Abstract
Epigenetic modifications, such as DNA cytosine methylation, contribute to the mechanisms underlying learning and memory by coordinating adaptive gene expression and neuronal plasticity. Transcription-dependent plasticity regulated by DNA methylation includes synaptic plasticity and homeostatic synaptic scaling. Memory-related plasticity also includes alterations in intrinsic membrane excitability mediated by changes in the abundance or activity of ion channels in the plasma membrane, which sets the threshold for action potential generation. We found that prolonged inhibition of DNA methyltransferase (DNMT) activity increased intrinsic membrane excitability of cultured cortical pyramidal neurons. Knockdown of the cytosine demethylase TET1 or inhibition of RNA polymerase blocked the increased membrane excitability caused by DNMT inhibition, suggesting that this effect was mediated by subsequent cytosine demethylation and de novo transcription. Prolonged DNMT inhibition blunted the medium component of the after-hyperpolarization potential, an effect that would increase neuronal excitability, and was associated with reduced expression of the genes encoding small-conductance Ca2+-activated K+ (SK) channels. Furthermore, the specific SK channel blocker apamin increased neuronal excitability but was ineffective after DNMT inhibition. Our results suggested that DNMT inhibition enables transcriptional changes that culminate in decreased expression of SK channel–encoding genes and decreased activity of SK channels, thus providing a mechanism for the regulation of neuronal intrinsic membrane excitability by dynamic DNA cytosine methylation. This study has implications for human neurological and psychiatric diseases associated with dysregulated intrinsic excitability.
INTRODUCTION
Mammalian learning and memory depend on activity-regulated neuronal plasticity. In response to external stimuli, neurons within memory-related networks in the adult central nervous system respond with patterns of activity that relay and encode information. Subsequently, alterations in synaptic strength and membrane excitability modify and shape neuronal connectivity and thus contribute to the integration and storage of experiential information (1–4). Fundamental to these processes is the adaptive regulation of gene expression (5, 6). In the context of development and differentiation, the cellular programs orchestrating the transcriptome are centered at the level of the nucleus, particularly in the regulation of chromatin structure through epigenetic modifications (7, 8). In the adult central nervous system, considerable evidence now supports a role for epigenetic modifications in learning and memory (9) and neuropsychiatric disorders (9, 10).
Once regarded as irreversible in postmitotic cells, DNA cytosine methylation has emerged as a dynamic epigenetic modification in adult neurons. In neurons, cytosines throughout the genome are differentially methylated and demethylated in response to activity and behavioral stimulation (9, 11–13). As in other cell types, the forward process of cytosine methylation is catalyzed by DNA methyltransferases (DNMTs) (14) and most commonly occurs at cytosines followed by a guanine residue, referred to as CpG sites (15). DNMTs, predominantly DNMT3A and
DNMT1, are abundant in postmitotic neurons. DNMT3A methylates cytosines on a single DNA strand, such as previously unmethylated CpG sites, and is thus called a de novo DNMT. DNMT1 recognizes methylated CpG sites on a single strand, subsequently methylates cytosines on the opposite strand, and is thus called “a maintenance” DNMT (16, 17). Experience stimulates DNMT-mediated conversion of cytosine to 5-methylcytosine, and this conversion is implicated in memory-related neuronal plasticity in brain regions such as hippocampus (12, 18–20), cortex (21), and midbrain (22, 23). Furthermore, pharmacological inhibition of DNMT activity (henceforth referred to as DNMTi) and genetic knockout of both Dnmt1 and Dnmt3a disrupt memory function in the adult rodent (19–22). The reverse process, DNA demethylation, occurs through an array of enzymatic reactions initiated by the ten-eleven translocation (TET) family–mediated oxidation of 5-methylcytosine to 5-hydroxymethylcytosine. Active DNA demethylation is also induced by experience and neuronal activity and is necessary for memory formation (13, 24–26). Collectively, these lines of evidence argue that in the behaving animal, DNA cytosine methylation represents a core epigenetic mechanism that promotes the incorporation of experiential information in a persistent yet flexible manner.
