Abstract
Defining pathways for amyloid assembly could impact therapeutic strategies for as many as 50 disease states. Here we show that amyloid assembly is subject to different forces regulating nucleation and propagation steps and provide evidence that the more global β-sheet/β-sheet facial complementarity is a critical determinant for amyloid nucleation and structural selection.
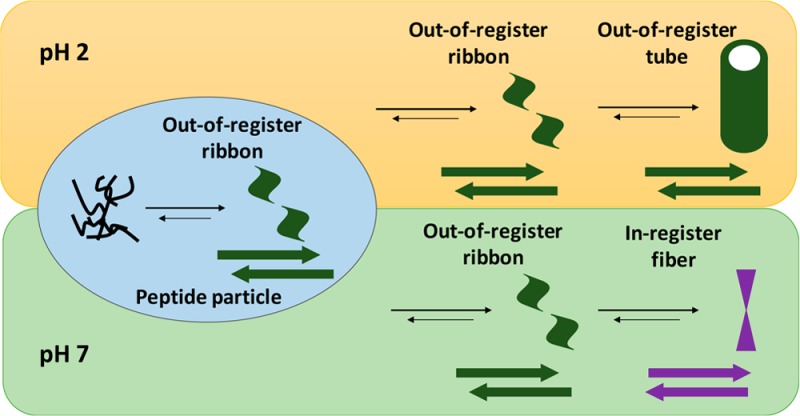
The prion hypothesis posits that protein condensates function as infectious agents underpinning many disease states.1,2 These amyloid assemblies can accumulate as fibers in brain plaques but can also access certain infective conformations that propagate through sustained Darwinian-like evolution3,4 for decades. The mechanism of how these structures arise, diversify, and propagate remains under active investigation.5−8 Ostwald’s rule of stages suggests that a less stable phase may appear first and accelerate the formation of more stable phases.9,10 Evidence obtained with simple peptides suggests that initial liquid–liquid transitions create metastable phases where peptides access β-sheets.11−16 The β-sheets may stack as laminates11,12 to create a stable cross-β nucleus capable of sustained template-directed propagation.13,14,16−20 This two-step nucleation21−23 would place assembly nucleation and propagation in distinct environments and diversify assembly.22−24 By defining the intermediates in a pH-dependent pathway, we now argue that in addition to electrochemical dynamics,13−15 a more global consideration of β-sheet facial complementarity11 contributes dominantly to assembly nucleation. Furthermore, template-directed propagation of the nucleus can lead to altered strand arrangements, which appear as propagation mutations. Together, these pathway dynamics can contribute significantly to the phase diversity accessible in these multistep processes.
The Aβ(16–22) peptide, Ac-KLVFFAE-NH2, is designated as the nucleating core25 of the Aβ peptide of Alzheimer’s disease (AD) and has a final assembled structure26 that is sensitive to environmental pH (Figure 1).27,28 At neutral pH, cross-strand pairing between the positively charged K16 and the deprotonated C-terminal E22 side-chain carboxylate stabilizes in-register β-sheet strand arrangements in fibers (Figures 1a,c and 2d).11,27 At acidic pH, the protonated E22 side chain weakens the K16–E22 salt bridge, and the strands shift out of register and give hollow nanotubes (Figures 1b,d and 2h).11 As highlighted in Figure 1b,d, the cross-strand packing of the bulky valine against the smaller alanine side chain29 stabilizes the antiparallel out-of-register strands. This arrangement places complementary charged side chains and positionally matched hydrophobic surfaces between the β-sheet faces (Figure 1d), achieving greater facial complementarity11 across neighboring β-sheets. This arrangement has previously been suggested to stabilize sheet lamination, allowing access to helical ribbons and ultimately the nanotubes (Figure 2h).11,18,30 The switch from antiparallel out-of-register strands to antiparallel in-register strands requires more than mere slippage, as the peptide must rotate 180° along its long axis, likely requiring dissociation from the β-sheet.
