE148K | ||
| ts(F)F67N | Yes | <10−5 | <10−5 | 0.8 | 0.5 | 0.1 |
| With cloned G gene expression | <10−5 | 0.1 | ||||
| ts(F)Y134Q | Yes | 10−5 | <10−5 | 0.7 | <10−4 | <10−4 |
| With cloned G gene expression | 10−5 | 0.05 | <10−4 | <10−4 | ||
| ts(F)Y134T | Yes | 10−5 | 10−4 | 1.0 | 10−4 | 10−5 |
| With cloned G gene expression | 10−5 | 0.07 | 10−4 | 10−5 | ||
| ts(F)Y134V | Yes | 10−5 | 10−4 | 0.9 | 10−5 | 10−4 |
| With cloned G gene expression | 10−5 | 10−4 | 10−5 | 10−4 | ||
| ts(F)F135I | Yes | <10−6 | 0.9 | 0.4 | 10−5 | 10−5 |
| With cloned G gene expression | <10−6 | 10−5 | 10−5 | |||
| ts(F)F67W | No | <10−4 | 1.0 | <10−5 | 1.0 | 1.0 |
| With cloned B gene expression | <10−4 | 10−3 | ||||
| With cloned G gene expression | <10−4 | <10−5 | ||||
| ts(F)Y134E | No | <10−6 | <10−2 | 10−4 | 0.9 | 10−4 |
| With cloned B gene expression | <10−6 | <10−2 | 1.2 | <10−2 | ||
| With cloned G gene expression | <10−5 | <10−2 | <10−2 | <10−2 | ||
| ts(F)F135H | No | <10−5 | <10−4 | 0.5pp | 10−4 | 10−4 |
| With cloned B gene expression | <10−5 | 0.4 | 0.7 | 0.2 | 0.6 | |
| With cloned G gene expression | <10−5 | 0.06 | ||||
| ts(F)F135W | No | <10−4 | 1.0 | 0.8 | <10−4 | |
| With cloned B gene expression | <10−4 | <10−4 | ||||
| With cloned G gene expression | <10−4 | <10−4 | ||||
Plating experiments were conducted at the restrictive and permissive temperatures of the parental mutant. Plating efficiency equals titer at elevated temperature/titer at lower temperature. EOP, efficiency of plating; pp, pinprick plaque morphology.
Indicates whether the parental mutant is rescued by the exogenous expression of a cloned B gene. No parental mutants were rescued by the exogenous expression of a cloned G gene.
If no rescue was observed in the above-described assay or in previous experiments utilizing exogenous cloned-gene expression, a second assay was performed. Restrictive su/ts(F) plaque formation was examined in cells expressing cloned B, D, G, or J genes. For example, the ts(F)F67W, ts(F)Y134E, ts(F)F135H, and ts(F)F135W mutants were not rescued by exogenous expression of cloned B gene, and eight of their corresponding su/tsF strains were not viable or plated very weakly at restrictive temperatures: suF-L49F/ts(F)F67W, suF-L49F/ts(F)Y134E, suB-B114V/ts(F)Y134E, suF-L49F/ts(F)F135H, suB-V114F/ts(F)F135H, suF-H203R/ts(F)F135H, suD-E148K/ts(F)F135H, and suF-H203R/ts(F)F135W. Thus, it was possible to determine whether the combination of the suppressor and exogenous B gene expression could restore high-temperature viability. Rescue was observed for six out of the eight su/ts(F) strains, indicating that the suppressors exhibited some level of activity (Table 3). Thus, the suppressor and exogenous B gene expression are necessary, but neither alone is sufficient for rescue. If a parental mutant was rescued by exogenous B gene expression, the su/ts(F) strain was also rescued.
Similar experiments were conducted with cells expressing cloned external scaffolding protein D, major spike protein G, or DNA binding protein J genes with all nonviable su/ts(F) strains. Four of the su/ts(F) mutants were weakly rescued by the overexpression of the cloned major spike G gene: su-B V114F/ts(F)F67N, su-B V114F/ts(F)Y134T, su-B V114F/ts(F)F135H, and su-B V114F/ts(F)Y134Q. The corresponding parental mutants did not form plaques under these conditions. Thus, the combination of the su-B V114F mutation and elevated spike protein levels in some genetic backgrounds can act synergistically. Increasing the intracellular levels of the external scaffolding protein D and DNA binding protein J did not restore viability to any nonviable su/ts(F) mutants.
The su-F L49F mutation appears to have the most relaxed allele specificity, exhibiting some degree of activity in all backgrounds. In some instances, it was both necessary and sufficient to suppress the ts phenotype, raising plating efficiencies to near 1.0 [e.g., ts(F)F67N, ts(F)Y134Q, ts(F)Y134T, ts(F)Y134V, ts(F)F135H, and ts(F)F135W]. However, in two backgrounds, ts(F)F67W and ts(F)Y134E, the suppressor required exogenous B gene expression to restore viability. Exogenous B gene expression did not rescue the last two parental mutants. No suppressor displayed strict allele specificity. Considering the various phenotypes of the parental mutants, this suggests that the suppressor and/or parental mutations confer pleiotropic effects, affecting more than one assembly step or interaction (see Discussion).
The suppressing internal scaffolding protein is dominant, whereas the suppressing external scaffolding protein is recessive.
To further characterize the extragenic suppressors, su-B V114F and su-D E148K, dominance assays were conducted with cells exogenously expressing cloned, wild-type internal or external scaffolding genes (Table 4). Extragenic suppressors rescued the parental mutants used in these assays, but exogenous wild-type B or D gene expression did not restore viability. Thus, these assays determine whether the presence of wild-type scaffolding proteins interferes with the activity of the suppressing proteins. The ability of the su-D E148K mutation to suppress was recessive. The exogenous expression of the wild-type external scaffolding protein D gene significantly lowered the plating efficiency of the su-D E148K/ts(F)F67N and su-D E148K/ts(F)F67W mutants. This effect was specific to the expression of gene D. No inhibition was observed when the su-D E148K/ts(F)F67W mutant was plated on cells expressing the cloned B gene. In contrast, the su-B V114F mutation appeared to be dominant. The plating efficiencies of the su-B V114F/ts(F)67W and su-B V114F/ts(F)F135W mutants were not affected by exogenous wild-type B gene expression. In one instance [su-B V114F/ts(F)F135H], the combination of increased wild-type B protein levels and the su-B V114F mutant appeared to act synergistically. In the absence of exogenous B gene expression, this mutant formed pinprick plaques, whereas exogenous B gene expression dramatically increased plaque size.
