Abstract
The recently discovered colistin resistance-encoding element, mcr-1, adds to the list of mobile resistance genes whose products rapidly erode the antimicrobial efficacy of not only the commonly used antibiotics, but also the last line agents of carbapenems and colistin. The relative prevalence of mcr-1-bearing strains in various ecological niches including 1,371 food samples, 480 animal faecal samples, 150 human faecal samples and 34 water samples was surveyed using a novel in-house method. Bacteria bearing mcr-1 were commonly detected in water (71% of samples), animal faeces (51%), food products (36%), and exhibited stable carriage in 28% of human subjects surveyed. Such strains, which exhibited variable antibiotic susceptibility profiles, belonged to various Enterobacteriaceae species, with Escherichia coli being the most dominant in each specimen type. The mcr-1 gene was detectable in the chromosome as well as plasmids of various sizes. Among these, two conjugative plasmids of sizes ca 33 and ca 60 kb were found to be the key vectors that mediated mcr-1 transmission in organisms residing in various ecological niches. The high mcr-1 carriage rate in humans found in this study highlights the importance of continued vigilance, careful antibiotic stewardship, and the development of new antimicrobials.
Keywords: mcr-1, Enterobacteriaceae, isolation method, food, water, animal, human, ecosystem, distribution
Introduction
The effectiveness of antibiotics to combat bacterial infections has diminished rapidly in the past decade due to incessant emergence of bacterial strains that exhibit novel and transmissible resistance mechanisms, such as carbapenem-resistant Enterobacteriaceae (CRE) strains that commonly cause untreatable and hard-to-treat infections among hospitalised patients. CRE are now considered an urgent public health threat according to reports by the European Centre for Disease Prevention and Control (ECDC) and the United States (US) Centers for Disease Control and Prevention (CDC) [1,2].
Colistin is currently considered a last-resort antibiotic that can be used to treat clinical CRE infections. Bacterial resistance to colistin was previously thought to be rare, and mainly attributed to chromosomal mutations leading to modification of lipid A or loss of lipopolysaccharide [3,4]. Recently, a new plasmid-encoded colistin resistance mechanism, mediated by the MCR-1 protein, a phosphoethanolamine transferase that modifies the phosphoethanolamine moiety of lipid A, has been reported [5]. Since its discovery in November 2015, the mcr-1 genehas been associated with a wide range of mobile elements in different bacterial species, suggesting that this resistance element is highly transmissible, thereby posing a huge challenge to the use of colistin as a reserved drug for treatment of CRE infections [6]. Currently there is a lack of studies comprehensively screening for mcr-1 positive bacteria in the environment, and the most frequent sources investigated for such bacteria consist of human clinical samples and veterinary specimens. Hence, information on the prevalence of mcr-1 gene in various ecological niches is not available to assess of the degree of mcr-1 contamination, which can potentially impact the clinical use of colistin. This is due to a lack of methods for specific isolation of mcr-1-positive bacteria, since many species of bacteria are intrinsically resistant to colistin, interfering with the isolation of mcr-1-positive organisms. In this study, we have developed a novel method that facilitates specific detection of mcr-1-positive bacteria. Using this method, we checked various environmental sources in China for the presence of mcr-1 positive bacteria. These included animal and human faecal samples, as well as sewage water, seawater and fresh water samples. Foods locally produced or imported from overseas were also tested.
Methods
Specific isolation of mcr-1-bearing bacteria
PCR screening for mcr-1-bearing organisms in food specimens
A PCR-based method was first developed to screen for the presence of mcr-1-bearing organisms in food products. A total of 25 g of each food sample was mixed with 75 mL alkaline peptone water and stomached for 30 s; 50 mL of the suspension (without sediment) were then incubated in a tube overnight at 37 °C without shaking. After incubation, the tube was inverted several times and left standing for 10 min for settlement of the food particles. To collect the bacteria, 1 mL of the suspension was centrifuged at 13,000 rpm, and the pellet was subsequently washed twice with phosphate buffered saline (PBS). The final pellet was resuspended in 100 µL of saline water and boiled at 100 °C for 5 min to release bacterial DNA. The sample was then subjected to centrifugation at 13,000 rpm for 10 min. The supernatant was then used as DNA template for PCR with the primers targeting mcr-1 as reported previously [5]. The genetic identity of all amplification products was confirmed by nt sequencing.
Isolation of Enterobacteriaceae containing mrc-1 in food
The samples that were positive for the mcr-1 gene were subjected to isolation of mcr-1-positive Enterobacteriaceae strains using the following procedure. Briefly, overnight-incubated food sample suspension as mentioned above was inverted several times to resuspend the bacteria, and then left standing for 10 min to facilitate settlement of large particles. 500 µL enrichment broth at the top of the tube were then removed and added into 5 mL Mossel Enterobacteria enrichment broth (MEE broth, Sigma-Aldrich) supplemented with 0, 2, 4, 8, 16, 32 and 64 µg/mL colistin E, followed by incubation at 37 °C overnight. A standard loop of overnight MEE culture was inoculated onto MacConkey agar plate and incubated at 37 °C overnight. Twenty colonies with different morphologies were selected for further purification and confirmation of the presence of mcr-1 by PCR as previously described [5]. The optimal concentration of colistin used in MEE broth, which could specifically select for mcr-1-bearing organisms, was determined. The mcr-1 negative samples as defined by the PCR assay were also subjected to isolation of mcr-1-bearing bacteria as described above.