Figure 1.
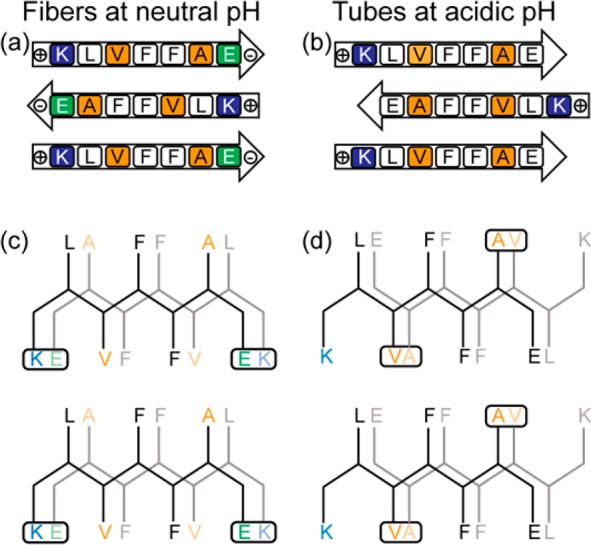
(a, b) Sequence alignment of (a) antiparallel in-register β-sheet and (b) antiparallel out-of-register β-sheet of Aβ(16–22) assemblies. (c, d) Projections down the H-bond axis, with the side chains for the front peptide drawn in black and those for the H-bonded second peptide in gray, highlighting cross-strand side-chain interactions of (c) antiparallel in-register β-sheets and (d) antiparallel out-of-register β-sheets. Positively charged side chains are indicated with blue and negatively charged side chains with green. V and A side chains are highlighted in orange, indicating the preferred packing of the valine side chain with the less bulky alanine side chain.
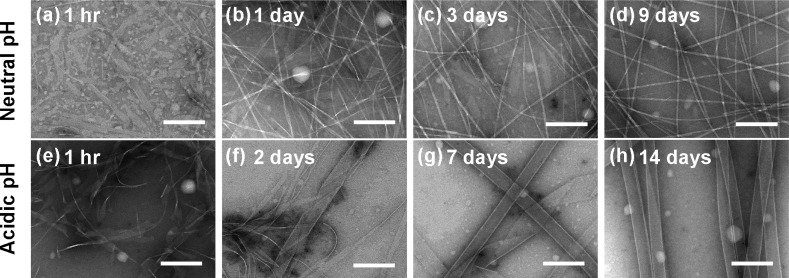
Figure 2.
TEM images of 1.0 mM [1-13C]F19 Aβ(16–22) in 40% acetonitrile in water at neutral pH for (a) 1 h, (b) 1 day, (c) 3 days, and (d) 9 days and at acidic pH for (e) 1 h, (f) 2 days, (g) 7 days, and (h) 14 days. Scale bars = 200 nm.
Despite these different registries, Aβ(16–22) assembly under these two pH conditions proceeds through a common structural intermediate. Transmission electron microscopy (TEM) images show that similar ribbon assemblies initially form under both neutral and acidic conditions (Figure 2a,e). Over time, the ribbons either transform into fibers (Figure 2a–d) or grow into nanotubes (Figure 2e–h), depending on the environmental pH.
Circular dichroism (CD) analyses provide evidence for the growth of β-sheet content in the assemblies. Under neutral conditions (Figure 3, black), the characteristic β-strand negative ellipticity at 217 nm develops almost immediately and plateaus after 5 days. In acidic environments, however (Figure 3, blue), the growing ellipticity is delayed for almost 4 days, then grows at a similar rate, and plateaus after 10 days with the significantly stronger molar ellipticity expected for the nanotubes.11 In both cases, the growth in β-sheet secondary structure is consistent with the morphological transitions observed by TEM.
Figure 3.
