Abstract
The p.R4810K (rs11273543, c.14429G > A) variant of the RNF213 gene is associated with increased risk of Moyamoya disease (MMD), which is an idiopathic progressive intracranial vascular steno-occlusive disease, in Asian populations. Numerous variant association studies for this MMD variant have been performed in Japan to date. Since another genetic study that utilized approximately 140,000 single nucleotide polymor (SNPs) has indicated that there still are genetic differences among mainland Japanese, there is a possibility that the variant distribution in patients with MMD and normal individuals varies between different Japanese regions. Additionally, the majority of variant association studies have used Sanger sequencing, which is labor-intensive, time-consuming, and costly. In this study, we analyzed the frequency of the variant genotype in patients with MMD and normal individuals in Kyushu using pyrosequencing, which is an accurate, cost-effective, and automated method. We found differences in the genotype frequencies in familial patients from Kyushu and normal populations in Tohoku compared with west Japan, which suggested that there were differences in the frequency of the variant among different regions in Japan.
Keywords: Moyamoya disease, RNF213, p.R4810K, rs112735431, pyrosequencing
Introduction
Moyamoya disease (MMD) is an idiopathic progressive intracranial vascular steno-occlusive disease that was first reported in 1957.1) MMD is characterized by the bilateral progressive occlusion of main arterial branches of the circle of Willis, i.e., the distal internal carotid artery (ICA), the proximal, middle cerebral arteries (MCA), and the anterior cerebral arteries (ACA). This results in the appearance of compensatory collateral vessels (moyamoya vessels) around the steno-occlusive regions.2) The moyamoya vessels appear as a “puff of smoke” in angiography.2)
The ring finger protein 213 gene (RNF213) has been identified as a disease-susceptibility gene for MMD,3,4) and numerous variant association studies for the p.R4810K (rs11273543, c.14429G > A) variant have been conducted around the world.5) The p.R4810K variant is associated with an increased risk of MMD in Asian populations, especially in Japanese and Korean populations, but much less so in Chinese populations.6) The Japanese Archipelago populations, especially the mainland Japanese population from Honshu, Shikoku, and Kyushu, are thought to be the result of admixture between the ancestral Yayoi and Jomon people, referred to as the dual-structure model.7) The dual-structure model was widely supported by genetic analyses that utilized more than one million single nucleotide polymorphisms (SNPs).8) However, another genetic study that utilized approximately 140,000 SNPs has indicated that there still are genetic differences among mainland Japanese.9) Although various Japanese regions were included in the above-mentioned association studies, there has not yet been a study on patients with MMD and normal individuals in Kyushu.
To date, the majority of studies investigating the p.R4810K variant have used Sanger sequencing for variant detection. Although Sanger sequencing is the gold standard for the analysis of sequence variation, it is labor-intensive, time-consuming, and costly. Pyrosequencing is a sequence-based detection technology that enables rapid and accurate quantification of sequence variation. Furthermore, it has the potential advantages of being accurate, flexible, and optimized for parallel processing, and can be easily automated.10)
In this study, we examined the practical utility of pyrosequencing for the detection of the RNF213 variant. Using pyrosequencing, we analyzed the variant genotype in patients with MMD and normal individuals in Kyushu, and compared the observed genotype frequencies with that of previous reports.
Materials and Methods
Patients
Ninety-one (70 sporadic and 21 familial) Japanese patients with MMD who visited the Department of Neurosurgery, Saga University Hospital participated in this study. All patients were born in Kyushu and were diagnosed based on the diagnostic criteria for MMD as described by the Research Committee on the Pathology and Treatment of Spontaneous Occlusion of the Circle of Willis.11) One hundred and eight healthy volunteers from Kyushu participated as normal controls. The sex ratio among sporadic patients, familial patients, and healthy controls was not statistically different (Table 1). Likewise, there were no statistical differences in the average age of sporadic and familial patients (Table 1). The average age of normal healthy controls could not be calculated because their personal information was de-identified. Genomic DNA was extracted from peripheral blood lymphocytes using the FlexiGene DNA Kit (Qiagen, Hilden, Germany).
