Abstract
Epidermal growth factor (EGF) activates the EGF receptor (EGFR) and stimulates its internalization and trafficking to lysosomes for degradation. However, a percentage of EGFR undergoes ligand‐independent endocytosis and is rapidly recycled back to the plasma membrane. Importantly, alterations in EGFR recycling are a common hallmark of cancer, and yet, our understanding of the machineries controlling the fate of endocytosed EGFR is incomplete. Intersectin‐s is a multi‐domain adaptor protein that is required for internalization of EGFR. Here, we discover that intersectin‐s binds DENND2B, a guanine nucleotide exchange factor for the exocytic GTPase Rab13, and this interaction promotes recycling of ligand‐free EGFR to the cell surface. Intriguingly, upon EGF treatment, DENND2B is phosphorylated by protein kinase D and dissociates from intersectin‐s, allowing for receptor targeting to degradation. Our study thus reveals a novel mechanism controlling the fate of internalized EGFR with important implications for cancer.
Keywords: DENN domain, endocytosis, exocytosis, growth signalling, intersectin‐s
Subject Categories: Membrane & Intracellular Transport, Signal Transduction
Introduction
In the absence of epidermal growth factor (EGF), the EGF receptor (EGFR) undergoes endocytosis, is dephosphorylated in perinuclear endosomes and rapidly recycles to the cell surface 1, 2. In contrast, EGF occupied EGFR recycles at a much slower rate, as it is ubiquitinated by the E3‐ligase Cbl and largely targeted to the lysosome for degradation 1, 2. While there is extensive understanding of the mechanisms targeting EGF‐activated receptor to lysosomes, relatively little is known regarding the mechanisms responsible for EGFR recycling 3. This is important as escape of EGFR from Cbl‐mediated lysosomal targeting, leading to enhanced recycling and signalling, is a reoccurring theme in cancer cell growth 3, 4. Thus, it is vital to better understand the mechanisms of EGFR recycling.
DENND2B is a member of the differentially expressed in normal and neoplastic cells (DENN) domain family 5, 6, 7. Through the DENN domain, these proteins function as guanine nucleotide exchange factors (GEFs) for Rab GTPases 7. DENND2B activates Rab13 selectively at the cell periphery and promotes the delivery of pro‐migratory cargo, and this activity has been linked to the growth and motility of cancer cells both in culture and in mouse models 8, 9, 10. DENND2B also stimulates the signalling of mitogen‐activated protein kinase (MAPK) downstream of EGFR, independent of its GEF activity 11, but the mechanism by which DENN2B activates MAPK, and the binding partners involved are unknown.
Intersectin (ITSN) is a multi‐domain endocytic protein that was originally identified based on its binding to proteins functioning in clathrin‐mediated endocytosis 12, 13. ITSN regulates the internalization and signalling of activated EGFR and has been implicated in the growth and migration of malignant gliomas 14, 15, 16. ITSN also functions in exocytosis and couples endocytosis to exocytosis in excitatory cells 17, 18. However, the role of ITSN in exocytosis has been largely attributed to its GEF activity towards Cdc42, and this activity resides in a protein domain present only in the ITSN‐long (ITSN‐l) isoform, which is expressed exclusively in excitatory cells such as neurons 17, 18, 19. Non‐neuronal cells express ITSN‐short (ITSN‐s), but a role for this isoform in coupling endocytosis to exocytosis has not been demonstrated.
Here, we identify ITSN‐s as a novel binding partner for DENND2B. We demonstrate that DENND2B is important for the recycling of EGFR to the cell surface, suggesting that ITSN‐s couples endocytosis to exocytosis of EGFR through DENND2B. Intriguingly, we discover that the ITSN‐s/DENND2B interaction is abolished upon phosphorylation of DENND2B by protein kinase D (PKD) in response to EGF stimulation. Thus, we propose that in the absence of EGF, ITSN‐s promotes recycling of internalized EGFR by recruiting DENND2B for activation of the Rab13‐dependent exocytic pathway. However, following EGF treatment, ITSN‐s no longer associates with DENND2B and EGFR proceeds towards a degradative pathway.
Results and Discussion
DENND2B binds ITSN‐s
Through its DENN domain, DENND2B functions enzymatically as a GEF for Rab13 8. Through regions outside of the DENN domain, DENND2B promotes MAPK signalling downstream of EGFR and binds to the SH3 domain‐containing adaptor protein Grb2, involved in regulating EGFR trafficking 11. Since DENND2B contains several putative SH3 binding sites, we wondered whether DENND2B interacts with additional SH3 containing proteins important for EGFR trafficking. We screened a panel of SH3 domain proteins known to regulate EGFR trafficking 20, 21, 22. In addition to GST‐Grb2, Flag‐DENND2B binds to GST‐ITSN SH3A, but not the SH3 domains of amphiphysin, endophilin or pacsin (Fig 1A). ITSN contains five tandem SH3 domains (Fig 3H), and binding is selective for the SH3A domain (Fig 1B). Binding of ITSN to DENND2B was confirmed by co‐immunoprecipitation (Fig 1C). Since both ITSN‐s and ITSN‐l contain the SH3A domain, these data suggest that DENND2B can interact with ITSN in both neuronal (ITSN‐l) and non‐neuronal (ITSN‐s) cells 13.
Figure 1. DENND2B binds ITSN.
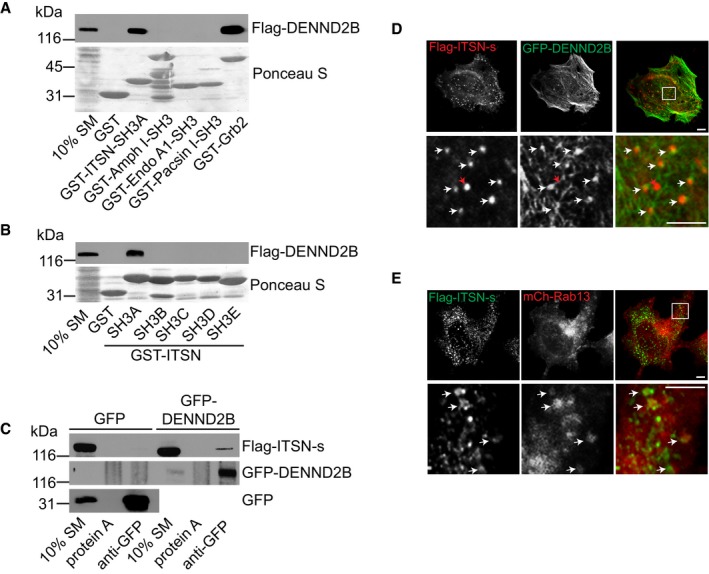
-
APurified GST‐SH3‐containing proteins were incubated with lysates of HEK‐293T cells expressing Flag‐DENND2B. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
-
BPurified GST‐SH3 domains of ITSN were incubated with lysates of HEK‐293T cells expressing Flag‐DENND2B. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
-
CLysates from HEK‐293T cells expressing Flag‐ITSN‐s and GFP or GFP‐DENND2B were incubated with protein A beads or protein A beads coupled to anti‐GFP. Bound proteins were detected by Western blot.
-
D, EHeLa cells expressing the indicated constructs. Flag‐ITSN‐s was visualized with anti‐Flag and AlexaFluor561 or AlexaFluor488 in (D and E), respectively. Scale bars: 5 μm. Boxed regions are magnified on the bottom. White arrows and red arrows point to colocalizing and non‐colocalizing structures, respectively.
Source data are available online for this figure.
Figure 3. 14‐3‐3 outcompetes ITSN for binding to phosphorylated DENND2B.
