Abstract
Understanding how cell fate decisions are regulated is a fundamental goal of developmental and stem cell biology. Most studies on the control of cell fate decisions address the contributions of changes in transcriptional programming, epigenetic modifications, and biochemical differentiation cues. However, recent studies have found that other aspects of cell biology also make important contributions to regulating cell fate decisions. These cues can have a permissive or instructive role and are integrated into the larger network of signaling, functioning both upstream and downstream of developmental signaling pathways. Here, we summarize recent insights into how cell fate decisions are influenced by four aspects of cell biology: metabolism, reactive oxygen species (ROS), intracellular pH (pHi), and cell morphology. For each topic, we discuss how these cell biological cues interact with each other and with protein‐based mechanisms for changing gene transcription. In addition, we highlight several questions that remain unanswered in these exciting and relatively new areas of the field.
Keywords: cell fate, cell morphology, intracellular pH, metabolism, reactive oxygen species
Subject Categories: Development & Differentiation, Stem Cells, Transcription
Glossary
- ECM
extracellular matrix
- ESCs
embryonic stem cells
- HSCs
hematopoietic stem cells
- iPSCs
induced pluripotent stem cells
- Metabolic reprogramming
Changes in metabolism that accompany and can sometimes be necessary or instructive for changes in cell fate
- MSCs
mesenchymal stem cells
- Niche
A specialized microenvironment in the tissue that maintains cells in the stem cell state
- NSCs
neural stem cells
- pH sensor
Selective proteins with post‐translational modification by protonation/deprotonation regulating activity or ligand binding.
- ROS
reactive oxygen species
- SAM
S‐adenosyl methionine
- Satellite cells
Stem cells of the skeletal muscle
Introduction
Cell fate decisions are tightly regulated by many layers of control. A change in cell fate is ultimately defined by the acquisition of new characteristics that come about largely through changes in transcription. Protein‐based signal transduction cascades leading to changes in transcription factor activity are the most direct causes of transcriptional changes and are among the most well‐studied aspects of the cell fate decision process. In contrast, much less is known about how other aspects of cell biology such as changes in metabolite concentration or mechanical forces contribute to cell fate decisions. This is due in part to the difficulty of studying cues that are not directly encoded in the genome. However, technological advances, including the generation of new biosensors that can be used for live cell imaging, improvements in quantitative fluorescence microscopy, and the development of more sensitive biochemical methods for detecting small molecules are making it easier to identify previously unrecognized control mechanisms. In this review, we discuss recent advances in understanding the role of metabolism, reactive oxygen species (ROS), intracellular pH (pHi), and cell morphology and adhesions to cell fate decisions, particularly during differentiation in adult, embryonic, and induced pluripotent stem cell lineages.
Metabolism
The metabolic state of a cell is the result of a complex array of inputs, including cell signaling, availability of nutrients and oxygen, energy needs, and biomass demands. These inputs and demands combine to influence the rate of ATP production from glycolysis versus oxidative phosphorylation, as well as the rate of side reactions that produce anabolic intermediates. As cells differentiate, the change in these inputs causes the metabolic state to shift. However, the metabolic state of the cell is not merely a consequence of differentiation. Instead, shifts in metabolism can have permissive and, in some cases, even instructive roles in promoting differentiation 1. This perspective positions metabolism as a key node in the regulation of cell fate transitions. In this section, we summarize the metabolic programs of cells at different stages of differentiation, briefly review some of the major cell signaling regulators of metabolic state, and discuss how changes in metabolic state contribute to cellular differentiation (Fig 1).
Figure 1. The connections between metabolism and cell fate decisions.
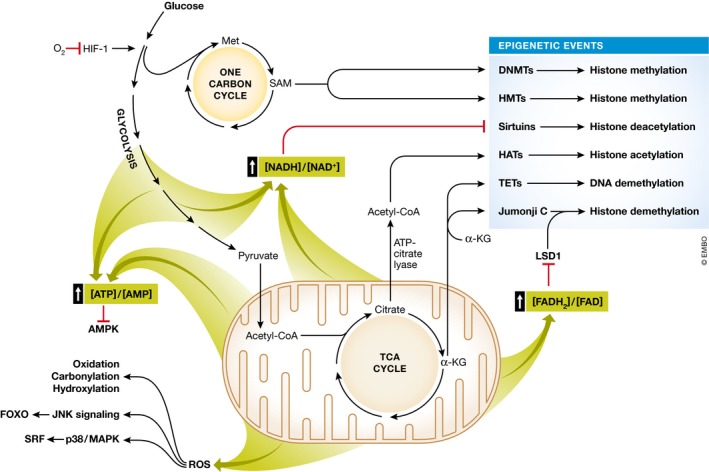
Metabolic inputs regulate epigenetics and cell signaling to promote changes in cell fate. Glycolysis produces metabolic intermediates that feed into the folate and one carbon metabolism cycle to produce S‐adenosylmethionine (SAM), which is a cofactor for DNA methyltransferases (DNMTs) and histone methyltransferases (HMTs). The energy released from glycolysis and oxidative phosphorylation also converts AMP to ATP and NAD+ to NADH. AMP stimulates AMPK activity, and NAD+ is a cofactor for sirtuins, so increased energy production decreases the activity of these enzymes. Glucose‐derived acetyl‐CoA enters the tricarboxylic acid (TCA) cycle to form citrate, which can be converted back to acetyl‐CoA by ATP‐citrate lyase. This source of acetyl‐CoA (but not acetyl‐CoA derived from fatty acid oxidation) contributes to the pool of nuclear acetyl‐CoA that is essential for histone acetylation by histone acetyltransferases (HATs). α‐ketoglutarate (α‐KG), which is produced in the TCA cycle and in the cytoplasm, is an essential cofactor for TET and Jumonji C enzymes, which demethylate DNA and histones, respectively. The energy released from oxidative phosphorylation converts FAD to FADH2, and FAD is a cofactor for lysine‐specific demethylase 1 (LSD1), so a reduction in FAD levels inhibits LSD1 activity. Increased oxidative phosphorylation also generates reactive oxygen species (ROS), which promote oxidation, carbonylation, and hydroxylation as well as increase the levels of JNK and p38/MAPK pathway activity. Low levels of oxygen (O2), for example in the HSC and satellite cell niches, increase the activity of the hypoxia inducible factor‐1 (HIF‐1), which promotes glycolysis.
Changes in metabolism, often collectively referred to as “metabolic reprogramming”, can shift the amount of energy and biomass produced by glycolysis versus oxidative phosphorylation to regulate changes in cell fate. In adult stem cell lineages, less active long‐term progenitors, such as quiescent hematopoietic stem cells (HSCs) or satellite cells (stem cells of skeletal muscle) utilize glycolysis over oxidative phosphorylation, whereas more actively growing and proliferating cells are bivalent and utilize both glycolysis and oxidative phosphorylation 2, 3, 4, 5. Embryonic stem cells (ESCs) transition through several metabolic states during differentiation. ESCs in the most undifferentiated, or “naive” state, have relatively high levels of oxidative phosphorylation 6, 7, 8, although these cells still consume high amounts of glucose and glutamine 6, 9. As ESCs differentiate toward the “primed” state, ATP production becomes decoupled from oxidative phosphorylation, and the metabolic program is shifted toward the use of glycolysis for energy and biomass production 6, 10 through a process that is regulated by the conserved RNA‐binding protein, LIN28 7. Energy production from oxidative phosphorylation then increases again as differentiation proceeds beyond the primed state. Likewise, the reprogramming of differentiated somatic cells into induced pluripotent stem cells (iPSCs) requires a shift from a bivalent metabolic program of glycolysis and oxidative phosphorylation toward a primarily glycolytic state that resembles the metabolism of primed ESCs 11, 12. Recent evidence indicates that this metabolic shift occurs prior to changes in gene expression, suggesting that it is a prerequisite for reprogramming rather than a consequence of the cell fate change 13.
Nonetheless, in most cases, metabolic changes are initiated by cell signaling molecules, including AMPK, HIF1α, AKT, and Myc. AMPK, which is activated by high [AMP]/[ATP] ratios that indicate low nutrient availability and metabolic stress, increases glycolytic energy production, activates FOXO proteins to promote the expression of antioxidants and autophagy genes, and restricts growth by inhibiting mTor 14, 15. This stress response program is important for maintaining cellular homeostasis in general, and thus functions during both self‐renewal and differentiation. HIF1α is an oxygen sensor that is stabilized by low oxygen levels and promotes a steady state level of energy production during periods of relatively low activity in quiescent and slowly dividing adult stem cells, such as HSCs 2, mesenchymal stem cells (MSCs) 16, and satellite cells 17. HIF1α shifts the metabolic program toward glycolysis over oxidative phosphorylation, which is conducive to the hypoxic environments of stem cell niches that maintain quiescent stem cells, and also minimizes the damage caused by ROS produced from mitochondrial respiration. In contrast, Akt and Myc promote an increase in energy production from oxidative phosphorylation and a switch in the utilization of glycolysis from a source of energy production to a source of anabolic intermediates. Akt activates mTor by inhibiting the Tsc complex, and several studies have found that this pathway promotes differentiation of adult stem cells including HSCs, NSCs, and ISCs 18, 19, 20. Akt signaling also increases ROS levels by inhibiting FOXO proteins, which has the effect of further promoting differentiation in some types of stem cells (see next section). Myc is also required for differentiation in the HSC and epidermal stem cell lineages 21, 22. In addition, Myc is an important factor for reprogramming into iPSCs, and inhibition of mTor or induced expression of metabolic enzymes can substitute for Myc in iPSC reprogramming 23, 24. Thus, shifts in metabolic state are a prerequisite for differentiation in cases where the shift is needed in order to meet the energetic and anabolic demands of the new cell state.