On the cellular level, activity-regulated DNA methylation contributes to memory-related synaptic plasticity. In rodents, strong membrane depolarization (27, 28) and synaptic activity (29) decrease promoter methylation and increase expression of the plasticity-promoting gene brain-derived neurotrophic factor (Bdnf ). In hippocampal (30) and cortical (31) slice preparations, DNMTi blocks the N-methyl-D-aspartate (NMDA) receptor–dependent, long-term potentiation of excitatory synapses and alters the methylation status of Bdnf and another plasticity-promoting gene, reelin (Reln). Additionally, hippocampal long-term potentiation is impaired and long-term depression is enhanced in mice with genetic forebrain deletion of Dnmt1 and Dnmt3a (20).
Active cytosine methylation also contributes to memory-related changes in synaptic strength by regulating synaptic scaling, a form of homeostatic plasticity involving cell-wide alterations in the abundance of postsynaptic receptors that occurs in response to chronic changes in neuronal activity (32–34). In cultured pyramidal neurons, inhibiting neuronal activity with tetrodotoxin (TTX) alters DNA cytosine methylation and differentially affects the expression of genes encoding proteins involved in demethylation (33, 34). Moreover, DNMTi and DNMT knockdown in cortical pyramidal cells (34) and TET3 knockdown in hippocampal pyramidal cells (33) induce a cell-wide increase in synaptic strength through the upscaling of glutamatergic synapses.
A functional link between DNA methylation and the excitability of the neuronal membrane remains an open question, however. Although synaptic activity mediates input of information within neuronal networks, the output of individual cells is ultimately conveyed by action potentials (APs). A neuron’s propensity for generating an AP from a given input is referred to as its intrinsic membrane excitability (IME). Both passive and active membrane properties determine IME. Passive properties, such as membrane capacitance, input resistance, and resting membrane potential, are primarily determined by cellular morphology and the presence or absence of ion channels in the plasma membrane that are open at rest, also known as leak channels. The active properties of the neuronal membrane are determined by the abundance, spatial distribution, and activity of voltage-gated and Ca2+-activated ion channels.
Memory-related changes in IME, termed intrinsic plasticity, predominantly occur through alterations in active neuronal membrane properties (4). Intrinsic plasticity occurs in response to a broad range of behavioral stimuli in neurons within the hippocampus (35–47) and cortex (48–52) and is thus activity-dependent. Additionally, intrinsic plasticity may contribute to learning-related processes as a form of homeostatic plasticity that occurs in response to chronic changes in neuronal activity (2, 53–55). Ultimately, intrinsic plasticity mediated by a behavioral experience is thought to subsequently modulate the acquisition of new memories (47, 50), a process known as metaplasticity (3, 56, 57). Although intrinsic plasticity is transcriptionally regulated (54, 55, 58, 59), it is unclear whether epigenetic mechanisms are involved.
Given the role of DNA methylation in learning and memory and the extent of genomic cytosines in neurons that may potentially be affected by activity-regulated methylation changes, DNA methylation may regulate adaptations in both synaptic strength and membrane excitability. In parallel to its regulatory contribution to synaptic plasticity and homeostatic synaptic scaling, DNA methylation might modulate the flow of information throughout memory-related networks by fine-tuning the IME of individual neurons. Therefore, we investigated whether there is a role of DNMT activity in regulating the IME of cortical pyramidal neurons.