TABLE 4.
Effects of exogenous scaffolding gene expression on the plating efficiency of extragenic second-site su/ts(F) strainsa
| Mutant | Scaffolding gene | Plating efficiency under indicated condition of exogenous scaffolding gene expressiona |
|
|---|---|---|---|
| Without | With | ||
| ts(F)F67N | External scaffolding D | <10−5 | <10−5 |
| su-D E148K/ts(F)67N | External scaffolding D | 0.1 | <10−5 |
| ts(F)F67W | External scaffolding D | <10−4 | <10−4 |
| su-D E148K/ts(F)67W | External scaffolding D | 1.0 | 10−2 |
| Internal scaffolding B | 1.0 | ||
| ts(F)F67W | Internal scaffolding B | <10−4 | <10−4 |
| su-B V114F/ts(F)67W | Internal scaffolding B | 1.0 | 0.8 |
| ts(F)F135W | Internal scaffolding B | 10−4 | <10−3 |
| su-B V114F/ts(F)F135W | Internal scaffolding B | 1.0 | 1.2 |
| ts(F)F135H | Internal scaffolding B | <10−5 | 10−5 |
| su-B V114F/ts(F)F135H | Internal scaffolding B | <10−5 | 0.4 |
Plating experiments were conducted at the restrictive and permissive temperatures of the parental mutant. Plating efficiency equals titer at elevated temperature/titer at lower temperature. The su-B V114F mutation resides in gene B, which encodes the internal scaffolding protein. The su-D E148K mutation resides in gene D, which encodes the external scaffolding protein.
DISCUSSION
Aromatic amino acids mediate most ϕX174 internal scaffolding-coat protein interactions. The most extensive contacts occur between internal scaffolding protein F120 and coat F protein residues F67, Y134, and F135 (37, 38). These four amino acids constitute the core of the B protein binding cleft (Fig. 1B). Nonaromatic substitutions for B protein F120 greatly reduce or totally eliminate coat-internal scaffolding interactions (16). In addition to F120, five other internal scaffolding aromatic residues are involved in contacts. However, these residues are not located in the core and interact with nonaromatic coat protein residues. Although mutationally altering these associations kinetically traps early assembly intermediates, coat-scaffolding interactions are maintained. In this study, the chemical requirements of the three core coat protein residues were determined. They appear to be less stringent than for F120 vis-à-vis aromaticity. This may reflect an inherent functional redundancy: the coat protein contributes three aromatic amino acids to the core, whereas the B protein contributes just one.
Phenotype and the geometry of the B protein-binding site.
The substitutions and the resulting phenotypes are summarized in Fig. 1D and Table 1 and in Table S1 in the supplemental material. Although every substitution was not recovered at all three sites, some limited correlations can be made. For all three sites the most conservative substitutions, Y→F or F→Y, resulted in wild-type phenotypes. In addition, methionine substitutions were well tolerated. Although the molecular basis is poorly understood, methionine residues are known to create stable interaction with aromatic rings (47).
Substitutions involving tryptophan, the largest aromatic amino acid, and histidine, a charged aromatic residue, at the three sites conferred different phenotypes. Y134→H or W mutations did not dramatically impair viability. In contrast, H and W substitutions at the other two locations produced more severe phenotypes. Within the context of the core's geometry, the orientation of the 134 side chain may more easily accommodate the bulkier tryptophan side chain. Moreover, charged residues were better tolerated at position 134. Unlike Y134, both F67 and F135 reside near charged residues, R239 and R290, respectively. Thus, the introduction of a partially protonated imidazole ring may be more consequential. Lastly, the size of the substituted side chain may also affect phenotype, at least at the Y134 site, for which the most mutations were isolated. For example, Y→S was less detrimental than Y→T. Similarly, Y→N was better tolerated than Y→Q.
Molecular and evolutionary implications of recessive phenotypes.
On the level of plaque formation, both frequency and size, all mutants were efficiently rescued by exogenous coat F gene expression. The plasmid- and genome-borne genes appeared to contribute equally to coat protein levels during infection (Fig. 2). Moreover, the expression of cloned lethal genes did affect wild-type plaque formation. Thus, the mutations exhibited a recessive phenotype. As most of the mutations do not appear to affect protein folding, two models may explain the recessive phenotypes. Heterogeneous pentamers, composed of mutant and wild-type coat proteins, may be productive assembly intermediates. Alternatively, coat proteins could be cotranslationally assembled into pentamers. Although cotranslational assembly of protein complexes has recently garnered much attention (48–50), it is technically difficult to demonstrate. Therefore, documented examples are sparse. If pentamer formation were a cotranslational phenomenon, mutant complexes would not interfere with wild-type pentamers, as they would not enter the assembly pathway. Evolutionarily this could promote speciation, allowing mutants to be maintained in a high-MOI environment, assisted by helper viruses, until compensatory mutations arise.
Unique versus pleiotropic defects, which reduce flux through the assembly pathway during multiple reactions.
Several ts(F) mutants were rescued by exogenous B gene expression (Table 1), which suggests a reduced affinity for the internal scaffolding protein. Thus, elevating the intracellular level of protein B may drive this reaction forward. In contrast, the lack of rescue is more difficult to interpret. Increasing B protein concentrations may not restore binding and/or the primary defect occurs after coat-internal scaffolding protein contact.