Table 1.
Background of the participants in the present study
| Headcount | Male:Female | P value* | Average age | P value** | |
|---|---|---|---|---|---|
| Sporadic MMD | 70 | 28:42 | 0.3370 | 29.7 ± 15.5 | 0.3849 |
| Familial MMD | 21 | 7:14 | 26.4 ± 14.9 | ||
| Control | 108 | 52:56 |
Comparison of sex ratio among the three cohorts using the Chi-square test.
Comparison of average age between sporadic and familial MMD using the Student’s t-test.
This study was approved by the Ethics Committee for Human Genome and Gene Analyses of the Faculty of Medicine, Saga University. Written informed consent was obtained from all study participants.
Pyrosequencing
Pyrosequencing was conducted using the PyroMark Q24 according to the manufacturer’s instructions (Qiagen). Primers for pyrosequencing were designed by PyroMark Assay Design 2.0 (Qiagen; Table 1). In brief, the genomic region encompassing the variant was amplified by polymerase chain reaction (PCR) with a forward primer (RNF213SNP-F: 5′-AAAGTTCCTGCCTGAGATTTTG-3′) and a biotinylated reverse primer (RNF213SNP-RB: 5′-AAATGCGGGACAGTCCTGGT-3′; Table 1). The PCR conditions were as follows: initial denaturation at 95°C for 2 min; 35 cycles of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 30 sec; followed by a final extension step at 72°C for 10 min. The size of the PCR products was verified by agarose gel electrophoresis. After denaturation, the biotinylated single-strand PCR products were purified using streptavidin-coated sepharose beads and used in the pyrosequencing reaction with a sequencing primer (RNF213SNP-Seq: 5′-CGTCCAGCAAGTTGA-3′). During the pyrosequencing reaction, dNTPs were added sequentially; for example, when dCTP was added, pyrophosphate (PPi) released as dCTP was incorporated by DNA polymerase and converted to ATP by sulfurylase. The ATP reacted with luciferin to generate visible light. The resulting light intensity was proportional to the amount of ATP, and was captured by a charge-coupled device (CCD) camera as raw data. Unincorporated dCTP and ATP were degraded by apyrase. After a sequence of these reactions, another dNTP was added, and the process repeated. The resulting pyrogram was automatically analyzed by PyroMark Q24 software (Qiagen) to provide quantitative results.
Sanger sequencing
The genomic region encompassing the variant was amplified by PCR, using the same primer pairs used for pyrosequencing (Table 1). The PCR products were subjected to sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Waltham, MA, U.S.A.) and the forward primer (RNF213SNP-F). The sequencing products were electrophoresed on an Applied Biosystems 3130 Genetic Analyzer and analyzed by Data Collection Software v3.0 (Applied Biosystems).
Statistical analysis
The Chi-square test and Student’s t-test were used to analyze differences in participants’ background characteristics. The Freeman-Halton extension of Fisher’s exact test and Steel’s test were used to compare the frequency of the p.R4810K genotype between groups. Statistical significance was set at P < 0.01.