Limit of PCR detection and isolation of mcr-1-bearing organisms in food samples
To determine the limit of detection and isolation of mcr-1-bearing bacteria for the above methods, three mcr-1 negative samples were selected for the following assays. 9.9 mL of pork suspension prepared as described above was inoculated with 10°, 101, 102, 103, 104 and 105 CFU of mcr-1-positive Escherichia coli isolates in 100 µL volume. Two E. coli isolates, SZM584, carrying a plasmid-borne mcr-1 gene, and SZM485, carrying the mcr-1 gene in the chromosome, were used. After overnight incubation at 37 °C, PCR template was prepared and subjected to PCR detection as described above. Similarly, pork suspensions inoculated with various amounts of mcr-1-positive E. coli, were subjected to isolation of mcr-1-bearing bacteria as described above, using an optimal concentration of colistin as determined above.
Isolation of mcr-1-bearing organisms from water and sewage
For isolation of mcr-1-positive Enterobacteriaceae strains from water and sewage, samples were collected at each site and filtered by vacuum filtration using a filter membrane (< 0.2 μm). Bacteria on the membrane was then suspended in 10 mL sterile 0.45% saline solution. 200 μL of the suspension were inoculated into 10 mL alkaline peptone water and incubated at 37 °C overnight. Subsequently, 500 µL of the later mixture were added to 5 mL MEE broth containing 2 µg/mL colistin E and incubated at 37 °C overnight. Bacterial isolation was then performed as described above.
Isolation of mcr-1-bearing organisms from faecal samples
For faecal samples, 5 g each were suspended in 15 mL alkaline peptone water and stomached for 30 s; 10 mL of the suspension were incubated overnight at 37 °C without shaking. After incubation, the enrichment broth was mixed and allowed to settle for 10 min. 500 µL of enrichment broth containing a positive sample was added into 5 mL MEE broth containing 2 µg/mL colistin E and incubated at 37 °C. The isolation procedure was the same as described above.
Surveillance of mcr-1-positive Enterobacteriaceae recovered from various niches in the ecosystem
Food samples including meat products (chicken, duck, pork, beef and mutton), seafood (shrimp, fish and shell fish), dairy products (yogurt, milk cheese and butter), fresh produce (lettuce, cabbage, broccoli, cauliflower etc.) and other food products (tofu etc.) were purchased from supermarkets and wet markets located in different districts of Shenzhen, mainland China (each of four different supermarkets and wet markets) and Hong Kong Special Administrative Region (each of three different supermarkets and wet markets). Meat and seafood products of overseas origin including Asian countries, Australia, Brazil, Canada, Denmark, New Zealand, Norway, and the US were collected from various stores of the three biggest chain supermarkets located in three districts of Hong Kong. The different districts chosen for this sampling in both Shenzhen and Hong Kong were all highly and densely populated residential areas. Water and sewage samples were collected from different bay areas, waste water treatment plants and a fresh water reservoir in Shenzhen. Human faecal samples were obtained from in- and outpatients in four different hospitals located in Shenzhen city and adjacent areas. Pet faecal samples were collected from three animal hospitals located in various geographical sites in Shenzhen city. Chicken and pig faecal samples were collected from farms located in the Chinese provinces of Fujian, Guangdong, Henan, Hubei, Jiangshu, Shandong, Shanxi and Zhejiang. All samples were collected during the period of December 2015 to May 2016. All samples were subjected to mcr-1-positive bacteria isolation using methods described above.
Antimicrobial susceptibility tests and detection of beta-lactamase genes
All Enterobacteriaceae isolates were subjected to antimicrobial susceptibility tests according to the standard agar dilution method described by the Clinical and Laboratory Standards Institute (CLSI) [7,8]. Sixteen antimicrobials were tested (as presented in the table shown in the result section). E. coli strain ATCC 25922 was used as a control. Beta-lactamase genes in cephalosporin-resistant E. coli isolates were determined by PCR as previously described [9].
Conjugation, S1-pulsed-field electrophoresis and Southern hybridisation
Conjugation experiments were carried out using the mixed broth method as previously described [10]. Pulsed-field gel electrophoresis (PFGE), S1-PFGE and Southern hybridisation were performed as previously described [11]. The chi-squared test was used to compare the conjugation rate among E. coli strains isolated from different sources.
Complete plasmid sequencing
One each of representative plasmids containing mcr-1, one of ca 33 kb and the other ca 60 kb in size, was recovered from transconjugants for sequencing. Plasmid sequencing was performed as previously described using the Illumina and PacBio RS II platforms [6]. All plasmid sequences were submitted to Rapid Annotation using Subsystem Technology (RAST) tool for annotations and modified manually by Basic Local Alignment Search Tool (BLAST) [12]. BLAST Ring Image Generator (BRIG) software was used to compare plasmids [13]. The nt sequences of the ca 33 kb plasmid (pECJS-B65–33) and the ca 60 kb plasmid (pECJS-61–63) were submitted to GenBank; the accession numbers were KX084392 and KX084393 respectively.
