Abstract
Background
Alzheimer’s disease (AD) is the most common type of dementia, affecting one in eight adults over 65 years of age. The majority of AD cases are sporadic, with unknown etiology, and only 5% of all patients with AD present the familial monogenic form of the disease. In the present study, our aim was to establish an in vitro cell model based on patient-specific human neurons to study the pathomechanism of sporadic AD.
Methods
We compared neurons derived from induced pluripotent stem cell (iPSC) lines of patients with early-onset familial Alzheimer’s disease (fAD), all caused by mutations in the PSEN1 gene; patients with late-onset sporadic Alzheimer’s disease (sAD); and three control individuals without dementia. The iPSC lines were differentiated toward mature cortical neurons, and AD pathological hallmarks were analyzed by RT-qPCR, enzyme-linked immunosorbent assay, and Western blotting methods.
Results
Neurons from patients with fAD and patients with sAD showed increased phosphorylation of TAU protein at all investigated phosphorylation sites. Relative to the control neurons, neurons derived from patients with fAD and patients with sAD exhibited higher levels of extracellular amyloid-β 1–40 (Aβ1–40) and amyloid-β 1–42 (Aβ1–42). However, significantly increased Aβ1–42/Aβ1–40 ratios, which is one of the pathological markers of fAD, were observed only in samples of patients with fAD. Additionally, we detected increased levels of active glycogen synthase kinase 3 β, a physiological kinase of TAU, in neurons derived from AD iPSCs, as well as significant upregulation of amyloid precursor protein (APP) synthesis and APP carboxy-terminal fragment cleavage. Moreover, elevated sensitivity to oxidative stress, as induced by amyloid oligomers or peroxide, was detected in both fAD- and sAD-derived neurons.
Conclusions
On the basis of the experiments we performed, we can conclude there is no evident difference except secreted Aβ1–40 levels in phenotype between fAD and sAD samples. To our knowledge, this is the first study in which the hyperphosphorylation of TAU protein has been compared in fAD and sAD iPSC-derived neurons. Our findings demonstrate that iPSC technology is suitable to model both fAD and sAD and may provide a platform for developing new treatment strategies for these conditions.
Electronic supplementary material
The online version of this article (doi:10.1186/s13195-017-0317-z) contains supplementary material, which is available to authorized users.
Keywords: Familial Alzheimer’s disease, Sporadic Alzheimer’s disease, Induced pluripotent stem cells, Amyloid β, TAU pathology, Hyperphosphorylation, GSK3B
Background
There are nearly 47 million people living with dementia worldwide, which is predicted to double every 20 years, increasing to more than 131 million by 2050. Alzheimer’s disease (AD) is the best characterized among them, and it accounts for 50–60% of all dementia cases [1]. This common neurodegenerative disease is clinically characterized by a progressive and gradual cognitive impairment, synapse loss, and substantial loss of neurons in later stages. Owing to the disease’s heterogeneity, the etiology of AD is still not very well understood. Most cases of early-onset AD are linked to autosomal dominant inherited mutations in the genes encoding amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2). These cases are referred to as familial Alzheimer’s disease (fAD) and are well characterized. In contrast, the etiology of the remaining 95% cases of late-onset AD, often referred to as sporadic Alzheimer’s disease (sAD), requires further investigation owing to the various factors involved in the pathology, including genetic and environmental exposures [2]. Moreover, cellular changes in the brain precede the first clinical symptoms by 10–15 years, and there is a lack of early diagnostic biomarkers for the prodromal stages of AD. Currently, there is no cure for AD, and the available medications can only slow down the progression of dementia and slightly improve the quality of life of the patients [3].
Two well-defined pathological hallmarks of AD have been described: the formation of extracellular amyloid plaques and the development of intracellular neurofibrillary tangles (NFTs) formed by aggregated hyperphosphorylated TAU protein [4]. The amyloid plaques are composed mainly of Aβ and form aggregates of regularly ordered amyloid fibers [5, 6]. Aβ peptides are derived from the sequential cleavage of APP by the proteolytic enzymes β- and γ-secretase. The cleavage of APP by β-secretase results in production of a large, secreted, derivative soluble amyloid precursor protein β protein and a 99-amino acid membrane-associated fragment (CT99) [7]. The γ-secretase complex cleaves within the transmembrane fragment of CT99 at various positions and generates peptides with differing lengths of the C-terminus (Aβ1–37, Aβ1–38, Aβ1–40, Aβ1–42, Aβ1–43; reviewed in [8]). Among these, Aβ1–40 is the most abundant isoform (~80–90% of the total Aβ peptide) [9], whereas the Aβ1–42 variant is the main isoform of the amyloid deposits found in AD [10, 11]. Aβ1–42 peptides have higher self-aggregating properties than the less hydrophobic, smaller Aβ species, and they have the ability to form oligomers. These diffusible, nonfibrillar, soluble, low-molecular-weight oligomers have neurotoxic effects [9]. It has been shown that soluble Aβ oligomers disrupt axonal transport [12] and cause synapse deterioration, reversal of spines, reduction of long-term potentiation (LTP), and induction of long-term depression (LTD), as well as altered mitochondrial distribution and dynamics in neurons [13]. Inhibition of LTP [14] and reversal of LTD [15] observed in the rat dentate gyrus are associated with impairment of cognitive function. Binding of Aβ oligomers to specific binding sites on neurons (reviewed in [16]) leads to redistribution of synaptic proteins and activation of ionotropic glutamate receptors [17]. Aβ oligomers inhibit LTP by increasing N-methyl-d-aspartate (NMDA) response through glutamate ionotropic receptor NMDA type subunit 2B. Furthermore, these molecules may cause a rapid depolarization of neurons and release of glutamate due to Ca2+ entry, leading to excitotoxicity and synapse loss [17].
Additionally, mutations in PSEN1, PSEN2, and APP cause imbalances between synthesis and degradation of Aβ. For instance, PSEN mutations lead to a partial loss of function in the γ-secretase complex and consequently to incomplete Aβ digestion [18], whereas most of the mutations in APP have been found to increase production of Aβ peptides [19]. The above-described gene aberrations result in many abnormal cellular responses, such as mitochondrial dysfunction, inflammation, activation of microglia, and neuronal loss [20].
The main mechanism in amyloid pathology in fAD is the increased production of Aβ species, whereas decreased Aβ clearance is postulated in sAD, which is modulated by the apolipoprotein E (APOE) genotype. APOE isoforms differentially regulate Aβ aggregation and clearance in the brain and have distinct functions in regulating brain lipid transport, glucose metabolism, neuronal signaling, neuroinflammation, and mitochondrial function (reviewed in [21]). There are at least three alleles of the APOE gene—ε2, ε3, and ε4—coding different isoforms of the protein. The most common allele (~60% in the population) is ε3. Individuals carrying the ε4 allele are at increased risk of AD compared with those carrying the ε3 allele, whereas the ε2 allele is known to decrease the risk.
The second characteristic of AD is TAU-based neurofibrillary pathology. TAU is a microtubule-associated protein (MAPT) required for stabilizing microtubules, the major component of the neuronal cytoskeleton. This protein is involved in neurite outgrowth, maintenance of neuronal polarity, and axonal transport (reviewed in [22]). Phosphorylation of TAU is regulated by kinases and phosphatases, and their actions on TAU molecules are required for proper neuronal growth. Site-specific phosphorylation of TAU plays a crucial role in microtubule stabilization, dynamic behavior, and spatial organization of microtubules in neurons and axonal transport regulation (reviewed in [23]).
In AD, hyperphosphorylated TAU (AD pTAU) spontaneously aggregates into paired helical filaments (PHFs) and form NFTs. Abnormal phosphorylation of TAU impairs microtubule-binding capacity and leads to microtubule destabilization [24]. These disturbances may cause improper anterograde and retrograde axonal transport and perturb intraneuronal signaling, including synaptic transmission (reviewed in [25]).
TAU dysfunction is also related to other forms of dementia, including frontotemporal lobar degeneration, commonly referred as frontotemporal dementia (FTD), which affects people before the age of 65 [26]. Mutations in several different genes can cause FTD; however, the granulin precursor (GRN) and MAPT genes are the most frequent harborers of such mutations. This genetic alteration induces TAU hyperphosphorylation and aberrant NFT formation [27, 28].
Studies on normal human brain tissue have revealed phosphorylation of TAU at several serine and threonine residues, whereas in AD more than 40 different phosphorylation sites have been identified [29]. Most of the sensitive TAU hyperphosphorylation sites are located at the microtubule-binding repeat domain in the proline-rich regions [30]. These regions include serine and threonine residues in serine-proline and threonine-proline motifs, and therefore they are targets of proline-directed protein kinases such as glycogen synthase kinase 3β (GSK3B) [31]. Studies have clearly demonstrated that phosphorylation of TAU by GSK3B and cyclin-dependent kinase 5 reduces the affinity of TAU to bind to microtubules [31], whereas phosphorylation of the serines within the KXGS motifs destructively affects TAU-microtubule interactions [32]. Recent data have shown that Aβ accumulated in the AD brain can activate kinases that promote TAU phosphorylation, including GSK3B [33]. GSK3B phosphorylates serine and threonine residues in TAU PHFs, and its activity corresponds with increased Aβ expression and Aβ-mediated neuronal death. Additionally, active forms of GSK3B were observed in cell lines with presenilin mutations [34].
Current understanding of AD pathogenesis is limited owing to the difficulties in obtaining and culturing human brain tissue and live neurons and inability to model the disease. The recently developed induced pluripotent stem cell (iPSC) technology may provide a new platform to create reliable human disease models for better understanding the pathological mechanisms of AD and for establishment new therapeutic strategies. An increasing number of studies have shown that neurons derived from patient-specific iPSCs can recapitulate the major cellular phenotypes of AD and hence may offer themselves for disease modeling or drug development [35]. In regard to AD, a few research groups have reported elevated levels of Aβ1–42 and increased Aβ1–42/Aβ1–40 ratio in neurons differentiated from iPSCs of patients with fAD [36–39], whereas authors of one report detected an increased secretion of Aβ1–42 in only one patient with sAD [40]. Additionally, AD neurons revealed elevated phosphorylation of TAU at Thr231 as well as an increased level of active GSK3B, and they accumulated large RAB5A-positive early endosomes [40].
We report the generation and neural differentiation of iPSCs derived from several patients with fAD and sAD, as well as from control individuals without dementia. The aim of our present study was to establish an in vitro cellular model that reveals major phenotypes of AD and enables investigation of the pathomechanisms of late-onset AD. Amyloid-β (Aβ) production, TAU phosphorylation, and GSK3B activation were investigated in all lines. Moreover, cellular responses to extracellular hydrogen peroxide and Aβ1–42 oligomers were analyzed in control and AD-derived neural cells. We demonstrate that both fAD and sAD pathology can be recapitulated and that TAU pathology can be investigated in an iPSC-based in vitro human cellular model.
Methods
The chemicals used were purchased from Sigma-Aldrich (St. Louis, MO, USA), and the cell culture reagents and culture plates were purchased from Thermo Fisher Scientific (Waltham, MA, USA), unless specified otherwise.
iPSC lines
iPSC lines derived from patients with AD used in this study were characterized and published earlier [41–46], as detailed in Table 1. The patients were clinically diagnosed and characterized by the Institute of Genomic Medicine and Rare Disorders, Semmelweis University, Budapest, Hungary, or at the Danish Dementia Research Centre, Rigshospitalet, University of Copenhagen, as described previously. Volunteers without dementia (assessed by clinical evaluation) were used as control subjects, from whom iPSC lines were established, characterized, and maintained under identical conditions as the AD iPSC lines. The hiPSC lines were maintained on Matrigel (BD Matrigel; STEMCELL Technologies, Vancouver, BC, Canada) in mTESR1 (STEMCELL Technologies) culture media. The media were changed daily, and the cells were passaged every 5–7 days using Gentle Cell Dissociation Reagent (STEMCELL Technologies) according to the manufacturer’s instructions.
Table 1.