Metabolic state can also influence cell fate decisions by affecting the availability of metabolites that are important for the epigenetic regulation of gene expression 24. Epigenetic regulation occurs primarily through the modification of histones and DNA, and histone acetylation and deacetylation as well as histone and DNA methylation and demethylation all can be regulated by metabolites. Histone acetyltransferases (HATs) use acetyl‐CoA, which is a key metabolic intermediate between glycolysis and the TCA cycle, as a substrate for histone acetylation. In the absence of sufficient acetyl‐CoA, global histone acetylation is reduced, and thus, the regulation of gene expression is impaired. This connection was clearly demonstrated in a study of in mouse adipocytes 25. The authors found that knockdown of ATP‐citrate lyase, which generates acetyl‐CoA from citrate, caused a decrease in histone acetylation and prevented the upregulation of genes such as glucose transporters that are required for differentiation. Likewise, deacetylation is also sensitive to acetyl‐CoA concentrations in the cell. For example, the addition of acetate (which increases acetyl‐CoA levels) to the culture media of human or mouse ESCs blocked histone deacetylation and delayed differentiation, whereas inhibition of glycolysis (which decreases acetyl‐CoA levels) accelerated differentiation 26. The effect of glycolysis inhibition could be reversed with the addition of acetate to the media, and pharmacological inhibition of the enzyme that produces acetyl‐CoA for histone acetylation produced a similar phenotype, but the effect on histone deacetylation was not tested directly. Deacetylation by sirtuins is also responsive to metabolic inputs 27. Sirtuins are deacetylases with a broad range of targets including histones and transcription factors. These enzymes are considered metabolic sensors because they use NAD+ as a cofactor and thus become more active when [NAD+]/[NADH] ratios are high. In addition, sirtuins both regulate and are regulated by AMPK 28. Sirtuin 1 (SIRT1) has been well studied during mammalian cell differentiation and may function through different mechanisms to both repress differentiation in some contexts and promote differentiation in others. For example, SIRT1 is highly expressed in ESCs, iPSCs, and early morula stage embryos, where it promotes pluripotency and is downregulated upon differentiation 29, 30. In contrast, genetic and pharmacological studies indicate that SIRT1 promotes differentiation in hematopoietic and neural lineages 31, 32.
The epigenetic regulators that catalyze the addition of methyl groups, DNA methyltransferases (DNMTs) and histone methyltransferases (HMTs), use S‐adenosyl methionine (SAM) as a substrate. The rates of histone methylation are different at active versus inactive promoters, and the concentration of intracellular SAM can directly influence these rates. SAM concentrations are relatively high in human and mouse ESCs and iPSCs, and SAM is required for histone methylation to maintain the pluripotent state in these cell types 33, 34, 35. In adult tissues, there is a well‐established role for SAM in the regulation of DNA and histone methylation during oncogenesis 36, and though less is known about the role of SAM in adult stem cell differentiation, many adult progenitors, including HSCs 37, ISCs 38, and epidermal progenitors 39 require DNMTs and HMTs 40. Thus, changes in the concentration of SAM influence cell fate transitions in many different cell types.
Likewise, enzymes that catalyze the removal of methyl groups from histones and DNA are sensitive to the availability of specific metabolites. For example, the Jumanji C family of histone demethylases and the TET‐family enzymes, which catalyze the first step of DNA demethylation, require both the TCA cycle intermediate α‐ketoglutarate (α‐KG) and the reduced (Fe2+) form of iron 41, 42. Iron is more commonly in the Fe3+ form but can be reduced to Fe2+ by vitamin C, and several recent studies revealed the importance of vitamin C for promoting the activity of Jumonji C or TET‐family enzymes in ESCs 43, 44, 45, 46, adult stem cells 47, 48, and during iPSC reprogramming 43, 49. Another important histone demethylase, lysine‐specific demethylase 1 (Lsd1), is also sensitive to metabolic changes as it relies on FAD as a cofactor 50. LSD1 is required in mouse ESCs (mESCs) to silence self‐renewal genes during differentiation 51, and the homologous gene, Su(var)3‐3, is also required in the somatic cells of the Drosophila ovary to promote germ cell differentiation 52, 53. Collectively, these findings demonstrate that metabolic processes can influence epigenetic regulation of gene expression at multiple levels.
In addition to the permissive roles for metabolism in cellular differentiation described above, metabolic cues can also be instructive, causing changes in cell signaling and gene expression sufficient to drive the change in cell fate. For example, in satellite cells, increased glycolysis during exit from quiescence causes a decrease in NAD+, which reduces SIRT activity and thus increases H4K16 acetylation, ultimately leading to the expression of key differentiation genes, such as MyoD 54. Another interesting example comes from a recent study that found that intestinal stem cells (ISCs) utilize lactate provided by the neighboring Paneth cells to sustain a high level of oxidative phosphorylation 55. Increased oxidative phosphorylation in ISCs causes an increase in reactive oxygen species (ROS), which activates the p38‐MAPK pathway (as discussed in the following section). Paneth cells are part of the ISC niche, so this suggests that metabolic cues can function as niche signals. Additional examples in which metabolic changes feed into signaling networks to instruct cell fate decisions involve mTOR, which is a master regulator of cell growth and proliferation. Several studies have demonstrated that mTOR is essential for the maintenance of pluripotency and the repression of differentiation genes in ESCs grown under standard conditions 56. In addition, a more recent study found that partial inhibition of mTOR in mESCs induces the cells to adopt a “paused” state resembling embryonic diapause 57. The mechanism of this effect is not fully understood, but the authors speculate that the paused state is induced by the combined effects of mTOR inhibition on transcription, translation, and metabolism. Lastly, in quiescent HSCs, activation of mTOR induces mitochondrial biogenesis, which activates proliferation and induces differentiation 58.
Two recent studies demonstrated that changes in pyruvate metabolism can contribute to the regulation of proliferation and differentiation in epidermal and intestinal cell lineages 59, 60. Pyruvate is the end product of glycolysis and can either enter be converted to lactate in the cytoplasm, or be transported into the mitochondria, where it is converted to acetyl‐CoA and oxidized in the TCA cycle. These studies provide evidence that hair follicle and intestinal stem cells are more glycolytic than their non‐stem cell progeny, and suggest that increased conversion of pyruvate to lactate drives stem cell proliferation whereas increased mitochondrial oxidation of pyruvate promotes differentiation. The downstream mechanism was not investigated, but both studies provide evidence suggesting that high levels of Myc in the stem cells may promote the shift toward lactate production. Interestingly, a separate study of intestinal differentiation in zebrafish found that Wnt signaling also regulates pyruvate metabolism 61. Wnt signaling is generally high in epithelial stem cells 62 and promotes Myc expression 63, 64, suggesting a model in which Wnt signaling, Myc, and pyruvate metabolism function together to promote epithelial stem cell identity.
Taken together, these studies demonstrate that changes in metabolism influence cell fate decisions in a variety of ways. In many cases, the link between the metabolic cue and the cell fate decision is reactive oxygen species as described in the next section.
Reactive oxygen species
Metabolic pathways can influence stem cell fate decisions through the activity of ROS (Fig 1). ROS, such as superoxide anion (O2 −), hydrogen peroxide (H2O2), and hydroxyl radicals (OH−), are formed by the reduction of molecular oxygen (O2). The toxic effects of these ROS have been studied extensively in the context of cell proliferation, DNA damage, and apoptosis. Additionally, ROS play a crucial role in regulating cellular processes like oxidative stress responses, aging, and stem cell fate decisions. In this section, we review recent advances in the understanding of the role of ROS in cell differentiation. ROS are commonly generated as by‐products of metabolic reactions occurring in the mitochondria, mainly in the electron transport chain. ROS levels are controlled by several proteins, such as NADPH oxidases, which have activity that results in formation of superoxides, superoxide dismutases (SOD), which reduce O2 − to H2O2, and other enzymes, including thioredoxins, glutathione peroxidases, and peroxiredoxins 65, 66.
Recent studies identified examples in which specific ROS regulators are necessary for stem cell differentiation. For example, Kim et al 67 found that peroxiredoxins, PrxI and Prx II, promote mouse embryonic stem cell differentiation into neurons by regulating ROS levels. In addition, Hochmuth et al 68 found that Nrf2, which controls transcription of antioxidant enzymes like thioredoxins and peroxidases, and Keap1, a negative regulator of Nrf2, regulate Drosophila intestinal stem cell proliferation by altering intracellular ROS. Other studies have focused on the downstream effects of changes to ROS levels, and in general, these studies find that increased ROS levels are associated with differentiation. During Drosophila testes germline stem cell (GSC) differentiation, GSCs maintain reduced levels of ROS, regulated by Keap1 and Nrf2 69. An increase in ROS in GSCs caused a decrease in the number of GSCs and promoted differentiation. In mammalian HSCs, an elevation in ROS levels occurs during differentiation into common myeloid progenitors 70. Likewise, quiescent multipotent hematopoietic progenitor cells in the Drosophila lymph gland have elevated levels of ROS, which promotes differentiation 71. In these studies, scavenging ROS by expressing antioxidant proteins like catalase in vivo or by the addition of N‐acetylcysteine, delayed differentiation, whereas increasing ROS by adding paraquat or mutating mitochondrial complex I proteins like ND75, promoted differentiation 69, 71, 72. During vascular smooth muscle cell differentiation, inhibition of ROS activity decreased the cellular expression of differentiation proteins, whereas an elevation in ROS activity increased expression of these differentiation markers 73. In contrast to this trend, two studies show that elevated ROS levels promote self‐renewing, proliferative neural and mouse spermatogonial stem cell fate 74, 75. Additionally, elevated ROS levels promote Drosophila intestinal stem cell proliferation 68.
Reactive oxygen species instruct stem cell fate decisions by regulating key signal transduction pathways. A mechanism by which ROS control signaling pathways that affect stem cell differentiation is by affecting post‐translational modifications of regulatory proteins, such as phosphatases. For example, ROS have been shown to mediate cysteine and methionine oxidation, protein carbonylation, and hydroxylation (reviewed by 66). Another mechanism by which ROS influences differentiation decisions is by directly affecting the activity of transcription factors and essential signaling pathway proteins responsible for activating genetic differentiation programs. The most commonly studied signaling pathways in this regard are the JNK and p38 MAPK pathways and an increase in ROS typically activates these pathways to promote differentiation 68, 71, 72, 73. For example, in Drosophila hematopoietic progenitor cells, elevated ROS levels stimulate the JNK pathway to promote differentiation by activating transcription factor FoxO and the derepression of polycomb activity 71. However, FoxO also increases antioxidant activity, which reduces ROS levels and thus creates a negative feedback loop that eventually brings FoxO activity back down. Likewise, in mammalian hematopoietic stem cells (HSCs), low levels of ROS are necessary for HSC self‐renewal whereas elevated levels of ROS promote differentiation by stimulating the activity of p38 and mTOR signaling pathways 76. During vascular smooth muscle differentiation and mouse spermatogonial stem cell self‐renewal, increased ROS levels activate p38 MAPK signaling pathway, which promotes the transcription of serum response factor (SRF), and ultimately increases the activity of differentiation proteins, such as α‐actin and calponin 73. Additionally, high levels of ROS in Drosophila GSCs promote differentiation by increasing the transcription of the epidermal growth factor receptor ligand, Spitz, thereby activating the MAPK signaling pathway 69. Collectively, these studies demonstrate that ROS concentrations are tightly controlled during cellular differentiation and that changes in ROS concentrations play important roles in the cell fate decision process.