Results
Method development
The PCR assay was able to detect the mcr-1 gene directly from various food samples. For isolation of mcr-1-positive bacteria from mcr-1 positive samples, 10 colonies with pink to red colour (suspicious for E. coli and Klebsiella pneumoniae) were picked from MacConkey plates spread with MEE broth supplemented with 2 µg/mL of colistin; all the 10 colonies were confirmed to be mcr-1 positive. Consistently, 10 colourless to yellow colonies (indicative of non-E. coli strains) collected from these plates were found to be mcr-1 negative. Eight of the 10 red colonies from MacConkey plates spread with MEE broth supplemented with 4 µg/mL of colistin were mcr-1 positive and all 10 yellow colonies were negative for mcr-1. However, MEE broth supplemented with 0, 1, 8, 16 and 32 µg/mL of colistin failed to isolate any mcr-1-bearing bacteria. All red or yellow colonies collected from MacConkey plates spread with these MEE broth were all negative for mcr-1. These data suggested that MEE broth supplemented with 2 µg/mL was optimal for specific isolation of mcr-1-bearing bacteria from food products. Using this method, the mcr-1 gene could be successfully detected by PCR in 9.9 mL of pork suspension inoculated with 103 CFU or more of mcr-1-positive E. coli strains, SZ584 or SZ485, which was equivalent to 104 CFU mcr-1-positive E. coli per 25 g of pork. For isolation of mcr-1-bearing organisms, E. coli strains SZ584 or SZ485 could be successfully isolated from 9.9 mL of pork suspension inoculated with 1×10° CFU or more of SZ584 or SZ485, which was equivalent to 101 CFU mcr-1-positive E. coli per 25 g of pork. Our data suggested that the isolation method was more sensitive than the PCR detection method. Therefore, the isolation method was used in subsequent surveillance experiments.
Prevalence of mcr-1-bearing bacteria in various food and environmental samples
Food samples
A total of 1,371 food samples obtained in Shenzhen and in Hong Kong, including 234 samples from overseas-imported foods, were subjected to screening of mcr-1-bearing organisms; 498 (36%) positive samples were identified.
Among the 620 food samples surveyed in Shenzhen, mcr-1-bearing bacteria isolates were isolated in 150 of 230 (65%) meat samples and 27 of 390 (7%) other food samples ( Table 1 ). Among the 230 meat samples, the mcr-1 isolation rate was 107/142 (75%), 29/43, 2/4, 2/6 and 10/35 for pork, chicken, duck, mutton and beef, respectively. The isolation rate in meat products purchased from supermarket (61/103, 59%) was slightly lower than that of meat purchased from wet market (89/127, 70%). In addition, seafood and vegetable products were also contaminated with mcr-1-bearing bacteria, with a rate of 9/63 (14%) and 18/271 (7%) respectively. All dairy products and other food such as tofu were negative for mcr-1-bearing bacteria.
Table 1. Prevalence of mcr-1-bearing bacteria in food, environmental, animal and human faecal samples, December 2015−May 2016.
| Specimen types | Number of specimens | Number of positive samples | % positive | Bacterial species recoverable (number of isolates) |
|---|---|---|---|---|
| Food | 1,371 | 498 | 36 | Escherichia coli (730), Klebsiella pneumoniae (3), Aeromonas veronii (4) |
| Shenzhen | 620 | 177 | 29 | |
| Meat | 230 | 150 | 65 | |
| Others | 390 | 27 | 7 | |
| Hong Kong | 517 | 215 | 42 | |
| Meat | 376 | 196 | 52 | |
| Others | 141 | 19 | 13 | |
| Overseas | 234 | 106 | 45 | |
| Meat | 222 | 104 | 47 | |
| Other | 12 | 2 | 17 | |
| Animal faeces | 480 | 243 | 51 | E. coli (576), K. pneumoniae (10), Enterobacter cloacae (4) |
| Piga | 245 | 124 | 51 | |
| Chickena | 180 | 113 | 63 | |
| Petb | 55 | 6 | 11 | |
| Humanc | 150 | 42 | 28 | E. coli (84), E. cloacae (5) |
| Inpatients | 85 | 22 | 26 | |
| Healthy individuals | 65 | 20 | 31 | |
| Water | 34 | 24 | 71 | E. coli (77), K. pneumoniae (4), K. variicola (3) |
| Sewage | 24 | 18 | 75 | E. coli (52), K. pneumoniae (4), K. variicola (3) |
| Seawater | 6 | 6 | 100 | E. coli (25) |
| Fresh water | 4 | 0 | 0 | None (0) |
a Chicken and pig faecal samples were collected from farms located in the Provinces of Fujian, Guangdong, Henan, Hubei, Jiangshu, Shandong, Shanxi and Zhejiang. The contamination rate of faecal samples collected from farms located in different provinces of China was 63% (25/40) in Fujian, 14% (8/58) in Guangdong, 55% (30/55) in Henan, 83% (75/90) in Hubei, 56% (28/50) in Jiangshu, 100% (42/42) in Shandong, 70% (28/40) in Shanxi, and 50% (25/50) in Zhejiang.
b Pet faecal samples were collected from three animal hospitals located in various geographical sites in Shenzhen city.
c Human faecal samples were obtained from in- and outpatients in four different hospitals located in Shenzhen city and adjacent areas.