Cell lines used in this study
| iPSC line name | Clone number | Identifier in the study | Disease | Mutation | Sex | Reference |
|---|---|---|---|---|---|---|
| BIOT-7183-PSEN1 | S1 | fAD-1 | Early-onset fAD | PSEN1 c.265G>C, p.V89L | F | [41] |
| S2 | fAD-2 | |||||
| H234 | C5 | fAD-3 | Early-onset fAD | PSEN1 c.449T>C, p.L150P | M | [42] |
| H235 | C6 | fAD-4 | Early-onset fAD | PSEN1 c.449T>C, p.L150P | M | – |
| BIOT-0904-LOAD | S2 | sAD-1 | Late-onset sAD | Unknown | M | [43] |
| S3 | sAD-2 | |||||
| BIOT-0630-LOAD | S4 | sAD-3 | Late-onset sAD | Unknown | F | [44] |
| BIOT-4828-LOAD | S1 | sAD-4 | Late-onset sAD | Unknown | F | [45] |
| S6 | sAD-5 | |||||
| BIOT-0726-LOAD | S3 | sAD-6 | Late-onset sAD | Unknown | F | [46] |
| CTRL1 | S9 | Ctrl-1 | Healthy | – | F | – |
| S11 | Ctrl-2 | |||||
| H250 | C16 | Ctrl-3 | Healthy | – | F | – |
| H256 | C6 | Ctrl-4 | Healthy | – | M | – |
Abbreviations: iPSC Induced pluripotent stem cell, LOAD Late-onset Alzheimer’s disease
Neural induction of iPSCs
Neural progenitor cells (NPCs) were generated from each of the human iPSCs by dual inhibition of SMAD signaling pathway using LDN193189 and SB431542 [47]. Neural induction was initiated upon reaching a desired confluence of iPSCs on Matrigel-coated dishes by addition of neural induction medium (NIM) (1:1 vol/vol mixture of DMEM/F12 and neurobasal medium, 1× N-2 supplement, 1× B-27 supplement, 1× nonessential amino acids [NEAA], 2 mM l-glutamine, 50 U/ml penicillin/streptomycin, 100 μM β-mercaptoethanol, 5 μg/ml insulin), which was supplemented with 5 ng/ml basic fibroblast growth factor (bFGF), 0.2 μM LDN193189 (Selleckchem, Houston, TX, USA), and 10 μM SB431542. The NIM was changed every day. At day 10, neural rosettes were picked manually, replated on poly-l-ornithine/laminin (POL/L) (Sigma-Aldrich)-coated dishes, and expanded in neural maintenance medium (NMM) (1:1 vol/vol mixture of DMEM/F12 and neurobasal medium, 1× N-2 supplement, 1× B-27 supplement, 1× NEAA, 2 mM l-glutamine, 50 U/ml penicillin/streptomycin), and supplemented with 10 ng/ml epidermal growth factor and 10 ng/ml bFGF.
Neural differentiation of NPCs
To generate human neurons, NPCs were plated on the POL/L-coated dishes and cultured in neural differentiation medium (1:1 vol/vol mixture of DMEM/F12 and neurobasal-A medium, 1× N-2 supplement, 1× B-27 supplement, 1× NEAA, 2 mM l-glutamine, 50 U/ml penicillin/streptomycin) supplemented with 0.2 mM ascorbic acid and 25 μM β-mercaptoethanol. For terminal differentiation (TD) into cortical neurons, the cells were plated on POL/L (0.002%/2 μg/cm2) at a seeding density of 40,000 cells/cm2 for immunocytochemistry (ICC) and 100,000 cells/cm2 for enzyme-linked immunosorbent assay (ELISA) and Western blotting experiments with NMM. The medium was changed every 3–4 days during the course of TD. The efficiency of TD was monitored by ICC staining and RT-qPCR for tubulin-β 3 class III (TUBB3) and microtubule-associated protein 2 (MAP2) expression at week 10. In the present study, NPCs from passage 9 to passage 10 were differentiated up to 10 weeks for ELISA and Western blotting experiments, whereas samples were collected at weekly intervals.
Immunocytochemistry
Cells were fixed in 4% paraformaldehyde (PFA) for 20 minutes at room temperature (RT), washed twice with PBS, and permeabilized with 0.2% Triton X-100 in PBS for 20 minutes. Then, cells were blocked with 3% bovine serum albumin (BSA) in the presence of 0.2% Triton X-100 in PBS for 60 minutes at RT. The respective primary antibodies were applied overnight at 4 °C (Additional file 1: Table S1). To detect the signal, cells were incubated for 60 minutes at RT with the appropriate secondary antibodies (Alexa Fluor 488 donkey anti-rabbit immunoglobulin G [IgG] [H + L], Alexa Fluor 594 donkey anti-mouse IgG [H + L], Alexa Fluor 488 donkey anti-mouse IgG [H + L], or Alexa Fluor 594 donkey anti-rabbit IgG [H + L]; Life Technologies, Carlsbad, CA, USA). Cell nuclei were visualized using VECTASHIELD Mounting Medium with 4′,6-diamidino-2-phenylindole (1.5 μg/ml; Vector Laboratories, Burlingame, CA, USA). Cells were analyzed under a fluorescence microscope equipped with a 3D imaging module (Axio Imager system with ApoTome; Carl Zeiss MicroImaging GmbH, Göttingen, Germany) controlled using AxioVision 4.8.1 software (Carl Zeiss MicroImaging GmbH).
Electron microscopy
To evaluate the neuronal cultures, a monolayer of 5-week-old neurons grown on POL/L-treated glass coverslips was fixed with a fixative solution containing 3.2% PFA, 0.2% glutaraldehyde, 1% sucrose, and 40 mM CaCl2 in 0.1 M cacodylate buffer for 24 h at 4 °C. Samples were rinsed for 2 days in cacodylate buffer, then postfixed in 1% ferrocyanide-reduced osmium tetroxide [48] for 1 h at RT. The samples were then treated with aqueous 1% uranyl-acetate for 30 minutes and embedded in Spurr low-viscosity epoxy resin medium (Sigma-Aldrich) according to the manufacturer’s instructions and cured for 24 h at 80 °C. Ultrathin sections were stained with Reynolds lead citrate for 2 minutes and examined under a JEOL JEM 1011 transmission electron microscope (JEOL, Peabody, MA, USA) operating at 60 kV. Photographs were taken using an Olympus Morada 11-megapixel camera and iTEM software (Olympus, Center Valley, PA, USA).
Flow cytometry
NPCs growing on POL/L-coated dishes were dissociated into single cells with Accutase (Sigma-Aldrich) and fixed with 4% PFA for 20 minutes at RT. Cells were permeabilized with 0.2% Triton X-100 in PBS for 5 minutes at RT and blocked with 10% FBS in PBS for 20 minutes at RT. Cells were stained for 1 h at RT with Alexa Fluor 647 mouse anti-Nestin and phycoerythrin mouse anti-human paired box 6 (PAX6) antibodies (BD Pharmingen, San Jose, CA, USA). Flow cytometric analysis was performed using a Cytomics FC 500 flow cytometer (Beckman Coulter, Brea, CA, USA). To detect Nestin (NES) and PAX6 expression in NPCs, an argon laser (488 nm) and a red solid laser (635 nm), respectively, were used. Flow cytometric data analysis was performed using FlowJo software (version 7.6.5; FlowJo, LLC, Ashland, OR, USA).
Immunoblotting
The cell cultures were lysed with RIPA Lysis and Extraction Buffer supplemented with Halt™ Protease and Phosphatase Inhibitor Cocktail and Pierce™ Universal Nuclease for Cell Lysis (all from Thermo Fisher Scientific). The protein extracts were derived from one well of a six-well plate of a single neuronal differentiation. Lysed samples were sonicated and incubated for 60 minutes on ice. Total protein concentration was determined using a Pierce BCA Protein Assay Kit. Cell lysates (2 μg) were separated on 10% sodium dodecyl sulfate-polyacrylamide gel and transferred to Immun-Blot® PVDF Membrane (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were blocked with Tris-buffered saline with Tween 20 (TBST; 20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween 20) containing 5% BSA and then incubated with the respective primary antibody solution overnight at 4 °C (Additional file 1: Table S1). After being washed with TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at RT. All the secondary antibodies were either goat anti-mouse or goat anti-rabbit (Sigma-Aldrich). All samples were analyzed at days 14, 28, 42, 56, and 70 of TD, and the amount of pTAU and pSer9GSK3B relative to total TAU and GSK3B in lysates was measured. Signals were detected with SuperSignal™ West Dura Extended Duration Substrate by KODAK Gel Logic 1500 Imaging System (Bruker, Billerica, MA, USA) and Kodak MI SE imaging software (Carestream, Atlanta, GA, USA). Densitometric measurement of protein band intensity was carried out using Image Studio™ Lite software (LI-COR Biosciences, Lincoln, NE, USA). Phosphorylation of TAU and GSK3B protein was calculated from densitometric data of phosphoproteins divided by densitometric data of nonphosphorylated proteins.
Measurement of Aβ1–40 and Aβ1–42 by ELISA
Conditioned medium was collected after 4 days of culture (without media change) at every week from one well of a six-well plate. To prevent protein degradation, 4-(2-aminomethyl)benzenesulfonyl fluoride hydrochloride was added to the medium. Extracellular Aβ1–40 and Aβ1–42 levels were measured using the Human β-Amyloid (1-40) ELISA Kit and Human β-Amyloid (1-42) ELISA Kit (Wako Chemicals, Neuss, Germany) according to the manufacturer’s instructions. The secreted Aβ levels determined (in picomolar concentrations) were normalized to total protein content of cell lysate. The signal was detected using a Varioskan Flash multimode reader (Thermo Fisher Scientific).
As a control value we used the average value (±SEM) of the four clones derived from healthy individuals (ctrl-1, ctrl-2, ctrl-3, ctrl-4) in all experiments. The individual expression levels are presented in Additional file 2: Figure S1.
RT-qPCR
Total RNA was isolated from differentiated neurons at different time points using the RNeasy Plus Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol. One microgram of RNA was transcribed using the SuperScript™ III VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific). The PCR conditions were subjected to 94 °C for 3 minutes as an initial denaturation step, followed by 40 cycles of 95 °C for 5 seconds for denaturation, 60 °C for 15 seconds for annealing, and 72 °C for 30 seconds for elongation. The amplification reactions were carried out in a total volume of 15 μl using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). RT-qPCR was run on the Rotor-Gene Q 5plex Platform (QIAGEN) using oligonucleotide primers detailed in Additional file 3: Table S2. Human glyceraldehyde 3-phosphate dehydrogenase GAPDH) was used as a reference gene. The data were analyzed using REST software (2009; version 2.0.13).
Library preparation and next-generation sequencing
Genomic DNA (gDNA) was isolated from peripheral blood mononuclear cells using the GenElute Mammalian Genomic DNA Miniprep Kit (Sigma-Aldrich). An ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used to determine the concentration and purity of the isolated gDNA. The gDNA integrity was evaluated by 0.8% agarose gel electrophoresis.
The multiplexed library preparation was performed by using the SureSelectQXT and SureSelectXT Human All Exon V6 Target Enrichment systems according to the manufacturer’s instructions (Agilent Technologies, Santa Clara, CA, USA). The probe set targeted > 99% of RefSeq (~20,000) genes with a total target size of 60 Mb. Pooled libraries were sequenced on the NextSeq 500 platform (Illumina, San Diego, CA, USA) in a 2 × 80-bp paired-end run. The average throughput was > 80 million reads (4 Gbp per sample). Primary data analysis, filtering, and adapter trimming were performed using NextSeq Control Software (version 1.4.1; Illumina). Alignment was done using the Burrows-Wheeler Aligner (Illumina) [49] to the GRCh37/hg19 reference genome, and variant calling was performed with the Genome Analysis Toolkit (GATK) [50]. Variants were annotated using the wANNOVAR web server [51, 52], and their phenotypic effects were evaluated using the American College of Medical Genetics and Genomics standards and guidelines [53].
Cell viability assay after H2O2 and Aβ treatment
Neurons growing on a 96-well plate at days 28 and 56 of TD were treated with 30 μM and 60 μM of hydrogen peroxide (H2O2) (Sigma-Aldrich) or 5 μM Aβ1–42 oligomer solution. For preparation of oligomer solution, iso-Aβ1–42 peptides were used as detailed elsewhere [54] (provided by University of Szeged, Hungary). Viability of the cultures following H2O2 and Aβ treatment was assessed at 24 h of stimulation using a CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI, USA). The luminescent signal was detected using the Varioskan Flash Multimode Reader. Neuronal survival was represented as a percentage of control.
Statistical analysis
All results were analyzed using Prism 5 (GraphPad Software, La Jolla, CA, USA) and Microsoft Office 2010 (Microsoft, Redmond, WA, USA) software. All data on the graphs represent the average of the triplicate measurements (n = 3). The “n” value corresponds to the number of replicates for each cell line. Analysis of data is presented as the mean ± SEM. Dunnett’s method was used to compare the individual groups with control subjects. In all cases, significance was noted at p < 0.05.