Intracellular pH
A long‐held view is that pHi is constitutively maintained between 7.2 and 7.4 in normal mammalian cells and only dysregulated from this narrow range in diseases, including being constitutively increased in cancer 77, 78 and decreased in neurodegenerative disorders 79, 80. However, emerging evidence indicates there are transient increases in pHi in normal mammalian cells during cell cycle progression 81, directional migration 82, 83, and differentiation 84, 85, 86, 87. Although the role of pHi dynamics in regulating cell fate decisions remains understudied, we highlight recent findings on this topic and emphasize questions that remain to be addressed (Fig 2).
Figure 2. Mechanisms by which pHi could regulate cell fate decisions.
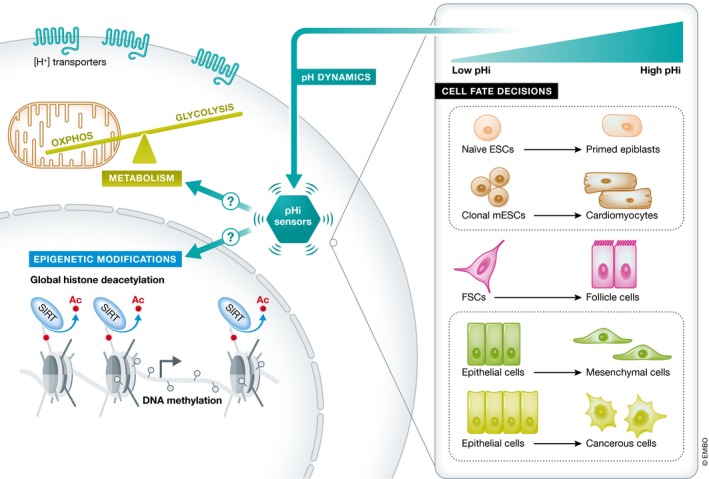
pHi increases during embryonic and adult stem cell differentiation, epithelial‐to‐mesenchymal transitions, and carcinoma transformations. Theoretically, pH‐sensitive proteins (“pH sensors”) that undergo protonation or deprotonation upon changes in pHi could regulate cell fate decisions by affecting proton transporter activity, cellular metabolism, and epigenetic modifications like histone deacetylation and DNA methylation. However, in most cases, the specific mechanisms by which pHi could regulate cell fate decisions are unknown.
Increasing evidence suggests that changes in pHi are necessary for embryonic stem cell differentiation. We recently showed a transient increase in pHi during differentiation of clonal naïve mESCs to primed epiblast‐like cells (EpiSC), which when prevented, blocks differentiation as indicated by attenuated expression of epiblast cell markers, including Pax6, Brachyury, and Fgf5, as well as the miRNA cluster mir‐302 84. The increased pHi from ~7.40 to ~7.65 occurs during the first 3 days of spontaneous differentiation and then returns to pHi values seen in naïve cells, which suggests that the higher pHi is necessary for the differentiation process but not for maintaining a differentiated state. Consistent with this prediction, an earlier study by Edwards et al 88 found that pHi increases from zygote to the morula stage. In a different embryonic cell model, Li et al 87 showed that inhibiting activity of the plasma membrane Na‐H exchanger‐1 (NHE1), markedly attenuates differentiation of CGR8 clonal mESCs into cardiomyocytes resulting in a decreased expression of the transcription factors Nkx2‐5 and Tbx5 and decreased abundance of α‐myosin heavy chain. Although changes in pHi during differentiation were not determined, inhibiting NHE1, which is an acid extruder, is predicted to lower pHi. This group also found that NHE1 activity potentiates differentiation of P19 embryonal carcinoma cells into neurons 89. In contrast, umbilical cord‐derived human mesenchymal stem cells (MSCs) have a higher pHi than differentiated cells. Lowering pHi of these cells by pharmacological inhibition of NHE1 promotes differentiation to an osteogenic lineage but has no effect on differentiation to an adipogenic lineage 90.
Recent studies, including studies from our laboratories, suggest that differentiation occurring during Drosophila adult epithelial follicle stem cell lineages requires changes in pHi. Using the genetically encoded pHi biosensor pHluorin, we showed a lower pHi in follicle stem cells of the adult Drosophila ovary compared with differentiated daughter cells. Preventing the increased pHi by loss of Dnhe2, the Drosophila ortholog of mammalian NHE1, inhibits differentiation, impairs germarium morphology, and results in infertility 84. Krüger and Bohrmann 91 also found an anteroposterior pHi‐gradient in follicle and nurse cells of the Drosophila ovary, although significance in oogenesis was not determined.
It remains to be determined whether the lower pHi in self‐renewing cells or the higher pHi in differentiating cells is an active process. In endometrial epithelial cells, LeftyA inhibits NHE1 to actively maintain a lower pHi 92. Likewise, in mESCs, our findings that the increased pHi with differentiation is transient and seen only during the first 72 h are consistent with an active regulation of pHi 84. Because increased pHi promotes proliferation, the decrease after 72 h may function to limit proliferation. As described below, a constitutively higher pHi is seen in most cancers and can induce hyperproliferation and dysplasia even in the absence of activated oncogenes 93.
Considered more broadly, a role for pHi dynamics in differentiation, epithelial plasticity, and morphogenesis remains understudied. Increased pHi is reported to enable or be necessary for the differentiation of CD4+ T helper 9 (Th9) cells 85, epithelial‐to‐mesenchymal transition (EMT) 86, and neural fates from ectoderm during Xenopus development 94. In contrast, expression of the Cl‐HCO3 exchanger AE2, which as an acid loader facilitating HCO3 efflux should lower pHi, is necessary for clonal mouse macrophages to differentiate into osteoclasts 95, although a role for pHi dynamics was not determined. Recent work shows that a glycolysis gradient in mouse and chick embryo tail bud generates a more acidic extracellular pH (pHe) in the tail bud, which when experimentally manipulated to be more alkaline, results in slower axis elongation 96. Additionally, decreased pHe from extracellular lactic acid generated by lactate dehydrogenase enables myofibroblast differentiation in an EMT‐like fibrosis by increasing acid‐induced activation of latent TGF‐β in the extracellular matrix 97, and an acidic pHe enables differentiation of MSCs into cancer‐associated fibroblasts through a mechanism involving a pH‐sensitive GPCR that regulates Yes‐associated protein (YAP) signaling 98. In normal adult tissues, pHe is ~7.4 and higher than pHi of ~7.2. In most cancers, this gradient is reversed, with pHe being ~7.0 or lower while pHi is ~7.6 and higher 77, 78. An intriguing prediction that remains to be verified is whether cancer initiating cells, analogous to stem cells, might have a lower pHe and higher pHi than differentiated cancer cells. In support of this prediction, an acidic pHe promotes self‐renewal of glioma stem cells by increasing stability of hypoxia inducible factor 2a 99.
The mechanisms by which pH dynamics regulates differentiation remain largely unknown. However, based on our previous findings on pHi‐dependent cell behaviors such as proliferation and migration, we speculate important roles for pH sensors, defined as selective proteins with post‐translational modification by protonation/deprotonation regulating activity or ligand binding 100. Our findings with pHi‐regulated Drosophila follicle stem cell differentiation suggest pH sensing by the hedgehog signaling pathway 84. Previous findings in Drosophila eye epithelium indicate pHi‐dependent Wnt signaling with a higher pHi enabling binding of disheveled to the plasma membrane and being necessary for planar cell polarity 101. With regard to pH‐dependent post‐translational modification, a decreased pHi is associated with global histone deacetylation 102. Epigenetic modifications such as histone modification and DNA methylation have established roles in cell differentiation by changing chromatin structure to activate or inhibit gene expression. Although unique for each stem cell lineage, in general, DNA silencing by methylation of CpG islands suppresses the expression of genes involved in cell cycle exit and terminal differentiation and hence preserves the progenitor self‐renewing state 103.
Metabolic reprogramming is another potential mechanism for pHi‐regulated epigenetic modifications 104. Recent findings show that a more acidic pHi promotes promiscuous enzymatic activity of lactate dehydrogenase to convert α‐ketoglutarate to the L enantiomer of 2‐hydroxyglutarate, compared with conventional lactate dehydrogenase conversion of pyruvate to lactate 105, 106. L‐ and D‐2‐hydroxyglutarate antagonize α‐ketoglutarate‐regulated chromatin modifications associated with differentiation and also stabilize expression of HIF‐1α 85, 86. Additionally, stabilized HIF‐1α promotes reprogramming to a glycolytic metabolism during the ESC to EpiSC transition 6. Hence, increased pHi during stem cell differentiation could enable reprogramming to a more glycolytic phenotype, which could be mediated by glycolytic enzymes that are pH sensors with increased activity at higher pH, such as phosphofructokinase‐1 77, 78. While studies have begun to uncover the integral role of pH dynamics in regulating cell fate changes, an important future direction is to identify the mechanisms mediating this effect.
Cell morphology and adhesion dynamics
Although differentiation often includes changes in cell shape and cell adhesion, including both cell–cell and cell‐matrix adhesion, we have an incomplete understanding of how these changes are regulated during differentiation and contribute to the differentiation process. Understanding the underlying cell biology of differentiation, especially during in vivo development, requires knowledge of how the cell interprets its niche through cell shape and adhesion‐derived mechanical forces. In this section, we review recent progress in how cell morphology and mechanical cues instruct cell fate decisions (Fig 3).
Figure 3. Mechanical and morphological cues regulate cell fate decisions through distinct signaling mechanisms.
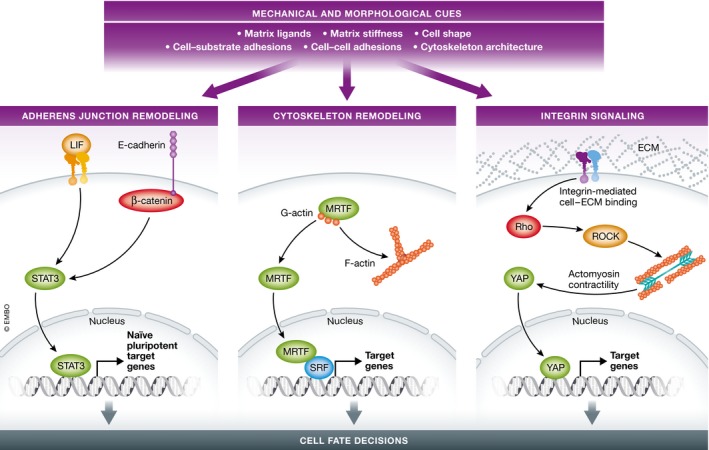
Cues provided by extracellular matrix (ECM) ligands, ECM stiffness, cell shape, cell‐substrate adhesion, cell–cell adhesion, and cytoskeleton architectures inform the cell of its surrounding niche (right panel). The naïve state of clonal embryonic stem cells is routinely maintained in medium supplemented with leukemia inhibitory factor (LIF), which activates STAT3 to induce expression of naïve pluripotent target genes. However, expression of E‐cadherin in pluripotent stem cells is sufficient to promote LIF‐independent self‐renewal by activating STAT3 to induce expression of naïve pluripotent target genes. This later effect requires the β‐catenin‐binding region of E‐cadherin (left panel). With increased actin polymerization, myocardin‐related transcription factor (MRTF), which is retained in the cytoplasm by binding to G‐actin, translocates to the nucleus where it binds the transcription factor serum response factor (SRF) to activate genes regulating differentiation programs (middle panel). In response to integrin‐mediated cell‐substrate adhesion, the low molecular weight GTPase Rho activates Rho‐associated protein kinase (ROCK) to generate actomyosin contractility, which results in nuclear translocation of yes‐associated protein (YAP) (right panel).