RESULTS
Prolonged DNMT inhibition increases IME
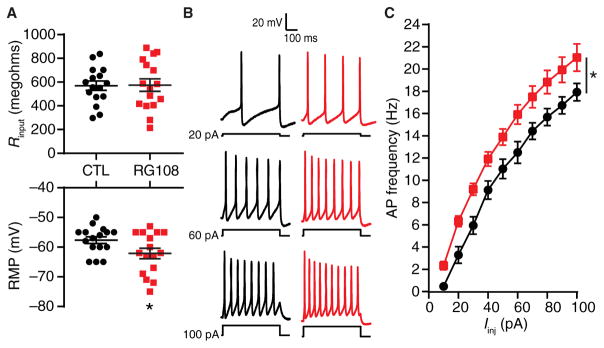
In our initial experiments, we used a pharmacological DNMTi, the small-molecule competitive antagonist RG108, to examine the role of DNMT activity in regulating the IME of cultured rat cortical pyramidal neurons. After a 24-hour exposure to 100 μM RG108, we performed whole-cell current-clamp electrophysiology of visually identified pyramidal neurons. Current-clamp recordings enabled the measurement of membrane potential both at rest and in response to intracellular injections of current. RG108 induced a hyperpolarization of the resting membrane potential (Fig. 1A and Table 1), suggesting a change in the ionic currents open at rest. However, RG108 did not affect whole-cell capacitance (Table 1), input resistance (Fig. 1A and Table 1), or the membrane time constant of recorded cells (Table 1), which indicated that passive neuronal properties were largely unaffected by this pharmacological DNMTi.
Fig. 1. Pharmacological DNMTi increases the membrane excitability of cortical pyramidal neurons.
(A) Input resistances (top) and resting membrane potentials (RMP; bottom) of pyramidal neurons after 24-hour vehicle control [CTL; dimethyl sulfoxide (DMSO)] and RG108 treatment. *P < 0.05, Student’s unpaired t test. (B) Representative recordings of control and RG108-treated neuronal membrane voltage responses to 500-ms-long intracellular current injections over a range of current intensities. (C) Mean AP firing rates evoked over a range of current intensities. Iinj, current injections. *P < 0.05, two-way repeated-measures analysis of variance (RM-ANOVA). (A and C) Graphs show individual cells as well as means ± SEM or means ± SEM from cells pooled from at least three experiments for each condition (CTL, n = 16 cells; RG108, n = 16 cells).
Table 1.
Passive membrane properties of the analyzed neurons exposed to various treatments.
| Figure | Treatment | n | Vm (mV) | Rinput (megohms) | τ(ms) | Cm (pF) |
|---|---|---|---|---|---|---|
| 1 | CTL | 16 | −57.6 ± 1.1 | 570 ± 39 | 62.7 ± 4.7 | 58.3 ± 2.6 |
| RG108 | 16 | −62.1 ± 1.7* | 575 ± 52 | 63.6 ± 5.9 | 66.7 ± 5.6 | |
| 2 | CTL | 7 | −60.1 ± 1.3 | 606 ± 75 | 63.3 ± 5.6 | 65.7 ± 4.4 |
| RG108 + APV | 5 | −60.4 ± 3.2 | 478 ± 80 | 66.4 ± 6.7 | 67.2 ± 8.3 | |
| RG108 + NBQX + APV | 7 | −58.4 ± 2.1 | 527 ± 50 | 67.9 ± 7.4 | 69.2 ± 6.3 | |
| RG108 + TTX | 7 | −58.7 ± 1.9 | 590 ± 96 | 77.9 ± 13.4 | 58.2 ± 5.8 | |
| 3 | CTL | 11 | −59.3 ± 1.2 | 589 ± 59 | 61.1 ± 4.7 | 60.9 ± 3.6 |
| RG108 + ActD | 4 | −59.3 ± 4.1 | 579 ± 153 | 64.1 ± 10.6 | 46.3 ± 2.0 | |
| Scrambled CTL | 6 | −57.3 ± 1.5 | 594 ± 30 | 62.6 ± 4.1 | 67.0 ± 5.4 | |
| Tet1 ASO + RG108 | 7 | −61.0 ± 1.7 | 619 ± 121 | 60.8 ± 5.2 | 52.7 ± 7.2 | |