Results
Neuronal differentiation capacity of iPSCs derived from patients with fAD and patients with sAD is similar
Samples from patients with fAD and patients with sAD were reprogrammed, and iPSC lines were generated and characterized by our teams as previously reported [41–46]. In the present study, we studied iPSC clones derived from three patients with fAD with mutation in PSEN1 (contributing with in total four lines: fAD-1, fAD-2, fAD-3, fAD-4), four patients with late-onset sAD (contributing with in total six lines: sAD-1, sAD-2, sAD-3, sAD-4, sAD-5, sAD-6), and three individuals without dementia as control subjects (contributing with in total four lines: ctrl-1, ctrl-2, ctrl-3, ctrl-4) (see also Table 1). In all sAD samples, next generation sequencing analysis revealed a lack of pathogenic mutations in APP, PSEN1, and PSEN2, indicating late-onset AD. Moreover, sequencing of MAPT and GRN excluded FTD as well. The APOE status of the cell lines included in the study is shown in Additional file 4: Table S3.
All AD iPSCs and control iPSCs were successfully converted into neuronal progenitor cells (NPCs) and expressed neuroepithelial markers PAX6 and NES (Fig. 1a). To quantify the efficiency of NPC formation, flow cytometry was applied, which showed that there were no significant differences between any of the cell lines with respect to generating NPCs including > 70% copositivity for PAX6 and NES (Fig. 1b).
Fig. 1.
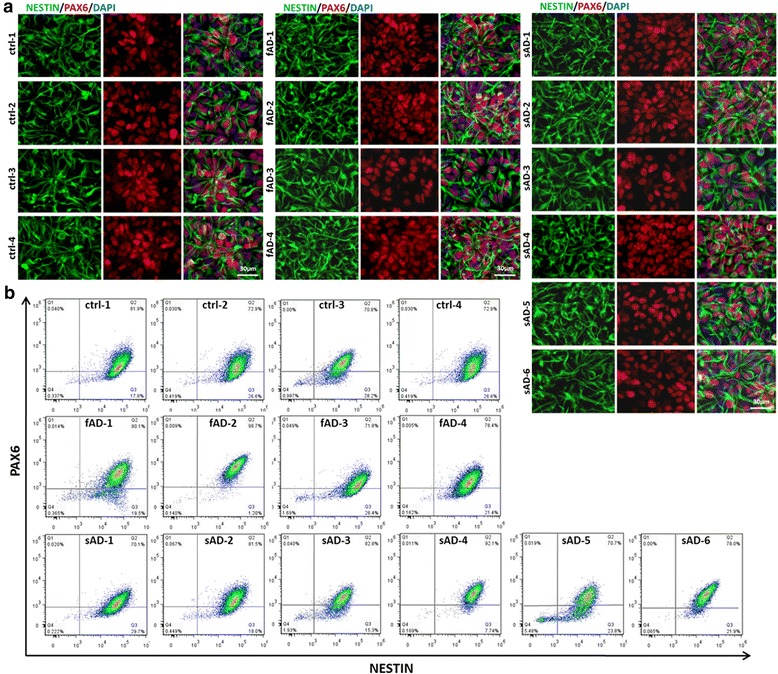
Immunofluorescence characterization of the induced pluripotent stem cells (iPSCs) derived neural progenitor cells. a Representative images of neural progenitor cells (NPCs) from healthy control individuals (ctrl-1–ctrl-4), patients with early-onset familial Alzheimer’s disease (fAD-1–fAD-4), and patients with late-onset sporadic Alzheimer’s disease (sAD-1–sAD-6) stained with Nestin (green), paired box 6 (PAX6; red), and 4′,6-diamidino-2-phenylindole (DAPI; blue). All three groups exhibited similar progenitor marker expression. scale bar = 30 μm. b Flow cytometric analysis of cultured NPCs. Dot plots demonstrate similar expression pattern of PAX6 and Nestin in control, fAD, and sAD iPSC line-derived NPC cultures with similar passage numbers (p6)
Next, the neuronal differentiation of fAD and sAD NPCs was investigated and compared with that of control lines at different time points (TD14, TD28, TD42, TD56, TD70). ICC labeling showed that the increasing expression of neuronal markers TUBB3 and MAP2 during neuronal differentiation was comparable among AD and control cell lines (Fig. 2a). More mature markers—TAU and neurofilament, heavy polypeptide 200 kDa (NF200)—appeared later during differentiation than MAP2 and TUBB3 (Fig. 2a). ICC analysis at day 70 of AD and control neural cultures revealed the presence of proteins related to distinct subtypes of mature neurons: markers of cholinergic neurons (vesicular acetylcholine transporter [VACHT]), dopaminergic neurons (tyrosine hydroxylase [TH]), GABAergic (glutamic acid decarboxylases 2 and 1 [GAD2/GAD1]), and glutamatergic neurons (vesicular glutamate transporter 1/2 [VGLUT1/2]) (Fig. 2b). In order to compare gene expression levels, RT-qPCR was performed on day 70 neurons. The analysis revealed that the AD and control lines expressed comparable levels of MAP2, sodium-dependent serotonin transporter (SLC6A4), NMDA receptor subunit NR1 (glutamate ionotropic receptor N-methyl-d-aspartate type subunit 1 [GRIN1]), marker of maturing neurons – RNA binding protein, fox-1 homolog 3 (RBFOX3), and GAD1 (Fig. 2c). Moreover, they also showed high expression levels of dopaminergic neuron marker (TH), astrocyte marker (glial fibrillary acidic protein [GFAP]), and an early oligodendrocyte marker (claudin 11 [CLDN11]) (Fig. 2c). Furthermore, the cells expressed TUBB3, choline O-acetyltransferase (CHAT), and postsynaptic density protein 95 (discs large MAGUK scaffold protein 4 [DLG4]). Importantly, no evident differences in gene expression levels were observed between control, fAD, and sAD lines. Additionally, electron microscopy of neural cell cultures at day 35 revealed the presence of synapses with docking of synaptic vesicles and synaptic clefts (Fig. 2d). These findings indicate that AD has no significant effect on the overall neural differentiation of iPSCs in the used system, as well as that NPCs can be differentiated into various neuronal and glial cell types with similar efficiency independent of their origin from patients with AD or healthy individuals.
Fig. 2.
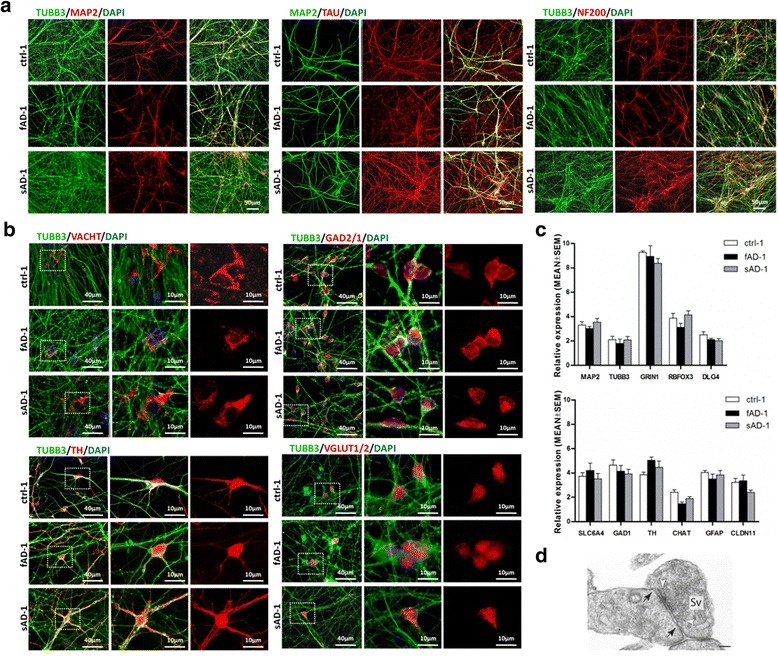
Neuronal differentiation from control and Alzheimer’s disease induced pluripotent stem cells (AD-iPSCs). a Representative immunofluorescence images show expression of neuronal markers at day 70 of terminal differentiation (TD70): tubulin β 3 class III (TUBB3; green, left panel), microtubule-associated protein 2 (MAP2; red, left panel), TAU (red, middle panel), and neurofilament, heavy polypeptide 200 kDa (NF200; red, right panel). Scale bar = 50 μm. b Differentiation of iPSCs into various neuronal subtypes at TD70 was confirmed by the presence of specific markers: vesicular acetylcholine transporter (VACHT) (cholinergic neurons), glutamic acid decarboxylases 2 and 1 (GAD2/1) (GABAergic neurons), TH (dopaminergic neurons), and vesicular glutamate transporter 1/2 (VGLUT1/2) (glutamatergic neurons). Scale bars = 40 μm and 10 μm as indicated. c Gene expression of neuronal markers from ctr-1, early-onset familial Alzheimer’s disease (fAD-1), and sporadic Alzheimer’s disease (sAD-1) lines at TD70 obtained by qPCR. The expression values were normalized to GAPDH and calculated as a relative amount of messenger RNA versus expression value of neural progenitor cells, which was set to 1. Data are reported as mean ± SEM of three independent measurements. AD-iPSC neurons demonstrate expression pattern similar to that of neuronal cells derived from control iPSCs. d Ultrastructure of a synapse at TD35 with synaptic vesicles (Sv) and synaptic cleft with synaptic junctions (tight junctions, black arrowheads). Docking synaptic vesicles are also observable (white arrowheads). Scale bar = 100 nm. DAPI 4′,6-Diamidino-2-phenylindole
Elevated level of Aβ was secreted by fAD and sAD neurons
To determine Aβ production from iPSC lines of patients with fAD and patients with sAD, secreted Aβ1–40 and Aβ1–42 levels were measured weekly by ELISA in the conditioned media of differentiated neural cell lines from the second to tenth weeks of differentiation. Elevated secreted Aβ1–40 and highly increased Aβ1–42 levels were observed in all fAD neural cultures compared with the mean level in the control lines at almost all time points (Fig. 3). The Aβ1–42/Aβ1–40 ratio was also increased up to twofold for the fAD-iPSC-derived neurons. The extracellular level of Aβ1–40 in all sAD neurons was significantly higher than in control and fAD neurons. The Aβ1–42 secretion in sAD neurons was also elevated compared with the average values of the control neurons; however, the levels did not reach the same levels as in fAD neurons. Interestingly, the measured Aβ1–42/Aβ1–40 ratio in all sAD-iPSC-derived neurons was similar to that in control neurons (Fig. 3c). In nonmature neurons up to day 14 (TD14), Aβ production was comparable among AD and control lines (Additional file 2: Figure S1). During neural differentiation, Aβ1–40 and Aβ1–42 levels gradually increased to reach the investigated maximum at day 70 of TD (endpoint of experiment; TD70), suggesting a maturation-dependent secretion of Aβ (Fig. 3, Additional file 2: Figure S1). The results at TD14 and TD28 indicated a transition period in which not all AD samples showed significant differences from the control samples, but from TD42, all AD samples displayed elevated amounts of Aβ. Therefore, we concentrated our further experimental analyses on the samples matured for a minimum of 6 weeks.
Fig. 3.

Characterization of amyloid-β (Aβ) secretion in control and Alzheimer’s disease induced pluripotent stem cell (AD iPSC)-derived neurons at terminal differentiation day 42 (TD42), TD56, and TD70. a The amount of secreted Aβ1–40 from control-, early-onset familial Alzheimer’s disease (fAD)- (fAD-1–fAD-4), and sporadic Alzheimer’s disease (sAD)- (sAD-1–sAD-6)-iPSC-derived neurons. b The amount of Aβ1–42 released from control- and AD-iPSC-derived neurons. c The ratio of Aβ1–42/Aβ1–40 from neurons derived from control and AD lines. Aβ1–40 and Aβ1–42 secreted from neural cells into the medium were measured at day 4 after the last medium change. The extracellular Aβ levels determined (in picomolar concentrations) were normalized to total protein content of cell lysates. Data represent mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control at the same time point of differentiation (*p < 0.05)
To evaluate the level of APP, the direct precursor of Aβ in neurons, we performed Western blot analysis. The results revealed that APP expression increased during neuronal differentiation and reached a maximum at day 70 (TD70) in all fAD and sAD neural cultures (Fig. 4, Additional file 5: Figure S2a). Expression of this protein was significantly increased in all AD cell lines from TD42. Subsequently, we quantified the amount of amyloid precursor protein carboxy-terminal fragment (APP-CTF) generated through APP cleavage by β-secretase. As shown in Fig. 4 and Additional file 5: Figure S2b, remarkably higher APP-CTF protein levels were detected in fAD and sAD neurons. No significant differences in the expression of APP and APP-CTF were observed between fAD and sAD neurons. In summary, both Aβ and APP levels were significantly elevated in fAD and sAD neural cultures compared with control cultures. These differences were detectable after 6 weeks (TD42) but were prominent with all sAD lines at week 10 (TD70).