The origin of food products surveyed in Hong Kong were categorised as either ‘Hong Kong’ or ‘overseas’. Among the 517 food samples tested in Hong Kong, which had not been imported from overseas, mcr-1-bearing bacteria isolates were isolated in 196 of 376 (52%) meat samples and 19 of 141 (13%) other food samples ( Table 1 ). The mcr-1-bearing bacteria isolation rate in different types of food samples was similar to that of Shenzhen except that vegetable products exhibited a higher contaminated rate (12/77, 15%) of mcr-1-bearing bacteria than those from Shenzhen. Food of overseas origin mainly included meat products and seafood (a total of 234 samples), in which the mcr-1-bearing bacteria isolation rate was 104 of 222 (47%) meat samples and 2 of 12 seafood samples respectively ( Table 1 ). The mcr-1-bearing bacteria isolation rates varied slightly between different countries in different regions of the world, with rates of 4/12, 37/69 (54%), 28/51 (55%), 27/72 (38%) and 11/18 recorded in food products originating from Asia, US/Canada, Brazil, Australia/New Zealand and Denmark/Norway respectively. Our data also indicated that meat products showed the highest contamination rate of mcr-1-bearing bacteria among all types of food products. Among the mcr-1 positive food samples recovered in both Shenzhen and Hong Kong, a total of 737 bacterial strains containing the mcr-1 element were isolated from the food samples tested, the majority of which (n = 730) being E. coli, followed by Aeromonas veronii (n = 4) and K. pneumoniae (n = 3)( Table 1 ).
Animal faecal samples
The mcr-1-bearing bacteria isolates were detected in 51% (124 of 245) and 63% (113 of 180) of pig and chicken faecal samples, respectively ( Table 1 ). The contamination rate of faecal samples collected from farms located in different provinces of China was 63% (25/40), 14% (8/58), 55% (30/55), 83% (75/90), 56% (28/50), 100% (42/42), 70% (28/40) and 50% (25/50), in Fujian, Guangdong, Henan, Hubei, Jiangshu, Shandong, Shanxi and Zhejiang respectively. In contrast, only six of 55 (11%) pet faecal samples (10 cats and 45 dogs) collected from three pet hospitals in Shenzhen were found to contain mcr-1 ( Table 1 ). A total of 590 strains containing the mcr-1 element were isolated from animal faecal samples, with the majority being E. coli (n = 576), followed by K. pneumoniae (n = 10) and Enterobacter cloacae (n = 4) ( Table 1 ).
Human faecal samples
Among the 150 human faecal samples tested, 85 were collected from inpatients and 65 were collected from healthy individuals who were admitted for physical examination. Twenty-two of the 85 faecal samples (26%) from inpatients and 20 of 65 samples (31%) from healthy individuals were found to contain strains that harboured the mcr-1 gene. The majority of the mcr-1-bearing bacterial strains were confirmed to be E. coli (n = 84) ( Table 1 ).
Water samples
Among the 24 sewage samples tested, six samples collected from the primary sedimentation tanks were negative, but all other 18 samples collected from other stages of water treatment were positive for mcr-1-bearing bacteria including water to be released to the sea after treatment ( Table 1 ). The mcr-1 gene was also detected in all six seawater samples collected from different locations in Shenzhen, but not in the four fresh water samples collected from the Meilin fresh water reservoir, a major source of fresh water in Shenzhen ( Table 1 ). Among the water samples, mcr-1-bearing E. coli was the only bacterial species isolated from seawater; in contrast, bacterial species such as K. pneumoniae and K. variicola were isolated from sewage, although E. coli remained the dominant species ( Table 1 ).
Antimicrobial susceptibility profiles of mcr-1-bearing Enterobacteriaceae strains recovered from various sources
Randomly selected mcr-1-positive bacterial strains collected from various sources, as shown in Table 2 , were subjected to assessment of their susceptibility to 16 antibiotics. Almost all of these strains exhibited a MIC of ≥ 4 µg/mL for colistin. Yet these colistin resistant strains were found to exhibit a diverse range of antibiotic susceptibility profiles, with co-resistance to antibiotics being a common phenomenon.
Table 2. Antimicrobial susceptibility of mcr-1-bearing Enterobacteriaceae strains isolated from different sources, Chinaa, December 2015−May 2016.
| Antibiotics | Resistance rate (%) | |||||
|---|---|---|---|---|---|---|
| Escherichia coli | Non-E. coli | |||||
| Animal (n = 400) | Non-imported food (n = 400) |
Overseas food (n = 100) | Human (84) |
Water (n = 50) |
(n = 34) | |
| AMC | 22 | 3 | 0 | 0 | 0 | 21 |
| AMP | 66 | 77 | 50 | 44 | 76 | 59 |
| CRO | 41 | 22 | 9 | 9 | 7 | 21 |
| CTX | 41 | 22 | 9 | 9 | 7 | 21 |
| MRP | 0 | 0 | 0 | 0 | 0 | 0 |
| CIP | 25 | 33 | 21 | 68 | 24 | 29 |
| NAL | 53 | 61 | 29 | 85 | 86 | 35 |
| CLS | 99 | 99 | 100 | 100 | 100 | 100 |
| AMK | 6 | 5 | 0 | 0 | 0 | 0 |
| STR | 72 | 53 | 59 | 29 | 17 | 29 |
| CHL | 49 | 73 | 38 | 0 | 69 | 41 |
| KAN | 63 | 55 | 21 | 12 | 76 | 41 |
| TET | 94 | 80 | 58 | 56 | 76 | 21 |
| SXT | 81 | 79 | 48 | 19 | 79 | 21 |
| TIG | 0 | 0 | 0 | 0 | 0 | 0 |
| FOS | 13 | 7 | 0 | 8 | 15 | 59 |
AMC: amoxicillin–clavulanic acid; AMK: amikacin; AMP: ampicillin; CHL: chloramphenicol; CIP: ciprofloxacin; CLS: colistin; CRO: ceftriaxone; CTX: ceftazidime; FOS: fosfomycin; KAN: kanamycin; MRP: meropenem; NAL: nalidixic acid; STR: streptomycin; SXT: trimethoprim–sulfamethoxazole; TET: tetracycline; TIG: tigecycline.
a Some strains isolated from foods imported to China from overseas were also tested, as indicated inside the Table.