Results
Genotyping of the RNF213 variant by pyrosequencing
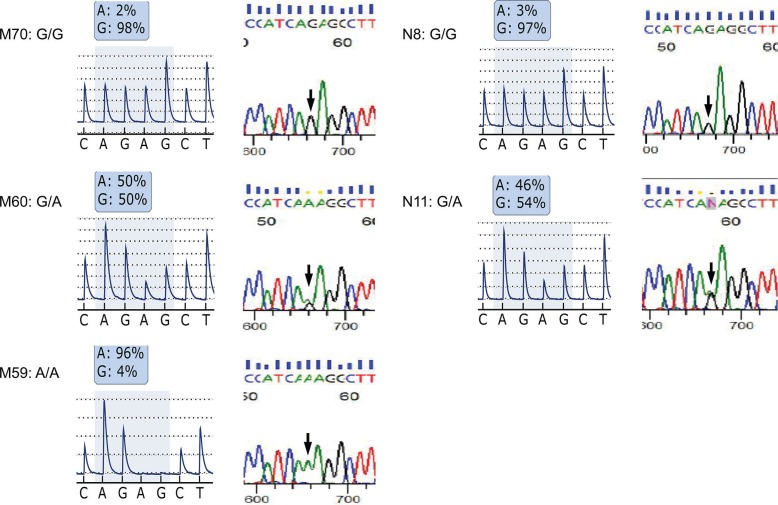
The pyrogram revealed that homozygotes (G/G or A/A) showed more than 95% of the G and A nucleotides, whereas heterozygotes showed approximately 50% of each nucleotide (Fig. 1). Therefore, each genotype was clearly distinguishable and easily identified by pyrosequencing. We confirmed the genotype of 27 subjects by Sanger sequencing, and found that the results obtained from Sanger sequencing were 100% identical to that of pyrosequencing (Fig. 1). The results demonstrated both the accuracy and reliability of pyrosequencing compared with Sanger sequencing.
Fig. 1.
Pyrogram data and Sanger sequencing data for each genotype. The results of pyrosequencing and Sanger sequencing were identical. M70, M60, and M59 were patients with MMD; N8 and N11 were normal controls.
Genotype frequencies of the RNF213 variant in patients with MMD
We identified 16 G/G, 74 G/A, and one A/A genotype in the MMD patient cohort (Table 2). The 70 sporadic patients consisted of 12 with the G/G genotype, 57 with the G/A genotype, and one with the A/A genotype, whereas the 21 familial patients consisted of four with the G/G genotype and 17 with the G/A genotype. There was no statistically significant difference in the genotype frequency of the RNF213 variant between sporadic and familial patients (Table 2). The genotype frequency was not significantly different between the sexes as previously reported12) (Freeman-Halton extension of the Fisher exact test, P = 0.7413).
Table 2.
Genotype frequencies of the RNF213 variant observed in the present study
| Genotype | G/G (%) | G/A (%) | A/A (%) | P value* |
|---|---|---|---|---|
| Patient | 16 (17.6) | 74 (81.3) | 1 (1.1) | 1.0000 |
| Sporadic | 12 (17.1) | 57 (81.4) | 1 (1.4) | |
| Familial | 4 (19.0) | 17 (81.0) | 0 (0.0) | |
| Control | 104 (96.3) | 4 (3.7) | 0 (0.0) |
Sporadic vs. familial using the Freeman-Halton extension of Fisher’s exact test.
We compared our results with three previous studies that analyzed the variant genotype in sporadic patients and familial patients separately.3,4,12) Patients in the previous studies were collected from all regions of the Japanese archipelago, with the likely exception of Kyushu, because no medical organization located in Kyushu was included in the studies.3,4,12) We found no statistical differences in the genotype frequency among sporadic patients. However, a statistically significant difference in familial patients was found between our results and those of Kamada et al.3) and Liu et al.4) (Table 3).
Table 3.
Comparison of genotype frequencies of the RNF213 variant in patients with MMD
| Study | G/G | G/A | A/A | P value* |
|---|---|---|---|---|
| Sporadic | ||||
| Present study | 12 | 57 | 1 | |
| Kamada et al.3) | 17 | 45 | 1 | 0.4029 |
| Liu et al.4) | 16 | 92 | 5 | 0.6620 |
| Miyatake et al.12) | 34 | 117 | 12 | 0.9936 |
| Familial | ||||
| Present study | 4 | 17 | 0 | |
| Kamada et al.3) | 0 | 39 | 3 | 0.0092 |
| Liu et al.4) | 0 | 43 | 5 | 0.0055 |
| Miyatake et al.12) | 2 | 36 | 3 | 0.0836 |
Previous vs. present study using Steel’s test.