Fig. 4.
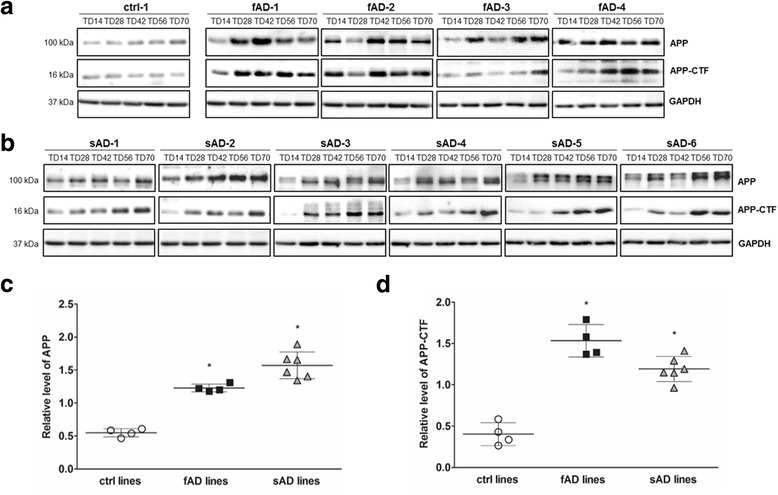
Characterization of amyloid precursor protein (APP) and amyloid precursor protein carboxy-terminal fragment (APP-CTF) expression during neuronal differentiation. Representative immunoblots of (a) APP and APP-CTF in control neurons (ctrl-1), early-onset familial Alzheimer’s disease (fAD)-derived neurons (fAD-1–fAD-4), and (b) sporadic Alzheimer’s disease (sAD)-derived neurons (sAD-1–sAD-6) at terminal differentiation day 14 (TD14), TD28, TD42, TD56, and TD70. Quantification of (c) APP and (d) APP-CTF signals at TD70 was normalized to GAPDH. Data are presented as mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05)
Epitope-specific TAU hyperphosphorylation was detected in fAD and sAD lines
One of the proposed mechanisms for the TAU protein pathomechanism in AD are posttranslational modifications through abnormal phosphorylation (hyperphosphorylation) (reviewed in [55]). The appearance and accumulation of abnormally phosphorylated TAU leads to mislocalization and aggregation of TAU as well as formation of NFTs [56]. Therefore, we focused on the alterations of TAU phosphorylation in our fAD and sAD neural cultures. TAU protein contains several phosphorylation sites (reviewed in [57]). We examined six different phosphorylation sites (Fig. 5a) in fAD, sAD, and control neural cultures at different time points of TD. We observed significantly higher phosphorylation of TAU at Ser262 (12E8 epitope) in all our AD neurons than in control neurons (Fig. 5b, Additional file 6: Figure S3a). Elevated phosphorylation was detected already at TD42 and reached the highest level between weeks 8 and 10 (TD56 and TD70) of TD. Further analysis revealed upregulated TAU phosphorylation at the Ser396 epitope in all fAD and sAD neural cultures (Fig. 5c, Additional file 6: Figure S3b). Western blot quantification showed an increase in TAU phosphorylation at Ser202/Thr205 (AT8 epitope) in the AD group (Fig. 5d, Additional file 6: Figure S3c). Furthermore, we observed higher phosphorylation of TAU at Thr181 and Ser400/Thr403/Ser404 in both fAD and sAD neurons (Fig. 5e and f, Additional file 6: Figure S3d and e). On the basis of these results, we can conclude that, although the phosphorylation level showed some clonal variations, the ratio of pTAU/TAU was significantly elevated in all AD neurons and that no major differences were observed between fAD- and sAD-derived neurons.
Fig. 5.

Western blot analysis of total TAU and phosphorylated TAU (pTAU) protein. a Schematic representation of human TAU isoform (441 amino acids) with the functional projection and microtubule-binding domains. Projection domains including a proline-rich region and N-terminal part interact with cytoskeletal elements and are involved in signal transduction. Microtubule-binding domains with a C-terminal part regulate the microtubule polymerization and bind to proteins such as presenilin 1 (PSEN1). Epitopes of pTAU antibodies analyzed in this study are indicated on the scheme. b–f Densitometric analysis of TAU phosphorylated at different epitopes: S262, S396, S202/T205, T181, and S400/T403/S404. All samples were analyzed at day 70 of terminal differentiation. The amount of pTAU relative to total TAU levels in the lysates was measured. GAPDH as the loading control was used to normalize the data. All values are the mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05)
GSK3B activation in AD-derived neurons
Scientific evidence suggests that GSK3B is involved in many pathological hallmarks of AD, including hyperphosphorylation of TAU [58], increased Aβ production [59], memory impairment, and neuronal loss [60]. To verify if AD neurons with elevated TAU phosphorylation have increased GSK3B activity, the percentage of the active form of GSK3B was calculated by assessing the ratio of inhibitory phosphorylation on Ser9 by immunoblotting. Our results demonstrate that fAD and sAD neurons in most cases exhibited significantly higher levels of active GSK3B than control neurons (Fig. 6a–c, Additional file 7: Figure S4a). During neuronal differentiation, the level of active GSK3B in control neurons remained constant, whereas in fAD and sAD neurons, the active GSK3B amount increased to reach its maximum at day 56 (sAD-2) or day 70 of TD for all remaining cell lines (Fig. 6c, Additional file 7: Figure S4a). Furthermore, higher phosphorylation of TAU at Thr231 (AT180 epitope), which can be catalyzed by GSK3B in vitro [61], was observed in all fAD and sAD neurons (Fig. 6d, Additional file 7: Figure S4b). These results indicate that activation of the GSK3B signaling in both fAD and sAD neurons contributes to the abnormal TAU phosphorylation pattern observed in these cell lines.
Fig. 6.

Analysis of glycogen synthase kinase 3β (GSK3B) and the active form of GSK3B in neuronal culture. a Representative immunoblotting shows the phosphorylation of GSK3B at Ser9 (inactive form of the kinase [102]) and GSK3B in control neurons (ctrl-1), early-onset familial Alzheimer’s disease (fAD) neurons (fAD-1–fAD-4), and (b) sporadic Alzheimer’s disease (sAD) neurons (sAD-1–sAD-6) at all time points of neuronal terminal differentiation (TD14–TD70). c Quantification of the active GSK3B form at TD70 was presented as a percentage of nonphosphorylated GSK3B at Ser9. d Densitometric analysis of TAU phosphorylated at the T231 epitope at TD70. All values were normalized to GAPDH and are presented as mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05)
Oxidative stress response was strongly affected in fAD and sAD neuronal cultures
Oxidative stress plays an important role in the pathogenesis of neurodegenerative disorders. The molecular mechanism of reactive oxygen species (ROS) action in the nervous system has been studied ([62, 63], reviewed in [64]). Despite this, the effect of oxidative stress on human iPSC-derived AD neurons is still not well studied. In our experiments, we used H2O2 in two different concentrations to examine neuronal response to a stressor. In AD and control neurons, treatment with increasing doses of H2O2 engendered a dose-dependent loss of cell viability; however, these two cell groups presented differential susceptibility to the stressor. The fAD and sAD neurons were more sensitive than control neurons to H2O2, and cell death caused by 30 μM and 60 μM H2O2 reached about 50% and 70%, respectively, in the AD cultures (Fig. 7a).
Fig. 7.
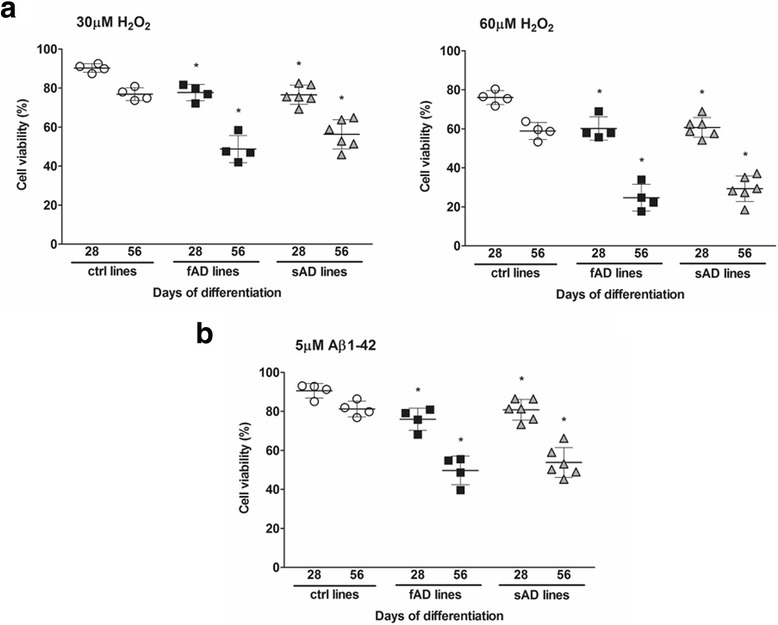
Effect of hydrogen peroxide (H2O2) and amyloid-β1–42 (Aβ1–42) oligomer treatment on neuronal viability. a Viability of induced pluripotent stem cell (iPSC)-derived neurons from control individuals (ctrl-1–ctrl-4), patients with early-onset familial Alzheimer’s disease (fAD) (fAD-1–fAD-4), and patients with sporadic Alzheimer’s disease (sAD) (sAD-1–sAD-6) at day 28 of terminal differentiation (TD28) and TD56 after 24 h of treatment with 30 μM H2O2 (left panel) and 60 μM H2O2 (right panel). b Neuronal survival of control and Alzheimer’s disease (AD) clones at TD28 and TD56 cultured 24 h in the presence of 5 μM Aβ1–42 oligomer solution. Neuronal survival was represented as a percentage of control. Viability of the cultures following H2O2 and Aβ treatment was assessed using the CellTiter-Glo® Luminescent Cell Viability Assay. Values are presented as the mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control at the same time point of differentiation (*p < 0.05)
Evidence of the toxicity of Aβ to neurons has been demonstrated in rodent models and neuronal cell lines (reviewed in [65, 66]). Owing to the limited studies on neurons generated from iPSCs, we examined the effect of synthetic Aβ1–42 oligomer solution on iPSC-derived neurons. Cells treated at days 28 and 56 of neural differentiation for 24 h with 5 μM Aβ1–42 oligomers showed reduced neuronal survival, depending on the neuronal maturation state (Fig. 7b). Relative to control samples, AD cultures had enhanced sensitivity to cell toxicity induced by Aβ1–42. The 28-day-old AD neurons displayed about 15–20% cell death compared with 10% cell loss in control individuals. Mature neurons at TD56 were more susceptible to cell death; almost 40% of neurons died in AD cultures, whereas cell loss in control neurons increased slightly to 15% (Fig. 7b). On the basis of our results, we can conclude that fAD and sAD neural cultures are more susceptible to Aβ1–42 oligomer-induced cell death than the control cultures.
Discussion
The present study shows that particular key disease phenotypes of the most common age-related neurological disorder, AD, can be modeled using patient-specific, iPSC-derived neural cells. Over the past few decades, primary neurons from animal models and immortalized neuronal cell lines have been used to study neurodegenerative diseases. Owing to many limitations, such as genetic and physiological differences between human and rodent brains, such studies of AD mechanisms have been controversial. Most animal models of AD are generated on the basis of modifications in three genes related to fAD (APP, PSEN1, and PSEN2), whereas modeling of sAD in rodents is currently not well examined. Therefore, there are huge expectations regarding iPSC technology, which allows for the generation of pluripotent cells from any individuals in the context of their own genetic identity, to provide new patient-specific in vitro disease models to study neurological disorders.
Modeling AD using iPSCs was initiated with fAD carrying mutations in PSEN1, PSEN2, and APP. Until now, only a few groups have reported generation of iPSC-derived neurons from patients with fAD, and sAD has been studied even less [36, 37, 40, 67]. In our present study, we analyzed samples and generated neurons from patients with fAD carrying pathogenic mutations in the PSEN1 gene (V89L and L150P; these mutations were first identified by Queralt et al. [68] and Wallon et al. [69], respectively) and patients with sAD (Table 1). Our findings demonstrate that iPSCs derived from patients with fAD and patients with sAD can be successfully induced into NPCs with very uniform expression of NES and PAX6 and that they can be further differentiated into neurons and glial cells. Gene expression and ICC analysis revealed the presence of various neuronal subtypes, including GABAergic, glutamatergic, cholinergic, and dopaminergic neurons and progenitor cells of astrocytes and oligodendrocytes in control and AD cultures. We therefore conclude that there is no prominent difference in the differentiation and maturation propensity or in marker gene expression between control and AD neural cells, which is in accordance with a previous report [37].