For example, up to 68% of colistin-resistant E. coli strains isolated from human faecal samples exhibited co-resistance to ciprofloxacin. Interestingly, organisms recovered from different sources also exhibited differential resistance profiles. In particular, E. coli strains recovered from food and animal faecal samples exhibited a higher rate of resistance to cephalosporins than those obtained from other sources ( Table 2 ). A total of 257 cephalosporin-resistant E. coli isolates were obtained, from which 100 isolates were randomly selected and subjected to screening for the presence of different beta-lactamases. Ninety-three of the 100 isolates carried different types of bla CTX-M genes, among which 75 belonged to bla CTX-M-1 group genes and the 18 remaining belonged to bla CTX-M-9 group genes (data not shown). For non-E. coli isolates, they were mainly K. pneumoniae isolates and showed a high resistance rate to cephalosporins and fosfomycin. Our data also showed that resistance to meropenem, amikacin and tigecycline remained extremely rare among mcr-1-positive strains, including those isolated from animals. Nevertheless, it should be noted that two mcr-1 positive E. coli strains that exhibited cross-resistance to meropenem and colistin, as well as most other tested antimicrobial drugs, had been recovered from animal faecal products (data not shown).
Genetic features of mcr-1
To investigate the genetic features of the mcr-1 gene harboured by strains of different bacterial species, randomly selected E. coli strains (maximum one from each sample) isolated from various sources, as shown in Table 3 , were subjected to assessment of their ability to undergo conjugative transfer of the mcr-1 gene to E. coli J53.
Table 3. Sizes and conjugation rate of plasmids harbouring the mcr-1 gene.
| Bacterial species / source of isolation | Number of isolates | Successful conjugation | Conjugation rate (%) | Unsuccessful conjugation (plasmid/chromosome) | ||||
|---|---|---|---|---|---|---|---|---|
| Number | Approximate plasmid size in kb (number of isolates harbouring this plasmid) |
Number | Approximate plasmid size range in kb (Number of isolates harbouring this plasmid) |
Number on chromosome | ||||
| Escherichia coli | Animal faeces | 60 | 31 | 60 (14), 33 (17) | 52 | 29 | 78−480 (19) | 10 |
| Food | 70 | 27 | 60 (14), 33 (13) | 39 | 43 | 78−480 (31) | 12 | |
| Water | 20 | 6 | 60 (3), 33 (3) | 30 | 14 | 120−250 (12) | 2 | |
| Human faeces | 50 | 38 | 60 (20), 33 (18) | 76 | 12 | 78−480 (10) | 2 | |
| Non-E. coli strains | 34 | 21 | 60 (8), 33 (13) | 62 | 38 | 78−250 (13) | 0 | |
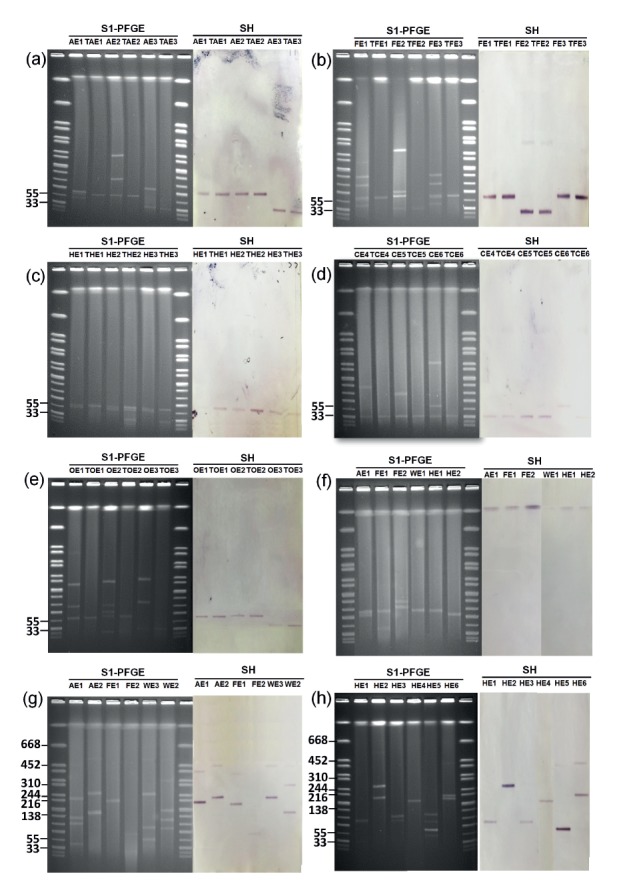
The transfer rate among E. coli strains isolated from different sources was significantly different (p = 0.0002) with E. coli strains obtained from human faeces and animals being the highest ( Table 3 ). S1-PFGE characterisation and Southern hybridisation were then performed on transconjugants as well as on their parental E. coli strains, using mcr-1-specific probes, to determine the range of transmissible and non-transmissible mcr-1-positive elements harboured by the test strains. Two major conjugative plasmids which contained the mcr-1 gene, with a size of ca 33 kb or 60 kb respectively, were detectable in most of the E. coli strains recovered from various sources and their corresponding transconjugants. On the other hand, the size of non-conjugative mcr-1-bearing plasmids in E. coli isolated from various sources varied (ca 78 kb − ca 480 kb). Interestingly, some E. coli strains, mostly in animal products, were found to harbour a chromosomal mcr-1 gene ( Figure 1 , Table 3 ). Apart from E. coli, 21 of 34 non-E. coli strains were found to successfully transfer their colistin resistance phenotypes to the recipient strains through the ca 33 kb or 60 kb plasmids, whereas the sizes of non-conjugative plasmids ranged from ca 78 kb − ca 250 kb ( Figure 1 , Table 3 ).