Genotype frequencies of the RNF213 variant in normal controls
We found 104 and four normal controls with the G/G and G/A genotype, respectively. No participants with the A/A genotype were found (Tables 2 and 4). In this study, nearly all of the subjects were from Kyushu; hence, we compared the frequency of the variant in the Kyushu population with other Japanese regions reported in previous studies (Table 4).13–15) With the exception of Noshiro (Akita prefecture) in Tohoku, the genotype frequency in Kyushu is similar to that in other Japanese regions. Because Nyugawa (Gifu prefecture) is located in Tokai-Hokuriku, we concluded that the genotype frequency in Kyushu was same as that in other Japanese districts, at least from the west of Tokai-Hokuriku. The results from Noshiro also suggest that the genotype frequency in the normal population varies across the Japanese archipelago.
Table 4.
Geographical distribution of genotype frequencies of the RNF213 variant in Japan
| Region | G/G | G/A | A/A | MAF (%) | P value* |
|---|---|---|---|---|---|
| Kyushu (present study) | 104 | 4 | 0 | 1.85 | |
| Okinawa13) | 98 | 2 | 0 | 1.00 | 0.8054 |
| Uji, Kyoto15) | 510 | 9 | 0 | 1.73 | 0.4079 |
| Nyugawa, Gifu14) | 959 | 23 | 2 | 2.74 | 0.8187 |
| Noshiro, Akita14) | 2432 | 11 | 0 | 0.45 | <0.0001 |
| Field14,15) | 857 | 23 | 1 | 2.84 | 0.9001 |
MAF: minor allele frequency,
Previous vs. present study using Steel’s test, Field study in west Japan.
Discussion
In this study, we examined the practical use of pyrosequencing to detect the p.R4810K RNF213 variant. Pyrosequencing could detect genotype variants with the same accuracy as Sanger sequencing. The entire pyrosequencing process took approximately four hours to complete, whereas more than eight hours were required for Sanger sequencing. Furthermore, pyrosequencing was far less costly compared to Sanger sequencing; according to our calculations, pyrosequencing cost approximately 190 Japanese yen (1.67 US dollars) per sample, whereas Sanger sequencing cost approximately 440 Japanese yen (3.87 US dollars) per sample. Our results indicate that pyrosequencing is reliable, and is simpler and more cost-effective than Sanger sequencing.
Although no differences in the genotype frequency in sporadic patients were found between this study and the three previous studies, there were statistically significant differences in familial patients between this study and two of the three studies (Table 3). As mentioned above, it is highly likely that no patient from Kyushu was included in the three previous studies.3,4,12) Therefore, there may be genetic differences in familial patients from Kyushu. However, since the number of familial patients in this study was relatively small, more patients from Kyushu and other many regions should be collected and genotyped to confirm the difference in genotype frequencies.
In addition, within the normal population, the genotype frequency in Noshiro (located in Tohoku) differed from other regions in west Japan, supporting the significantly low minor allele frequency (MAF) in Noshiro reported by Koizumi et al. (Table 4).14) Genetic differences even within Honshu Island have been reported, especially between Tohoku and other regions in Honshu Island, using numerous SNPs.9) Thus, the significantly low MAF in Noshiro might be attributed to such genetic differences.
In conclusion, we confirmed the practical utility of pyrosequencing for the detection of the RNF213 variant, and found differences in the genotype frequencies in familial patients from Kyushu and normal populations in Tohoku compared with west Japan. Further investigation of the genotype frequency in patients and the normal population should be conducted region-by-region throughout the Archipelago.
Acknowledgement
We thank all the patients for participating in this study, and Prof. Atsushi Kawaguchi (Center for Comprehensive Community Medicine, Faculty of Medicine, Saga University) for his support on statistical analyses. This work was supported in part by a Grant for Research on Intractable Diseases from the Ministry of Health, Labor, and Welfare (H26-nanchitou(nan)-ippan-035); a Grant for Child Health and Development from the National Center for Child Health and Development (26–13); a Grant-in-Aid for Challenging Exploratory Research (26670169); a Grant-in-Aid for Scientific Research (C) from the Japan Society for the Promotion of Science (25461554); and the Practical Research Project for Rare/Intractable Diseases from Japan Agency for Medical Research and Development (AMED).