Initially, our main question was to evaluate if sAD can be modeled through iPSC-derived neural cultures and represent a suitable model system for neuropathological investigations and drug development studies. Therefore, we first analyzed the relevant in vivo pathological hallmarks of the disease in our in vitro system. The accumulation of Aβ into extracellular aggregates is one of the pathological signs of AD in the human brain. Alterations in the level of Aβ peptides are often presented as the ratios between different isoforms. In PSEN mutant mouse models, elevated Aβ production can be detected [70–72]. In humans, elevated Aβ production was revealed in iPSC-derived neural lines from patients with fAD [36]. However, another study demonstrated that Aβ secretion in sAD-derived neurons is not consistently altered [37]. Our study demonstrated increased extracellular Aβ1–40 and Aβ1–42 levels in neurons derived from all fAD and sAD lines in a maturation-dependent manner. Interestingly, Aβ1–40 secretion was approximately twofold higher in sAD neurons than in fAD, whereas Aβ1–42 levels were similar. Moreover, we observed an elevated ratio of Aβ1–42 to Aβ1–40, one of the AD hallmarks, in fAD-derived neurons, whereas the Aβ1–42/Aβ1–40 ratio in sAD lines remained unchanged and comparable to that of non-AD control lines. Additionally, we observed upregulated expression of APP and APP-CTF in all AD-derived cell lines, which is in line with the increased Aβ levels we measured. Some groups also detected an increased level of Aβ1–40, Aβ1–42, and Aβ1–42/Aβ1–40 ratio in fAD cell lines with mutations in PSEN1, PSEN2, and APP [36, 37, 40]. On the basis of the above results, we can conclude that mutations in PSEN1 may change the metabolism of Aβ peptides and drive amyloidosis in patients with fAD. Our findings also indicate the possible heterogeneity of fAD and sAD. AD-iPSC lines with PSEN1 mutations and sAD do not always recapitulate the same phenotypes [37]. In fAD, genetic factors modify the clinical phenotype of the disease, whereas mechanisms underlying the pathogenesis of sAD are still not well understood and combine multiple genetic and environmental risk factors. It is possible that underlying mutations that have not yet been discovered may play an important role in sAD, reflecting the inherent variability of iPSCs. Thus, more cell lines have to be analyzed to reveal the broad heterogeneity of AD phenotypes.
We have demonstrated that another pathological hallmark characteristic of AD, TAU hyperphosphorylation, could be detected in AD neurons. Conformational changes and misfolded protein structure result in aberrant aggregation of TAU into neurofibrillary structures, NFTs [73]. TAU phosphorylation at various sites affects TAU activity, as well as its biological function and pathogenic role. Studies on the physiological properties of TAU have revealed that phosphorylation of Ser262 significantly diminishes the ability of TAU to bind microtubules [74]. Others have reported that phosphorylation of few KXGS motifs, including Ser262 and Ser356, reduces TAU binding capacity to microtubules and thus increases the dynamics of microtubules, which plays an important role in neurite growth and the development of neuronal polarity [75]. Phosphorylation at Thr231 leads to decreased ability of TAU to bind microtubules, reduces the level of acetylated tubulin, and consequently leads to microtubule destabilization [33]. Moreover, the increased TAU phosphorylation at Ser396 and Ser404 impairs microtubule assembly by detachment of TAU molecules from microtubules [76]. Elevated phosphorylation on Ser262, Thr231, and Ser396 residues can be detected early in the AD disease process [77, 78].
Quantitative in vitro data have revealed a negative impact of TAU phosphorylation at many epitopes on TAU activity and microtubule stability [31, 32, 79]. Previous studies on iPSC-derived, patient-specific fAD and sAD neurons have been limited to detecting mostly only one TAU phosphorylation site at Thr231 [40, 67]. We analyzed TAU phosphorylation in iPSC-derived neurons from patients with fAD and patients with sAD at six different phosphorylation sites. As a novel finding, we demonstrated increased TAU phosphorylation at all examined epitopes (Fig. 5a): Ser262, Ser202/Thr205, Ser396, Ser400/Thr403/Ser404, Thr181, and Thr231 in both fAD and sAD neurons. According to the literature, phosphorylation of TAU at Ser262 and Thr231 greatly diminishes its ability to bind microtubules, by 35% and 25%, respectively [74], whereas Ser396 and Ser404 phosphorylation generates more fibrillogenic TAU in vitro [78], which shows an increased propensity to aggregate [80]. Moreover, phosphorylation of the AT8 epitope results in a decrease in TAU microtubule nucleation activity, leading to microtubule depolymerization and destabilization [81]. TAU phosphorylation increased during neural differentiation, which is in agreement with in vivo studies on mice with APP mutations [82].
Furthermore, we have shown higher levels of active GSK3B in our AD cultures. This observation correlates with previously described findings for which researchers showed increased levels of active GSK3B measured in fAD and sAD neurons in vitro [40] and in transgenic animal models [83]. Pathological activation of GSK3B establishes a feedforward loop that contributes to abnormal APP processing [84], enhanced apoptosis in hippocampal neurons, TAU hyperphosphorylation, and synaptic failure in rodent models of AD [85, 86]. Our findings revealed that activation of GSK3B might contribute to TAU misregulation and abnormal phosphorylation. This is in accordance with previous reports confirming the important role of GSK3B in regulating TAU phosphorylation mostly on Thr231 and Ser199, Ser396, Ser400, Ser404, and Ser413 [87]. Consistent with this, restoring the normal level of GSK3B has been shown to reduce TAU hyperphosphorylation, decrease Aβ production and neuronal death in AD murine models [88], and decrease Aβ-induced neurotoxicity in cultured mouse primary neurons in vitro [89].
In our study, we evaluated cell viability after hydrogen peroxide and extracellular Aβ1–42 exposure in neurons at different maturation stages. Exposure to stress agents such as H2O2 induces ROS production and toxicity in many different cell types [90–92]. ROS show high reactivity with macromolecules and play an important role as signaling molecules (reviewed in [93]). Oxidative damage is linked with mitochondrial abnormalities and is catalyzed by the presence of Fe and Cu ions. In our results, we demonstrated a significant H2O2 dose-related decrease in the survival of fAD and sAD neurons. More mature neurons (TD56) showed a greater sensitivity to H2O2 than younger neuronal cultures (TD28). These observations indicate that H2O2 may provoke an antioxidant stress response resulting in increased level of ROS and may lead to subsequent cell death. Additionally, we observed that treatment with Aβ1–42 oligomers induced cell death in both fAD and sAD neurons. According to the literature, Aβ treatment may lead to activation of glutamate receptors and inhibition of glutamate transporters that leads to abnormal release of glutamate and disturbances in glutamatergic neurotransmission. Aβ treatment induces NMDA receptor-mediated cellular events in neurons and astrocytes, leading to synaptic damage and spine loss [94]. Chronic stimulation of NMDA receptor results in Ca2+ influx, which activates apoptotic pathways and increased glutamate excitotoxicity, leading to generation of ROS and to neuronal damage and cell death [95]. In cortical neurons, accumulation of glutamate and NMDA receptors promotes H2O2-mediated neurotoxicity and oxidative damage in DNA.
Furthermore, increased influx of Ca2+ mediated by Aβ treatment activates mitochondrial permeability transition pore, leading to deregulation of respiratory chain enzymes and ROS overproduction, and consequently to neurotoxicity [96]. On the basis of the above data, we can speculate that the neuronal death observed in our cultures upon synthetic Aβ treatment may be a consequence of mitochondrial stress and higher ROS production.
It has previously been reported that accumulated Aβ oligomers induce endoplasmic reticulum (ER) stress and ROS production [97], which lead to membrane lipid peroxidation and impairment of membrane protein function (reviewed in [98]). Furthermore, gene analysis of APP E693Δ neurons revealed upregulation of the oxidative stress-related markers [37]. Additionally, in vivo studies showed increased protein oxidation and lipid peroxidation in PSEN1 mutant brains [99, 100], leading to destruction of spine morphology and impaired synaptic plasticity [101]. Thus, we speculate that our fAD and sAD neurons may also exhibit increased levels of stress-related gene expression, suggesting ER and Golgi abnormalities. It is worth considering that mutations in PSEN1 and pathological changes in sAD, combined with neuronal aging, can upregulate ROS production, leading to mitochondrial damage that may contribute to neurodegenerative processes and AD progression.
Our results provide insight into the molecular basis of AD and provide patient cell models that are relevant for further AD investigations. In all fAD and sAD lines, we observed higher Aβ1–40 and Aβ1–42 secretion, increased activation of GSK3B, and hyperphosphorylation of TAU at six different epitopes. We showed that both fAD and sAD neurons revealed AD phenotypes, suggesting that sAD, with an unknown disease etiology, resembles the fAD phenotype.
The results of this study provide strong evidence that iPSC technology can be used in modeling the most common age-related neurodegenerative disorders, such as AD. This new strategy will make it possible to study pathological mechanisms on the molecular level and analyze patient-derived neurons in vitro from individuals who are still alive. Potentially, iPSCs from complex, unknown genetic background diseases, such as sAD, may be equally useful and able to provide suitable models such as the monogenic forms of AD carrying mutations in PSEN1, PSEN2, and APP.
Although iPSC-derived models afford the opportunity to identify factors associated with disease phenotypes, there are still many uncertainties that should be resolved in the near future. The most crucial limitations that must be addressed in future studies include development of reliable protocols for more rapid neuronal differentiation and optimization of culture conditions as well as current differentiation methods to eliminate clonal variability and heterogeneity of differentiated neuronal cells.
Several preclinical studies using fAD and sAD models have established new methods to gather insight into the molecular mechanisms, the role of different cell types, and patient-specific drug response. We believe that iPSCs have great potential for improving the model systems used in preclinical and clinical applications in neurodegenerative diseases.
Conclusions
Our findings demonstrate that increased TAU phosphorylation occurs in fAD and sAD iPSC-derived mature neurons. To our knowledge, this is the first study involving comparison of TAU protein hyperphosphorylation in an iPSC-based fAD and sAD model. We also present evidence of elevated Aβ1–40 and Aβ1–42 levels in fAD and sAD samples; however, the Aβ1–42/Aβ1–40 ratios are significantly different from control cell lines in fAD cases, whereas the ratio of sAD cell lines not differ significantly. Moreover, we have shown the more sensitive oxidative stress response and higher susceptibility to exogenously added synthetic Aβ1–42 peptide solution of AD lines. Our findings demonstrate that the iPSC technology is suitable to model both fAD and sAD and may provide a platform for developing new treatment strategies for these conditions.
Additional files
Antibodies used in this study. (TIF 141 kb)
Characterization of Aβ secretion in control and AD iPSC-derived neurons at TD14 and TD28. a The amount of secreted Aβ1–40 and (b) Aβ1–42 from control-, fAD- (fAD-1–fAD-4), and sAD- (sAD-1–sAD-6)-iPSC-derived neurons. c The ratio of Aβ1–42/Aβ1–40 from neurons derived from control and AD lines. Aβ1–40 and Aβ1–42 secreted from neural cells into the medium were measured at day 4 after the last medium change. The extracellular Aβ levels determined (in picomolar concentrations) were normalized to total protein content. Data represent mean ± SEM (n = 3). Because the detected levels of Aβ1–40 and Aβ1–42 were not significantly different between the four healthy individual-derived clones (ctrl-1, ctrl-2, ctrl-3, ctrl-4), the average value (±SEM) as a control value was used in all graphs. Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 396 kb)
PCR primers used in this study. (TIF 109 kb)
Polymorphism of AD- and FTD-related genes and APOE status of cell lines used in this study. Footnote: Cell lines not detailed in the table were not permitted to be involved in whole exome sequencing; therefore, only clinical mutation identification was available. (TIF 130 kb)
Characterization of APP and APP-CTF expression during neuronal differentiation. Densitometric analysis of (a) APP and (b) APP-CTF expression in control neurons (ctrl-1), fAD-derived neurons (fAD-1–fAD-4), and sAD-derived neurons (sAD-1–sAD-6) at TD42, TD56, and TD70. Quantification of APP and APP-CTF signals was normalized to GAPDH. Data are presented as mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 1295 kb)
Western blot analysis of total TAU and pTAU protein. a–e Densitometric analysis of TAU phosphorylated at different epitopes: S262, S396, S202/T205, T181, and S400/T403/S404. All samples were analyzed at days 42, 56, and 70 of terminal differentiation. The amount of pTAU relative to total TAU levels in the lysates was measured. GAPDH as the loading control was used to normalize the data. All values are the mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 804 kb)
Analysis of GSK3B and active form of GSK3B in neuronal culture. a Quantification of active GSK3B form in control neurons (ctrl-1), fAD neurons (fAD-1–fAD-4), and sAD neurons (sAD-1–sAD-6) at days 42, 56, and 70 of terminal differentiation was presented as a percentage of nonphosphorylated GSK3B at Ser9 (inactive form of the kinase [102]). b Densitometric analysis of TAU phosphorylated at T231 epitope at days 42, 56, and 70 of terminal differentiation. All values were normalized to GAPDH and are presented as mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 384 kb)
Acknowledgements
We are grateful for Dr. J. M. Molnár and her team at the Institute of Genomic Medicine and Rare Disorders, Semmelweis University, Budapest (Hungary), for organizing the sampling and consenting of the Hungarian patients for iPSC generation. We also thank C. Nemes and E. Varga (BioTalentum Ltd.), C. Pires (Department of Veterinary Clinical and Animal Science, University of Copenhagen, Denmark), and A. Tubsuwan (Stem Cell Research Group, Institute of Molecular Biosciences, Mahidol University, Thailand) for providing iPSCs for the experiments. In addition, we thank L. Fülöp (Department of Medical Chemistry, University of Szeged, Hungary) for providing Aβ1–42 peptides, M. Truszka for the preparation of samples for electron microscopy, and K. Szczesna (BioTalentum Ltd.) for valuable support.