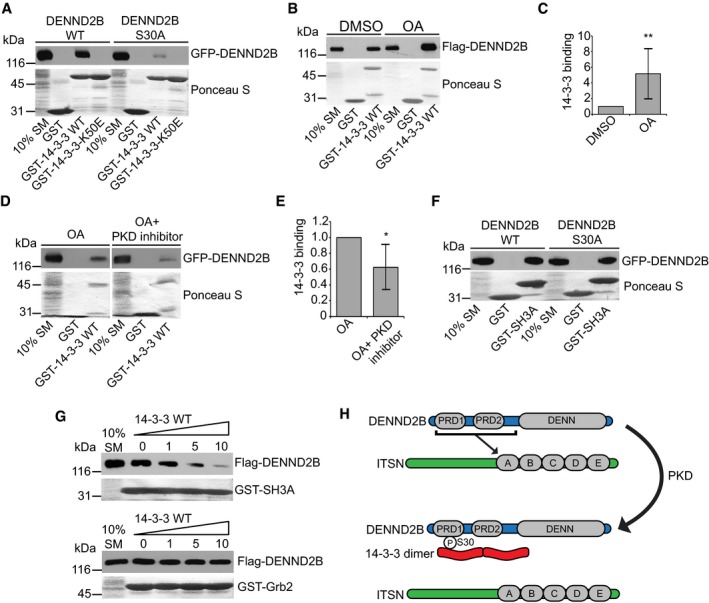
- Purified GST‐14‐3‐3 WT or K50E was incubated with lysates of HEK‐293T cells expressing GFP‐DENND2B WT or S30A. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
- HEK‐293T cells expressing Flag‐DENND2B were treated with DMSO or 250 nM OA, and cell lysates were incubated with GST‐14‐3‐3. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
- Quantification of (B) where bound DENND2B is normalized to the SM. Mean ± SD. Welch's t‐test **P = 0.006. n = 8 for both treatment groups from five independent experiments.
- HEK‐293T cells expressing Flag‐DENND2B were treated with 250 nM OA alone or with 50 μm PKD inhibitor CID755673, and cell lysates were incubated with GST‐14‐3‐3. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
- Quantification of (D) where bound DENND2B is normalized to the SM. Mean ± SD. Welch's t‐test *P = 0.040. n = 5 for both treatment groups from three independent experiments.
- GST‐SH3A was incubated with lysates of HEK‐293T cells expressing GFP‐DENND2B WT or S30A. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
- Lysates of HEK‐293T cells expressing Flag‐DENND2B were pre‐incubated with increasing concentrations of purified 14‐3‐3 WT and subsequently incubated with GST‐SH3A (top panel) or GST‐Grb2 (bottom panel). Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
- Regulation of the ITSN‐DENND2B interaction. In the absence of phosphorylation, DENND2B binds the SH3A (A) domain of ITSN. Phosphorylation of DENND2B by PKD occurs on Ser30 residue within the first proline‐rich domain (PRD). Phosphorylation of Ser30 recruits 14‐3‐3 which outcompetes ITSN binding.
Source data are available online for this figure.
Since DENND2B and ITSN‐s interact biochemically, we predicted that they would co‐localize. DENND2B localizes to the cell periphery and on filamentous actin, while ITSN‐s localizes on puncta throughout the cell 8, 19. When co‐expressed, GFP‐DENND2B and Flag‐ITSN‐s co‐localize on punctate structures seemingly associated with the cortical actin pool of DENND2B (Fig 1D, white arrows). However, not all ITSN‐s‐positive puncta co‐localize with DENND2B (Fig 1D, red arrow). This is expected if ITSN‐s traffics on vesicles that are not in contact with the plasma membrane. We previously found that Rab13 traffics on vesicles from endosomes to the cell periphery where it is activated locally by DENND2B 8. Interestingly, Flag‐ITSN‐s also co‐localizes on a subset of mCh‐Rab13‐positive vesicles (Fig 1E, white arrows). Therefore, a portion of ITSN‐s appears to traffic on Rab13‐positive recycling endosomes to the cell periphery where it then associates with DENND2B.
DENND2B binding to ITSN‐s is phosphorylation dependent
We next examined whether binding of ITSN‐s to DENND2B is regulated. Other DENN domain‐containing proteins are regulated by phosphorylation. For example, phosphorylation of DENND1A by Akt relieves its auto‐inhibition, promoting GEF activity 23, and phosphorylation of DENND3 by ULK activates its GEF activity towards Rab12 24. We therefore wondered whether DENND2B is also phosphorylated and if so, does this influence ITSN‐s binding. By Western blot, we observed a dose‐dependent upward mobility shift in Flag‐DENND2B from cells treated with the serine/threonine phosphatase inhibitor okadaic acid (OA), indicative of phosphorylation (Fig 2A). To test whether this potential phosphorylation event on DENND2B influenced ITSN binding, we performed GST pull‐down assays following OA treatment. While Flag‐DENND2B binding to Grb2 was unaffected by OA treatment, ITSN‐SH3A binding was strongly reduced (Fig 2B). Therefore, it appears that phosphorylation of DENND2B negatively regulates binding to ITSN.
Figure 2. DENND2B phosphorylation by PKD disrupts ITSN interaction.
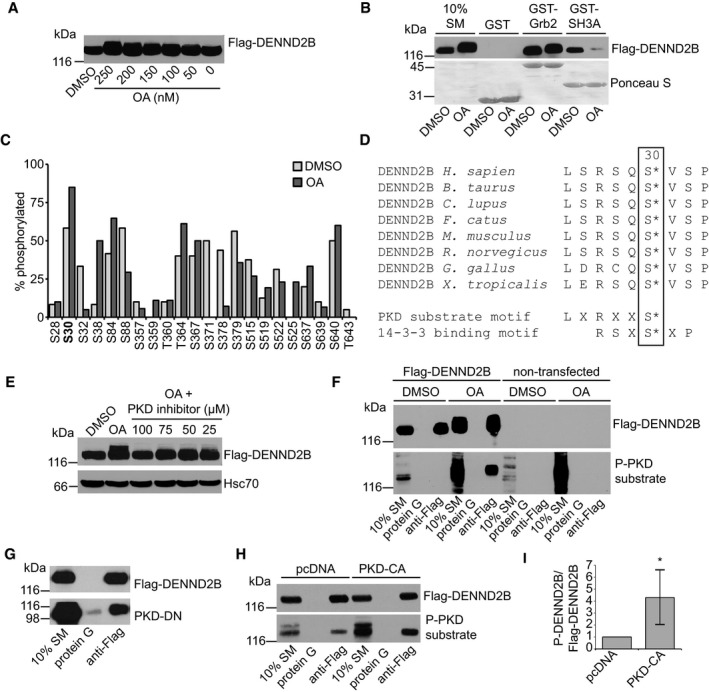
- HEK‐293T cells were treated with DMSO or okadaic acid (OA), lysed and analysed by Western blot.
- HEK‐293T cells expressing Flag‐DENND2B were treated with DMSO or 250 nM OA, and cell lysates were incubated with GST proteins. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
- Percentage of phosphorylated peptides following DMSO or OA treatment as determined by mass spectrometry. Low abundance peptides not included. Refer to Table EV1 for a complete peptide list.
- Alignment of DENND2B sequence from different species flanking the Ser30 residue. PKD and 14‐3‐3 consensus motifs are shown on the bottom.
- HEK‐293T cells expressing Flag‐DENND2B were treated with DMSO, 250 nM OA or with increasing concentrations of PKD inhibitor CID755673. Cell lysates were analysed by Western blot.
- HEK‐293T cells expressing Flag‐DENND2B or control cells were treated with 250 nM OA or DMSO, incubated with protein G beads coupled to anti‐Flag and analysed by Western blot.
- Lysates from HEK‐293T cells expressing Flag‐DENND2B and PKD‐dominant negative (DN) were incubated with protein G beads or protein G beads coupled to anti‐Flag. Bound proteins were detected by Western blot.
- HEK‐293T cells co‐expressing Flag‐DENND2B with pcDNA3 or PKD‐constitutively active (CA) were incubated with protein G beads or protein G beads coupled to anti‐Flag and bound proteins were detected by Western blot.