| 5 | CTL | 10 | −59.0 ± 1.3 | 621 ± 53 | 62.2 ± 4.7 | 55.6 ± 5.1 |
| RG108 | 13 | −62.6 ± 1.2 | 599 ± 59 | 62.9 ± 6.1 | 60.7 ± 3.7 | |
| 6 (A to E) | Scrambled CTL | 12 | −56.8 ± 1.1 | 516 ± 32 | 59.5 ± 3.9 | 55.5 ± 4.2 |
| Dnmt1 ASO | 9 | −58.9 ± 1.2 | 613 ± 83 | 61.0 ± 4.6 | 56.9 ± 7.4 | |
| Dnmt3a ASO | 7 | −60.7 ± 2.0 | 626 ± 61 | 66.6 ± 7.2 | 50.4 ± 3.5 | |
| 6 (F to H) | Scrambled CTL | 8 | −57.0 ± 1.0 | 541 ± 35 | 56.8 ± 4.1 | 60.8 ± 4.8 |
| Dnmt1 ASO | 5 | −58.4 ± 2.0 | 507 ± 22 | 55.0 ± 4.5 | 65.4 ± 11.6 | |
| Dnmt3a ASO | 5 | −63.4 ± 2.0 | 670 ± 71 | 70.3 ± 9.3 | 55.2 ± 8.1 | |
| 7 (A and B, and E and F) | CTL | 8 | −56.4 ± 1.0 | 634 ± 54 | 63.0 ± 5.8 | 50.4 ± 5.0 |
| CTL + apamin | 5 | −58.4 ± 3.0 | 704 ± 41 | 73.3 ± 4.3 | 54.9 ± 5.3 | |
| RG108 | 10 | −62.0 ± 1.4 | 610 ± 68 | 61.6 ± 6.5 | 56.5 ± 2.8 | |
| RG108 + apamin | 4 | −59.8 ± 4.0 | 683 ± 110 | 67.8 ± 5.7 | 58.1 ± 4.0 | |
| 7 (C and D, and G and H) | CTL | 18 | −58.1 ± 1.1 | 570 ± 38 | 62.3 ± 4.2 | 60.3 ± 2.7 |
| CTL + apamin | 5 | −58.4 ± 3.0 | 704 ± 41 | 73.3 ± 4.3 | 54.9 ± 5.3 | |
| RG108 | 19 | −62.7 ± 1.5 | 575 ± 48 | 63.5 ± 5.4 | 67.1 ± 5.1 | |
| RG108 + apamin | 4 | −59.8 ± 4.0 | 683 ± 110 | 67.8 ± 5.7 | 58.1 ± 4.0 |
All values are means ± SEM.
P < 0.05, Student’ t test. n, number of cells; Vm, resting membrane potential; Rinput, input resistance; τ, membrane time constant; Cm, membrane capacitance; ActD, actinomycin D.
To assess active neuronal membrane properties, we injected 500-ms-long depolarizing current pulses of increasing amplitude into the neuronal soma. The membrane potential for all neurons was adjusted to −60 mV between each 500-ms current pulse to control for alterations in resting membrane potential induced by RG108. In cells from both the control and RG108-exposed groups, stronger depolarization resulted in an increased number of evoked APs and an increased rate of AP firing (Fig. 1B). However, neurons exposed to RG108 displayed an increased IME as indicated by an increased evoked AP frequency over the range of delivered current injections (Fig. 1C). These results indicated that inhibition of ongoing DNA cytosine methylation affects the active membrane properties that regulate the input-output relationships and repetitive AP firing behavior of individual cortical pyramidal neurons. The increased IME observed with DNMTi is consistent with the hypothesis that DNA cytosine methylation regulates the abundance, membrane distribution, or activity of voltage-gated or Ca2+-activated ion channels, either through direct or indirect transcriptional control of the genes encoding the ion channel subunits or by changing the expression of genes encoding enzymes that alter ion channel activity through posttranslational modifications.