Cell‐substrate adhesion
Extracellular matrix (ECM) interactions with integrins and the changes in cell shape and tensional forces they generate provide instructive cues in stem cell fate decisions for both embryonic and adult stem cells, although downstream pathways result in divergent outcomes depending on the cellular context. Variable matrix elasticity directs MSC lineage specification with a greater selectivity than through biochemical cues and generates cellular fate memory that persists after cells are removed from a given matrix 107. Furthermore, pre‐committing naive MSCs on a matrix stiffness that most closely recapitulates in vivo niche stiffness improves microenvironment adaptation upon implantation 107. The effect of cell shape on MSC fate decisions has also been shown by plating cells on small fibronectin islands, which reveals that cells with a rounded morphology differentiate to adipogenic lineages, while cells with a flattened cell shape differentiate to osteogenic lineages 108. This morphology‐driven differentiation is dependent on activity of the low molecular weight GTPase RhoA, indicating that the mechanical cues of cell shape and contractility contribute to lineage commitment in MSCs. In support of this finding, McBeath and colleagues 108 suggest that changes in cell shape sensed through integrin binding of ECM ligands, which provide tensional forces, can drive signaling cascades that result in altered gene expression in MSCs.
In contrast to these MSCs that respond to integrin signaling with self‐renewal, ECM–integrin interactions facilitate differentiation in mouse ESCs 109. Teasing apart the roles of mechanical forces resulting from integrin‐mediated cell–ECM adhesion versus E‐cadherin‐mediated cell–cell adhesion, Uda et al 110 found that force via integrins but not E‐cadherins decreases Oct3/4 expression in mouse ESCs, suggesting mechanical forces from distinct force transduction pathways can play divergent roles in embryonic stem cell biology.
Divergent roles for cadherin‐mediated and integrin‐mediated force transduction pathways may occur in the stem cell niches present in Drosophila melanogaster gonad development. DE‐cadherin, the Drosophila melanogaster homolog of E‐cadherin, mediates cell–cell adhesion between germline stem cells and other cells within the Drosophila ovary niche for both proper recruitment and anchoring 111. Somatic stem cells within the Drosophila ovary generate follicle progenitor cells and several differentiated cells within the chamber. These epithelial stem cells are similarly anchored to the surrounding niche by DE‐cadherin in order to prevent differentiation 112. In addition to this cadherin‐mediated cell–cell adhesion, integrins also enable follicle stem cells in the Drosophila ovary to adhere to surrounding basal lamina in the niche, anchoring them in position to respond to cues regulating their differentiation 113. During gonad morphogenesis in the Drosophila testis, germline stem cells contact hub cells in the niche. Integrin‐dependent adhesion but not DE‐cadherin‐dependent adhesion positions the hub cells such that ECM surrounding the gonads anchors the niche and the germline stem cells 114, 115. Somatic stem cells within the Drosophila testis must also contact hub cells to maintain self‐renewal and proliferation, but these contacts are DE‐cadherin‐mediated 115. Though dependent on distinct anchoring mechanisms, positioning of both germline stem cells and somatic stem cells along hub cells within the Drosophila testis allows cooperation during gametogenesis as both cell types respond in different ways to local JAK‐STAT signaling within the niche 116. A recent study suggests that DE‐cadherin affects signaling in the Drosophila ISCs through a feedback loop that couples enterocyte cell death to ISC divisions 117. In this tissue, β‐catenin is typically sequestered at the adherens junctions in enterocytes, but enterocyte cell death disrupts these junctions and thus causes the release of β‐catenin. β‐catenin then translocates to the nucleus where it activates the expression of rhomboid, which promotes the secretion of EGF ligands and ultimately leads to increased ISC proliferation.
We highlight here merely some advances in our understanding of how cell–ECM interactions and cell shape contribute to stem cell fate decisions. For more comprehensive discussions, we refer readers to reviews on ECM, integrins, and growth factors directing stem cell fate 118, nanoscale features of integrin–matrix interactions, matrix stiffness and 2D versus 3D cultures 119, and integrin‐ and cadherin‐mediated adhesion in maintaining a supportive niche for stem cell anchoring, self‐renewal, and differentiation 120, 121, 122.
Cell–cell adhesion
The role of cadherin‐mediated cell–cell adhesion in pluripotent cells is currently an area of active investigation. In mouse embryos, the adherens junction protein E‐cadherin is highly expressed until gastrulation, when E‐cadherin is downregulated as epithelial epiblasts undergo an epithelial‐to‐mesenchymal transition (EMT) and germ layers are specified. Animals null for E‐cadherin are unable to complete embryogenesis beyond this point 123, 124, which may be due in part to the lack of mechanical forces at adherens junctions 125. However, heterozygous loss of E‐cadherin combined with N‐cadherin knock‐in results in normal embryonic development 126. Whether the in vivo role for E‐cadherin is similar for differentiation of embryonic stem cells in vitro remains controversial. Spencer et al 127 found that mouse ESC differentiation involves traditional markers of EMT such as an E‐cadherin to N‐cadherin switching, increased expression of the E‐cadherin repressors Snail and Slug, and increased cell motility. Also in support of a pluripotent self‐renewal promoting role for cell–cell adhesion, E‐cadherin‐mediated cell–cell contacts promote mouse ESC self‐renewal and induced pluripotent stem cell (iPSC) generation 128, 129. In agreement with this proposed role, mouse ESCs null for E‐cadherin have a transcriptional profile that more closely resembles differentiated epiblast stem cells than self‐renewing naïve ESCs 130. Interestingly, genes most differentially expressed in self‐renewing ESCs from E‐cadherin−/− compared with WT mice are not limited to cell adhesion and motility but also includes transcripts related to metabolic processes, catabolism, and apoptosis 130. A comprehensive evaluation of the roles for E‐cadherin in embryonic stem cells, pluripotency, and self‐renewal is beyond the scope of our discussion of lesser‐studied regulators of stem cell biology, but we refer the reader to several excellent reviews on this topic 131, 132, 133.
Like E‐cadherin, the role of β‐catenin in stem cell self‐renewal and differentiation is currently controversial, despite consensus on the importance of repressive transcriptional activity Tcf3 downstream of canonical Wnt signaling, as described more completely in recent reviews on embryonic 134 and adult 135 stem cells. For embryonic stem cells, conflicting findings may result from distinct β‐catenin functions as an adherens junction protein and a signaling molecule in the Wnt pathway, with perhaps a cell–cell adhesion function being more critical. In brief, for embryonic stem cells, one view is that β‐catenin is not necessary for the self‐renewal and expansion of naïve mESCs, but its absence eliminates the self‐renewal response to Gsk3 inhibition 136. Another non‐contradictory view is that a complex of β‐catenin, E‐cadherin and Oct 4 but not β‐catenin transcriptional activity is necessary for pluripotency 137. Additionally, β‐catenin may be necessary for subsequent differentiation stages because mesendodermal germ layer formation and neuronal differentiation are defective in β‐catenin‐null mESCs 138. Redundancy between catenins may also explain conflicting findings because in β‐catenin‐null mESCs, loss of γ‐catenin promotes exit from pluripotency 139, which further suggests the importance of the adherens junctions but not signaling function of β‐catenin in embryonic stem cell self‐renewal and differentiation.
Actin filaments
Although actin filament dynamics regulate cell‐substrate adhesion, cell–cell adhesion, and cell morphology, we have limited understanding of its direct role in stem cell differentiation and lineage specification. Moreover, how actin cytoskeleton dynamics might regulate transcriptional programs in cell differentiation is incompletely understood, although current evidence implicates roles for YAP, transcriptional activator with PDZ‐binding motif (TAZ), and myocardin‐related transcription factor (MRTF), which are transcriptional regulators responding to mechanical force or actin remodeling. YAP and TAZ, transcriptional cofactors in the Hippo signaling pathway, are both required for early mouse embryo development 140. In response to mechanical cues, YAP and TAZ translocate from the cytoplasm to the nucleus where they bind the transcription factor TEAD and other promoter‐specific transcription factors (reviewed in 141). Higher stiffness of the surrounding extracellular matrix results in nuclear YAP/TAZ localization by an unclear mechanism that senses cell tension 142. Multiple types of mouse stem and progenitor cells, including ESCs, are characterized by upregulated YAP expression, suggesting that Hippo signaling promotes pluripotency‐related pathways 143. Additionally, Yorkie, the Drosophila homolog of Yap, causes increased ISC proliferation in response to intestinal epithelia damage 144, 145 and also functions downstream of hedgehog signaling to promote proliferation of follicle stem cells 146. In the mouse intestine, Yap activity contributes to the downregulation Wnt signaling, which is the key ISC self‐renewal signal, and overexpression of Yap causes ISC loss whereas knockout of Yap causes an increase in the number of ISCs and Paneth cells. In contrast, overexpression of Yap in the epidermis has the opposite effect, causing an expansion of the stem cell pool and the formation of squamous cell‐like carcinomas. However, knockout of the upstream negative regulator, Mst1/2, does not have the same effect, suggesting that Yap is activated by a non‐canonical mechanism in this tissue.
Myocardin‐related transcription factor is another link between actin remodeling and transcriptional regulation. In contrast to nuclear translocation of YAP/TAZ in response to cell‐substrate signals, MRTF is translocated from the cytosol to the nucleus in response to increased actin polymerization 147, 148. In the nucleus, MRTF is a cofactor for transcriptional regulation by SRF to induce expression of over 200 transcripts, mostly related to actin dynamics, cell motility, muscle‐specific genes, and miRNAs (reviewed in 149). Although a role for MRTF in ESCs remains undetermined, it is important for adult MSC differentiation. Specifically, the degree of cell spreading in a precursor of the adipogenic and osteogenic lineages increases actin polymerization, and MRTF is translocated to the nucleus to promote osteogenic gene expression programs 150, 151, 152, 153, 154.