Footnotes
Conflicts of Interest Disclosure
The authors declare no conflicts of interest.
References
- 1). Takeuchi K, Shimizu K: [Hypoplasia of the bilateral internal carotid arteries]. Brain Nerve 9: 37– 43, 1957. (Japanese) [Google Scholar]
- 2). Suzuki J, Takaku A: Cerebrovascular “moyamoya” disease. Disease showing abnormal net-like vessels in base of brain. Arch Neurol 20: 288– 299, 1969. [DOI] [PubMed] [Google Scholar]
- 3). Kamada F, Aoki Y, Narisawa A, et al. : A genome-wide association study identifies RNF213 as the first moyamoya disease gene. J Hum Genet 56: 34– 40, 2011. [DOI] [PubMed] [Google Scholar]
- 4). Liu W, Morito D, Takashima S, et al. : Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS ONE 6: e22542, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5). Shoemaker LD, Clark MJ, Patwardhan A, et al. : Disease variant landscape of a large multiethnic population of moyamoya patients by exome sequencing. G3 (Bethesda) 6: 41– 49, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6). Sun XS, Wen J, Li JX, et al. : The association between the ring finger protein 213 (RNF213) polymorphisms and moyamoya disease susceptibility: a meta-analysis based on case-control studies. Mol Genet Genomics 291: 1193– 1203, 2016. [DOI] [PubMed] [Google Scholar]
- 7). Jinam TA, Kanzawa-Kiriyama H, Saitou N: Human genetic diversity in the Japanese Archipelago: dual structure and beyond. Genes Genet Syst 90: 147– 152, 2015. [DOI] [PubMed] [Google Scholar]
- 8). Japanese Archipelago Human Population Genetics Consortium. Jinam T, Nishida N, Hirai M, et al. : The history of human populations in the Japanese Archipelago inferred from genome-wide SNP data with a special reference to the Ainu and the Ryukyuan populations. J Hum Genet 57: 787– 795, 2012. [DOI] [PubMed] [Google Scholar]
- 9). Yamaguchi-Kabata Y, Nakazono K, Takahashi A, et al. : Japanese population structure, based on SNP genotypes from 7003 individuals compared to other ethnic groups: effects on population-based association studies. Am J Hum Genet 83: 445– 456, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10). Ronaghi M: Pyrosequencing sheds light on DNA sequencing. Genome Res 11: 3– 11, 2001. [DOI] [PubMed] [Google Scholar]
- 11). Research Committee on the Pathology and Treatment of Spontaneous Occlusion of the Circle of Willis; Health Labour Sciences Research Grant for Research on Measures for Infractable Diseases : Guidelines for diagnosis and treatment of moyamoya disease (spontaneous occlusion of the circle of Willis). Neurol Med Chir (Tokyo) 52: 245– 266, 2012. [DOI] [PubMed] [Google Scholar]
- 12). Miyatake S, Miyake N, Touho H, et al. : Homozygous c.14576G>A variant of RNF213 predicts early-onset and severe form of moyamoya disease. Neurology 78: 803– 810, 2012. [DOI] [PubMed] [Google Scholar]
- 13). Liu W, Hitomi T, Kobayashi H, Harada KH, Koizumi A: Distribution of moyamoya disease susceptibility polymorphism p.R4810K in RNF213 in East and Southeast Asian populations. Neurol Med Chir (Tokyo) 52: 299– 303, 2012. [DOI] [PubMed] [Google Scholar]
- 14). Koizumi A, Kobayashi H, Liu W, et al. : P.R4810K, a polymorphism of RNF213, the susceptibility gene for moyamoya disease, is associated with blood pressure. Environ Health Prev Med 18: 121– 129, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15). Cao Y, Kobayashi H, Morimoto T, Kabata R, Harada KH, Koizumi A: Frequency of RNF213 p.R4810K, a susceptibility variant for moyamoya disease, and health characteristics of carriers in the Japanese population. Environ Health Prev Med 21: 387– 390, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]