DNMTi-enhanced IME requires signaling through NMDA receptors
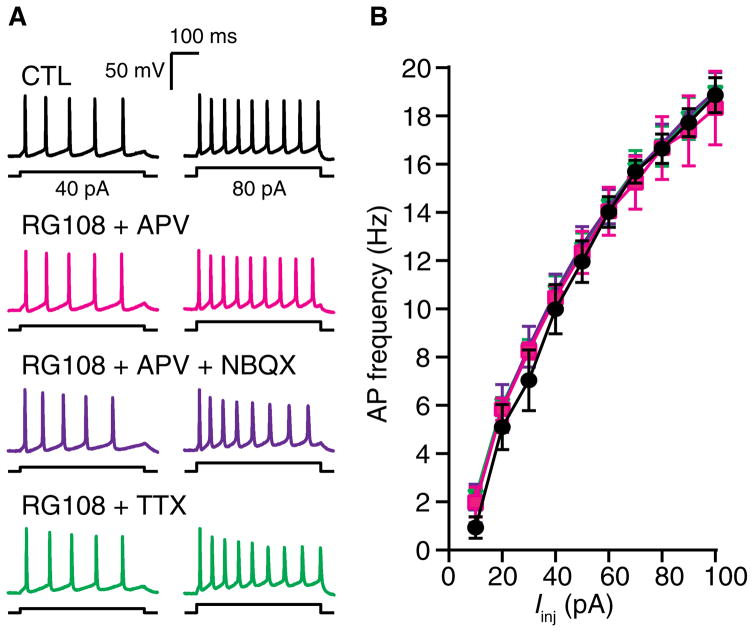
Experience-induced and homeostatic intrinsic plasticity depends on signaling through the NMDA type of glutamate receptors (54, 55, 60–64). Therefore, we examined the underlying neuronal activity required for the increased IME caused by DNMTi. We used 2,3-dioxo-6-nitro-1,2,3,4-tetrahydrobenzo[f]quinoxaline-7-sulfonamide (NBQX) to antagonize AMPA-type glutamate receptors and D,L-2-amino-5-phosphonovaleric acid (APV) NMDA-type glutamate receptors; we used TTX to suppress Na+ channel–dependent AP generation. Cultured cortical neurons were exposed to RG108 in the presence of APV, APV + NBQX, or TTX. At 24 hours, we measured neuronal passive membrane properties in the absence of pharmacological blockers. Exposure to RG108 + APV, RG108 + APV + NBQX, or RG108 + TTX did not affect passive membrane properties (Table 1) or AP firing frequencies over a range of somatic current injections (Fig. 2, A and B). Together, these results indicated that the enhanced IME driven by inhibition of DNA cytosine methylation requires neuronal activity. Furthermore, because the inhibition of NMDA receptors alone with APV blocked DNMTi-mediated changes in IME, our results indicated a role for glutamatergic signaling through NMDA receptors and subsequent Ca2+ influx.
Fig. 2. DNMTi-enhanced excitability requires neuronal activity and NMDA receptor activity.
(A) Representative recordings of voltage responses from pyramidal neurons after a 24-hour exposure to CTL, RG108 + APV, RG108 + NBQX + APV, or RG108 + TTX. (B) Mean AP firing rates evoked over a range of current intensities. Graph shows means ± SEM from cells pooled from at least three experiments for each condition (CTL, n = 7 cells; RG108 + APV, n = 5 cells; RG108 + NBQX + APV, n = 7 cells; RG108 + TTX, n = 7 cells).