The results of the physiological, genetic, and second-site genetic analyses suggest that a specific defect cannot be easily assigned to the various mutant proteins. Many mutants could be rescued via (i) independently elevating the level of more than one coat-interacting protein, (ii) extragenic and/or intragenic suppressors, or (iii) a combination of the two mechanisms. The mutant assembly pathways discussed herein are diagrammed in Fig. 5. ts(F)Y134Q is rescued by increased intracellular levels of the internal scaffolding protein B, indicating that one defect may be inefficient internal scaffolding protein binding. In addition, increased intracellular levels of the external scaffolding protein D rescued this mutant. This implies that during the course of a normal infection (no exogenous gene expression), the mutant protein can form 12S* particles. However, the concentration of these particles may be too low and/or they have a reduced affinity for the external scaffolding D protein. The latter barrier can be overcome by increasing the intracellular D protein concentration. Thus, the parental mutation appeared to have pleiotropic effects, reducing the flux through the pathway at a minimum of two points. Rescue via increased intracellular D protein concentrations was considerably weaker than rescue by increased B protein concentrations. This is consistent with the temporal steps of the assembly pathway. The number of assembly intermediates that can be driven through morphogenesis will decrease with each successive step. Similarly, ts(F)F67N was efficiently rescued by cloned B gene expression, suggesting a B protein binding defect. It is also rescued, albeit more weakly, by the su-D E148K mutation, which resides in the external scaffolding protein and likely affected the procapsid elongation reaction.
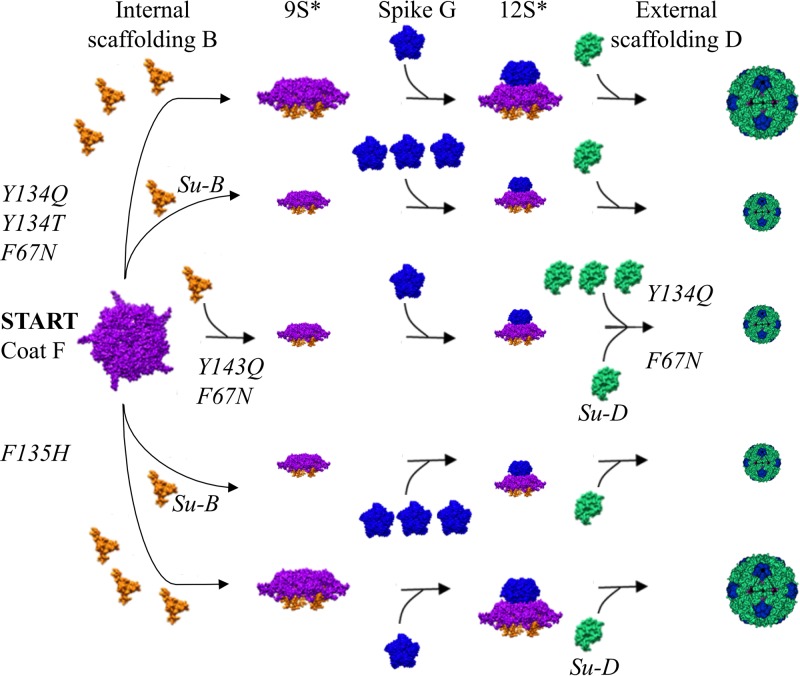
FIG 5.
Mutant assembly pathways. The assembly pathways diagram the various ways ts(F) mutants can be rescued via (i) elevating wild-type internal scaffolding, external scaffolding, and major spike protein levels, (ii) extragenic suppressors, or (iii) a combination of the two mechanisms. Multiple copies of internal scaffolding, external scaffolding, and major spike proteins represent elevated protein levels, which were achieved by the exogenous expression of a cloned gene. The smaller size of the depicted assembly 9S* and 12S* assembly intermediates represents a hypothesized concentration decrease within the reduced flux model described in the text. The size of procapsid conveys the efficiency of rescue. Small procapsids represent weak rescue as described in Table 3. Su-B and Su-D, second-site suppressors in the internal B and external D scaffolding proteins, respectively.
Interpreting results was more complex for the su/ts(F) double mutants. However, for strains with extragenic suppressors, the data suggest that flux through the assembly pathway was affected at multiple steps. For example, ts(F)F135H was rescued neither by exogenous internal scaffolding B gene expression nor by the extragenic suppressor located in the external scaffolding protein, suD-E148K. The external scaffolding protein enters the assembly pathway after the formation of the pentameric 12S* assembly intermediate. In contrast, su-D E148K/ts(F)F135H was efficiently rescued by exogenous B gene expression. Interpreted within the reduced flux model, the ts(F)F135H protein does not efficiently bind the B protein, the first defect. If the reaction is driven forward by increasing B protein concentration, the resulting 12S* particle cannot participate in productive assembly, the second defect, unless the external scaffolding protein contains the su-D E148K suppressor.
Lastly, the su-B V114F suppressor, in some mutant backgrounds, conferred the ability to be rescued by the exogenous expression of the major spike G gene. In these instances, the parental mutations were efficiently rescued by cloned B gene expression [ts(F)F67N, ts(F)Y134Q, and ts(F)Y134T] or rescued required both the su-B V114F mutation and cloned B gene expression [ts(F)F135H] (Table 3). Rescue by exogenous G gene expression was considerably less robust than rescue by exogenous B gene expression. Again, this is consistent with the temporal steps of the assembly pathway and a reduced flux model. Coat protein pentamers interact with the internal scaffolding protein before interacting with the G protein spikes.
Possible mechanisms of suppression.
When the suppressing mutations are mapped onto the atomic structure of the procapsid, some, but not all, provide mechanistic insights (Fig. 6). The suppressor in protein B may strengthen and/or correct defective internal scaffolding protein interactions. Coat-internal scaffolding contacts are primarily mediated by aromatic amino acids (15, 16). The internal scaffolding suppressor confers a V→F substitution at position 114. The orientation of this side chain could position an aromatic ring to interact with the other rings, including B protein residue F120 (Fig. 1C), thereby stabilizing the association.