Figure 1.
S1-PFGE and Southern hybridisation (SH) analysis of mcr-1-bearing conjugative and non-conjugative plasmids harboured by strains of E. coli or other Enterobacteriaceae species isolated from various sources, December 2015−May 2016
PFGE: pulsed-field gel electrophoresis.
(a) Representative E. coli strains recovered from animal faeces (AE), transconjugants which have acquired plasmids recovered from animal E. coli isolates (TAE); (b) Representative E. coli strains recovered from food specimens (FE), transconjugants which have acquired plasmids recovered from food-borne E. coli strains (TFE); (c) Representative E. coli strains recovered from human faeces (HE), transconjugants which have acquired plasmids recovered from human faecal E. coli isolates (THE); (d) Representative E. coli strains recovered from human clinical specimens (CE), transconjugants which have acquired plasmids recovered from human clinical E. coli isolates (TCE); (e) Other Enterobacteriaceae species recovered from various sources (OE), transconjugants which have acquired plasmids recovered from strains of other Enterobacteriaceae species (TOE); (f) E. coli from various sources carrying mcr-1 on the chromosome; E. coli strains recovered from water samples (WE); (g,h) Representative non-conjugative plasmids recovered from E. coli in animal/ food / water and human faecal specimens.
Genetic features of the most common transmissible mcr-1-positive plasmids
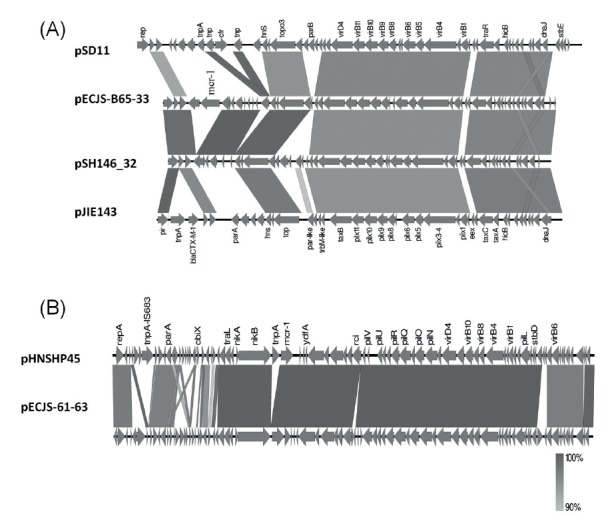
One representative ca 33 kb conjugative plasmid (pECJS-B65–33) was sequenced and shown to belong to the IncX4 type, with a size of 33,298 bp and 41.85% GC content ( Figure 2 ). Several similar plasmids have been reported from various parts of the world. One representative ca 60 kb plasmid (pECJS-61–63) was also sequenced and shown to belong to the IncI2 type, with a size of 63,656 bp and 42.64% GC content. It shares 99% similarity and 88% coverage with the pHNSHP45 plasmid except that ISApI1 is missing in the upstream region of in mcr-1 ( Figure 2 ) [5].
Figure 2.
Comparison of the ca 33 kb and ca 60 kb mcr-1-bearing conjugative plasmids found in this study with other previously reported plasmids
(A) Comparison of pECJS-B65–33 with other IncX4 plasmids with similar backbone structure (plasmid name and the corresponding GenBank accession number): pSD11(KM212169) carrying cfr was isolated from an animal E. coli strain in China; pSH146_32(JX258655), which was known to contain a backbone highly similar to pECJS-B65–33 but without any antimicrobial resistance gene, was isolated from a Salmonella Heidelberg strain in the United States; pJIE143(JN194214) carrying bla CTX-M-15 was recovered from a clinical E. coli ST131 strain isolated in Sydney, Australia. The gene labels follow that of the original annotations in GenBank. Gene alignment was performed by EasyFigure 2.1 [22]. (B) Genetic comparison of pECJS-61–63 with pHNSHP45(KP347127), the first mcr-1-bearing IncI2 plasmid reported in the literature. The upstream region of mcr-1 in pECJS-61–63 was found to lack ISApI1 when compared with pHNSHP45.
Discussion
Just a few months after the discovery of the plasmid-mediated colistin resistance gene, mcr-1, a flood of information regarding this gene was reported in the literature [5,14-17]. However, these studies failed to provide comprehensive understanding on the distribution of mcr-1-like elements in various ecological niches in order to assess the potential impact of dissemination of this novel colistin resistance element on current antimicrobial treatment protocols, especially the use of colistin to treat infections due to CRE.
In this study, we developed a sensitive and specific method for the isolation of mcr-1-bearing bacteria from various sources and used it to investigate the prevalence of mcr-1 in various sample types collected from different settings. It should be noted that the traditional selective isolation method using colistin-containing agar plates is not suited for isolation of mcr-1-positive bacteria due to the prevalence of organisms that are intrinsically resistant to colisin, such as the Proteus, Morganella, Neisseria, Providencia and Serratia spp., some of which are normal flora of animals and human, rendering measurement of the true mcr-1 positive rate among Enterobacteriaceae species challenging. In this work, we tried to focus on isolation of Enterobacteriaceae strains harbouring the mcr-1 gene. To eliminate non-Enterobacteriaceae strains before plating, we added another selective step by diluting peptone water enrichment broth in MEE broth (selective for Enterobacteriaceae) supplemented with 2 µg/mL colistin, which was proven to be optimal in selecting strains harbouring mcr-1. Using this in-house method, we were able to isolate different species of mcr-1-positive bacteria including Enterobacteriaceae and other species exhibiting intrinsic resistance to colistin. During the course of this study, two new variant of the plasmid-mediated colistin resistance gene, namely mcr-2 and mcr-3 were also discovered [18,19]. We screened all isolates that were positive for mcr-1 for the presence of these two variants, but none of these isolates contained any of these two variants (data not shown).