Funding
This work was supported by grants from European Union Seventh Framework Programme for Research and Technological Development projects (STEMMAD, PIAPP-GA-2012-324451; EpiHealth, HEALTH-2012-F2-278418; EpiHealthNet, PITN-GA-2012-317146), a Richter Gedeon research grant (RG-IPI-2013_TP7/026), and Innovation Fund Denmark (BrainStem 4108-00008B and NeuroStem 4096-00001B).
Availability of data and materials
Additional files are available online along with this article.
Abbreviations
- Aβ1–40
Amyloid-β1–40
- Aβ1–42
Amyloid-β1–42
- AD
Alzheimer’s disease
- AD pTAU
Alzheimer’s disease hyperphosphorylated TAU
- APP
Amyloid precursor protein
- APP-CTF
Amyloid precursor protein carboxy-terminal fragment
- bFGF
Basic fibroblast growth factor
- BSA
Bovine serum albumin
- CHAT
Choline acetyltransferase
- CLDN11
Claudin 11
- DLG4
Discs large MAGUK scaffold protein 4
- ELISA
Enzyme-linked immunosorbent assay
- ER
Endoplasmic reticulum
- fAD
Early-onset familial Alzheimer’s disease
- FTD
Frontotemporal dementia
- GAD1
Glutamic acid decarboxylase 1
- GAD2
Glutamic acid decarboxylase 2
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- gDNA
Genomic DNA
- GFAP
Glial fibrillary acidic protein
- GRIN1
Glutamate ionotropic receptor N-methyl-d-aspartate type subunit 1
- GRN
Granulin precursor
- GSK3B
Glycogen synthase kinase 3β
- ICC
Immunocytochemistry
- IgG
Immunoglobulin G
- iPSC
Induced pluripotent stem cell
- LOAD
Late-onset Alzheimer’s disease
- LTD
Long-term depression
- LTP
Long-term potentiation
- MAP2
Microtubule-associated protein 2
- MAPT
Microtubule-associated protein TAU
- NEAA
Nonessential amino acids
- NES
Nestin
- NF200
Neurofilament, heavy polypeptide 200 kDa
- NFT
Neurofibrillary tangle
- NIM
Neural induction medium
- NMDA
N-methyl-d-aspartate
- NMM
Neural maintenance medium
- NPC
Neural progenitor cell
- PAX6
Paired box 6
- PFA
Paraformaldehyde
- PHF
Paired helical filament
- POL/L
Poly-l-ornithine/laminin
- PSEN1
Presenilin 1
- PSEN2
Presenilin 2
- RBFOX3
RNA binding protein, fox-1 homolog 3
- RT
Room temperature
- sAD
Sporadic Alzheimer’s disease
- SLC6A4
Sodium-dependent serotonin transporter
- TBST
Tris-buffered saline with Tween 20
- TD
Terminal differentiation
- TH
Tyrosine hydroxylase
- TUBB3
Tubulin β 3 class III
- VACHT
Vesicular acetylcholine transporter
- VGLUT1/2
Vesicular glutamate transporter 1/2
Authors’ contributions
AO performed experimental design; implemented the experiments; performed flow cytometry, immunocytochemistry, Western blot experiments, ELISA, and RT-qPCR; analyzed the data; participated in the interpretation of the results; and wrote the manuscript. BM and HXA designed the experiments and participated in the implementation of experiments and interpretation of the results. AC and AT were involved in cell culturing. IB participated in whole exome sequencing and performed the data analysis. MLG performed Western blot experiments. ZT performed iPSC establishment and maintenance. KM and LL performed and analyzed electron microscopy data. JEN organized the sampling and consenting of the Danish patients. BH, KF, and PH participated in the implementation of experiments and interpretation of the results. JK was involved in the experimental design and participated in data analysis, interpretation of the results, and manuscript preparation. AD initiated the study and read and approved the manuscript. All authors read and approved the final manuscript.
Authors’ information
Not applicable.
Ethics approval and consent to participate
Written informed consent was obtained from the subjects who provided samples for iPSC derivation. Ethical approval was obtained from the competent authority to establish and maintain hiPSC lines (in Hungary: Medical Research Council (in Hungarian: Egészségügyi Tudományos Tanács - Tudományos és Kutatásetikai Bizottság; ETT-TUKEB) ETT-TUKEB 834/PI/09, 8-333/2009-1018EKU; In Denmark: De Videnskabsetiske Komiteer for Region Hovedstaden; StemMad and BrainStem: Approval: H-4-2011-157).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13195-017-0317-z) contains supplementary material, which is available to authorized users.
Contributor Information
Anna Ochalek, Email: anna.ochalek@biotalentum.hu.
Balázs Mihalik, Email: balazs.mihalik@biotalentum.hu.
Hasan X. Avci, Email: hasan.avci@uni-tuebingen.de
Abinaya Chandrasekaran, Email: abinaya.c@biotalentum.hu.
Annamária Téglási, Email: annamaria.teglasi@biotalentum.hu.
István Bock, Email: istvan.bock@biotalentum.hu.
Maria Lo Giudice, Email: maria.lo@biotalentum.hu.
Zsuzsanna Táncos, Email: zsuzsanna.tancos@biotalentum.hu.
Kinga Molnár, Email: kinga.molnar@ttk.elte.hu.
Lajos László, Email: laszlo@elte.hu.
Jørgen E. Nielsen, Email: joergen.erik.nielsen.01@regionh.dk
Bjørn Holst, Email: bho@bioneer.dk.
Kristine Freude, Email: kkf@sund.ku.dk.
Poul Hyttel, Email: poh@sund.ku.dk.
Julianna Kobolák, Email: julianna.kobolak@biotalentum.hu.
András Dinnyés, Phone: +36-28-526-159, Email: manuscript.dinnyes@biotalentum.hu.
References
- 1.Prince M, Comas-Herrera A, Knapp M, Guerchet M, Karagiannidou M, World Alzheimer report 2016 . Improving health care for people living with dementia: coverage, quality and costs now and in the future. London: Alzheimer’s Disease International; 2016. [Google Scholar]
- 2.Borenstein AR, Copenhaver CI, Mortimer JA. Early-life risk factors for Alzheimer disease. Alzheimer Dis Assoc Disord. 2006;20(1):63–72. doi: 10.1097/01.wad.0000201854.62116.d7. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D. The therapeutics of Alzheimer’s disease: where we stand and where we are heading. Ann Neurol. 2013;74(3):328–336. doi: 10.1002/ana.24001. [DOI] [PubMed] [Google Scholar]
- 4.Murphy MP, LeVine H., 3rd Alzheimer’s disease and the β-amyloid peptide. J Alzheimers Dis. 2010;19(1):311–323. doi: 10.3233/JAD-2010-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt M, Sachse C, Richter W, Xu C, Fändrich M, Grigorieff N. Comparison of Alzheimer Aβ1–40 and Aβ1–42 amyloid fibrils reveals similar protofilament structures. Proc Natl Acad Sci U S A. 2009;106(47):19813–19818. doi: 10.1073/pnas.0905007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R, Hu X, Khant H, Ludtke SJ, Chiu W, Schmid MF, Frieden C, Lee JM. Interprotofilament interactions between Alzheimer’s Aβ1–42β peptides in amyloid fibrils revealed by cryoEM. Proc Natl Acad Sci U S A. 2009;106(12):4653–4658. doi: 10.1073/pnas.0901085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi XP, Tugusheva K, Bruce JE, Lucka A, Wu GX, Chen-Dodson E, Price E, Li Y, Xu M, Huang Q, Sardana MK, Hazuda DJ. β-Secretase cleavage at amino acid residue 34 in the amyloid β peptide is dependent upon γ-secretase activity. J Biol Chem. 2003;278(23):21286–21294. doi: 10.1074/jbc.M209859200. [DOI] [PubMed] [Google Scholar]
- 8.De Strooper B, Aizenstein H, Nebes R. Proteases and proteolysis in Alzheimer disease: a multifactorial view on the disease process. Physiol Rev. 2010;90(2):465–494. doi: 10.1152/physrev.00023.2009. [DOI] [PubMed] [Google Scholar]
- 9.Gu L, Guo Z. Alzheimer’s Aβ42 and Aβ40 peptides form interlaced amyloid fibrils. J Neurochem. 2013;126(3):305–311. doi: 10.1111/jnc.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roher AE, Lowenson JD, Clarke S, Woods AS, Cotter RJ, Gowing E, Ball MJ. β-Amyloid-(1–42) is a major component of cerebrovascular amyloid deposits: implications for the pathology of Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90(22):10836–10840. doi: 10.1073/pnas.90.22.10836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwatsubo T, Saido TC, Mann DM, Lee VM, Trojanowski JQ. Full-length amyloid-β(1–42(43)) and amino-terminally modified and truncated amyloid-β 42(43) deposit in diffuse plaques. Am J Pathol. 1996;149(6):1823–1830. [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Perry G, Smith MA, Zhu X. Amyloid-β-derived diffusible ligands cause impaired axonal transport of mitochondria in neurons. Neurodegener Dis. 2010;7(1-3):56–59. doi: 10.1159/000283484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-β overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci U S A. 2008;105(49):19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416(6880):535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 15.Wang HW, Pasternak JF, Kuo H, Ristic H, Lambert MP, Chromy B, Viola KL, Klein WL, Stine WB, Krafft GA, Trommer BL. Soluble oligomers of β amyloid (1–42) inhibit long-term potentiation but not long-term depression in rat dentate gyrus. Brain Res. 2002;924(2):133–140. doi: 10.1016/S0006-8993(01)03058-X. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira ST, Lourenco MV, Oliveira MM, De Felice FG. Soluble amyloid-β oligomers as synaptotoxins leading to cognitive impairment in Alzheimer’s disease. Front Cell Neurosci. 2015;9:191. doi: 10.3389/fncel.2015.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li S, Jin M, Koeglsperger T, Shepardson N, Shankar G, Selkoe D. Soluble Aβ oligomers inhibit long-term potentiation through a mechanism involving excessive activation of extrasynaptic NR2B-containing NMDA receptors. J Neurosci. 2011;31(18):6627–6638. doi: 10.1523/JNEUROSCI.0203-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Strooper B. Loss‐of‐function presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8(2):141–146. doi: 10.1038/sj.embor.7400897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou L, Brouwers N, Benilova I, Vandersteen A, Mercken M, van Laere K, van Damme P, Demedts D, van Leuven F, Sleegers K, et al. Amyloid precursor protein mutation E682K at the alternative β-secretase cleavage β′-site increases Aβ generation. EMBO Mol Med. 2011;3:291–302. doi: 10.1002/emmm.201100138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Jiao B, Zhou M, Zhou T, Shen L. Modeling Alzheimer’s disease with induced pluripotent stem cells: current challenges and future concerns. Stem Cells Int. 2016;2016:7828049. doi: 10.1155/2016/7828049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu CC, Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9(2):106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guzmán-Martinez L, Farías GA, Maccioni RB. Tau oligomers as potential targets for Alzheimer’s diagnosis and novel drugs. Front Neurol. 2013;4(10):167. doi: 10.3389/fneur.2013.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mi K, Johnson GVW. The role of tau phosphorylation in the pathogenesis of Alzheimer’s disease. Curr Alzheimer Res. 2006;3(5):449–463. doi: 10.2174/156720506779025279. [DOI] [PubMed] [Google Scholar]
- 24.Alonso AC, Zaidi T, Grundke-Iqbal I, Iqbal K. Role of abnormally phosphorylated tau in the breakdown of microtubules in Alzheimer disease. Proc Natl Acad Sci U S A. 1994;91(12):5562–5566. doi: 10.1073/pnas.91.12.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaPointe NE, Morfini G, Pigino G, Gaisina IN, Kozikowski AP, Binder LI, Brady ST. The amino terminus of tau inhibits kinesin-dependent axonal transport: Implications for filament toxicity. J Neurosci Res. 2009;87(2):440–451. doi: 10.1002/jnr.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bang J, Spina S, Miller BL. Frontotemporal dementia. Lancet. 2015;386(10004):1672–1682. doi: 10.1016/S0140-6736(15)00461-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hosokawa M, Arai T, Masuda-Suzukake M, Kondo H, Matsuwaki T, Nishihara M, Hasegawa M, Akiyama H. Progranulin reduction is associated with increased tau phosphorylation in P301L tau transgenic mice. J Neuropathol Exp Neurol. 2015;74(2):158–165. doi: 10.1097/NEN.0000000000000158. [DOI] [PubMed] [Google Scholar]