- Quantification of (H) where PKD‐CA treatment is normalized to pcDNA3 control. Mean ± SD. Welch's t‐test *P = 0.016, n = 6 for both treatment groups from three independent experiments.
Source data are available online for this figure.
DENND2B is phosphorylated by protein kinase D
We next sought to identify the kinase responsible for DENND2B phosphorylation and disruption of ITSN binding. A previous large‐scale proteomic screen revealed that DENND2B is phosphorylated on Ser30 25. Using mass spectrometry, we confirmed that Ser30 is phosphorylated upon OA treatment (Fig 2C and Table EV1). Interestingly, not only are the residues flanking Ser30 conserved across vertebrates, but Ser30 is part of a protein kinase D (PKD) consensus motif (Fig 2D). Similar to DENND2B and ITSN, PKD has been implicated in signalling downstream of EGFR 26. We therefore questioned whether DENND2B is phosphorylated by PKD.
First, we found that the upward mobility shift of Flag‐DENND2B induced by OA was prevented by a PKD inhibitor (Fig 2E). Moreover, a phospho‐PKD substrate antibody that recognizes PKD consensus motifs upon phosphorylation 27 recognizes Flag‐DENND2B purified by immunoprecipitation following OA treatment but not following treatment with vehicle control (Fig 2F). We did not detect any signal when we performed immunoprecipitation in non‐transfected cells, further supporting that the antibody specifically recognizes phosphorylated DENND2B (Fig 2F). We next tested whether DENND2B interacts biochemically with PKD using a dominant‐negative PKD mutant. The dominant‐negative mutation renders the protein kinase dead and creates a substrate trap with stable binding of the enzyme to its substrate 28, 29. Indeed, dominant‐negative PKD co‐immunoprecipitates with DENND2B (Fig 2G). Next, using the phospho‐PKD substrate antibody, we found that expression of a constitutively active PKD mutant increased the phosphorylation of purified Flag‐DENND2B (Fig 2H and I). Collectively, these data indicate that DENND2B is a substrate for PKD.
DENND2B phosphorylation recruits 14‐3‐3 to outcompete ITSN binding
DENND2B was identified in a proteomic analysis of 14‐3‐3 binding partners 30, and interestingly, Ser30 is part of a 14‐3‐3 binding motif (Fig 2C). 14‐3‐3 binds to phosphorylated serine or threonine residues to regulate protein function in numerous cellular processes including protein trafficking and signal transduction 31. We thus hypothesized that 14‐3‐3 binds to Ser30 to regulate DENND2B function. Using GST pull‐down assays, we observed that Flag‐DENND2B binds wild‐type 14‐3‐3 but does not interact with 14‐3‐3 K50E, a binding defective mutant (Fig 3A). Moreover, binding to wild‐type 14‐3‐3 was strongly reduced with a S30A DENND2B mutation (Fig 3A). Further, we observed an increase in 14‐3‐3 binding to DENND2B from cells treated with OA compared to the control (Fig 3B and C). We also observed a decrease in 14‐3‐3 binding in cells that were treated with OA in the presence of a PKD inhibitor compared to OA alone (Fig 3D and E). Together these results indicate that phosphorylation of DENND2B on Ser30 by PKD promotes 14‐3‐3 binding.
Although the Ser30 mutation on DENND2B does not affect ITSN binding (Fig 3F), we wondered whether 14‐3‐3 binding to DENND2B disrupts ITSN binding as 14‐3‐3 binding to substrate proteins can prevent interactions with other molecular partners by steric hindrance. Using competition assays, we found that increasing amounts of purified 14‐3‐3 outcompetes GST‐SH3A for binding to DENND2B (Fig 3G top panels). In contrast, 14‐3‐3 did not compete with GST‐Grb2 for binding to DENND2B (Fig 3G bottom panels). These data show that 14‐3‐3 binds DENND2B thereby disrupting binding to ITSN.
Altogether, our data reveal that DENND2B binds specifically to the SH3A domain of ITSN and that phosphorylation of DENND2B on Ser30 recruits 14‐3‐3 to disrupt binding to ITSN (Fig 3H). While Ser30 is clearly an important site in this process, it is possible that PKD phosphorylates DENND2B at additional sites. Furthermore, 14‐3‐3 proteins typically bind their substrates as dimers, and therefore, additional phosphorylation events could influence the interaction between ITSN and DENND2B. Finally, although 14‐3‐3 binding occurs on Ser30, this does not necessarily mean that ITSN binding occurs in the same region. Depending on how DENND2B is folded and where other 14‐3‐3 binding sites are located, 14‐3‐3 could outcompete ITSN binding at a distant location by inducing a conformation change in DENND2B or by steric hindrance.
DENND2B regulates EGFR recycling
We next sought to determine the physiological relevance of the regulated interaction between DENND2B and ITSN‐s. One emerging commonality of both ITSN‐s and DENND2B is the ability to promote tumorigenicity. ITSN‐s is overexpressed in several cancers 32, and activation of Rab13 by DENND2B drives cancer cell growth and metastasis 8. Interestingly, both ITSN‐s and DENND2B enhance mitogenic signalling downstream of EGFR by promoting MAPK activation 11, 14. We confirmed these findings by expressing Flag‐DENND2B or Flag‐ITSN‐s in HEK‐293T cells, which each caused an increase in phosphorylated MAPK compared to cells expressing Flag alone (Fig EV1).
Figure EV1. DENND2B and ITSN activate MAPK.
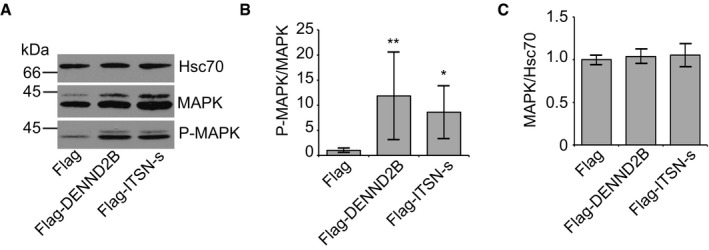
- HEK‐293T cells expressing Flag, Flag‐DENND2B or Flag‐ITSN‐s were analysed by Western blot.
- Quantification of (A) where surface P‐MAPK is normalized to MAPK. Mean ± SD, ANOVA with Dunnett's post hoc test **P = 0.005 and *P = 0.045. n = 7 for all treatment groups from three independent experiments.
- Quantification of (A) where MAPK is normalized to Hsc70. Mean ± SD, n = 8 for all treatment groups from three independent experiments.
Source data are available online for this figure.
We next sought to determine how these proteins promote mitogenic signalling. Following EGFR activation, ITSN is necessary for the internalization of EGFR by clathrin‐mediated endocytosis 14, 33, 34 and ITSN can promote MAPK activation independent of its role in EGFR endocytosis 35. However, ITSN can also recruit Cbl to stimulate ubiquitination and subsequent degradation of EGFR thereby dampening EGFR signalling 36. DENND2B also interacts with proteins involved in EGFR trafficking, notably those involved in facilitating receptor endocytosis and early trafficking steps, such as Grb2 interaction 37, 38, and those involved in attenuating receptor degradation, such as c‐abl 11, 39. Therefore, we questioned whether EGFR trafficking, and consequently its signalling, could be influenced by ITSN's interaction with DENND2B.
To explore a role for DENND2B in EGFR trafficking, we first examined for potential localization of EGFR to Rab13‐positive vesicles. Rab13 traffics on vesicles where it is activated locally by DENND2B to drive vesicle fusion with the plasma membrane 8, 40, 41, and we observed co‐localization of EGFR with Rab13 on vesicles (Fig 4A, arrows) and the plasma membrane (Fig 4A, arrowheads). This is consistent with previous studies showing the importance of Rab13 in regulating EGFR trafficking 42, 43. Moreover, a role for Rab13 in endocytosis has been excluded as Rab13 does not localize on early endosomes where EGFR is initially found upon internalization 40, 41, 44. Thus, EGFR appears to use Rab13 vesicles for resurfacing following endocytosis. Consistent with a role in recycling, we find co‐localization of EGFR with DENND2B on the plasma membrane (Fig 4B, arrowheads).