It became difficult to determine the origin of mcr-1 since it has been disseminated to various species of bacteria. However, comprehensive data generated by this study suggested that the mcr-1 gene may originate from E. coli in animal gastrointestinal (GI) tract due to prolonged usage of colistin in livestock. Pet animals which are rarely exposed to colistin exhibited a much lower level of prevalence of mcr-1-positive organisms than pigs and chickens. Additional evidence supporting this hypothesis includes findings that E. coli is the predominant species among mcr-1-bearing Enterobacteriaceae strains and that mcr-1 was detectable in E. coli isolated from animals during the 1980s [20], a date much earlier than that of the first detection of this gene in human (2008) in a retrospective study [14]. Enterobacteriaceae strains carrying mcr-1 in the animal GI tract can cause contamination of their meat products and the environment as evidenced by the highly prevalent mcr-1- positive Enterobacteriaceae strains in food, waste water and seawater, but not in a fresh water reservoir that is not contaminated by faeces. Findings of this work also confirmed that mcr-1 is an extremely common mobile element detectable worldwide, and commonly recoverable from food products originating from different parts of the world, including Australia, the most geographically isolated continent.
One limitation in investigating food samples from overseas in our study was however the uncertainty in the sources of contamination. The food products could have been contaminated in the country of origin or during the re-packaging process after they were imported into Hong Kong. Another limitation of the current study is that it involved non-probability sampling. Hence, while many samples were investigated, and new information on mcr-1-bearing Enterobacteriaceae in various ecological niches was provided, the extent of data representativeness of the areas under study is difficult to derive.
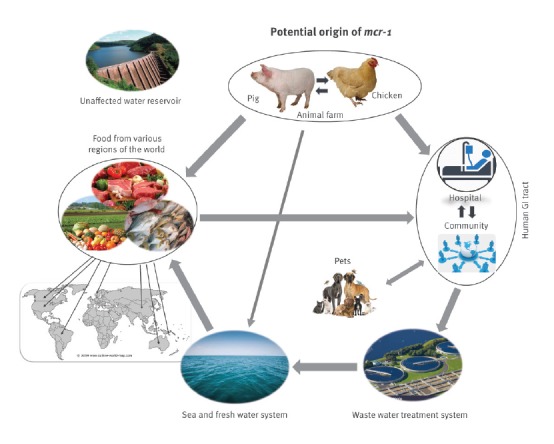
Based on the molecular epidemiology data in this study, we propose a potential mcr-1 transmission route. The mcr-1 gene may have evolved from animal GI tract with the prolonged use of colistin as a growth promoter in livestock. The mcr-1 gene might have then been transmitted to humans through the food chain or direct contact between animals and humans, as well as through contamination of the fresh and seawater system, which in turn lead to contamination of vegetables and seafood. The persistence of mcr-1 in the human GI tract microflora can cause further contamination of our water systems through improper disposal of waste water. A fresh water reservoir that is outside these transmission routes maintained clear of mcr-1 contamination ( Figure 3 ).
Figure 3.
Potential transmission route of mcr-1 in the ecosystem
The mcr-1 gene may undergo evolutionary changes in the animal GI tract upon prolonged usage of colistin as growth promoter. The gene is then transmitted to human through the food chain or direct human contact with animals, as well as through contamination of the fresh and seawater system, which in turn cause contamination of vegetables and seafood. The persistence of mcr-1 in the human GI tract microflora can cause further contamination of the water systems through disposal of waste water containing human faeces. Fresh water systems outside these transmission routes remain clear of mcr-1 contamination.
Genetic characterisation of mcr-1-bearing plasmids revealed that the gene may reside in both chromosome and plasmids, but most commonly on two conjugative elements of ca 33 and 60 kb in size. A much higher proportion of Enterobacteriaceae strains in the human GI tract and clinical specimens was found to carry these two conjugative plasmids when compared with Enterobacteriaceae strains recovered from other sources, suggesting that the prevalence of mcr-1 among human Enterobacteriaceae strains is mainly due to the transmission of mcr-1-bearing conjugative plasmids to the human GI tract microflora. The fact that mcr-1-bearing organisms recoverable from human faecal samples and clinical specimens exhibit highly different antibiotic susceptibility and PFGE profiles (data not shown) from those of other sources also supports the idea that plasmids may play an important role in mcr-1 transmission to humans. Sequence analysis revealed the presence of mcr-1-negative plasmids with a backbone similar to that of the 33 and 60 kb elements in Enterobacteriaceae strains isolated from animals, thus further supporting the theory that these two conjugative plasmids originated from animals [16]. The finding that these two plasmids are highly conjugative (conjugation efficiency at 10 − 1 level) [21] and stably inherited in Enterobacteriaceae strains in the human GI tract without colistin selective pressure suggests that they may severely compromise efforts to control dissemination of the mcr-1 among bacterial pathogens. It is apparently too late to eradicate organisms harbouring mcr-1. Upon approval of clinical use of colistin in China and other regions of the world this year or in the near future, the two colistin resistance-encoding plasmids described in our study may potentially spread in the hospital environment within a short period. The use of colistin to treat CRE infections may result in rapid selection of organisms that exhibit resistance to both carbapenems and colistin. This highlights the importance of vigilance and antimicrobial stewardship. Development of effective inhibitors for MCR-1 or intervention measures to disrupt the transmission of these two plasmids may be an effective strategy to prolong the use of colistin as a last-line antibiotic to treat life-threatening bacterial infections.