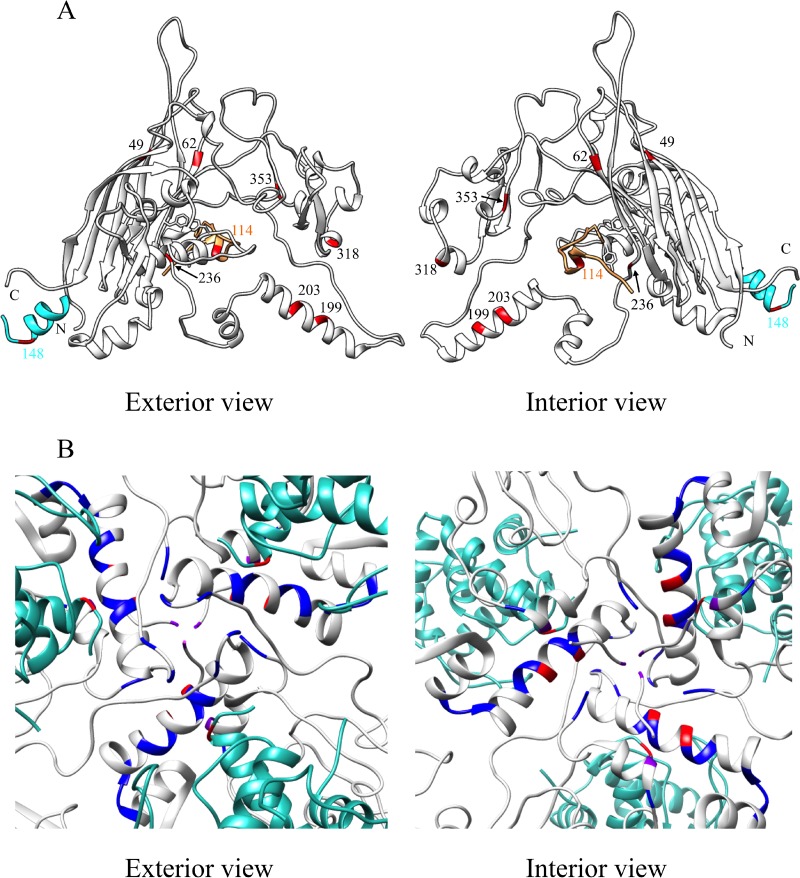
FIG 6.
Locations of the second-site suppressors. (A) The viral coat protein is depicted gray, the internal scaffolding protein in orange, and the D4 external scaffolding protein unit in cyan. There are four D protein subunits per asymmetric unit. Only a portion of the scaffolding proteins are depicted. The residues in which the suppressors reside are depicted in red. The color of the numerical labels indicates the affected protein and the suppressor's location in the primary structure. The side chains of residues F67, Y134, and F135 in F protein and F120 in B protein are included to identify the core of the binding cleft. (B) The 3-fold axis of symmetry. The blue, red, and purple colors identify the locations of the suppressors. Red indicates suppressors identified in this study. Blue indicates suppressors of defective external scaffolding protein function isolated in previous studies. Purple indicates those suppressors identified in this study that were identical and independently isolated in previous studies.
The suppressors located in the coat protein confer changes in the large insertions loops or in the terminal amino acids of the β-strands that compose the protein's β-barrel core (Fig. 6A). Most assembly suppressors that correct defects associated with mutant and/or foreign internal scaffolding B, external scaffolding D, spike G and host cell rep helicase proteins also reside in these regions (15–18, 20, 39, 45, 46, 51–57). This suggests that loop regions provide the requisite flexibility for conformational switches and protein-protein interactions during assembly. Thus, once folded, the contribution of the β-barrel core to morphogenesis is minimal.
Many of the suppressors of ts(F)F67W mutation are located in a large α-helix at the 3-fold axis of symmetry (Fig. 6B). The suppressing substitutions are very similar or identical to previously characterized mutations that correct defective external scaffolding function (17, 18, 45). This is consistent with the biochemical characterization of the mutant assembly pathway. After nucleation, 12S* particles did not appear to be incorporated during the procapsid elongation reaction (Fig. 4C and D). Indeed, one of the second-site suppressors mapped to the external scaffolding D gene. The altered amino acid also resides at the 3-fold axis.
Although trends were apparent when the allele specificity of the suppressors was examined, no strict correlations emerged. The allele specificity of the su-F H203R and su-D E148K mutations illustrates this point. As defined by rescue via exogenous B gene expression, the parental mutations genetically fell into two classes. In general, these suppressors did not display any activity when combined with parental mutations rescued by elevating B protein levels (Table 3). However, they rescued ts(F)F67N. Biochemically, at least on a superficial level, the parental mutants displayed two phenotypes vis-à-vis the solubility of coat protein containing intermediates. Yet some suppressors were active in both types of parental backgrounds. If parental mutations confer pleiotropic defects during assembly, then they may be rescued by multiple mechanisms. Alternatively, the suppressors may be pleiotropic, able to suppress several types of defects, and/or some parental mutations may confer related defects that cannot be discerned by standard assays. The elongation defect associated with ts(F)F67W was the easiest to discern; the assembly products remain soluble. Other mutations may also confer elongation defects but the resulting products are insoluble. Thus, the suppressors could act by inhibiting defective off-pathway elongation reactions, regardless of the solubility of the off-pathway product.
Pleiotropic mutations are quite common in viruses. However, they are usually documented as tradeoffs, secondary phenotypes, or defects associated with suppressing, adaptive, or resistance mutations that lower fitness when selective pressures are removed (40, 58–60). Moreover, the molecular bases of the secondary defects are rarely characterized. The mutations described herein appear to affect multiple steps within a single pathway, thus affecting the overall morphogenetic flux. While this likely occur in other viruses, the phenomenon may be more difficult to define without the extensive biochemical and genetic tools available for the ϕX174 system.