Figure 4. DENND2B promotes EGFR recycling.
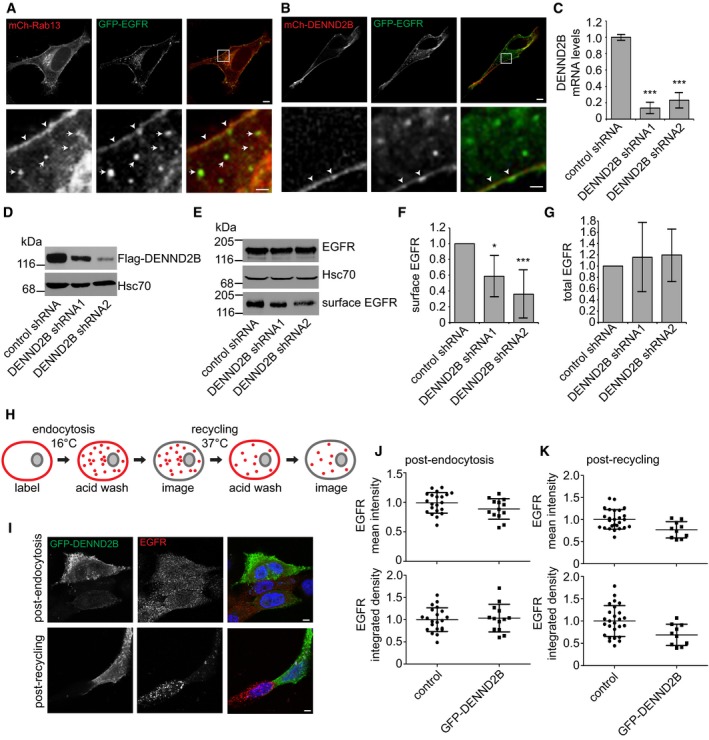
-
A, BHeLa cells expressing the indicated constructs. Boxed regions are magnified on the bottom. Scale bars: 5 μm in the top panel and 1 μm in the bottom panel. White arrows and arrowheads point to co‐localization on vesicles and plasma membrane, respectively.
-
CTransduced MCF10A cells were quantified by real‐time PCR. Mean ± SD, ANOVA with Dunnett's post hoc test ***P = 0.000 for both comparisons, n = 3 from three independent experiments.
-
DTransduced HEK‐293T cells expressing Flag‐DENND2B were analysed by Western blot.
-
ETransduced MCF10A cells were biotinylated and cell lysates were incubated with streptavidin beads. Bound proteins were detected by Western blot.
-
FQuantification of (E) where surface EGFR is normalized to total EGFR. Mean ± SD, ANOVA with Dunnett's post hoc test *P = 0.013 and ***P = 0.000. n = 6 for all treatment groups from four independent experiments.
-
GQuantification of (E) where total EGFR is normalized to Hsc70. Mean ± SD, n = 3 for all treatment groups from three independent experiments.
-
HSchematic of trafficking assay. Surface EGFR is labelled and internalized at 16°C or recycled at 37°C. Acid washing removes labelled surface pools and allows analysis of the internal pool of EGFR originating from the surface.
-
IMCF10A cells expressing GFP‐DENND2B were labelled for surface EGFR on ice, incubated at 16°C to allow internalization, acid washed to remove remaining labelled‐EGFR and processed for immunofluorescence to visualize the internal pool of EGFR (post‐endocytosis, top panel) or incubated at 37°C to allow recycling, acid washed again and processed for immunofluorescence to visualize the remaining internal pool of EGFR (post‐recycling, bottom panel). Scale bars: 5 μm.
-
JQuantification of experiment in (I), top panel, internal EGFR post‐endocytosis. Mean fluorescence per cell ± SD, n = 13 for GFP‐DENND2B and n = 21 for control cells pooled from two independent experiments.
-
KQuantification of experiment in (I), bottom panel, internal EGFR post‐recycling. Mean fluorescence per cell ± SD, n = 10 for GFP‐DENND2B and n = 26 for control cells pooled from 2 independent experiments.
Source data are available online for this figure.
To explore the role of DENND2B in EGFR resurfacing, we first examined the cell surface levels of the receptor following DENND2B knockdown. We used lentivirus to deliver two independent shRNAmiRs to reduce DENND2B mRNA (Fig 4C) and prevent protein expression (Fig 4D) compared to a non‐targeting shRNAmiR control. We then performed surface biotinylation assays to measure the surface levels of EGFR. Indeed, DENND2B knockdown led to decreased surface levels of EGFR (Fig 4E and F) with no change in total levels of the receptor (Fig 4E and G).
Since alterations in surface levels of EGFR could be due to changes in endocytosis or recycling, we tested the effects of DENND2B expression on EGFR trafficking. Using an antibody that recognizes the extracellular domain of EGFR, we labelled the surface pool of EGFR at 4°C, shifted the cells to 16°C to allow internalization but inhibit recycling, stripped the remaining surface pool by acid wash and examined the amount of EGFR that was internalized (Fig 4H). We observed no difference in the amount of endocytosed EGFR with DENND2B overexpression (Fig 4I and J). Following internalization, we shifted the cells to 37°C to allow recycling to the plasma membrane, stripped the surface pool and examined the amount of internal EGFR remaining (Fig 4H). Here, we observed a decrease in the remaining pool of EGFR with enhanced DENND2B expression following recycling (Fig 4I and K). Cumulatively, our data suggest that EGFR recycles to the plasma membrane in Rab13‐positive vesicles and requires DENND2B for resurfacing. Thus, by binding DENND2B, ITSN‐s can couple endocytosis with exocytosis of EGFR in non‐neuronal cells. Future studies are needed to determine whether DENND2B and ITSN play a similar role in facilitating the recycling of proteins in addition to EGFR.
It is interesting to note that EGFR is inactivated in recycling endosomes 2, suggesting that Grb2 would dissociate from these vesicles before reaching DENND2B on the plasma membrane. Therefore, it remains unclear under what circumstances DENND2B binds Grb2. While we did not detect any influence of DENND2B on EGFR endocytosis in our assays, it is still possible that DENND2B plays an unknown role in early endosomal trafficking.
EGF treatment dissociates ITSN‐s and DENND2B
As EGF stimulation targets EGFR to a degradative pathway, while EGFR internalized in a ligand‐independent manner is recycled back to the plasma membrane, we wondered whether EGF plays a role in regulating the interaction between DENND2B and ITSN‐s. Since we identified PKD as the kinase responsible for phosphorylating DENND2B, we first tested whether PKD is activated in response to EGF treatment. We treated serum‐starved cells with EGF and analysed the phosphorylation of PKD on Ser916, an autophosphorylation event that accompanies activation of PKD kinase activity 45. Indeed, we observed increased PKD activation following EGF treatment (Fig 5A and B).
Figure 5. EGF treatment dissociates DENND2B from ITSN.
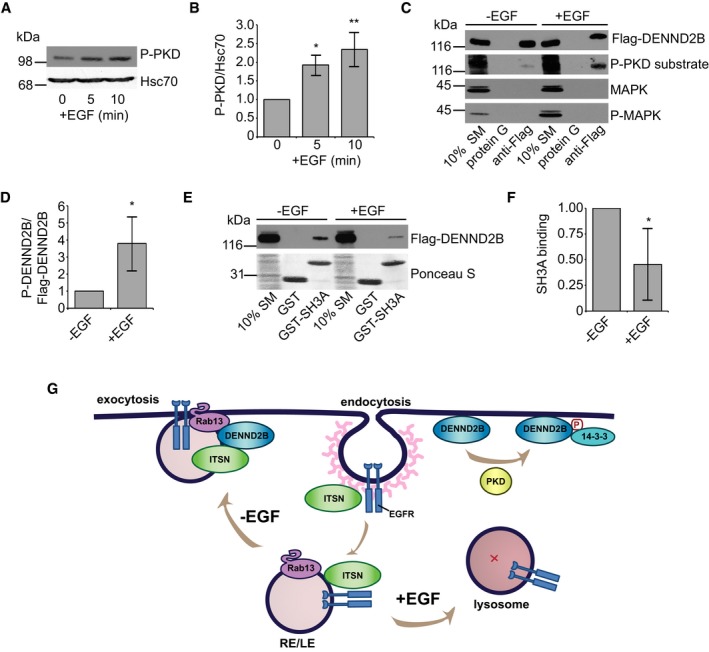
- HEK‐293T cells were serum‐starved, treated with 100 ng/ml EGF for the indicated times, and cell lysates were analysed by Western blot.