Despite recent advances, further understanding of how cell shape, adhesion, and actin filament dynamics contribute to stem cell differentiation is needed to inform how directed in vitro differentiation protocols are optimized for regenerative medicine applications. For example, Gilbert et al 155 showed that differentiating muscle cells reorganizes their actin cytoskeleton to match their cultured substrate stiffness, significantly improving the cell's ability to engraft and properly heal after implantation when the cultured substrate stiffness matched that of the in vivo niche. Additionally, Myers et al 156 found that cell colony geometry is a driver of stem cell fate decisions in 2D culture systems: patterning of colonies according to uniform size, density, and shape resulted in improved homogeneity and yield of human iPSC‐derived cardiomyocytes. Zoldan et al 125 found that culturing hESCs on variable scaffold stiffnesses was sufficient to induce lineage‐specific gene expression. As the field of regenerative medicine continues to develop in vitro‐derived cell replacement therapeutics, knowledge of the underlying cell biology of stem cell shape and adhesion as it pertains to both in vivo development and in vitro differentiation will greatly inform future studies.
Conclusion
Our review highlights the diversity of mechanisms used to regulate cell fate decisions. Assuming that a more robust cell fate determination process provides an evolutionary advantage, it seems likely that different cells are regulated by multiple and sometimes distinct cues. Extracellular chemical and mechanical cues integrate with intracellular protein‐ and metabolite‐based signaling for the complex control of cell fate decisions (Fig 4). Changes in metabolic state directly impact protein‐based regulation of cell fate decisions by, for example, shifting the availability of metabolites that are required for epigenetic modifications and regulating metabolic sensors, such as AMPK and sirtuins 24. Metabolic changes are also the primary causes of changes in ROS concentrations, which contribute to cell fate decisions through the JNK and p38 MAPK pathways 66. Increased ROS levels feedback to regulate metabolism by activating transcription factors such as FoxO family members that regulate metabolism, and can promote the activity of Rho‐associated protein kinase (ROCK), which regulates cytoskeletal‐associated proteins such as myosin, talin, and cofilin 157. Additionally, Hippo signaling, which responds to mechanical cues, can induce changes in metabolism 158, and metabolism can affect cell shape and cytoskeletal dynamics through the effects of AMPK on cell polarity proteins and myosin regulatory light chain 159, 160, 161. Metabolic changes that shift in the balance of energy and biomass production from glycolysis versus oxidative phosphorylation also affect pHi by changing the redox state of the cell. Changes in pHi can also feedback to regulate metabolism by, for example, affecting the activity of pH‐sensitive enzymes such as phosphofructokinase‐1 162, 163. Finally, changes in pHi can also impact cell shape and mechanical cues through effects on pH‐sensing actin regulatory proteins, such as cofilin and talin 164, 165.
Figure 4. Network of cell biological cues that instruct cell fate decisions.
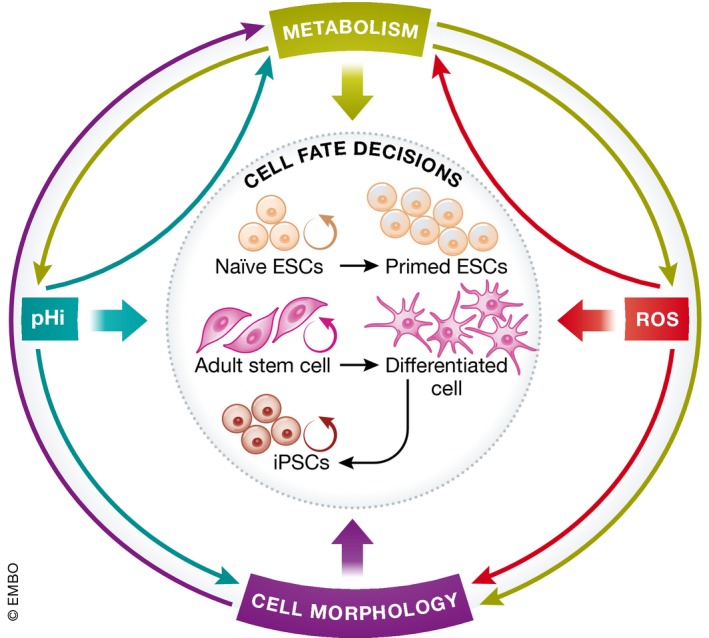
Examples of cell fate decisions include naïve embryonic stem cells (ESC) undergoing self‐renewal (curved arrow) or differentiating into a primed ESC (straight arrow); adult stem cells self‐renewing (curved arrow) or becoming differentiated cells (straight arrow); and reprogramming of differentiated cells to form induced pluripotent stem cells (iPSCs). Cell fate decisions are directly influenced by cell biological cues like metabolism, intracellular pH (pHi), reactive oxygen species (ROS), and cell morphology (short colored arrows). Additionally, since these cell biological cues can affect each other (long colored arrows), these cues also affect cell fate decisions indirectly. Metabolism can affect pHi, ROS and cell morphological changes. pHi and ROS can influence cellular metabolism and morphology. Cell morphological changes can also affect metabolic changes. Therefore, the interaction between these cell biological cues forms a network of cues that instruct cell fate decisions.
Collectively, the studies summarized here demonstrate the extensive contribution of cell biological regulators to the mechanisms that govern cell fate decisions. Nevertheless, many open questions remains (Box 1). However, the field is still relatively new, and the increasing interest combined with new methods for studying the cell biology of cell fate decisions in vivo is likely to lead to more insights into this significant area of developmental biology.
Box 1:In need of answers.
What roles do metabolic pathways other than glycolysis and the TCA cycle play in cellular differentiation?
Do changes in S‐adenosyl methionine concentrations regulate adult stem cell self‐renewal and/or differentiation?
Do the ROS signals that contribute to the regulation of cellular differentiation promote aging?
What are the key pH‐sensing proteins that mediate effects of pHi dynamics in regulating cell fate?
Do changes in pHi affect cellular differentiation by influencing metabolism, reactive oxygen species, or the cytoskeleton?
Despite considerable work, there is still a lack of a comprehensive understanding about the role of adherens junctions in ESC pluripotency.
How do changes in actin dynamics, cell shape, and cell adhesion regulate cellular differentiation, particularly with regard to epigenetic and transcriptional effects?
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was funded by a National Institute of Health grant GM116384 to D. L. Barber and T. G. Nystul. In addition, Francesca Aloisio is supported by the Howard Hughes Medical Institute Gilliam Fellowship, UCSF Moritz‐Heyman Discovery Fellowship, and an NIH T32 grant GM008568.
EMBO Reports (2017) 18: 2105–2118
See the Glossary for abbreviations used in this article.
References
- 1. Shyh‐Chang N, Daley GQ, Cantley LC (2013) Stem cell metabolism in tissue development and aging. Development 140: 2535–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, Sadek HA (2010) The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7: 380–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Suda T, Takubo K, Semenza GL (2011) Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9: 298–310 [DOI] [PubMed] [Google Scholar]
- 4. Almada AE, Wagers AJ (2016) Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat Rev Mol Cell Biol 17: 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rocheteau P, Gayraud‐Morel B, Siegl‐Cachedenier I, Blasco MA, Tajbakhsh S (2012) A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148: 112–125 [DOI] [PubMed] [Google Scholar]
- 6. Zhou W, Choi M, Margineantu D, Margaretha L, Hesson J, Cavanaugh C, Blau CA, Horwitz MS, Hockenbery D, Ware C et al (2012) HIF1α induced switch from bivalent to exclusively glycolytic metabolism during ESC‐to‐EpiSC/hESC transition. EMBO J 31: 2103–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang J, Ratanasirintrawoot S, Chandrasekaran S, Wu Z, Ficarro SB, Yu C, Ross CA, Cacchiarelli D, Xia Q, Seligson M et al (2016) LIN28 regulates stem cell metabolism and conversion to primed pluripotency. Cell Stem Cell 19: 66–80 [DOI] [PubMed] [Google Scholar]
- 8. Takashima Y, Guo G, Loos R, Nichols J, Ficz G, Krueger F, Oxley D, Santos F, Clarke J, Mansfield W et al (2014) Resetting transcription factor control circuitry toward ground‐state pluripotency in human. Cell 158: 1254–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carey BW, Finley LWS, Cross JR, Allis CD, Thompson CB (2015) Intracellular α‐ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 518: 413–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang J, Khvorostov I, Hong JS, Oktay Y, Vergnes L, Nuebel E, Wahjudi PN, Setoguchi K, Wang G, Do A et al (2011) UCP2 regulates energy metabolism and differentiation potential of human pluripotent stem cells. EMBO J 30: 4860–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Folmes CDL, Dzeja PP, Nelson TJ, Terzic A (2012) Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell 11: 596–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Folmes CDL, Nelson TJ, Martinez‐Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez‐Terzic C, Terzic A (2011) Somatic oxidative bioenergetics transitions into pluripotency‐dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14: 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J (2012) Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep 2: 1579–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mihaylova MM, Shaw RJ (2011) The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol 13: 1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ito K, Suda T (2014) Metabolic requirements for the maintenance of self‐renewing stem cells. Nat Rev Mol Cell Biol 15: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Palomäki S, Pietilä M, Laitinen S, Pesälä J, Sormunen R, Lehenkari P, Koivunen P (2013) HIF‐1α is upregulated in human mesenchymal stem cells. Stem Cells 31: 1902–1909 [DOI] [PubMed] [Google Scholar]
- 17. Liu W, Wen Y, Bi P, Lai X, Liu XS, Liu X, Kuang S (2012) Hypoxia promotes satellite cell self‐renewal and enhances the efficiency of myoblast transplantation. Development 139: 2857–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamazaki S, Iwama A, Takayanagi S‐I, Morita Y, Eto K, Ema H, Nakauchi H (2006) Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. EMBO J 25: 3515–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yilmaz ÖH, Katajisto P, Lamming DW, Gültekin Y, Bauer‐Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M et al (2012) mTORC1 in the Paneth cell niche couples intestinal stem‐cell function to calorie intake. Nature 486: 490–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou J, Shrikhande G, Xu J, McKay RM, Burns DK, Johnson JE, Parada LF (2011) Tsc1 mutant neural stem/progenitor cells exhibit migration deficits and give rise to subependymal lesions in the lateral ventricle. Genes Dev 25: 1595–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson A, Murphy MJ, Oskarsson T, Kaloulis K, Bettess MD, Oser GM, Pasche A‐C, Knabenhans C, Macdonald HR, Trumpp A (2004) c‐Myc controls the balance between hematopoietic stem cell self‐renewal and differentiation. Genes Dev 18: 2747–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waikel RL, Kawachi Y, Waikel PA, Wang XJ, Roop DR (2001) Deregulated expression of c‐Myc depletes epidermal stem cells. Nat Genet 28: 165–168 [DOI] [PubMed] [Google Scholar]