- 28.Hosokawa M, Kondo H, Serrano GE, Beach TG, Robinson AC, Mann DM, Akiyama H, Hasegawa M, Arai T. Accumulation of multiple neurodegenerative disease-related proteins in familial frontotemporal lobar degeneration associated with granulin mutation. Sci Rep. 2017;7:1513. doi: 10.1038/s41598-017-01587-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gong CX, Liu F, Grundke-Iqbal I, Iqbal K. Post-translational modifications of tau protein in Alzheimer’s disease. J Neural Transm. 2005;112(6):813–838. doi: 10.1007/s00702-004-0221-0. [DOI] [PubMed] [Google Scholar]
- 30.Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297(Pt 2):249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner U, Utton M, Gallo JM, Miller CC. Cellular phosphorylation of tau by GSK-3β influences tau binding to microtubules and microtubule organisation. J Cell Sci. 1996;109(Pt 6):1537–1543. doi: 10.1242/jcs.109.6.1537. [DOI] [PubMed] [Google Scholar]
- 32.Drewes G, Ebneth A, Preuss U, Mandelkow EM, Mandelkow E. MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell. 1997;89(2):297–308. doi: 10.1016/S0092-8674(00)80208-1. [DOI] [PubMed] [Google Scholar]
- 33.Cho JH, Johnson GVW. Primed phosphorylation of tau at Thr231 by glycogen synthase kinase 3β (GSK3β) plays a critical role in regulating tau’s ability to bind and stabilize microtubules. J Neurochem. 2004;88(2):349–358. doi: 10.1111/j.1471-4159.2004.02155.x. [DOI] [PubMed] [Google Scholar]
- 34.Kang DE, Yoon IS, Repetto E, Busse T, Yermian N, Ie L, Koo EH. Presenilins mediate phosphatidylinositol 3-kinase/AKT and ERK activation via select signaling receptors: selectivity of PS2 in platelet-derived growth factor signaling. J Biol Chem. 2005;280(36):31537–31547. doi: 10.1074/jbc.M500833200. [DOI] [PubMed] [Google Scholar]
- 35.Boissart C, Poulet A, Georges P, Darville H, Julita E, Delorme R, Bourgeron T, Peschanski M, Benchoua A. Differentiation from human pluripotent stem cells of cortical neurons of the superficial layers amenable to psychiatric disease modeling and high-throughput drug screening. Transl Psychiatry. 2013;3:e294. doi: 10.1038/tp.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Modeling familial Alzheimer’s disease with induced pluripotent stem cells. Hum Mol Genet. 2011;20(23):4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- 37.Kondo T, Asai M, Tsukita K. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell. 2013;12(4):487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 38.Sproul AA, Jacob S, Pre D, Kim SH, Nestor MW, Navarro-Sobrino M, Santa-Maria I, Zimmer M, Aubry S, Steele JW, et al. Characterization and molecular profiling of PSEN1 familial Alzheimer’s disease iPSC-derived neural progenitors. PLoS One. 2014;9(1):e84547. doi: 10.1371/journal.pone.0084547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muratore CR, Rice HC, Srikanth P, Callahan DG, Shin T, Benjamin LN, Walsh DM, Selkoe DJ, Young-Pearse TL. The familial Alzheimer’s disease APPV717I mutation alters APP processing and Tau expression in iPSC-derived neurons. Hum Mol Genet. 2014;23(13):3523–3536. doi: 10.1093/hmg/ddu064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, van Gorp S, Nazor KL, Boscolo FS, et al. Probing sporadic and familial Alzheimer’s disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemes C, Varga E, Tancos Z, Bock I, Francz B, Kobolak J, Dinnyes A. Establishment of PSEN1 mutant induced pluripotent stem cell (iPSC) line from an Alzheimer’s disease (AD) female patient. Stem Cell Res. 2016;17(1):69–71. doi: 10.1016/j.scr.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 42.Tubsuwan A, Pires C, Rasmussen MA, Schmid B, Nielsen JE, Hjermind LE, Hall V, Nielsen TT, Waldemar G, Hyttel P, et al. Generation of induced pluripotent stem cells (iPSCs) from an Alzheimer’s disease patient carrying a L150P mutation in PSEN-1. Stem Cell Res. 2016;16(1):110–112. doi: 10.1016/j.scr.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 43.Táncos Z, Varga E, Kovács E, Dinnyés A, Kobolák J. Establishment of induced pluripotent stem cell (iPSC) line from an 84-year old patient with late onset Alzheimer’s disease (LOAD) Stem Cell Res. 2016;17(1):75–77. doi: 10.1016/j.scr.2016.05.016. [DOI] [PubMed] [Google Scholar]
- 44.Táncos Z, Varga E, Kovács E, Dinnyés A, Kobolák J. Establishment of induced pluripotent stem cell (iPSC) line from a 75-year old patient with late onset Alzheimer’s disease (LOAD) Stem Cell Res. 2016;17(1):81–83. doi: 10.1016/j.scr.2016.05.013. [DOI] [PubMed] [Google Scholar]
- 45.Ochalek A, Nemes C, Varga E, Táncos Z, Kobolák J, Dinnyés A. Establishment of induced pluripotent stem cell (iPSC) line from a 57-year old patient with late onset Alzheimer’s disease (LOAD) Stem Cell Res. 2016;17(1):72–74. doi: 10.1016/j.scr.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 46.Chandrasekaran A, Varga E, Nemes C, Táncos Z, Kobolák J, Dinnyés A. Establishment of induced pluripotent stem cell (iPSC) line from a 63-year old patient with late onset Alzheimer’s disease (LOAD) Stem Cell Res. 2016;17(1):78–80. doi: 10.1016/j.scr.2016.05.014. [DOI] [PubMed] [Google Scholar]
- 47.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White DL, Mazurkiewicz JE, Barrnett RJ. A chemical mechanism for tissue staining by osmium tetroxide-ferrocyanide mixtures. J Histochem Cytochem. 1979;27(7):1084–1091. doi: 10.1177/27.7.89155. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49(7):433–436. doi: 10.1136/jmedgenet-2012-100918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10(10):1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bozso Z, Penke B, Simon D, Laczko I, Juhasz G, Szegedi V, Kasza A, Soos K, Hetenyi A, Weber E, Tohati H, Csete M. Controlled in situ preparation of Aβ1–42 oligomers from the isopeptide ‘iso-Aβ1–42’, physicochemical and biological characterization. Peptides. 2010;31(2):248–256. doi: 10.1016/j.peptides.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Johnson GV, Stoothoff WH. Tau phosphorylation in neuronal cell function and dysfunction. J Cell Sci. 2004;117(Pt 24):5721–5729. doi: 10.1242/jcs.01558. [DOI] [PubMed] [Google Scholar]
- 56.Zempel H, Thies E, Mandelkow E, Mandelkow EM. Aβ oligomers cause localized Ca2+ elevation, missorting of endogenous Tau into dendrites, Tau phosphorylation, and destruction of microtubules and spines. J Neurosci. 2010;30(36):11938–11950. doi: 10.1523/JNEUROSCI.2357-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sergeant N, Delacourte A, Buée L. Tau protein as a differential biomarker of tauopathies. Biochim Biophys Acta. 2005;1739(2):179–197. doi: 10.1016/j.bbadis.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 58.Rankin CA, Sun Q, Gamblin TC. Tau phosphorylation by GSK-3β promotes tangle-like filament morphology. Mol Neurodegener. 2007;2:12. doi: 10.1186/1750-1326-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Phiel CJ, Wilson CA, Lee VMY, Klein PS. GSK-3α regulates production of Alzheimer’s disease amyloid-β peptides. Nature. 2003;423(2):435–439. doi: 10.1038/nature01640. [DOI] [PubMed] [Google Scholar]
- 60.Hooper C, Killick R, Lovestone S. The GSK3 hypothesis of Alzheimer’s disease. J Neurochem. 2008;104(6):1433–1439. doi: 10.1111/j.1471-4159.2007.05194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cho JH, Johnson GVW. Glycogen synthase kinase 3β phosphorylates tau at both primed and unprimed sites: differential impact on microtubule binding. J Biol Chem. 2003;278(1):187–193. doi: 10.1074/jbc.M206236200. [DOI] [PubMed] [Google Scholar]
- 62.Fukui K, Takatsu H, Koike T, Urano S. Hydrogen peroxide induces neurite degeneration: prevention by tocotrienols. Free Radic Res. 2011;45(6):681–691. doi: 10.3109/10715762.2011.567984. [DOI] [PubMed] [Google Scholar]
- 63.Watson SN, Nelson MA, Wildering WC. Redox agents modulate neuronal activity and reproduce physiological aspects of neuronal aging. Neurobiol Aging. 2012;33(1):149–161. doi: 10.1016/j.neurobiolaging.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 64.Tönnies E, Trushina E. Oxidative stress, synaptic dysfunction, and Alzheimer’s disease. J Alzheimers Dis. 2017;57(4):1105–1121. doi: 10.3233/JAD-161088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol Med. 2016;8(6):595–608. doi: 10.15252/emmm.201606210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foley AM, Ammar ZM, Lee RH, Mitchell CS. Systematic review of the relationship between amyloid-β levels and measures of transgenic mouse cognitive deficit in Alzheimer’s disease. J Alzheimers Dis. 2015;44(3):787–795. doi: 10.3233/JAD-142208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hossini AM, Megges M, Prigione A, Lichtner B, Toliat MR, Wruck W, Schröter F, Neurnberg P, Kroll H, Makrantonaki E, et al. Induced pluripotent stem cell-derived neuronal cells from a sporadic Alzheimer’s disease donor as a model for investigating AD-associated gene regulatory networks. BMC Genomics. 2015;16:84. doi: 10.1186/s12864-015-1262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Queralt R, Ezquerra M, Lleo A, Castellvi M, Gelpi J, Ferrer I, Acarin N, Pasarin L, Blesa R, Oliva R. A novel mutation (V89L) in the presenilin 1 gene in a family with early onset Alzheimer’s disease and marked behavioural disturbances. J Neurol Neurosurg Psychiatry. 2002;72(2):266–269. doi: 10.1136/jnnp.72.2.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallon D, Rousseau S, Rovelet-Lecrux A, Quillard-Muraine M, Guyant-Marechal L, Martinaud O, Pariente J, Puel M, Rollin-Sillaire A, Croisile B, et al. The French series of autosomal dominant early onset Alzheimer’s disease cases: mutation spectrum and cerebrospinal fluid biomarkers. J Alzheimers Dis. 2012;30(4):847–856. doi: 10.3233/JAD-2012-120172. [DOI] [PubMed] [Google Scholar]
- 70.Duff K, Eckman C, Zehr C, Yu X, Prada CM, Perez-tur J, Hutton M, Buee L, Harigaya Y, Yager D, et al. Increased amyloid-β42(43) in brains of mice expressing mutant presenilin 1. Nature. 1996;383(6602):710–713. doi: 10.1038/383710a0. [DOI] [PubMed] [Google Scholar]
- 71.Huang XG, Yee BK, Nag S, Chan STH, Tang F. Behavioral and neurochemical characterization of transgenic mice carrying the human presenilin-1 gene with or without the leucine-to-proline mutation at codon 235. Exp Neurol. 2003;183(2):673–681. doi: 10.1016/S0014-4886(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 72.Dewachter I, Ris L, Croes S, Borghgraef P, Devijver H, Voets T, Nilius B, Godaux E, van Leuven F. Modulation of synaptic plasticity and Tau phosphorylation by wild-type and mutant presenilin1. Neurobiol Aging. 2008;29(5):639–652. doi: 10.1016/j.neurobiolaging.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 73.Grundke-Iqbal I, Iqbal K, Tung YC, Quinlan M, Wisniewski HM, Binder LI. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc Natl Acad Sci U S A. 1986;83(13):4913–4917. doi: 10.1073/pnas.83.13.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Biernat J, Gustke N, Drewes G, Mandelkow E, Mandelkow EM. Phosphorylation of Ser262 strongly reduces binding of tau to microtubules: distinction between PHF-like immunoreactivity and microtubule binding. Neuron. 1993;11(1):153–163. doi: 10.1016/0896-6273(93)90279-Z. [DOI] [PubMed] [Google Scholar]