- Quantification of (A) where P‐PKD is normalized to Hsc70. Mean ± SD. ANOVA with Dunnett's post‐test *P = 0.015 and **P = 0.003, n = 3 from three independent experiments.
- HEK‐293T cells were serum‐starved, treated with 100 ng/ml EGF for 5 min, and cell lysates were incubated with protein G beads or protein G beads coupled to anti‐Flag. Bound proteins were detected by Western blot.
- Quantification of (B) where P‐PKD substrate is normalized to Flag‐DENND2B. Mean ± SD, Welch's t‐test *P = 0.036, n = 4 from three independent experiments.
- HEK‐293T cells were serum‐starved, treated with 100 ng/ml EGF for 5 min, and cell lysates were incubated with GST‐SH3A. Total proteins and bound proteins were detected by Ponceau S staining and Western blot, respectively.
- Quantification of (E) where bound Flag‐DENND2B is normalized to SM. Mean ± SD, Welch's t‐test *P = 0.012, n = 6 from three independent experiments.
- Schematic of EGFR trafficking. ITSN promotes EGFR internalization upon activation. EGF‐independent activation of EGFR: EGFR traffics on Rab13‐positive recycling or late endosomes (RE/LE) to the plasma membrane where ITSN binds the DENND2B to facilitate EGFR exocytosis. EGF‐dependent activation of EGFR: DENND2B is phosphorylated by PKD, binds 14‐3‐3 and loses its affinity for ITSN and EGFR is targeted towards lysosomal degradation.
Source data are available online for this figure.
Next, we wondered whether EGF treatment enhanced DENND2B phosphorylation. Using the phospho‐PKD substrate antibody that recognizes PKD consensus motifs upon phosphorylation, we found an increase in the phosphorylation of purified Flag‐DENND2B from cells treated with EGF (Fig 5C and D). Therefore, DENND2B is phosphorylated upon EGF treatment. Finally, we tested whether EGF treatment influenced the binding of DENND2B to ITSN‐s. We performed GST pull‐down assays following EGF treatment and discovered diminished binding of Flag‐DENND2B to ITSN‐SH3A following EGF treatment (Fig 5E and F). EGF treatment thus enhances phosphorylation of DENND2B by PKD, reducing its ability to bind ITSN. In addition to EGF, EGFR can be activated by other ligands that influence receptor trafficking fate. Heparin‐binding EGF‐like growth factor and beta‐cellulin target EGFR for degradation while TGF‐alpha, epiregulin and amphiregulin all promote receptor recycling 46. Therefore, future work is needed to determine whether ITSN and DENND2B also play a role in EGFR trafficking downstream of different ligands.
Overall, our study uncovers the importance of DENND2B in EGFR recycling. We propose that ITSN‐s couples EGF‐independent EGFR internalization with its recycling by binding DENND2B. However, upon EGF treatment, DENND2B is phosphorylated by PKD, the interaction of DENND2B with ITSN‐s is abolished, and EGFR is no longer recycled to the surface but instead traffics to the lysosome for degradation (Fig 5G). The exact mechanisms by which this interaction controls EGFR trafficking remain unknown. One possibility is that DENND2B influences ubiquitination of EGFR as this modification is crucial for targeting EGFR to the lysosome for degradation. In fact, EGF‐activated EGFR is recycled back to the plasma membrane in the absence of ubiquitination 47. Previous studies have shown that ITSN activates the ubiquitin ligase Cbl by binding to Spry2 and disrupting the inhibitory Spry2/Cbl interaction 36. Interestingly, Spry2 also binds to ITSN's SH3A domain 36. Therefore, it is possible that in the absence of EGF, DENND2B binding to ITSN outcompetes Spry2 causing Cbl to remain inactive, while EGF treatment releases ITSN to enhance EGFR ubiquitination and subsequent degradation. Future studies are needed to test these and other potential mechanisms by which the interaction between ITSN and DENND2B control EGFR trafficking.
Materials and Methods
Antibodies and DNA constructs
Mouse monoclonal Flag (M2) antibody was from Sigma‐Aldrich. Mouse monoclonal EGFR (ab30) recognizing extracellular domain was from Abcam. Rat monoclonal Hsc70 (1B5) antibody was from Enzo LifeSciences. Rabbit polyclonal GFP (A6455), Alexa Fluor®647‐conjugated Phalloidin, Alexa Fluor®488 and Alexa Fluor®561‐conjugated anti‐mouse were from Invitrogen. Rabbit monoclonal EGFR (D38B1) and rabbit polyclonal phospho‐PKD/PKCμ (Ser916), PKD/PKCμ, phospho‐(Ser/Thr) PKD substrate antibodies and mouse monoclonal p44/42 MAPK (3A7) and phospho‐p44/42 MAPK (Thr202/Tyr204) (E10) antibodies were from Cell Signalling Technology. HRP‐conjugated anti‐mouse and anti‐rabbit were obtained from Jackson ImmunoResearch. Alexa Fluor 488 and Alexa Fluor 561‐conjugated anti‐mouse were purchased from Invitrogen.
Flag‐DENND2B, mCh‐DENND2B, EGFP‐DENND2B and mCh‐Rab13 were previously described 8. GST‐14‐3‐3 WT and GST‐14‐3‐3 K50E were previously described 23. EGFP‐DENND2B S30A was generated using the QuikChange II site‐directed mutagenesis kit (Agilent Technologies) with 5′‐GGACTCTGAGCAGGTCTCAGGCAGTCTCTCCACCTCCAGTTCTC and 3′‐GAGAACTGGAGGTGGAGAGACTGCCTGAGACCTGCTCAGAGTCC primers. GST‐tagged intersectin SH3 domains A‐E, amphiphysin, endophilin A1, pacsin 1 and Grb2 were previously described 5, 48. Flag‐intersectin was previously described 49. EGFR‐GFP was a gift from Alexander Sorkin 50. PKD‐dominant negative (D733A) and PKD‐constitutively active (S744E/S748E) in pcDNA3 were gifts from Teresa Iglesias (Instituto de Investigaciones Biomédicas “Alberto Sols” CSIC‐UAM, Madrid, Spain).
Lentivirus production and DENND2B knockdown
Lentivirus‐mediated knockdown of DENND2B was performed as previously described 8. In brief, shRNAmiR sequences for DENND2B and non‐targeting control were first cloned into pcDNA6.2/GW‐emGFP–miR cassette. The emGFP‐miR cassette was then PCR‐amplified and subcloned into the pRRLsinPPT viral expression vector (Invitrogen). The following sequences for shRNAmiR were used: non‐targeting control AATTCTCCGAACGTGTCACGT, DENND2B shRNA1 TGCTGTGAGGTTTCACTATCACTGAGGTTTTGGCCACTGACTGACCTCAGTGAGTGAAACCTCA and DENND2B shRNA2 TGCTGCTTGGATGAAGCCAGCAAACAGTTTTGGCCACTGACTGACTGTTTGCTCTTCATCCAAG. Viral particles were produced in HEK‐293T cells. The media containing viral particles was collected and filtered with a 0.45‐μm filter to remove cell debris and purified by ultracentrifugation. HEK‐293T and MCF10A cells were plated the day of transduction. Cells were transduced using an MOI of 5, and the media was replaced with fresh culture media after 24 h. All experiments were performed 5–7 days post‐transduction.