- 23. Cao Y, Guo W‐T, Tian S, He X, Wang X‐W, Liu X, Gu K‐L, Ma X, Huang D, Hu L et al (2015) miR‐290/371‐Mbd2‐Myc circuit regulates glycolytic metabolism to promote pluripotency. EMBO J 34: 609–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ryall JG, Cliff T, Dalton S, Sartorelli V (2015) Metabolic Reprogramming of Stem Cell Epigenetics. Cell Stem Cell 17: 651–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB (2009) ATP‐citrate lyase links cellular metabolism to histone acetylation. Science 324: 1076–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moussaieff A, Rouleau M, Kitsberg D, Cohen M, Levy G, Barasch D, Nemirovski A, Shen‐Orr S, Laevsky I, Amit M et al (2015) Glycolysis‐mediated changes in acetyl‐CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab 21: 392–402 [DOI] [PubMed] [Google Scholar]
- 27. Correia M, Perestrelo T, Rodrigues AS, Ribeiro MF, Pereira SL, Sousa MI, Ramalho‐Santos J (2017) Sirtuins in metabolism, stemness and differentiation. Biochim Biophys Acta 1861: 3444–3455 [DOI] [PubMed] [Google Scholar]
- 28. Ruderman NB, Xu XJ, Nelson L, Cacicedo JM, Saha AK, Lan F, Ido Y (2010) AMPK and SIRT1: a long‐standing partnership? Am J Physiol Endocrinol Metab 298: E751–E760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Calvanese V, Lara E, Suárez‐Alvarez B, Abu Dawud R, Vázquez‐Chantada M, Martínez‐Chantar ML, Embade N, López‐Nieva P, Horrillo A, Hmadcha A et al (2010) Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc Natl Acad Sci USA 107: 13736–13741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Saunders LR, Sharma AD, Tawney J, Nakagawa M, Okita K, Yamanaka S, Willenbring H, Verdin E (2010) miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging 2: 415–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hisahara S, Chiba S, Matsumoto H, Tanno M, Yagi H, Shimohama S, Sato M, Horio Y (2008) Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci USA 105: 15599–15604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Peled T, Shoham H, Aschengrau D, Yackoubov D, Frei G, Rosenheimer GN, Lerrer B, Cohen HY, Nagler A, Fibach E et al (2012) Nicotinamide, a SIRT1 inhibitor, inhibits differentiation and facilitates expansion of hematopoietic progenitor cells with enhanced bone marrow homing and engraftment. Exp Hematol 40:342–355.e1 [DOI] [PubMed] [Google Scholar]
- 33. Shyh‐Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H et al (2013) Influence of threonine metabolism on S‐adenosylmethionine and histone methylation. Science 339: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Teslaa T, Teitell MA (2015) Pluripotent stem cell energy metabolism: an update. EMBO J 34: 138–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S (2014) Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 19: 780–794 [DOI] [PubMed] [Google Scholar]
- 36. Hirschey MD, DeBerardinis RJ, Diehl AME, Drew JE, Frezza C, Green MF, Jones LW, Ko YH, Le A, Lea MA et al (2015) Dysregulated metabolism contributes to oncogenesis. Semin Cancer Biol 35(Suppl): S129–S150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bröske A‐M, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R et al (2009) DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet 41: 1207–1215 [DOI] [PubMed] [Google Scholar]
- 38. Sheaffer KL, Kim R, Aoki R, Elliott EN, Schug J, Burger L, Schübeler D, Kaestner KH (2014) DNA methylation is required for the control of stem cell differentiation in the small intestine. Genes Dev 28: 652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sen GL, Reuter JA, Webster DE, Zhu L, Khavari PA (2010) DNMT1 maintains progenitor function in self‐renewing somatic tissue. Nature 463: 563–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Avgustinova A, Benitah SA (2016) Epigenetic control of adult stem cell function. Nat Rev Mol Cell Biol 17: 643–658 [DOI] [PubMed] [Google Scholar]
- 41. Tsukada Y‐I, Fang J, Erdjument‐Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y (2006) Histone demethylation by a family of JmjC domain‐containing proteins. Nature 439: 811–816 [DOI] [PubMed] [Google Scholar]
- 42. Yang H, Lin H, Xu H, Zhang L, Cheng L, Wen B, Shou J, Guan K, Xiong Y, Ye D (2014) TET‐catalyzed 5‐methylcytosine hydroxylation is dynamically regulated by metabolites. Cell Res 24: 1017–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hore TA, von Meyenn F, Ravichandran M, Bachman M, Ficz G, Oxley D, Santos F, Balasubramanian S, Jurkowski TP, Reik W (2016) Retinol and ascorbate drive erasure of epigenetic memory and enhance reprogramming to naïve pluripotency by complementary mechanisms. Proc Natl Acad Sci USA 113: 12202–12207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. D'Aniello C, Habibi E, Cermola F, Paris D, Russo F, Fiorenzano A, Di Napoli G, Melck DJ, Cobellis G, Angelini C et al (2017) Vitamin C and l‐proline antagonistic effects capture alternative states in the pluripotency continuum. Stem Cell Reports 8: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. D'Aniello C, Cermola F, Patriarca EJ, Minchiotti G (2017) Vitamin C in stem cell biology: impact on extracellular matrix homeostasis and epigenetics. Stem Cells Int 2017: 8936156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Blaschke K, Ebata KT, Karimi MM, Zepeda‐Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A et al (2013) Vitamin C induces Tet‐dependent DNA demethylation and a blastocyst‐like state in ES cells. Nature 500: 222–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Agathocleous M, Meacham CE, Burgess RJ, Piskounova E, Zhao Z, Crane GM, Cowin BL, Bruner E, Murphy MM, Chen W et al (2017) Ascorbate regulates haematopoietic stem cell function and leukaemogenesis. Nature 549: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He X‐B, Kim M, Kim S‐Y, Yi S‐H, Rhee Y‐H, Kim T, Lee E‐H, Park C‐H, Dixit S, Harrison FE et al (2015) Vitamin C facilitates dopamine neuron differentiation in fetal midbrain through TET1‐ and JMJD3‐dependent epigenetic control manner. Stem Cells 33: 1320–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G et al (2011) The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin‐C‐dependent manner. Cell Stem Cell 9: 575–587 [DOI] [PubMed] [Google Scholar]
- 50. Forneris F, Binda C, Vanoni MA, Mattevi A, Battaglioli E (2005) Histone demethylation catalysed by LSD1 is a flavin‐dependent oxidative process. FEBS Lett 579: 2203–2207 [DOI] [PubMed] [Google Scholar]
- 51. Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA (2012) Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 482: 221–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Eliazer S, Shalaby NA, Buszczak M (2011) Loss of lysine‐specific demethylase 1 nonautonomously causes stem cell tumors in the Drosophila ovary. Proc Natl Acad Sci USA 108: 7064–7069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Eliazer S, Palacios V, Wang Z, Kollipara RK, Kittler R, Buszczak M (2014) Lsd1 restricts the number of germline stem cells by regulating multiple targets in escort cells. PLoS Genet 10: e1004200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ryall JG, Dell'Orso S, Derfoul A, Juan A, Zare H, Feng X, Clermont D, Koulnis M, Gutierrez‐Cruz G, Fulco M et al (2015) The NAD+‐dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16: 171–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rodríguez‐Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras‐Raves M, Sacchetti A, Hornsveld M, Oost KC, Snippert HJ et al (2017) Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543: 424–427 [DOI] [PubMed] [Google Scholar]
- 56. Yu JSL, Cui W (2016) Proliferation, survival and metabolism: the role of PI3K/AKT/mTOR signalling in pluripotency and cell fate determination. Development 143: 3050–3060 [DOI] [PubMed] [Google Scholar]
- 57. Bulut‐Karslioglu A, Biechele S, Jin H, Macrae TA, Hejna M, Gertsenstein M, Song JS, Ramalho‐Santos M (2016) Inhibition of mTOR induces a paused pluripotent state. Nature 540: 119–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Vannini N, Girotra M, Naveiras O, Nikitin G, Campos V, Giger S, Roch A, Auwerx J, Lutolf MP (2016) Specification of haematopoietic stem cell fate via modulation of mitochondrial activity. Nat Commun 7: 13125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Flores A, Schell J, Krall AS, Jelinek D, Miranda M, Grigorian M, Braas D, White AC, Zhou JL, Graham NA et al (2017) Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat Cell Biol 19: 1017–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schell JC, Wisidagama DR, Bensard C, Zhao H, Wei P, Tanner J, Flores A, Mohlman J, Sorensen LK, Earl CS et al (2017) Control of intestinal stem cell function and proliferation by mitochondrial pyruvate metabolism. Nat Cell Biol 19: 1027–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sandoval IT, Delacruz RGC, Miller BN, Hill S, Olson KA, Gabriel AE, Boyd K, Satterfield C, Remmen HV, Rutter J et al (2017) A metabolic switch controls intestinal differentiation downstream of Adenomatous polyposis coli (APC). Elife 6: e22706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Clevers H, Loh KM, Nusse R (2014) Stem cell signaling. An integral program for tissue renewal and regeneration: Wnt signaling and stem cell control. Science 346: 1248012 [DOI] [PubMed] [Google Scholar]
- 63. Muncan V, Sansom OJ, Tertoolen L, Phesse TJ, Begthel H, Sancho E, Cole AM, Gregorieff A, de Alboran IM, Clevers H et al (2006) Rapid loss of intestinal crypts upon conditional deletion of the Wnt/Tcf‐4 target gene c‐Myc. Mol Cell Biol 26: 8418–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cordero JB, Stefanatos RK, Scopelliti A, Vidal M, Sansom OJ (2012) Inducible progenitor‐derived Wingless regulates adult midgut regeneration in Drosophila . EMBO J 31: 3901–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nathan C, Cunningham‐Bussel A (2013) Beyond oxidative stress: an immunologist's guide to reactive oxygen species. Nat Rev Immunol 13: 349–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bigarella CL, Liang R, Ghaffari S (2014) Stem cells and the impact of ROS signaling. Development 141: 4206–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kim S‐U, Park Y‐H, Kim J‐M, Sun H‐N, Song I‐S, Huang SM, Lee S‐H, Chae J‐I, Hong S, Sik Choi S et al (2014) Dominant role of peroxiredoxin/JNK axis in stemness regulation during neurogenesis from embryonic stem cells. Stem Cells 32: 998–1011 [DOI] [PubMed] [Google Scholar]