MATERIALS AND METHODS
Phage plating, media, buffers, and stock preparation.
Plating, media, buffers, and stock preparation have been described previously (51).
Bacterial strains and plasmids.
The Escherichia coli C strains C122 and BAF30 (recA) have been described previously (51, 61). RY7211 contains a mutation in the mraY gene, conferring resistance to ϕX174-mediated lysis (62). The construction of the complementing plasmids for gene B (63), gene D (20), and gene G (39) is reported in the literature. All three genes are induced by isopropyl β-d-1-thiogalactopyranoside (IPTG). The rhamnose-induced complementing clone of gene F (64) was a gift from D. Endy and P. Jaschke. To obtain a complementing clone of gene J, the J gene was amplified by PCR with primers that introduce an upstream NcoI site and a downstream HindIII site. After amplification, the purified fragment was digested with NcoI and HindIII and ligated into pSE420 (Invitrogen) digested with the same enzymes. The gene is under IPTG induction.
ssDNA isolation and purification.
Viral single-stranded DNA (ssDNA) was isolated by the OLT (Our Little Trick) protocol. Twenty-five milliliters, or a multiple thereof, of lysis-resistant cells was grown to a concentration of 1.0 × 108 cells/ml in TKY medium (1.0% tryptone, 0.5% KCl, 0.5% yeast extract). Medium was supplemented with 10 mM MgCl2 and 5.0 mM CaCl2 at the time of infection. Cells were infected at a multiplicity of infection (MOI) of 3. As ssDNA biosynthesis and packaging are concurrent processes (65), infections must occur at a permissive temperature to ensure virion production and cells cannot contain plasmid DNA. After a 3-h incubation at 37°C, 4-h incubation 33°C, or 6-h incubation at 24°C, cells were concentrated by centrifugation. Each 25-ml volume is treated as one sample in a Wizard plus SV miniprep kit (Promega) by following the manufacturer's instructions but with one critical modification. The incubation time with the alkaline protease is extended to 30 min. Typical ssDNA yields are approximately 100 ng/μl. Copurification of replicative-form double-stranded DNA is negligible. However, the presence of plasmid DNA considerably reduces ssDNA yield and purity.
Generation of mutations in the F67, Y134, and F135 codons of gene F.
Amber mutants were separately generated at the F67, Y134, and F135 codons by site-directed mutagenesis as previously described (45). Two approaches were taken to generate missense mutants with temperature sensitive (ts), cold-sensitive (cs), and silent phenotypes. In the first approach, amber revertants were directly selected by plating on E. coli C122 (sup0). In the second approach, amber mutant DNA was mutagenized with randomizing primers in PCRs with Q5 DNA polymerase (New England BioLabs) as previously described (66). For both protocols, wild-type cells (sup0) were used and progeny were recovered at a variety of temperatures: 24°C, 33°C, and 42°C. The resulting plaques were then stabbed into indicator lawns incubated at 24°C, 33°C, and 42°C and assigned preliminary phenotypes: cs, ts, or resembling the wild type, i.e., no temperature restrictions. The isolation and the initial screening temperatures (24°C, 33°C, and 42°C) were chosen to span a broad range of growth conditions. These may differ from the temperatures listed in Table 1, which were refined when phenotypes were further tested in plating assays. To generate lethal mutants, wild-type DNA was mutagenized, as described above, using codon-randomizing primers or primers designed to introduce codons for aspartic and glutamic acid (GAG/C) or lysine and arginine (AG/AA). Mutagenized DNA was transfected into cells harboring a complementing F clone at 37°C. Progeny were screened for complementation-dependent phenotypes by stabbing plaques into indicator lawns seeded with the wild-type cells with and without the complementing F plasmid.
Isolation of second-site suppressors (su) and the construction of su/ts(F) mutants for the cross-suppressor analysis.
Second-site suppressors were selected by plating approximate 1.0 × 106 particles on E. coli C122 at restrictive temperatures. DNA from revertant plaques was first sequenced in gene F to distinguish between same-site and second-site events. The entire genomes of the su/ts(F) strains were sequenced to verify the identity of the suppressing mutation and to ensure that it was both necessary and sufficient to confer the suppressing phenotype. Suppressors were moved into ts(F) backgrounds in which they were not isolated by Q5 DNA polymerase PCRs as previously described (66). ts(F) mutant DNA served as the templates in these reactions, and the primers introduced the second-site suppressors.
Rate zonal sedimentation and protein electrophoresis.
The protocols for ssDNA, double-stranded replicative form (RF) DNA isolation and purification, rate zonal sedimentation, and protein electrophoresis protocols have been previously described (18, 52). To determine the relative ratio of viral proteins, densitometry was performed on digitized gels using the ImageJ (NIH) program.
Propagation of mutants.
Although it was possible to generate ts(F) mutant stocks with high enough titers (>1010) and low enough reversion frequencies (<10−4) by standard protocols (51) at permissive temperatures, suitable stocks of the lethal mutants could not be generated by this technique, which typically resulted in very low titers and/or very high reversion frequencies, ∼10−2 to 10−1. Presumably, high reversion frequencies arose from recombination with the F gene plasmid. To circumvent this problem, lethal mutants were propagated by successive infections in lysis-resistant cells bearing the complementing plasmid. To begin, small plaques were picked and resuspended in 100 μl of HFB buffer (0.06 M NH4Cl, 0.09 M NaCl, 0.1 M KCl, 0.1 M Tris-HCl [pH 7.4], 1.0 mM MgSO4, 1.0 mM CaCl2) and titers were determined to ensure that the reversion frequency was below 10−4. Ten milliliters of plasmid-containing cells was then grown to a concentration of 1 × 108 cells/ml in TKY medium. Ten minutes before infection, medium was supplemented with 100 mM rhamnose, 10 mM MgCl2, and 5 mM CaCl2. In the first round of infection, cells were typically infected at a multiplicity of infection of 10−5 and incubated for 3 h at 37°C. Cells were concentrated by centrifugation, resuspended in 200 μl of HFB buffer, and lysed overnight at 4°C by the addition of egg white lysozyme (1.0 mg/ml). The protocol was repeated, typically two times at increasing MOI, until titers were over 1010 virions/ml, the concentration required to characterize the assembly pathway in vivo. Reversion frequencies typically ranged between 10−3 and 10−2, a background level low enough for biochemical analyses but still too high for genetic analyses.