To validate protein knockdown: HEK‐293T cells were transduced for 7 days with control shRNA or two shRNAs targeting DENND2B. Cells were transfected with Flag‐DENND2B using jetPRIME® transfection reagent (Polyplus) according to the manufacturer's instructions. 24 h post‐transfection, cells were lysed in HEPES buffer (20 mM HEPES pH 7.4, 100 mM NaCl, 1 mM dithiothreitol) supplemented with protease inhibitors (0.83 mM benzamidine, 0.25 mM phenylmethylsulphonyl fluoride, 0.5 μg/ml aprotinin and 0.5 μg/ml leupeptin), resolved by SDS–PAGE and processed for Western blotting.
To validate mRNA knockdown: MCF10A cells were transduced for 7 days, and total RNA was extracted using an RNeasy extraction kit (Qiagen, Germany). An aliquot of total RNA (800 ng) was used for cDNA synthesis using iScript Reverse Transcription Supermix (Bio‐Rad). Real‐time PCR was performed with SsoFast™ EvaGreen Supermix (Bio‐Rad) on Bio‐Rad CFX Connect Real‐Time PCR Detection System. The reaction was performed in triplicate for each sample. TATA‐binding protein (TBP) and beta‐2‐microglobulin (B2M) were used as endogenous control. The following primers were used: DENND2B‐F GTCTCCTACCAGTTTCCCAAG, DENND2B‐R AAAAGGTCTCACTGCTATACTCTG, TBP‐F GAACCACGGCACTGATTTTC, TBP‐R CCCCACCATGTTCTGAATCT, B2M‐F ACTGAATTCACCCCCACTGA and B2M‐R CCTCCATGATGCTGCTTACA.
Pull‐down assays
Various GST‐tagged proteins were expressed in Escherichia coli BL21 and purified in HEPES buffer supplemented with protease inhibitors and purified on glutathione‐Sepharose beads. HEK‐293T cells were transfected with Flag‐DENND2B constructs. At 18 h post‐transfection, cells were washed with phosphate‐buffered saline, scraped into HEPES buffer, sonicated and Triton X‐100 was added to 1% final concentration. After 15 min of incubation at 4°C, the lysates were centrifuged at 305,000 g for 15 min at 4°C. The supernatants were incubated with 2.5–40 μg of GST fusion proteins pre‐coupled to glutathione‐Sepharose beads for 2 h at 4°C. For competition assays, GST‐14‐3‐3 wild type or K50E was purified on glutathione‐Sepharose beads, cleaved with precision protease overnight at 4°C and concentrated with Amicon 10‐kDa ultracentrifugation filters. Cell lysates containing Flag‐DENND2B were incubated with increasing amounts of purified and cleaved 14‐3‐3 rocking for 1.5 h at 4°C followed by incubation with GST‐SH3A or GST‐Grb2 rocking for an additional 1.5 h at 4°C. Samples were washed three times with HEPES buffer, eluted in SDS–PAGE sample buffer, resolved by SDS–PAGE and processed for Western blotting. For all pull‐down assays, aliquots of starting material (SM) equal to 10% of that added to the beads were analysed in parallel.
Phosphorylation assays
HEK‐293T cells were transfected with Flag‐DENND2B overnight and treated with DMSO or okadaic acid (OA) with or without PKD inhibitor CID755673 (Tocris Biosciences) for 2 h or serum‐starved for 4 h and treated with or without 100 ng/ml EGF (Cedarlane) for 5 min. Cells were washed twice in PBS and collected in phospho‐lysis buffer (20 mM HEPES, 125 mM NaCl, 1 mM dithiothreitol, 10% Triton X pH 7.4, 5 mM sodium pyrophosphate, 500 nM OA, 1 mM Na3VO4, 10 mM NaF), supplemented with protease inhibitors (0.83 mM benzamidine, 0.20 mM phenylmethylsulfonyl fluoride, 0.5 mg/ml aprotinin and 0.5 mg/ml leupeptin). Cell lysates were centrifuged at 21,000 g for 15 min, and supernatant was resolved by SDS–PAGE and processed for Western blotting.
Immunoprecipitation assays
For immunoprecipitation of phosphorylated DENND2B, HEK‐293T cells were transfected with Flag‐DENND2B overnight and treated with either DMSO or OA for 2 h, or serum‐starved for 4 h and treated with or without 100 ng/ml EGF (Cedarlane) for 5 min. For PKD kinase assay, HEK‐293T cells were co‐transfected with Flag‐DENND2B and pcDNA3 or PKD‐CA overnight. Cells were washed twice in PBS and collected in phospho‐lysis buffer. For co‐immunoprecipitation assays, HEK‐293T cells were transfected with indicated constructs overnight, washed twice in PBS and lysed in HEPES buffer. Lysates were centrifuged at 10,000 g for 15 min to remove cell debris, and supernatants were incubated with either Protein A‐Sepharose beads with anti‐GFP or Protein G‐Sepharose beads with anti‐Flag for 2 h at 4°C. Samples were washed three times with the same buffer, eluted in SDS–PAGE sample buffer, resolved by SDS–PAGE and processed for Western blotting. For all immunoprecipitation assays, aliquots of starting material (SM) equal to 10% of that added to the beads were analysed in parallel.
Phospho‐mass spectrometry
HEK‐293T cells were transfected with Flag‐DENND2B overnight and treated with either DMSO or 250 nM OA for 2 h; cells were washed twice in PBS and lysed in phospho‐lysis buffer. Lysates were centrifuged at 10,000 g for 15 min to remove cell debris, and supernatants were incubated with Protein G‐Sepharose beads with anti‐Flag for 2 h at 4°C. Samples were washed three times with the same buffer, eluted in SDS–PAGE sample buffer and resolved by SDS–PAGE, and Flag‐DENND2B bands were excised following staining with Coomassie blue. The gel slices were treated with trypsin, and the phosphorylated peptides were enriched using titanium dioxide and analysed as described 51.
Biotinylation assays
MCF10A cells were transduced with shRNAmiR constructs targeting DENND2B or non‐targeting control. Cells were washed three times in ice‐cold wash buffer (PBS containing 1 mM MgCl2 and 0.1 mM CaCl2) then incubated with 0.5 mg/ml EZ‐link sulfo‐NHS‐LC‐biotin (Pierce, Thermo Scientific) in wash buffer for 30 min on ice. Cells were subsequently washed two times in wash buffer and incubated in wash buffer containing 10 mM glycine for 2 min on ice. Cells were lysed in 20 mM HEPES pH 7.4, 200 mM NaCl, 1% Triton X‐100, 0.5% deoxycholate, 0.1% SDS, 5% glycerol and protease inhibitors and centrifuged at 205,000 g at 4°C for 15 min. Protein lysates (300 μg) were incubated with streptavidin‐coupled agarose beads (Pierce, Thermo Scientific) rocking at 4°C for 1 h, washed three times with lysis buffer, resolved by SDS–PAGE and processed for Western blotting.
Trafficking assays
MCF10A cells were transfected with GFP‐DENND2B constructs using jetPRIME® transfection reagent (Polyplus) according to the manufacturer's instructions. Cells were starved in DMEM/F12 for 2 h, chilled on ice for 2 min, incubated with anti‐EGFR on ice for 2 h and washed with PBS. To monitor endocytosis, cells were incubated at 16°C for 2 h in serum‐free media, and remaining surface‐bound antibody was removed by acid wash (0.5 M NaCl, 0.2 M acetic acid, pH 2.5) followed by a PBS wash. To monitor recycling, cells were incubated at 16°C for 2 h and surface‐bound antibody was removed by acid wash followed by a PBS wash. Cells were then incubated at 37°C for 30 min, and the recycled surface‐bound antibody was removed by acid wash. Cells were fixed, permeabilized in 0.2% Triton X‐100 and processed for immunofluorescence using an Alexa Fluor 561‐conjugated secondary antibody. Imaging was performed using a 710 laser scanning confocal microscope equipped with a plan‐apochromat 63× oil objective (NA = 1.4) (Zeiss). Acquisition was performed using ZEN 11.0 software (Zeiss), and analysis was done using ImageJ 1.43 m. Internal EGFR levels were quantified by measuring both the mean fluorescence and the integrated density to control for changes in cell size (ImageJ). EGFR levels were normalized to non‐transfected control cells in the same field of view.