- 75.Biernat J, Wu YZ, Timm T, Zheng-Fischhofer Q, Mandelkow E, Meijer L, Mandelkow EM. Protein kinase MARK/PAR-1 is required for neurite outgrowth and establishment of neuronal polarity. Mol Biol Cell. 2002;13(11):4013–4028. doi: 10.1091/mbc.02-03-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Evans DB, Rank KB, Bhattacharya K, Thomsen DR, Gurney ME, Sharma SK. Tau phosphorylation at serine 396 and serine 404 by human recombinant tau protein kinase II inhibits tau’s ability to promote microtubule assembly. J Biol Chem. 2000;275(32):24977–24983. doi: 10.1074/jbc.M000808200. [DOI] [PubMed] [Google Scholar]
- 77.Augustinack JC, Schneider A, Mandelkow EM, Hyman BT. Specific tau phosphorylation sites correlate with severity of neuronal cytopathology in Alzheimer’s disease. Acta Neuropathol. 2002;103(1):26–35. doi: 10.1007/s004010100423. [DOI] [PubMed] [Google Scholar]
- 78.Abraha A, Ghoshal N, Gamblin TC, Cryns V, Berry RW, Kuret J, Binder LI. C-terminal inhibition of tau assembly in vitro and in Alzheimer’s disease. J Cell Sci. 2000;113:3737–3745. doi: 10.1242/jcs.113.21.3737. [DOI] [PubMed] [Google Scholar]
- 79.Sengupta A, Kabat J, Novak M, Wu Q, Grundke-Iqbal I, Iqbal K. Phosphorylation of tau at both Thr 231 and Ser 262 is required for maximal inhibition of its binding to microtubules. Arch Biochem Biophys. 1998;357(2):299–309. doi: 10.1006/abbi.1998.0813. [DOI] [PubMed] [Google Scholar]
- 80.Haase C, Stieler J, Arendt T, Holzer M. Pseudophosphorylation of tau protein alters its ability for self-aggregation. J Neurochem. 2004;88(6):1509–1520. doi: 10.1046/j.1471-4159.2003.02287.x. [DOI] [PubMed] [Google Scholar]
- 81.Wada Y, Ishiguro K, Itoh TJ, Uchida T, Hotani H, Saito T, Kishimoto T, Hisanaga S. Microtubule-stimulated phosphorylation of tau at Ser202 and Thr205 by cdk5 decreases its microtubule nucleation activity. J Biochem. 1998;124(4):738–746. doi: 10.1093/oxfordjournals.jbchem.a022174. [DOI] [PubMed] [Google Scholar]
- 82.Masliah E, Sisk A, Mallory M, Games D. Neurofibrillary pathology in transgenic mice overexpressing V717F β-amyloid precursor protein. J Neuropathol Exp Neurol. 2001;60(4):357–368. doi: 10.1093/jnen/60.4.357. [DOI] [PubMed] [Google Scholar]
- 83.Ryan KA, Pimplikar SW. Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J Cell Biol. 2005;171(2):327–335. doi: 10.1083/jcb.200505078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Deng Y, Xiong Z, Chen P, Wei J, Chen S, Yan Z. β-Amyloid impairs the regulation of N-methyl-D-aspartate receptors by glycogen synthase kinase 3. Neurobiol Aging. 2014;35(3):449–459. doi: 10.1016/j.neurobiolaging.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fuster-Matanzo A, Llorens-Martin M, de Barreda E, Avila J, Hernandez F. Different susceptibility to neurodegeneration of dorsal and ventral hippocampal dentate gyrus: a study with transgenic mice overexpressing GSK3β. PLoS One. 2011;6(11):e27262. doi: 10.1371/journal.pone.0027262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Llorens-Martín M, Fuster-Matanzo A, Teixeira CM, Jurado-Arjona J, Ulloa F, deFelipe J, Rabano A, Hernandez F, Soriano E, Avila J. GSK-3β overexpression causes reversible alterations on postsynaptic densities and dendritic morphology of hippocampal granule neurons in vivo. Mol Psychiatry. 2013;18(4):451–460. doi: 10.1038/mp.2013.4. [DOI] [PubMed] [Google Scholar]
- 87.Kolarova M, García-Sierra F, Bartos A, Ricny J, Ripova D. Structure and pathology of tau protein in Alzheimer disease. Int J Alzheimers Dis. 2012;2012:731526. doi: 10.1155/2012/731526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Engel T, Hernández F, Avila J, Lucas JJ. Full reversal of Alzheimer’s disease-like phenotype in a mouse model with conditional overexpression of glycogen synthase kinase-3. J Neurosci. 2006;26(19):5083–5090. doi: 10.1523/JNEUROSCI.0604-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Koh SH, Noh MY, Kim SH. Amyloid-β-induced neurotoxicity is reduced by inhibition of glycogen synthase kinase-3. Brain Res. 2008;1188(1):254–262. doi: 10.1016/j.brainres.2007.10.064. [DOI] [PubMed] [Google Scholar]
- 90.Lee Y, Ha M, YK K. H2O2-induced cell death in human glioma cells: role of lipid peroxidation and PARP activation. Neurochem Res. 2001;26(4):337–43. [DOI] [PubMed]
- 91.Kim M, Chung J, Jang J, Chung S, Kwag N, Yoo J. Hydrogen peroxide-induced cell death in a human retinal pigment epithelial cell line, ARPE-19. Korean J Ophthalmol. 2003;17(1):19–28. doi: 10.3341/kjo.2003.17.1.19. [DOI] [PubMed] [Google Scholar]
- 92.Tochigi M, Inoue T, Suzuki-Karasaki M, Ochiai T, Ra C, Suzuki-Karasaki Y. Hydrogen peroxide induces cell death in human TRAIL-resistant melanoma through intracellular superoxide generation. Int J Oncol. 2013;42(3):863–872. doi: 10.3892/ijo.2013.1769. [DOI] [PubMed] [Google Scholar]
- 93.Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative stress and neurodegenerative diseases: a review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol. 2009;7(1):65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Talantova M, Sanz-Blasco S, Zhang X, Xia P, Akhtar MW, Okamoto S, Dziewczapolski G, Nakamura T, Cao G, Pratt AE, et al. Aβ induces astrocytic glutamate release, extrasynaptic NMDA receptor activation, and synaptic loss. Proc Natl Acad Sci U S A. 2013;110:E2518–E2527. doi: 10.1073/pnas.1306832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mailly F, Marin P, Israël M, Glowinski J, Prémont J. Increase in external glutamate and NMDA receptor activation contribute to H2O2-induced neuronal apoptosis. J Neurochem. 1999;73(3):1181–1188. doi: 10.1046/j.1471-4159.1999.0731181.x. [DOI] [PubMed] [Google Scholar]
- 96.Morkuniene R, Cizas P, Jankeviciute S, Petrolis R, Arandarcikaite O, Krisciukaitis A, Borutaite V. Small Aβ1–42 oligomer-induced membrane depolarization of neuronal and microglial cells: role of N-methyl-D-aspartate receptors. J Neurosci Res. 2015;93(3):475–486. doi: 10.1002/jnr.23510. [DOI] [PubMed] [Google Scholar]
- 97.Nishitsuji K, Tomiyama T, Ishibashi K, Ito K, Teraoka R, Lambert MP, Klein WL, Mori H. The E693Δ mutation in amyloid precursor protein increases intracellular accumulation of amyloid β oligomers and causes endoplasmic reticulum stress-induced apoptosis in cultured cells. Am J Pathol. 2009;174(3):957–969. doi: 10.2353/ajpath.2009.080480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Butterfield DA. Amyloid β-peptide [1-42]-associated free radical-induced oxidative stress and neurodegeneration in Alzheimer’s disease grain: Mechanisms and consequences. Curr Med Chem. 2003;10(24):2651–2659. doi: 10.2174/0929867033456422. [DOI] [PubMed] [Google Scholar]
- 99.Mohmmad Abdul H, Wenk GL, Gramling M, Hauss-Wegrzyniak B, Butterfield DA. APP and PS-1 mutations induce brain oxidative stress independent of dietary cholesterol: implications for Alzheimer’s disease. Neurosci Lett. 2004;368(2):148–150. doi: 10.1016/j.neulet.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 100.Schuessel K, Frey C, Jourdan C, Keil U, Weber CC, Muller-Spahn F, Muller WE, Eckert A. Aging sensitizes toward ROS formation and lipid peroxidation in PS1M146L transgenic mice. Free Radic Biol Med. 2006;40(5):850–862. doi: 10.1016/j.freeradbiomed.2005.10.041. [DOI] [PubMed] [Google Scholar]
- 101.Auffret A, Gautheron V, Repici M, Kraftsik R, Mount HT, Mariani J, Rovira C. Age-dependent impairment of spine morphology and synaptic plasticity in hippocampal CA1 neurons of a presenilin 1 transgenic mouse model of Alzheimer’s disease. J Neurosci. 2009;29(32):10144–10152. doi: 10.1523/JNEUROSCI.1856-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. Crystal structure of glycogen synthase kinase 3β: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell. 2001;105(6):721–732. doi: 10.1016/S0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antibodies used in this study. (TIF 141 kb)
Characterization of Aβ secretion in control and AD iPSC-derived neurons at TD14 and TD28. a The amount of secreted Aβ1–40 and (b) Aβ1–42 from control-, fAD- (fAD-1–fAD-4), and sAD- (sAD-1–sAD-6)-iPSC-derived neurons. c The ratio of Aβ1–42/Aβ1–40 from neurons derived from control and AD lines. Aβ1–40 and Aβ1–42 secreted from neural cells into the medium were measured at day 4 after the last medium change. The extracellular Aβ levels determined (in picomolar concentrations) were normalized to total protein content. Data represent mean ± SEM (n = 3). Because the detected levels of Aβ1–40 and Aβ1–42 were not significantly different between the four healthy individual-derived clones (ctrl-1, ctrl-2, ctrl-3, ctrl-4), the average value (±SEM) as a control value was used in all graphs. Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 396 kb)
PCR primers used in this study. (TIF 109 kb)
Polymorphism of AD- and FTD-related genes and APOE status of cell lines used in this study. Footnote: Cell lines not detailed in the table were not permitted to be involved in whole exome sequencing; therefore, only clinical mutation identification was available. (TIF 130 kb)
Characterization of APP and APP-CTF expression during neuronal differentiation. Densitometric analysis of (a) APP and (b) APP-CTF expression in control neurons (ctrl-1), fAD-derived neurons (fAD-1–fAD-4), and sAD-derived neurons (sAD-1–sAD-6) at TD42, TD56, and TD70. Quantification of APP and APP-CTF signals was normalized to GAPDH. Data are presented as mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 1295 kb)
Western blot analysis of total TAU and pTAU protein. a–e Densitometric analysis of TAU phosphorylated at different epitopes: S262, S396, S202/T205, T181, and S400/T403/S404. All samples were analyzed at days 42, 56, and 70 of terminal differentiation. The amount of pTAU relative to total TAU levels in the lysates was measured. GAPDH as the loading control was used to normalize the data. All values are the mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 804 kb)
Analysis of GSK3B and active form of GSK3B in neuronal culture. a Quantification of active GSK3B form in control neurons (ctrl-1), fAD neurons (fAD-1–fAD-4), and sAD neurons (sAD-1–sAD-6) at days 42, 56, and 70 of terminal differentiation was presented as a percentage of nonphosphorylated GSK3B at Ser9 (inactive form of the kinase [102]). b Densitometric analysis of TAU phosphorylated at T231 epitope at days 42, 56, and 70 of terminal differentiation. All values were normalized to GAPDH and are presented as mean ± SEM (n = 3). Dunnett’s test was performed to evaluate the significance of groups compared with control (*p < 0.05). (TIF 384 kb)
Data Availability Statement
Additional files are available online along with this article.