Supplementary Material
ACKNOWLEDGMENTS
We thank D. Endy and P. Jaschke for providing the complementing clone of gene F, and M.D.L. Johnson for discussions.
This research was supported by National Science Foundation grant MCB-1408217 (B.A.F.) and The BIO5 Institute at the University of Arizona.
We also acknowledge the support of programs in which undergraduate and high school students participated: the undergraduate biology research program, the Steps 2 STEM program, and the advanced math/science research programs at Berkshire School and Flowing Wells High School. Phage systems are ideal tools for teaching fundamental genetic concepts. Accordingly, the four high school students (A.R.P., R.H.V., C.J.J., and E.C.T.) conducted the extensive reversion and cross-suppressor suppressor analyses under the supervision of their teachers (M.H. and A.D.B.) at their schools or laboratory personnel (A.P.R. and B.A.F.) at the University of Arizona.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01384-17.
REFERENCES
- 1.Fane BA, Prevelige PE Jr. 2003. Mechanism of scaffolding-assisted viral assembly. Adv Protein Chem 64:259–299. doi: 10.1016/S0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 2.Prevelige PE, Fane BA. 2012. Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Adv Exp Med Biol 726:325–350. doi: 10.1007/978-1-4614-0980-9_14. [DOI] [PubMed] [Google Scholar]
- 3.Zlotnick A, Fane BA. 2010. Mechanisms of icosahedral virus assembly, p 180–202. In Agbandje-McKenna M, McKenna R (ed), Structural virology. Royal Society of Chemistry, London, United Kingdom. [Google Scholar]
- 4.Moore SD, Prevelige PE Jr. 2002. Bacteriophage P22 portal vertex formation in vivo. J Mol Biol 315:975–994. doi: 10.1006/jmbi.2001.5275. [DOI] [PubMed] [Google Scholar]
- 5.Parker MH, Casjens S, Prevelige PE Jr. 1998. Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol 281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- 6.Botstein D, Waddell CH, King J. 1973. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. I. Genes, proteins, structures and DNA maturation. J Mol Biol 80:669–695. [DOI] [PubMed] [Google Scholar]
- 7.Kikuchi Y, King J. 1975. Genetic control of bacteriophage T4 baseplate morphogenesis. III. Formation of the central plug and overall assembly pathway. J Mol Biol 99:695–716. [DOI] [PubMed] [Google Scholar]
- 8.Tonegawa S, Hayashi M. 1970. Intermediates in the assembly of ϕX174. J Mol Biol 48:219–242. doi: 10.1016/0022-2836(70)90158-0. [DOI] [PubMed] [Google Scholar]
- 9.Moore SD, Prevelige PE Jr. 2002. A P22 scaffold protein mutation increases the robustness of head assembly in the presence of excess portal protein. J Virol 76:10245–10255. doi: 10.1128/JVI.76.20.10245-10255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tuma R, Tsuruta H, French KH, Prevelige PE. 2008. Detection of intermediates and kinetic control during assembly of bacteriophage P22 procapsid. J Mol Biol 381:1395–1406. doi: 10.1016/j.jmb.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D'Lima NG, Teschke CM. 2015. A molecular staple: D-loops in the I domain of bacteriophage P22 coat protein make important intercapsomer contacts required for procapsid assembly. J Virol 89:10569–10579. doi: 10.1128/JVI.01629-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suhanovsky MM, Teschke CM. 2015. Nature's favorite building block: deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 479–480:487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suhanovsky MM, Parent KN, Dunn SE, Baker TS, Teschke CM. 2010. Determinants of bacteriophage P22 polyhead formation: the role of coat protein flexibility in conformational switching. Mol Microbiol 77:1568–1582. doi: 10.1111/j.1365-2958.2010.07311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suhanovsky MM, Teschke CM. 2011. Bacteriophage P22 capsid size determination: roles for the coat protein telokin-like domain and the scaffolding protein amino-terminus. Virology 417:418–429. doi: 10.1016/j.virol.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gordon EB, Fane BA. 2013. The effects of an early conformational switch defect during ϕX174 morphogenesis are belatedly manifested late in the assembly pathway. J Virol 87:2518–2525. doi: 10.1128/JVI.02839-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon EB, Knuff CJ, Fane BA. 2012. Conformational switch-defective ϕX174 internal scaffolding proteins kinetically trap assembly intermediates before procapsid formation. J Virol 86:9911–9918. doi: 10.1128/JVI.01120-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchiyama A, Chen M, Fane BA. 2007. Characterization and function of putative substrate specificity domain in microvirus external scaffolding proteins. J Virol 81:8587–8592. doi: 10.1128/JVI.00301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchiyama A, Fane BA. 2005. Identification of an interacting coat-external scaffolding protein domain required for both the initiation of ϕX174 procapsid morphogenesis and the completion of DNA packaging. J Virol 79:6751–6756. doi: 10.1128/JVI.79.11.6751-6756.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherwa JE Jr, Tyson J, Bedwell GJ, Brooke D, Edwards AG, Dokland T, Prevelige PE, Fane BA. 2017. ϕX174 procapsid assembly: the effects of an inhibitory external scaffolding protein and resistant coat proteins in vitro. J Virol 91:e01878-16. doi: 10.1128/JVI.01878-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherwa JE Jr, Uchiyama A, Fane BA. 2008. Scaffolding proteins altered in the ability to perform a conformational switch confer dominant lethal assembly defects. J Virol 82:5774–5780. doi: 10.1128/JVI.02758-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortines JR, Weigele PR, Gilcrease EB, Casjens SR, Teschke CM. 2011. Decoding bacteriophage P22 assembly: identification of two charged residues in scaffolding protein responsible for coat protein interaction. Virology 421:1–11. doi: 10.1016/j.virol.