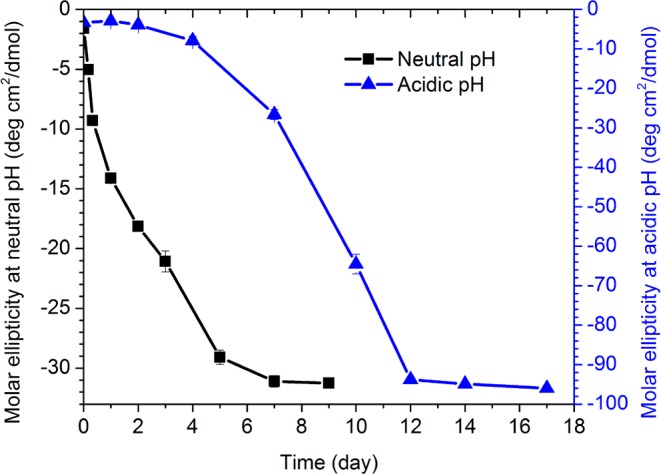
Molar ellipticities at 217 nm measured by CD spectroscopy for 1 mM [1-13C]F19 Aβ(16–22) solution in 40% acetonitrile at (blue, right y axis) acidic pH (pH = 2) and (black, left y axis) neutral pH (pH = 6) over time.
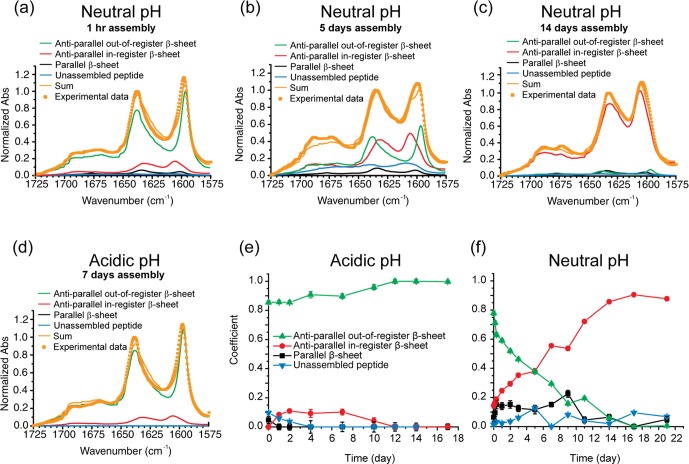
Amide-I stretching transitions provide a direct assessment of strand registry in β-sheets via extended normal mode coupling (Figure S1).31−35Figure 4 contains fits to the isotope-edited infrared (IE-IR) spectra of the enriched [1-13C]F19 Aβ(16–22), Ac-16KLV[1-13C]FFA22E-NH2. With a linear combination of spectra corresponding to mature assemblies with (i) parallel, (ii) in-register antiparallel, and (iii) out-of-register antiparallel β-sheets and (iv) unassembled peptide (Figures 4a–d and S1),15 the fits allow us to argue that the antiparallel out-of-register β-sheets predominate initially under both pH conditions (Figures 4, S2, and S3). While the absolute concentrations cannot be determined without the molar absorptivity for peptide orientations, a slight but significant strengthening in normal mode delocalization occurs from day 4 to day 10 under acidic conditions (Figures 4e and S2), consistent with the transition to nanotube morphology. At neutral pH, the early antiparallel out-of-register sheets transition completely to the in-register strands of fibers over 2 weeks (Figures 4f and S3).18,36,37
Figure 4.
(a–d) Representative fits of the IR spectra for [1-13C]F19 Aβ(16–22) incubated at (a–c) neutral pH for (a) 1 h, (b) 5 days, and (c) 14 days and (d) at acidic pH for 7 days. Linear combinations of spectra corresponding to mature assemblies with parallel (black), in-register antiparallel (red), and out-of-register antiparallel (green) β-sheets and unassembled peptide (blue) are presented as well as the resulting sums (orange lines), which are compared to the experimental spectra (orange squares). (e, f) IR spectral coefficients derived from fits to the IR spectra of assembled [1-13C]F19 Aβ(16–22) under (e) acidic and (f) neutral pH assembly conditions.