- 68. Hochmuth CE, Biteau B, Bohmann D, Jasper H (2011) Redox regulation by Keap1 and Nrf2 controls intestinal stem cell proliferation in Drosophila . Cell Stem Cell 8: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tan SWS, Lee QY, Wong BSE, Cai Y, Baeg GH (2017) Redox homeostasis plays important roles in the maintenance of the Drosophila testis germline stem cells. Stem Cell Reports 9: 342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo‐Kallanian S, Williams IR, Sears C et al (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128: 325–339 [DOI] [PubMed] [Google Scholar]
- 71. Owusu‐Ansah E, Banerjee U (2009) Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461: 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pervaiz S, Taneja R, Ghaffari S (2009) Oxidative stress regulation of stem and progenitor cells. Antioxid Redox Signal 11: 2777–2789 [DOI] [PubMed] [Google Scholar]
- 73. Su B, Mitra S, Gregg H, Flavahan S, Chotani MA, Clark KR, Goldschmidt‐Clermont PJ, Flavahan NA (2001) Redox regulation of vascular smooth muscle cell differentiation. Circ Res 89: 39–46 [DOI] [PubMed] [Google Scholar]
- 74. Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, Wu H, Kornblum HI (2011) Proliferative neural stem cells have high endogenous ROS levels that regulate self‐renewal and neurogenesis in a PI3K/Akt‐dependant manner. Cell Stem Cell 8: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Morimoto H, Iwata K, Ogonuki N, Inoue K, Atsuo O, Kanatsu‐Shinohara M, Morimoto T, Yabe‐Nishimura C, Shinohara T (2013) ROS are required for mouse spermatogonial stem cell self‐renewal. Cell Stem Cell 12: 774–786 [DOI] [PubMed] [Google Scholar]
- 76. Jang Y‐Y, Sharkis SJ (2007) A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low‐oxygenic niche. Blood 110: 3056–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Webb BA, Chimenti M, Jacobson MP, Barber DL (2011) Dysregulated pH: a perfect storm for cancer progression. Nat Rev Cancer 11: 671–677 [DOI] [PubMed] [Google Scholar]
- 78. White KA, Grillo‐Hill BK, Barber DL (2017) Cancer cell behaviors mediated by dysregulated pH dynamics at a glance. J Cell Sci 130: 663–669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Harguindey S, Reshkin SJ, Orive G, Arranz JL, Anitua E (2007) Growth and trophic factors, pH and the Na+/H+ exchanger in Alzheimer's disease, other neurodegenerative diseases and cancer: new therapeutic possibilities and potential dangers. Curr Alzheimer Res 4: 53–65 [DOI] [PubMed] [Google Scholar]
- 80. Wolfe DM, Lee J‐H, Kumar A, Lee S, Orenstein SJ, Nixon RA (2013) Autophagy failure in Alzheimer's disease and the role of defective lysosomal acidification. Eur J Neurosci 37: 1949–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Putney LK, Barber DL (2003) Na‐H exchange‐dependent increase in intracellular pH times G2/M entry and transition. J Biol Chem 278: 44645–44649 [DOI] [PubMed] [Google Scholar]
- 82. Denker SP, Barber DL (2002) Cell migration requires both ion translocation and cytoskeletal anchoring by the Na‐H exchanger NHE1. J Cell Biol 159: 1087–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Stock C, Schwab A (2009) Protons make tumor cells move like clockwork. Pflugers Arch 458: 981–992 [DOI] [PubMed] [Google Scholar]
- 84. Ulmschneider B, Grillo‐Hill BK, Benitez M, Azimova DR, Barber DL, Nystul TG (2016) Increased intracellular pH is necessary for adult epithelial and embryonic stem cell differentiation. J Cell Biol 215: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Singh Y, Zhou Y, Shi X, Zhang S, Umbach AT, Salker MS, Lang KS, Lang F (2016) Alkaline cytosolic ph and high sodium hydrogen exchanger 1 (NHE1) activity in Th9 cells. J Biol Chem 291: 23662–23671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Amith SR, Wilkinson JM, Fliegel L (2016) Na+/H+ exchanger NHE1 regulation modulates metastatic potential and epithelial‐mesenchymal transition of triple‐negative breast cancer cells. Oncotarget 7: 21091–21113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Li X, Karki P, Lei L, Wang H, Fliegel L (2009) Na+/H+ exchanger isoform 1 facilitates cardiomyocyte embryonic stem cell differentiation. Am J Physiol Heart Circ Physiol 296: H159–H170 [DOI] [PubMed] [Google Scholar]
- 88. Edwards LJ, Williams DA, Gardner DK (1998) Intracellular pH of the mouse preimplantation embryo: amino acids act as buffers of intracellular pH. Hum Reprod 13: 3441–3448 [DOI] [PubMed] [Google Scholar]
- 89. Wang H, Singh D, Fliegel L (1997) The Na+/H+ antiporter potentiates growth and retinoic acid‐induced differentiation of P19 embryonal carcinoma cells. J Biol Chem 272: 26545–26549 [DOI] [PubMed] [Google Scholar]
- 90. Gao W, Zhang H, Chang G, Xie Z, Wang H, Ma L, Han Z, Li Q, Pang T (2014) Decreased intracellular pH induced by cariporide differentially contributes to human umbilical cord‐derived mesenchymal stem cells differentiation. Cell Physiol Biochem 33: 185–194 [DOI] [PubMed] [Google Scholar]
- 91. Krüger J, Bohrmann J (2015) Bioelectric patterning during oogenesis: stage‐specific distribution of membrane potentials, intracellular pH and ion‐transport mechanisms in Drosophila ovarian follicles. BMC Dev Biol 15: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Salker MS, Zhou Y, Singh Y, Brosens J, Lang F (2015) LeftyA sensitive cytosolic pH regulation and glycolytic flux in Ishikawa human endometrial cancer cells. Biochem Biophys Res Commun 460: 845–849 [DOI] [PubMed] [Google Scholar]
- 93. Grillo‐Hill BK, Choi C, Jimenez‐Vidal M, Barber DL (2015) Increased H+ efflux is sufficient to induce dysplasia and necessary for viability with oncogene expression. Elife 4: e03270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Uzman JA, Patil S, Uzgare AR, Sater AK (1998) The role of intracellular alkalinization in the establishment of anterior neural fate in Xenopus. Dev Biol 193: 10–20 [DOI] [PubMed] [Google Scholar]
- 95. Wu J, Glimcher LH, Aliprantis AO (2008) HCO3‐/Cl‐ anion exchanger SLC4A2 is required for proper osteoclast differentiation and function. Proc Natl Acad Sci US A 105: 16934–16939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Oginuma M, Moncuquet P, Xiong F, Karoly E, Chal J, Guevorkian K, Pourquié O (2017) A gradient of glycolytic activity coordinates FGF and Wnt signaling during elongation of the body axis in amniote embryos. Dev Cell 40: 342–353.e10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kottmann RM, Trawick E, Judge JL, Wahl LA, Epa AP, Owens KM, Thatcher TH, Phipps RP, Sime PJ (2015) Pharmacologic inhibition of lactate production prevents myofibroblast differentiation. Am J Physiol Lung Cell Mol Physiol 309: L1305–L1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Zhu H, Guo S, Zhang Y, Yin J, Yin W, Tao S, Wang Y, Zhang C (2016) Proton‐sensing GPCR‐YAP signalling promotes cancer‐associated fibroblast activation of mesenchymal stem cells. Int J Biol Sci 12: 389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hjelmeland AB, Wu Q, Heddleston JM, Choudhary GS, MacSwords J, Lathia JD, McLendon R, Lindner D, Sloan A, Rich JN (2011) Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ 18: 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Schönichen A, Webb BA, Jacobson MP, Barber DL (2013) Considering protonation as a posttranslational modification regulating protein structure and function. Annu Rev Biophys 42: 289–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Simons M, Gault WJ, Gotthardt D, Rohatgi R, Klein TJ, Shao Y, Lee H‐J, Wu A‐L, Fang Y, Satlin LM et al (2009) Electrochemical cues regulate assembly of the Frizzled/Dishevelled complex at the plasma membrane during planar epithelial polarization. Nat Cell Biol 11: 286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. McBrian MA, Behbahan IS, Ferrari R, Su T, Huang T‐W, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB et al (2013) Histone acetylation regulates intracellular pH. Mol Cell 49: 310–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Iglesias‐Bartolome R, Callejas‐Valera JL, Gutkind JS (2013) Control of the epithelial stem cell epigenome: the shaping of epithelial stem cell identity. Curr Opin Cell Biol 25: 162–169 [DOI] [PubMed] [Google Scholar]
- 104. Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H et al (2015) The metabolome regulates the epigenetic landscape during naive‐to‐primed human embryonic stem cell transition. Nat Cell Biol 17: 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Nadtochiy SM, Schafer X, Fu D, Nehrke K, Munger J, Brookes PS (2016) Acidic pH is a metabolic switch for 2‐hydroxyglutarate generation and signaling. J Biol Chem 291: 20188–20197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Intlekofer AM, Wang B, Liu H, Shah H, Carmona‐Fontaine C, Rustenburg AS, Salah S, Gunner MR, Chodera JD, Cross JR et al (2017) L‐2‐Hydroxyglutarate production arises from noncanonical enzyme function at acidic pH. Nat Chem Biol 13: 494–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126: 677–689 [DOI] [PubMed] [Google Scholar]
- 108. McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS (2004) Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell 6: 483–495 [DOI] [PubMed] [Google Scholar]
- 109. Hayashi Y, Furue MK, Okamoto T, Ohnuma K, Myoishi Y, Fukuhara Y, Abe T, Sato JD, Hata R‐I, Asashima M (2007) Integrins regulate mouse embryonic stem cell self‐renewal. Stem Cells 25: 3005–3015 [DOI] [PubMed] [Google Scholar]
- 110. Uda Y, Poh Y‐C, Chowdhury F, Wu DC, Tanaka TS, Sato M, Wang N (2011) Force via integrins but not E‐cadherin decreases Oct3/4 expression in embryonic stem cells. Biochem Biophys Res Commun 415: 396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Song X, Zhu C‐H, Doan C, Xie T (2002) Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296: 1855–1857 [DOI] [PubMed] [Google Scholar]
- 112. Song X, Xie T (2002) DE‐cadherin‐mediated cell adhesion is essential for maintaining somatic stem cells in the Drosophila ovary. Proc Natl Acad Sci USA 99: 14813–14818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. O'Reilly AM, Lee H‐H, Simon MA (2008) Integrins control the positioning and proliferation of follicle stem cells in the Drosophila ovary. J Cell Biol 182: 801–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tanentzapf G, Devenport D, Godt D, Brown NH (2007) Integrin‐dependent anchoring of a stem‐cell niche. Nat Cell Biol 9: 1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Voog J, D'Alterio C, Jones DL (2008) Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature 454: 1132–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Issigonis M, Tulina N, de Cuevas M, Brawley C, Sandler L, Matunis E (2009) JAK‐STAT signal inhibition regulates competition in the Drosophila testis stem cell niche. Science 326: 153–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Liang J, Balachandra S, Ngo S, O'Brien LE (2017) Feedback regulation of steady‐state epithelial turnover and organ size. Nature 548: 588–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Brizzi MF, Tarone G, Defilippi P (2012) Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr Opin Cell Biol 24: 645–651 [DOI] [PubMed] [Google Scholar]