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padilla-Meier GP, Gilcrease EB, Weigele PR, Cortines JR, Siegel M, Leavitt JC, Teschke CM, Casjens SR. 2012. Unraveling the role of the C-terminal helix turn helix of the coat-binding domain of bacteriophage P22 scaffolding protein. J Biol Chem 287:33766–33780. doi: 10.1074/jbc.M112.393132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parent KN, Suhanovsky MM, Teschke CM. 2007. Polyhead formation in phage P22 pinpoints a region in coat protein required for conformational switching. Mol Microbiol 65:1300–1310. doi: 10.1111/j.1365-2958.2007.05868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon CL, King J. 1993. Temperature-sensitive mutations in the phage P22 coat protein which interfere with polypeptide chain folding. J Biol Chem 268:9358–9368. [PubMed] [Google Scholar]
- 25.Siden EJ, Hayashi M. 1974. Role of the gene beta-product in bacteriophage ϕX174 development. J Mol Biol 89:1–16. doi: 10.1016/0022-2836(74)90159-4. [DOI] [PubMed] [Google Scholar]
- 26.Chen M, Uchiyama A, Fane BA. 2007. Eliminating the requirement of an essential gene product in an already very small virus: scaffolding protein B-free ϕX174, B-free. J Mol Biol 373:308–314. doi: 10.1016/j.jmb.2007.07.064. [DOI] [PubMed] [Google Scholar]
- 27.Cherwa JE Jr, Young LN, Fane BA. 2011. Uncoupling the functions of a multifunctional protein: the isolation of a DNA pilot protein mutant that affects particle morphogenesis. Virology 411:9–14. doi: 10.1016/j.virol.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 28.Ruboyianes MV, Chen M, Dubrava MS, Cherwa JE Jr, Fane BA. 2009. The expression of N-terminal deletion DNA pilot proteins inhibits the early stages of ϕX174 replication. J Virol 83:9952–9956. doi: 10.1128/JVI.01077-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cherwa JE Jr, Organtini LJ, Ashley RE, Hafenstein SL, Fane BA. 2011. In vitro assembly of the ϕX174 procapsid from external scaffolding protein oligomers and early pentameric assembly intermediates. J Mol Biol 412:387–396. doi: 10.1016/j.jmb.2011.07.070. [DOI] [PubMed] [Google Scholar]
- 30.Prevelige PE Jr, Thomas D, King J. 1993. Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys J 64:824–835. doi: 10.1016/S0006-3495(93)81443-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang JR, Spilman MS, Rodenburg CM, Dokland T. 2009. Functional domains of the bacteriophage P2 scaffolding protein: identification of residues involved in assembly and protease activity. Virology 384:144–150. doi: 10.1016/j.virol.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fu CY, Morais MC, Battisti AJ, Rossmann MG, Prevelige PE Jr. 2007. Molecular dissection of ϕ29 scaffolding protein function in an in vitro assembly system. J Mol Biol 366:1161–1173. doi: 10.1016/j.jmb.2006.11.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oien NL, Thomsen DR, Wathen MW, Newcomb WW, Brown JC, Homa FL. 1997. Assembly of herpes simplex virus capsids using the human cytomegalovirus scaffold protein: critical role of the C terminus. J Virol 71:1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tuma R, Parker MH, Weigele P, Sampson L, Sun Y, Krishna NR, Casjens S, Thomas GJ Jr, Prevelige PE Jr. 1998. A helical coat protein recognition domain of the bacteriophage P22 scaffolding protein. J Mol Biol 281:81–94. doi: 10.1006/jmbi.1998.1916. [DOI] [PubMed] [Google Scholar]
- 35.Weigele PR, Sampson L, Winn-Stapley D, Casjens SR. 2005. Molecular genetics of bacteriophage P22 scaffolding protein's functional domains. J Mol Biol 348:831–844. doi: 10.1016/j.jmb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Burch AD, Fane BA. 2000. Efficient complementation by chimeric Microviridae internal scaffolding proteins is a function of the COOH-terminus of the encoded protein. Virology 270:286–290. doi: 10.1006/viro.2000.0306. [DOI] [PubMed] [Google Scholar]
- 37.Dokland T, Bernal RA, Burch A, Pletnev S, Fane BA, Rossmann MG. 1999. The role of scaffolding proteins in the assembly of the small, single-stranded DNA virus ϕX174. J Mol Biol 288:595–608. doi: 10.1006/jmbi.1999.2699. [DOI] [PubMed] [Google Scholar]
- 38.Dokland T, McKenna R, Ilag LL, Bowman BR, Incardona NL, Fane BA, Rossmann MG. 1997. Structure of a viral procapsid with molecular scaffolding. Nature 389:308–313. doi: 10.1038/38537. [DOI] [PubMed] [Google Scholar]
- 39.Doore SM, Fane BA. 2015. The kinetic and thermodynamic aftermath of horizontal gene transfer governs evolutionary recovery. Mol Biol Evol 32:2571–2584. doi: 10.1093/molbev/msv130. [DOI] [PubMed] [Google Scholar]
- 40.Aramli LA, Teschke CM. 1999. Single amino acid substitutions globally suppress the folding defects of temperature-sensitive folding mutants of phage P22 coat protein. J Biol Chem 274:22217–22224. doi: 10.1074/jbc.274.32.22217. [DOI] [PubMed] [Google Scholar]
- 41.Aramli LA, Teschke CM. 2001. Alleviation of a defect in protein folding by increasing the rate of subunit assembly. J Biol Chem 276:25372–25377. doi: 10.1074/jbc.M101759200. [DOI] [PubMed] [Google Scholar]
- 42.Luria SE, Delbruck M. 1943. Mutations of bacteria from virus sensitivity to virus resistance. Genetics 28:491–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McKenna R, Ilag LL, Rossmann MG. 1994. Analysis of the single-stranded DNA bacteriophage ϕX174, refined at a resolution of 3.0 A. J Mol Biol 237:517–543. [DOI] [PubMed] [Google Scholar]