The initial ordering of Aβ(16–22) strands at neutral pH into antiparallel out-of-register β-sheets was unexpected. Our operating hypothesis was that Aβ(16–22) nucleation at neutral pH would be dominated by electrostatic forces in the particles14,15 and that the Aβ(16–22) peptides would initially adopt antiparallel in-register β-sheets stabilized by the cross-strand paired K/E residues (Figure 1a,c). In a related study on Aβ(16–22)E22Q, Ac-KLVFFAQ-NH2,14,15 which has no C-terminal charge, also forms antiparallel out-of-register strands initially and then transitions to parallel strands as a result of Q cross-strand pairing. These analyses were consistent with charge repulsion in the particle phase driving antiparallel assembly, but here the electrostatic dynamics do not explain the out-of-register selection, suggesting other contributing factors.
As shown in Figure 1b,d, both the polar and nonpolar side chains in out-of-register antiparallel β-sheets are positioned for stabilizing electrostatic and hydrophobic stacking, suggesting that the limiting constraint on nucleation arises from global β-sheet stacking and facial complementarity in the desolvated particle.11 The hydrophobic effect in the desolvated particle remains to be quantified, as does the number of stacks needed to stabilize a persistently propagating nucleus.18,37
These results provide insight into how the selected nucleus serves as a template for diverse peptide phase propagation.38 Differential solvation during propagation is expected to modulate electrostatic energetics, allowing salt bridge cross-strand pairing to lock the incoming peptide into a conformation distinct from the template. For Aβ(16–22) at neutral pH, a simple shift of registry is insufficient (Figure 1b,d), as the peptide must rotate to access the K/E salt bridge and lock the new strand into the final antiparallel in-register sheet (Figure 1a,c). Subsequent propagation of this new template might drive the transition from ribbons to fibers.14 Such stepwise processes are consistent with previous modeling studies39 and may explain many of the anomalies observed in earlier studies on different model peptides. For example, previous IE-IR experiments13 suggested two assembly mechanistic pathways to give a single final structure and that concentration differences could differentially impact the rates of nucleation and propagation much as in polymer synthesis. These conformational transitions, mediated specifically by environmental changes,20 are not only expected to diversify the range of structures available to model peptide amyloid assemblies but also should be far richer in the complex cellular matrix where so many amyloid diseases are initiated. The nature of the initial nucleation phase may well depend on different cellular protein and membrane surfaces, and understanding the nucleation and propagation mechanisms in cellular environments will become increasingly critical to defining disease etiology. Moreover, these assembly mechanisms in multistep processes have already been used to extend the design and construction of new self-assembling mesoscale materials.40
Acknowledgments
The authors thank the McDonnell Foundation (21st Century Science Initiative Grant on Studying Complex Systems, 220020271), the NSF (Grants CHE-1507932 and NSF/DMR-BSF 1610377), and the NIH Alzheimer’s Disease Research Center (P50AG025688) for financial support. The authors also thank the Robert P. Apkarian Integrated Electron Microscopy Core at Emory University for assistance with TEM measurements.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b09362.