Acknowledgements
We thank members of Sheng’s lab for their help in sample collection and bacterial isolation. This work was supported by the Chinese National Key Basic Research and Development (973) Program (2013CB127200) and Collaborative Research Fund from Research Grant Council (C7038-15G and C5026-16G).
Conflict of interest: None declared.
Authors’ contributions: KCC performed the experiments; EWCC designed the study and performed the experiments; MMX performed the experiments; LWY performed the experiments; ND helped with the strain collection and plasmid sequencing; SC designed the study and supervised the whole project.
References
- 1.Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2013.Atlanta: CDC; 2013. Available from: https://www.cdc.gov/drugresistance/threat-report-2013/index.html
- 2.European Centre for Disease Control Prevention and Control (ECDC). Rapid risk assessment: Carbapenem-resitant Enterobacteriaceae – 8 April 2016. Stockholm: ECDC; 2016. Available from: https://ecdc.europa.eu/sites/portal/files/media/en/publications/Publications/carbapenem-resistant-enterobacteriaceae-risk-assessment-april-2016.pdf
- 3. Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, et al. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob Agents Chemother. 2011;55(12):5761-9. 10.1128/AAC.05391-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beceiro A, Moreno A, Fernández N, Vallejo JA, Aranda J, Adler B, et al. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother. 2014;58(1):518-26. 10.1128/AAC.01597-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- 6. Li R, Xie M, Zhang J, Yang Z, Liu L, Liu X, et al. Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J Antimicrob Chemother. 2017;72(2):393-401. 10.1093/jac/dkw411 [DOI] [PubMed] [Google Scholar]
- 7.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-fifth informational supplement. Wayne, PA: CLSI; 2015. CLSI document M100-S25. [Google Scholar]
- 8.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-sixth informational supplement. Wayne, PA: CLSI; 2016. CLSI document M100-S26. [Google Scholar]
- 9. Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490-5. 10.1093/jac/dkp498 [DOI] [PubMed] [Google Scholar]
- 10. Borgia S, Lastovetska O, Richardson D, Eshaghi A, Xiong J, Chung C, et al. Outbreak of carbapenem-resistant enterobacteriaceae containing blaNDM-1, Ontario, Canada. Clin Infect Dis. 2012;55(11):e109-17. 10.1093/cid/cis737 [DOI] [PubMed] [Google Scholar]
- 11. Wang X, Chen G, Wu X, Wang L, Cai J, Chan EW, et al. Increased prevalence of carbapenem resistant Enterobacteriaceae in hospital setting due to cross-species transmission of the bla NDM-1 element and clonal spread of progenitor resistant strains. Front Microbiol. 2015;6:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014;42(Database issue):D206-14. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. 2016;21(9):30155. 10.2807/1560-7917.ES.2016.21.9.30155 [DOI] [PubMed] [Google Scholar]
- 15. Zhi C, Lv L, Yu LF, Doi Y, Liu JH. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(3):292-3. 10.1016/S1473-3099(16)00063-3 [DOI] [PubMed] [Google Scholar]
- 16. Stoesser N, Mathers AJ, Moore CE, Day NP, Crook DW. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis. 2016;16(3):285-6. 10.1016/S1473-3099(16)00010-4 [DOI] [PubMed] [Google Scholar]
- 17. Zhang R, Huang Y, Chan EW, Zhou H, Chen S. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis. 2016;16(3):291-2. 10.1016/S1473-3099(16)00062-1 [DOI] [PubMed] [Google Scholar]
- 18. Xavier BB, Lammens C, Ruhal R, Kumar-Singh S, Butaye P, Goossens H, et al. Identification of a novel plasmid-mediated colistin-resistance gene, mcr-2, in Escherichia coli, Belgium, June 2016. Euro Surveill. 2016;21(27):30280. 10.2807/1560-7917.ES.2016.21.27.30280 [DOI] [PubMed] [Google Scholar]
- 19. Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel Plasmid-Mediated Colistin Resistance Gene mcr-3 in Escherichia coli. MBio. 2017;8(3):e00543-17. 10.1128/mBio.00543-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shen Z, Wang Y, Shen Y, Shen J, Wu C. Early emergence of mcr-1 in Escherichia coli from food-producing animals. Lancet Infect Dis. 2016;16(3):293. 10.1016/S1473-3099(16)00061-X [DOI] [PubMed] [Google Scholar]
- 21. Quesada A, Ugarte-Ruiz M, Iglesias MR, Porrero MC, Martínez R, Florez-Cuadrado D, et al. Detection of plasmid mediated colistin resistance (MCR-1) in Escherichia coli and Salmonella enterica isolated from poultry and swine in Spain. Res Vet Sci. 2016;105:134-5. 10.1016/j.rvsc.2016.02.003 [DOI] [PubMed] [Google Scholar]
- 22. Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27(7):1009-10. 10.1093/bioinformatics/btr039 [DOI] [PMC free article] [PubMed] [Google Scholar]