Cellular imaging
HeLa cells were transfected with DNA constructs using jetPRIME® transfection reagent (Polyplus) according to the manufacturer's instructions. Cells were washed in PBS and fixed for 10 min in 3% PFA at 37°C. Imaging was performed using an 880 laser scanning confocal microscope equipped with a plan‐apochromat 63× oil objective (NA = 1.4) (Zeiss). Acquisition was performed using ZEN 11.0 software (Zeiss), and analysis was done using ImageJ 1.43 m.
Statistics
Statistics were analysed using SPSS version 17 or GraphPad Prism 5. Mean ± standard deviation (SD) was used to determine significant differences between pairs. Comparisons were performed using an unpaired two‐tailed Student's t‐test or Welch's t‐test for two samples. One‐way analysis of variance (ANOVA) and Dunnett's post‐test were used for multiple comparisons.
Author contributions
MSI and PSM conceived the experiments. MSI, GK, MF, JJM, CH, ST, NN, TH and EM performed experiments. MSI and PSM wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Expanded View
Acknowledgements
We also thank Jacynthe Philie for excellent technical assistance and the Proteomics Platform of the McGill University Health Centre Research Institute for the phospho‐mass spectrometry analysis. This work was supported by grant MOP‐62684 from the CIHR to PSM. MSI was supported by a Jeanne Timmins‐Costello Fellowship and an Anne and Richard Sievers Award in Neuroscience. NN was supported by a CIHR Canada Graduate Scholarship, the Max E. Binz Fellowship and a Jeanne Timmins‐Costello studentship. GK was supported by a fellowship from le Fonds de recherche du Québec–Santé and a Jeanne Timmins‐Costello Fellowship. PSM is a James McGill Professor and Fellow of the Royal Society of Canada.
EMBO Reports (2017) 18: 2119–2130
Contributor Information
Maria S Ioannou, Email: maria.ioannou@mail.mcgill.ca.
Peter S McPherson, Email: peter.mcpherson@mcgill.ca.
References
- 1. Herbst JJ, Opresko LK, Walsh BJ, Lauffenburger DA, Wiley HS (1994) Regulation of postendocytic trafficking of the epidermal growth factor receptor through endosomal retention. J Biol Chem 269: 12865–12873 [PubMed] [Google Scholar]
- 2. Baumdick M, Bruggemann Y, Schmick M, Xouri G, Sabet O, Davis L, Chin JW, Bastiaens PI (2015) EGF‐dependent re‐routing of vesicular recycling switches spontaneous phosphorylation suppression to EGFR signaling. Elife 4: e12223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tomas A, Futter CE, Eden ER (2014) EGF receptor trafficking: consequences for signaling and cancer. Trends Cell Biol 24: 26–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peschard P, Park M (2003) Escape from Cbl‐mediated downregulation: a recurrent theme for oncogenic deregulation of receptor tyrosine kinases. Cancer Cell 3: 519–523 [DOI] [PubMed] [Google Scholar]
- 5. Allaire PD, Ritter B, Thomas S, Burman JL, Denisov AY, Legendre‐Guillemin V, Harper SQ, Davidson BL, Gehring K, McPherson PS (2006) Connecdenn, a novel DENN domain‐containing protein of neuronal clathrin‐coated vesicles functioning in synaptic vesicle endocytosis. J Neurosci 26: 13202–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA (2010) Family‐wide characterization of the DENN domain Rab GDP‐GTP exchange factors. J Cell Biol 191: 367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Marat AL, Dokainish H, McPherson PS (2011) DENN domain proteins: regulators of Rab GTPases. J Biol Chem 286: 13791–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ioannou MS, Bell ES, Girard M, Chaineau M, Hamlin JN, Daubaras M, Monast A, Park M, Hodgson L, McPherson PS (2015) DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J Cell Biol 208: 629–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nishikimi A, Ishihara S, Ozawa M, Etoh K, Fukuda M, Kinashi T, Katagiri K (2014) Rab13 acts downstream of the kinase Mst1 to deliver the integrin LFA‐1 to the cell surface for lymphocyte trafficking. Sci Signal 7: ra72 [DOI] [PubMed] [Google Scholar]
- 10. Wu C, Agrawal S, Vasanji A, Drazba J, Sarkaria S, Xie J, Welch CM, Liu M, Anand‐Apte B, Horowitz A (2011) Rab13‐dependent trafficking of RhoA is required for directional migration and angiogenesis. J Biol Chem 286: 23511–23520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Majidi M, Hubbs AE, Lichy JH (1998) Activation of extracellular signal‐regulated kinase 2 by a novel Abl‐binding protein, ST5. J Biol Chem 273: 16608–16614 [DOI] [PubMed] [Google Scholar]
- 12. Yamabhai M, Hoffman NG, Hardison NL, McPherson PS, Castagnoli L, Cesareni G, Kay BK (1998) Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem 273: 31401–31407 [DOI] [PubMed] [Google Scholar]
- 13. Hussain NK, Yamabhai M, Ramjaun AR, Guy AM, Baranes D, O'Bryan JP, Der CJ, Kay BK, McPherson PS (1999) Splice variants of intersectin are components of the endocytic machinery in neurons and nonneuronal cells. J Biol Chem 274: 15671–15677 [DOI] [PubMed] [Google Scholar]
- 14. Martin NP, Mohney RP, Dunn S, Das M, Scappini E, O'Bryan JP (2006) Intersectin regulates epidermal growth factor receptor endocytosis, ubiquitylation, and signaling. Mol Pharmacol 70: 1643–1653 [DOI] [PubMed] [Google Scholar]
- 15. Ma Y, Wang B, Li W, Liu X, Wang J, Ding T, Zhang J, Ying G, Fu L, Gu F (2011) Intersectin 1‐s is involved in migration and invasion of human glioma cells. J Neurosci Res 89: 1079–1090 [DOI] [PubMed] [Google Scholar]
- 16. Gu F, Zhang H, Qin F, Liu X, Li W, Fu L, Ying G, Li B, Zhang M, Ma Y (2015) Intersectin 1‐S, a multidomain adapter protein, is essential for malignant glioma proliferation. Glia 63: 1595–1605 [DOI] [PubMed] [Google Scholar]
- 17. Malacombe M, Ceridono M, Calco V, Chasserot‐Golaz S, McPherson PS, Bader MF, Gasman S (2006) Intersectin‐1L nucleotide exchange factor regulates secretory granule exocytosis by activating Cdc42. EMBO J 25: 3494–3503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Momboisse F, Ory S, Calco V, Malacombe M, Bader MF, Gasman S (2009) Calcium‐regulated exocytosis in neuroendocrine cells: intersectin‐1L stimulates actin polymerization and exocytosis by activating Cdc42. Ann N Y Acad Sci 1152: 209–214 [DOI] [PubMed] [Google Scholar]
- 19. Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, Antonarakis SE, Kay BK, Stossel TP, Lamarche‐Vane N et al (2001) Endocytic protein intersectin‐l regulates actin assembly via Cdc42 and N‐WASP. Nat Cell Biol 3: 927–932 [DOI] [PubMed] [Google Scholar]
- 20. Wigge P, Vallis Y, McMahon HT (1997) Inhibition of receptor‐mediated endocytosis by the amphiphysin SH3 domain. Curr Biol 7: 554–560 [DOI] [PubMed] [Google Scholar]
- 21. Kaneko T, Maeda A, Takefuji M, Aoyama H, Nakayama M, Kawabata S, Kawano Y, Iwamatsu A, Amano M, Kaibuchi K (2005) Rho mediates endocytosis of epidermal growth factor receptor through phosphorylation of endophilin A1 by Rho‐kinase. Genes Cells 10: 973–987 [DOI] [PubMed] [Google Scholar]