Synthetic and experimental details, including peptide synthesis and purification; TEM, CD, and isotope-edited FTIR results; and the construction of the basis set for deconvolution of IR spectra (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Li J.; Browning S.; Mahal S. P.; Oelschlegel A. M.; Weissmann C. Science 2010, 327, 869. 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M.; Walker L. C. Nature 2015, 525, 193. 10.1038/525193a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C. Trends Biochem. Sci. 2011, 36, 151. 10.1016/j.tibs.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders D. W.; Kaufman S. K.; Holmes B. B.; Diamond M. I. Neuron 2016, 89, 433. 10.1016/j.neuron.2016.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles T. P. J.; Waudby C. A.; Devlin G. L.; Cohen S. I. A.; Aguzzi A.; Vendruscolo M.; Terentjev E. M.; Welland M. E.; Dobson C. M. Science 2009, 326, 1533. 10.1126/science.1178250. [DOI] [PubMed] [Google Scholar]
- Qiang W.; Kelley K.; Tycko R. J. Am. Chem. Soc. 2013, 135, 6860. 10.1021/ja311963f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell A. K. Int. Rev. Cell Mol. Biol. 2017, 329, 187. 10.1016/bs.ircmb.2016.08.014. [DOI] [PubMed] [Google Scholar]
- Aleksis R.; Oleskovs F.; Jaudzems K.; Pahnke J.; Biverstal H. Biochimie 2017, 140, 176. 10.1016/j.biochi.2017.07.011. [DOI] [PubMed] [Google Scholar]
- Levin A.; Mason T. O.; Adler-Abramovich L.; Buell A. K.; Meisl G.; Galvagnion C.; Bram Y.; Stratford S. A.; Dobson C. M.; Knowles T. P. J.; Gazit E. Nat. Commun. 2014, 5, 5219. 10.1038/ncomms6219. [DOI] [PubMed] [Google Scholar]
- Nyvlt J. Cryst. Res. Technol. 1995, 30, 443. 10.1002/crat.2170300402. [DOI] [Google Scholar]
- Mehta A. K.; Lu K.; Childers W. S.; Liang Y.; Dublin S. N.; Dong J.; Snyder J. P.; Pingali S. V.; Thiyagarajan P.; Lynn D. G. J. Am. Chem. Soc. 2008, 130, 9829. 10.1021/ja801511n. [DOI] [PubMed] [Google Scholar]
- Eisenberg D. S.; Sawaya M. R. Annu. Rev. Biochem. 2017, 86, 69. 10.1146/annurev-biochem-061516-045104. [DOI] [PubMed] [Google Scholar]
- Petty S. A.; Decatur S. M. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 14272. 10.1073/pnas.0502804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C.; Ni R.; Smith J. E.; Childers W. S.; Mehta A. K.; Lynn D. G. J. Am. Chem. Soc. 2014, 136, 15146. 10.1021/ja508621b. [DOI] [PubMed] [Google Scholar]
- Smith J. E.; Liang C.; Tseng M.; Li N.; Li S.; Mowles A. K.; Mehta A. K.; Lynn D. G. Isr. J. Chem. 2015, 55, 763. 10.1002/ijch.201500012. [DOI] [Google Scholar]
- Hsieh M.-C.; Lynn D. G.; Grover M. A. J. Phys. Chem. B 2017, 121, 7401. 10.1021/acs.jpcb.7b03085. [DOI] [PubMed] [Google Scholar]
- Rha A. K.; Das D.; Mehta A. K.; Taran O.; Ke Y.; Lynn D. G.. 2017, submitted.