DNMTi-enhanced IME depends on gene expression and TET1-mediated DNA demethylation
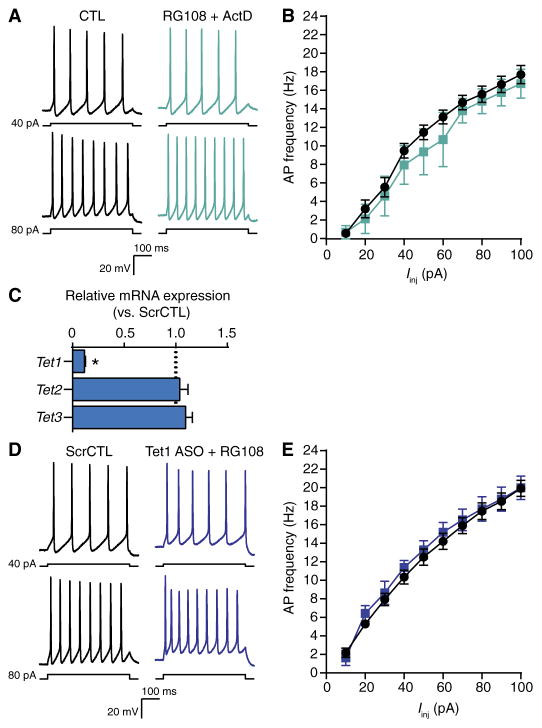
NMDA receptor–mediated Ca2+-dependent signaling is critical for the transcriptional regulation of both learning-related synaptic plasticity and intrinsic plasticity (6, 54, 65–68). Furthermore, pharmacological DNMTi blocks NMDA receptor–dependent changes in gene expression and promoter methylation in slice preparations (30, 31) and in the behaving animal (19, 22, 69). Because DNMTi-enhanced IME required NMDA receptor–mediated signaling (Fig. 2) and because DNA methylation regulates gene expression (12, 70, 71), we hypothesized that gene expression is necessary for IME. To initially address this question, we exposed cultured cortical neurons to RG108 in the presence of the broadly acting transcriptional inhibitor actinomycin D. In pyramidal cells, combined exposure to RG108 and actinomycin D did not affect passive membrane properties (Table 1) or the rate of repetitive AP firing evoked by somatic current pulses (Fig. 3, A and B). These results showed that DNMTi-enhanced IME requires de novo transcription.
Fig. 3. Gene expression and DNA demethylation mediated by TET1 are necessary for DNMTi-enhanced excitability.
(A) Representative recordings of voltage responses from pyramidal neurons after 24-hour exposure to CTL or RG108 + actinomycin D. (B) Mean AP firing rates evoked from increasing current injections. Graph shows means ± SEM from cells pooled from at least three experiments for each condition (CTL, n = 11 cells; RG108 + actinomycin D, n = 4 cells). (C) Relative expression versus scrambled control of Tet1, Tet2, and Tet3 mRNA after treatment with a Tet1-targeting ASO. Data are normalized to the abundance in the scrambled control and are means ± SEM from three experiments (ScrCTL, n = 12 biological replicates; Tet1 ASO, n = 9 biological replicates). *P < 0.05, Mann-Whitney test. (D) Representative evoked membrane voltage responses of cells exposed to scrambled control ASO or Tet1 ASO + 24-hour RG108. (E) Mean AP firing rates evoked with 500-ms current pulses of increasing intensity. Bar graph shows means ± SEM from cells pooled from at least three experiments for each condition (scrambled control, n = 6 cells; Tet1 ASO + RG108, n = 7 cells).
DNA methylation, particularly within gene promoter regions, is considered suppressive of transcription (70). Therefore, we examined whether DNMTi changed gene expression and IME through cytosine demethylation using an antisense oligonucleotide (ASO)–targeted approach to knock down the expression of the Tet1 gene. The dioxygenase TET1 mediates neuronal activity–induced cytosine demethylation and associated alterations in gene expression (13, 24, 26). Previously, we used ASOs delivered to cultured neurons without transfection reagents and showed specific transcript knockdown that persisted a week after a single dose (34). Here, we found a selective ASO-mediated knockdown of Tet1 mRNA, with no effect on Tet2 or Tet3 mRNA abundance (Fig. 3C). Six to 7 days after a single ASO dose, cultured cortical neurons were exposed for 24 hours to RG108. In neurons with the Tet1-specific ASO, RG108 did not affect passive membrane properties (Table 1) and did not change evoked AP firing rates (Fig. 3, D and E). These results indicated that the effect of DNMTi on IME depends on secondary TET1-mediated cytosine demethylation. Together, our results implied that changes in cytosine methylation are sufficient to regulate the expression of voltage- and Ca2+-dependent channels or their associated modifiers and control IME.