- 119. Dalby MJ, Gadegaard N, Oreffo ROC (2014) Harnessing nanotopography and integrin‐matrix interactions to influence stem cell fate. Nat Mater 13: 558–569 [DOI] [PubMed] [Google Scholar]
- 120. Raymond K, Deugnier M‐A, Faraldo MM, Glukhova MA (2009) Adhesion within the stem cell niches. Curr Opin Cell Biol 21: 623–629 [DOI] [PubMed] [Google Scholar]
- 121. Xi R (2009) Anchoring stem cells in the niche by cell adhesion molecules. Cell Adh Migr 3: 396–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Chen S, Lewallen M, Xie T (2013) Adhesion in the stem cell niche: biological roles and regulation. Development 140: 255–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Larue L, Ohsugi M, Hirchenhain J, Kemler R (1994) E‐cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc Natl Acad Sci USA 91: 8263–8267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Stephenson RO, Yamanaka Y, Rossant J (2010) Disorganized epithelial polarity and excess trophectoderm cell fate in preimplantation embryos lacking E‐cadherin. Development 137: 3383–3391 [DOI] [PubMed] [Google Scholar]
- 125. Zoldan J, Karagiannis ED, Lee CY, Anderson DG, Langer R, Levenberg S (2011) The influence of scaffold elasticity on germ layer specification of human embryonic stem cells. Biomaterials 32: 9612–9621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Kan NG, Stemmler MP, Junghans D, Kanzler B, de Vries WN, Dominis M, Kemler R (2007) Gene replacement reveals a specific role for E‐cadherin in the formation of a functional trophectoderm. Development 134: 31–41 [DOI] [PubMed] [Google Scholar]
- 127. Spencer HL, Eastham AM, Merry CLR, Southgate TD, Perez‐Campo F, Soncin F, Ritson S, Kemler R, Stern PL, Ward CM (2007) E‐cadherin inhibits cell surface localization of the pro‐migratory 5T4 oncofetal antigen in mouse embryonic stem cells. Mol Biol Cell 18: 2838–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Chen T, Yuan D, Wei B, Jiang J, Kang J, Ling K, Gu Y, Li J, Xiao L, Pei G (2010) E‐cadherin‐mediated cell‐cell contact is critical for induced pluripotent stem cell generation. Stem Cells 28: 1315–1325 [DOI] [PubMed] [Google Scholar]
- 129. Bedzhov I, Alotaibi H, Basilicata MF, Ahlborn K, Liszewska E, Brabletz T, Stemmler MP (2013) Adhesion, but not a specific cadherin code, is indispensable for ES cell and induced pluripotency. Stem Cell Res 11: 1250–1263 [DOI] [PubMed] [Google Scholar]
- 130. Soncin F, Mohamet L, Ritson S, Hawkins K, Bobola N, Zeef L, Merry CLR, Ward CM (2011) E‐cadherin acts as a regulator of transcripts associated with a wide range of cellular processes in mouse embryonic stem cells. PLoS One 6: e21463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Pieters T, van Roy F (2014) Role of cell‐cell adhesion complexes in embryonic stem cell biology. J Cell Sci 127: 2603–2613 [DOI] [PubMed] [Google Scholar]
- 132. Mohamet L, Hawkins K, Ward CM (2011) Loss of function of e‐cadherin in embryonic stem cells and the relevance to models of tumorigenesis. J Oncol 2011: 352616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Soncin F, Ward CM (2011) The function of e‐cadherin in stem cell pluripotency and self‐renewal. Genes 2: 229–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ying Q‐L, Smith A (2017) The art of capturing pluripotency: creating the right culture. Stem Cell Reports 8: 1457–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kretzschmar K, Clevers H (2017) Wnt/β‐catenin signaling in adult mammalian epithelial stem cells. Dev Biol 428: 273–282 [DOI] [PubMed] [Google Scholar]
- 136. Wray J, Kalkan T, Gomez‐Lopez S, Eckardt D, Cook A, Kemler R, Smith A (2011) Inhibition of glycogen synthase kinase‐3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat Cell Biol 13: 838–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Faunes F, Hayward P, Descalzo SM, Chatterjee SS, Balayo T, Trott J, Christoforou A, Ferrer‐Vaquer A, Hadjantonakis A‐K, Dasgupta R et al (2013) A membrane‐associated β‐catenin/Oct4 complex correlates with ground‐state pluripotency in mouse embryonic stem cells. Development 140: 1171–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lyashenko N, Winter M, Migliorini D, Biechele T, Moon RT, Hartmann C (2011) Differential requirement for the dual functions of β‐catenin in embryonic stem cell self‐renewal and germ layer formation. Nat Cell Biol 13: 753–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Mahendram S, Kelly KF, Paez‐Parent S, Mahmood S, Polena E, Cooney AJ, Doble BW (2013) Ectopic γ‐catenin expression partially mimics the effects of stabilized β‐catenin on embryonic stem cell differentiation. PLoS One 8: e65320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Morin‐Kensicki EM, Boone BN, Howell M, Stonebraker JR, Teed J, Alb JG, Magnuson TR, O'Neal W, Milgram SL (2006) Defects in yolk sac vasculogenesis, chorioallantoic fusion, and embryonic axis elongation in mice with targeted disruption of Yap65. Mol Cell Biol 26: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Halder G, Dupont S, Piccolo S (2012) Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol 13: 591–600 [DOI] [PubMed] [Google Scholar]
- 142. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S et al (2011) Role of YAP/TAZ in mechanotransduction. Nature 474: 179–183 [DOI] [PubMed] [Google Scholar]
- 143. Ramalho‐Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA (2002) ‘Stemness’: transcriptional profiling of embryonic and adult stem cells. Science 298: 597–600 [DOI] [PubMed] [Google Scholar]
- 144. Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, Tapon N (2010) The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137: 4147–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Staley BK, Irvine KD (2010) Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr Biol 20: 1580–1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Huang J, Kalderon D (2014) Coupling of Hedgehog and Hippo pathways promotes stem cell maintenance by stimulating proliferation. J Cell Biol 205: 325–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. McGee KM, Vartiainen MK, Khaw PT, Treisman R, Bailly M (2011) Nuclear transport of the serum response factor coactivator MRTF‐A is downregulated at tensional homeostasis. EMBO Rep 12: 963–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Miralles F, Posern G, Zaromytidou A‐I, Treisman R (2003) Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113: 329–342 [DOI] [PubMed] [Google Scholar]
- 149. Olson EN, Nordheim A (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 11: 353–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Wang D‐Z, Li S, Hockemeyer D, Sutherland L, Wang Z, Schratt G, Richardson JA, Nordheim A, Olson EN (2002) Potentiation of serum response factor activity by a family of myocardin‐related transcription factors. Proc Natl Acad Sci USA 99: 14855–14860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Parmacek MS (2007) Myocardin‐related transcription factors: critical coactivators regulating cardiovascular development and adaptation. Circ Res 100: 633–644 [DOI] [PubMed] [Google Scholar]
- 152. Nobusue H, Onishi N, Shimizu T, Sugihara E, Oki Y, Sumikawa Y, Chiyoda T, Akashi K, Saya H, Kano K (2014) Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. Nat Commun 5: 3368 [DOI] [PubMed] [Google Scholar]
- 153. McDonald ME, Li C, Bian H, Smith BD, Layne MD, Farmer SR (2015) Myocardin‐related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell 160: 105–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Bian H, Lin JZ, Li C, Farmer SR (2016) Myocardin‐related transcription factor A (MRTFA) regulates the fate of bone marrow mesenchymal stem cells and its absence in mice leads to osteopenia. Mol Metab 5: 970–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Gilbert PM, Havenstrite KL, Magnusson KEG, Sacco A, Leonardi NA, Kraft P, Nguyen NK, Thrun S, Lutolf MP, Blau HM (2010) Substrate elasticity regulates skeletal muscle stem cell self‐renewal in culture. Science 329: 1078–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Myers FB, Silver JS, Zhuge Y, Beygui RE, Zarins CK, Lee LP, Abilez OJ (2013) Robust pluripotent stem cell expansion and cardiomyocyte differentiation via geometric patterning. Integr Biol 5: 1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Muliyil S, Narasimha M (2014) Mitochondrial ROS regulates cytoskeletal and mitochondrial remodeling to tune cell and tissue dynamics in a model for wound healing. Dev Cell 28: 239–252 [DOI] [PubMed] [Google Scholar]
- 158. Irvine KD, Harvey KF (2015) Control of organ growth by patterning and hippo signaling in Drosophila . Cold Spring Harb Perspect Biol 7: a019224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Jansen M, ten Klooster JP, Offerhaus GJ, Clevers H (2009) LKB1 and AMPK family signaling: the intimate link between cell polarity and energy metabolism. Physiol Rev 89: 777–798 [DOI] [PubMed] [Google Scholar]
- 160. Lee JH, Koh H, Kim M, Kim Y, Lee SY, Karess RE, Lee S‐H, Shong M, Kim J‐M, Kim J et al (2007) Energy‐dependent regulation of cell structure by AMP‐activated protein kinase. Nature 447: 1017–1020 [DOI] [PubMed] [Google Scholar]
- 161. Mirouse V, Swick LL, Kazgan N, St Johnston D, Brenman JE (2007) LKB1 and AMPK maintain epithelial cell polarity under energetic stress. J Cell Biol 177: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 162. Trivedi B, Danforth WH (1966) Effect of pH on the kinetics of frog muscle phosphofructokinase. J Biol Chem 241: 4110–4112 [PubMed] [Google Scholar]
- 163. Erecińska M, Deas J, Silver IA (1995) The effect of pH on glycolysis and phosphofructokinase activity in cultured cells and synaptosomes. J Neurochem 65: 2765–2772 [DOI] [PubMed] [Google Scholar]
- 164. Srivastava J, Barreiro G, Groscurth S, Gingras AR, Goult BT, Critchley DR, Kelly MJS, Jacobson MP, Barber DL (2008) Structural model and functional significance of pH‐dependent talin‐actin binding for focal adhesion remodeling. Proc Natl Acad Sci USA 105: 14436–14441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Frantz C, Barreiro G, Dominguez L, Chen X, Eddy R, Condeelis J, Kelly MJS, Jacobson MP, Barber DL (2008) Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J Cell Biol 183: 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]