- Childers W. S.; Anthony N. R.; Mehta A. K.; Berland K. M.; Lynn D. G. Langmuir 2012, 28, 6386. 10.1021/la300143j. [DOI] [PubMed] [Google Scholar]
- Anthony N. R.; Mehta A. K.; Lynn D. G.; Berland K. M. Soft Matter 2014, 10, 4162. 10.1039/C4SM00361F. [DOI] [PubMed] [Google Scholar]
- Chen C.; Tan J.; Hsieh M.-C.; Pan T.; Goodwin J. T.; Mehta A. K.; Grover M. A.; Lynn D. G. Nat. Chem. 2017, 9, 799. 10.1038/nchem.2737. [DOI] [PubMed] [Google Scholar]
- Vorontsova M. A.; Maes D.; Vekilov P. G. Faraday Discuss. 2015, 179, 27. 10.1039/C4FD00217B. [DOI] [PubMed] [Google Scholar]
- Peng Y.; Wang F.; Wang Z.; Alsayed A. M.; Zhang Z.; Yodh A. G.; Han Y. Nat. Mater. 2015, 14, 101. 10.1038/nmat4083. [DOI] [PubMed] [Google Scholar]
- Auer S.; Ricchiuto P.; Kashchiev D. J. Mol. Biol. 2012, 422, 723. 10.1016/j.jmb.2012.06.022. [DOI] [PubMed] [Google Scholar]
- Schiener A.; Magerl A.; Krach A.; Seifert S.; Steinruck H. G.; Zagorac J.; Zahn D.; Weihrich R. Nanoscale 2015, 7, 11328. 10.1039/C5NR01602A. [DOI] [PubMed] [Google Scholar]
- Williams A. D.; Shivaprasad S.; Wetzel R. J. Mol. Biol. 2006, 357, 1283. 10.1016/j.jmb.2006.01.041. [DOI] [PubMed] [Google Scholar]
- Meinke J. H.; Hansmann U. H. E. J. Chem. Phys. 2007, 126, 014706. 10.1063/1.2423013. [DOI] [PubMed] [Google Scholar]
- Balbach J. J.; Ishii Y.; Antzutkin O. N.; Leapman R. D.; Rizzo N. W.; Dyda F.; Reed J.; Tycko R. Biochemistry 2000, 39, 13748. 10.1021/bi0011330. [DOI] [PubMed] [Google Scholar]
- Lu K.; Jacob J.; Thiyagarajan P.; Conticello V. P.; Lynn D. G. J. Am. Chem. Soc. 2003, 125, 6391. 10.1021/ja0341642. [DOI] [PubMed] [Google Scholar]
- Liang Y.; Pingali S. V.; Jogalekar A. S.; Snyder J. P.; Thiyagarajan P.; Lynn D. G. Biochemistry 2008, 47, 10018. 10.1021/bi801081c. [DOI] [PubMed] [Google Scholar]
- Guo Q.; Mehta A. K.; Grover M. A.; Chen W.; Lynn D. G.; Chen Z. Appl. Phys. Lett. 2014, 104, 211901. 10.1063/1.4878941. [DOI] [Google Scholar]
- Krimm S.; Bandekar J. Adv. Protein Chem. 1986, 38, 181. 10.1016/S0065-3233(08)60528-8. [DOI] [PubMed] [Google Scholar]
- Halverson K. J.; Sucholeiki I.; Ashburn T. T.; Lansbury P. T. J. Am. Chem. Soc. 1991, 113, 6701. 10.1021/ja00017a068. [DOI] [Google Scholar]
- Moran S. D.; Zanni M. T. J. Phys. Chem. Lett. 2014, 5, 1984. 10.1021/jz500794d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch W. R. W.; Kubelka J.; Keiderling T. A. J. Phys. Chem. B 2013, 117, 10343. 10.1021/jp4056126. [DOI] [PubMed] [Google Scholar]
- Sarroukh R.; Goormaghtigh E.; Ruysschaert J.-M.; Raussens V. Biochim. Biophys. Acta, Biomembr. 2013, 1828, 2328. 10.1016/j.bbamem.2013.04.012. [DOI] [PubMed] [Google Scholar]
- Lin D. D.; Luo Y.; Wu S.; Ma Q. Q.; Wei G. H.; Yang X. J. Langmuir 2014, 30, 3170. 10.1021/la4048165. [DOI] [PubMed] [Google Scholar]
- Liang Y.; Lynn D. G.; Berland K. M. J. Am. Chem. Soc. 2010, 132, 6306. 10.1021/ja910964c. [DOI] [PubMed] [Google Scholar]
- Nguyen P. H.; Li M. S.; Stock G.; Straub J. E.; Thirumalai D. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 111. 10.1073/pnas.0607440104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace J. A.; Shen J. K. Biochemistry 2010, 49, 5290. 10.1021/bi100137y. [DOI] [PubMed] [Google Scholar]
- Rengifo R. F.; Li N. X.; Sementilli A.; Lynn D. G. Org. Biomol. Chem. 2017, 15, 7063. 10.1039/C7OB01170A. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.