- 22. Wasiak S, Quinn CC, Ritter B, de Heuvel E, Baranes D, Plomann M, McPherson PS (2001) The Ras/Rac guanine nucleotide exchange factor mammalian Son‐of‐sevenless interacts with PACSIN 1/syndapin I, a regulator of endocytosis and the actin cytoskeleton. J Biol Chem 276: 26622–26628 [DOI] [PubMed] [Google Scholar]
- 23. Kulasekaran G, Nossova N, Marat AL, Lund I, Cremer C, Ioannou MS, McPherson PS (2015) Phosphorylation‐dependent regulation of connecdenn/DENND1 guanine nucleotide exchange factors. J Biol Chem 290: 17999–18008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu J, Fotouhi M, McPherson PS (2015) Phosphorylation of the exchange factor DENND3 by ULK in response to starvation activates Rab12 and induces autophagy. EMBO Rep 16: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huttlin EL, Jedrychowski MP, Elias JE, Goswami T, Rad R, Beausoleil SA, Villen J, Haas W, Sowa ME, Gygi SP (2010) A tissue‐specific atlas of mouse protein phosphorylation and expression. Cell 143: 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hurd C, Rozengurt E (2001) Protein kinase D is sufficient to suppress EGF‐induced c‐Jun Ser 63 phosphorylation. Biochem Biophys Res Commun 282: 404–408 [DOI] [PubMed] [Google Scholar]
- 27. Doppler H, Storz P, Li J, Comb MJ, Toker A (2005) A phosphorylation state‐specific antibody recognizes Hsp27, a novel substrate of protein kinase D. J Biol Chem 280: 15013–15019 [DOI] [PubMed] [Google Scholar]
- 28. Mendenhall MD, Richardson HE, Reed SI (1988) Dominant negative protein kinase mutations that confer a G1 arrest phenotype. Proc Natl Acad Sci USA 85: 4426–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McClendon CL, Kornev AP, Gilson MK, Taylor SS (2014) Dynamic architecture of a protein kinase. Proc Natl Acad Sci USA 111: E4623–E4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Benzinger A, Muster N, Koch HB, Yates JR III, Hermeking H (2005) Targeted proteomic analysis of 14‐3‐3 sigma, a p53 effector commonly silenced in cancer. Mol Cell Proteomics 4: 785–795 [DOI] [PubMed] [Google Scholar]
- 31. Morrison DK (2009) The 14‐3‐3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol 19: 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hunter MP, Russo A, O'Bryan JP (2013) Emerging roles for intersectin (ITSN) in regulating signaling and disease pathways. Int J Mol Sci 14: 7829–7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Simpson F, Hussain NK, Qualmann B, Kelly RB, Kay BK, McPherson PS, Schmid SL (1999) SH3‐domain‐containing proteins function at distinct steps in clathrin‐coated vesicle formation. Nat Cell Biol 1: 119–124 [DOI] [PubMed] [Google Scholar]
- 34. Thomas S, Ritter B, Verbich D, Sanson C, Bourbonniere L, McKinney RA, McPherson PS (2009) Intersectin regulates dendritic spine development and somatodendritic endocytosis but not synaptic vesicle recycling in hippocampal neurons. J Biol Chem 284: 12410–12419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tong XK, Hussain NK, Adams AG, O'Bryan JP, McPherson PS (2000) Intersectin can regulate the Ras/MAP kinase pathway independent of its role in endocytosis. J Biol Chem 275: 29894–29899 [DOI] [PubMed] [Google Scholar]
- 36. Okur MN, Ooi J, Fong CW, Martinez N, Garcia‐Dominguez C, Rojas JM, Guy G, O'Bryan JP (2012) Intersectin 1 enhances Cbl ubiquitylation of epidermal growth factor receptor through regulation of Sprouty2‐Cbl interaction. Mol Cell Biol 32: 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Di Guglielmo GM, Baass PC, Ou WJ, Posner BI, Bergeron JJ (1994) Compartmentalization of SHC, GRB2 and mSOS, and hyperphosphorylation of Raf‐1 by EGF but not insulin in liver parenchyma. EMBO J 13: 4269–4277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fortian A, Sorkin A (2014) Live‐cell fluorescence imaging reveals high stoichiometry of Grb2 binding to the EGF receptor sustained during endocytosis. J Cell Sci 127: 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tanos B, Pendergast AM (2006) Abl tyrosine kinase regulates endocytosis of the epidermal growth factor receptor. J Biol Chem 281: 32714–32723 [DOI] [PubMed] [Google Scholar]
- 40. Ioannou MS, Girard M, McPherson PS (2016) Rab13 traffics on vesicles independent of prenylation. J Biol Chem 291: 10726–10735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nokes RL, Fields IC, Collins RN, Folsch H (2008) Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J Cell Biol 182: 845–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Abou‐Zeid N, Pandjaitan R, Sengmanivong L, David V, Le PG, Salamero J, Zahraoui A (2011) MICAL‐like1 mediates epidermal growth factor receptor endocytosis. Mol Biol Cell 22: 3431–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zahraoui A (2014) MICAL‐like1 in endosomal signaling. Methods Enzymol 535: 419–437 [DOI] [PubMed] [Google Scholar]
- 44. Morimoto S, Nishimura N, Terai T, Manabe S, Yamamoto Y, Shinahara W, Miyake H, Tashiro S, Shimada M, Sasaki T (2005) Rab13 mediates the continuous endocytic recycling of occludin to the cell surface. J Biol Chem 280: 2220–2228 [DOI] [PubMed] [Google Scholar]
- 45. Matthews SA, Rozengurt E, Cantrell D (1999) Characterization of serine 916 as an in vivo autophosphorylation site for protein kinase D/Protein kinase Cmu. J Biol Chem 274: 26543–26549 [DOI] [PubMed] [Google Scholar]
- 46. Roepstorff K, Grandal MV, Henriksen L, Knudsen SL, Lerdrup M, Grovdal L, Willumsen BM, van DB (2009) Differential effects of EGFR ligands on endocytic sorting of the receptor. Traffic 10: 1115–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Duan L, Miura Y, Dimri M, Majumder B, Dodge IL, Reddi AL, Ghosh A, Fernandes N, Zhou P, Mullane‐Robinson K et al (2003) Cbl‐mediated ubiquitinylation is required for lysosomal sorting of epidermal growth factor receptor but is dispensable for endocytosis. J Biol Chem 278: 28950–28960 [DOI] [PubMed] [Google Scholar]
- 48. Tong XK, Hussain NK, de Heuvel E, Kurakin A, Abi‐Jaoude E, Quinn CC, Olson MF, Marais R, Baranes D, Kay BK et al (2000) The endocytic protein intersectin is a major binding partner for the Ras exchange factor mSos1 in rat brain. EMBO J 19: 1263–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jenna S, Hussain NK, Danek EI, Triki I, Wasiak S, McPherson PS, Lamarche‐Vane N (2002) The activity of the GTPase‐activating protein CdGAP is regulated by the endocytic protein intersectin. J Biol Chem 277: 6366–6373 [DOI] [PubMed] [Google Scholar]
- 50. Carter RE, Sorkin A (1998) Endocytosis of functional epidermal growth factor receptor‐green fluorescent protein chimera. J Biol Chem 273: 35000–35007 [DOI] [PubMed] [Google Scholar]
- 51. Thingholm TE, Jorgensen TJ, Jensen ON, Larsen MR (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat Protoc 1: 1929–1935 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expanded View Figures PDF
Table EV1
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Expanded View