- 44.McKenna R, Xia D, Willingmann P, Ilag LL, Krishnaswamy S, Rossmann MG, Olson NH, Baker TS, Incardona NL. 1992. Atomic structure of single-stranded DNA bacteriophage ϕX174 and its functional implications. Nature 355:137–143. doi: 10.1038/355137a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fane BA, Shien S, Hayashi M. 1993. Second-site suppressors of a cold-sensitive external scaffolding protein of bacteriophage ϕX174. Genetics 134:1003–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherwa JE Jr, Sanchez-Soria P, Wichman HA, Fane BA. 2009. Viral adaptation to an antiviral protein enhances the fitness level to above that of the uninhibited wild type. J Virol 83:11746–11750. doi: 10.1128/JVI.01297-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Valley CC, Cembran A, Perlmutter JD, Lewis AK, Labello NP, Gao J, Sachs JN. 2012. The methionine-aromatic motif plays a unique role in stabilizing protein structure. J Biol Chem 287:34979–34991. doi: 10.1074/jbc.M112.374504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shieh YW, Minguez P, Bork P, Auburger JJ, Guilbride DL, Kramer G, Bukau B. 2015. Operon structure and cotranslational subunit association direct protein assembly in bacteria. Science 350:678–680. doi: 10.1126/science.aac8171. [DOI] [PubMed] [Google Scholar]
- 49.Natan E, Wells JN, Teichmann SA, Marsh JA. 2017. Regulation, evolution and consequences of cotranslational protein complex assembly. Curr Opin Struct Biol 42:90–97. doi: 10.1016/j.sbi.2016.11.023. [DOI] [PubMed] [Google Scholar]
- 50.Kiho Y, Rich A. 1964. Induced enzyme formed on bacterial polyribosomes. Proc Natl Acad Sci U S A 51:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fane BA, Hayashi M. 1991. Second-site suppressors of a cold-sensitive prohead accessory protein of bacteriophage ϕX174. Genetics 128:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burch AD, Ta J, Fane BA. 1999. Cross-functional analysis of the Microviridae internal scaffolding protein. J Mol Biol 286:95–104. doi: 10.1006/jmbi.1998.2450. [DOI] [PubMed] [Google Scholar]
- 53.Burch AD, Fane BA. 2000. Foreign and chimeric external scaffolding proteins as inhibitors of Microviridae morphogenesis. J Virol 74:9347–9352. doi: 10.1128/JVI.74.20.9347-9352.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burch AD, Fane BA. 2003. Genetic analyses of putative conformation switching and cross-species inhibitory domains in Microviridae external scaffolding proteins. Virology 310:64–71. doi: 10.1016/S0042-6822(03)00076-X. [DOI] [PubMed] [Google Scholar]
- 55.Cherwa JE Jr, Fane BA. 2009. Complete virion assembly with scaffolding proteins altered in the ability to perform a critical conformational switch. J Virol 83:7391–7396. doi: 10.1128/JVI.00479-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doore SM, Schweers NJ, Fane BA. 2017. Elevating fitness after a horizontal gene exchange in bacteriophage ϕX174. Virology 501:25–34. doi: 10.1016/j.virol.2016.10.029. [DOI] [PubMed] [Google Scholar]
- 57.Ekechukwu MC, Oberste DJ, Fane BA. 1995. Host and ϕX174 mutations affecting the morphogenesis or stabilization of the 50S complex, a single-stranded DNA synthesizing intermediate. Genetics 140:1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pepin KM, Samuel MA, Wichman HA. 2006. Variable pleiotropic effects from mutations at the same locus hamper prediction of fitness from a fitness component. Genetics 172:2047–2056. doi: 10.1534/genetics.105.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreno-Pérez MG, Garcia-Luque I, Fraile A, Garcia-Arenal F. 2016. Mutations that determine resistance breaking in a plant RNA virus have pleiotropic effects on its fitness that depend on the host environment and on the type, single or mixed, of infection. J Virol 90:9128–9137. doi: 10.1128/JVI.00737-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldhill DH, Turner PE. 2014. The evolution of life history trade-offs in viruses. Curr Opin Virol 8:79–84. doi: 10.1016/j.coviro.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 61.Fane BA, Head S, Hayashi M. 1992. Functional relationship between the J proteins of bacteriophages phi X174 and G4 during phage morphogenesis. J Bacteriol 174:2717–2719. doi: 10.1128/jb.174.8.2717-2719.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernhardt TG, Struck DK, Young R. 2001. The lysis protein E of ϕX174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J Biol Chem 276:6093–6097. doi: 10.1074/jbc.M007638200. [DOI] [PubMed] [Google Scholar]
- 63.Novak CR, Fane BA. 2004. The functions of the N terminus of the ϕX174 internal scaffolding protein, a protein encoded in an overlapping reading frame in a two scaffolding protein system. J Mol Biol 335:383–390. doi: 10.1016/j.jmb.2003.09.050. [DOI] [PubMed] [Google Scholar]
- 64.Jaschke PR, Lieberman EK, Rodriguez J, Sierra A, Endy D. 2012. A fully decompressed synthetic bacteriophage ϕX174 genome assembled and archived in yeast. Virology 434:278–284. doi: 10.1016/j.virol.2012.09.020. [DOI] [PubMed] [Google Scholar]
- 65.Fujisawa H, Hayashi M. 1976. Viral DNA-synthesizing intermediate complex isolated during assembly of bacteriophage ϕX174. J Virol 19:409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roznowski AP, Fane BA. 2016. Structure-function analysis of the ϕX174 DNA-piloting protein using length-altering mutations. J Virol 90:7956–7966. doi: 10.1128/JVI.00914-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.