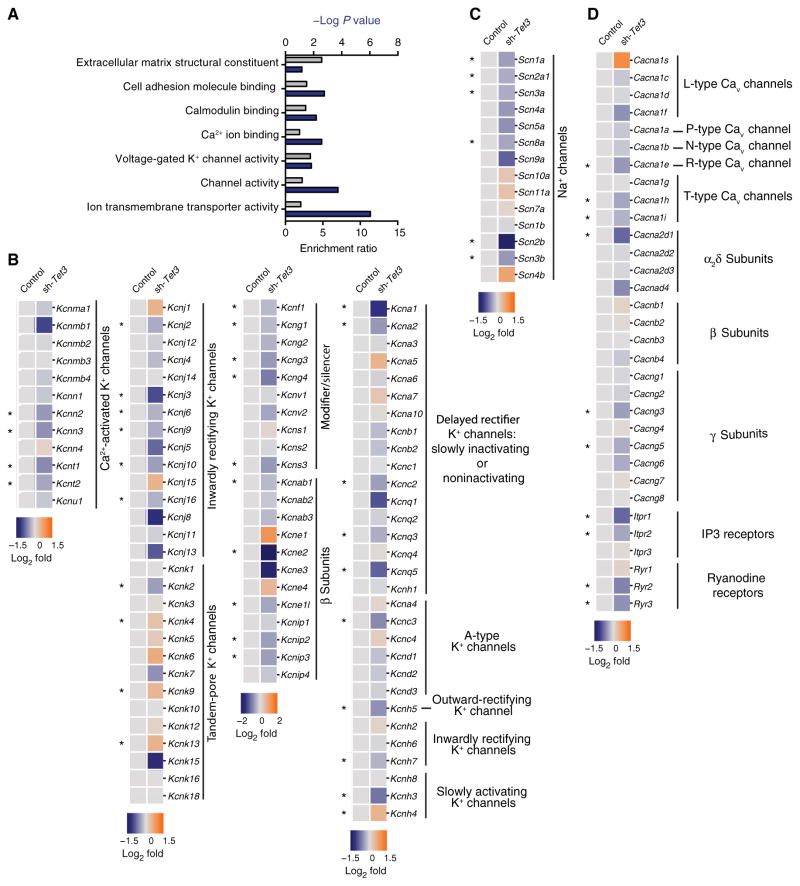
To further support this interpretation, we preformed post hoc analyses of publicly available RNA sequencing (RNA-seq) data from a study that used short hairpin RNA (shRNA)–mediated TET3 knockdown in cultured mouse hippocampal cells (33). These data showed a role for DNA demethylation in the transcriptional regulation of genes encoding proteins involved in synapse structure and synaptic activity. However, whether TET3 knockdown altered the expression of genes encoding ion channels related to IME was not specifically examined. Here, we hypothesized that TET3 knockdown–induced changes in DNA methylation would also alter the expression of genes encoding voltage-gated and Ca2+-activated ion channels. Notably, a gene set enrichment analysis (72–75) of differentially repressed genes in control versus TET3 knockdown neurons revealed a significant overrepresentation of genes ascribed with ion channel activity, specifically voltage-gated K+ channels (Fig. 4A). Furthermore, all subfamilies of K+ channels had specific differentially expressed ion channel–encoding genes (Fig. 4B), including, but not limited to, those encoding Ca2+-activated K+ channels, delayed rectifier K+ channels, and the α and β subunits of voltage-gated K+ channels, both of which regulate the activity of their cognate pore-forming subunits. In addition, TET3 knockdown altered the expression of genes encoding both voltage-gated Na+ and voltage-gated Ca2+ channels (Fig. 4, C and D). These observations highlight the transcriptional potential of cytosine methylation to broadly regulate the expression of genes encoding ion channels that govern neuronal IME.
Fig. 4. Altered DNA methylation status from shRNA-mediated TET3 knockdown regulates the expression of genes associated with ion channel activity.
(A) Gene ontology analysis of differentially down-regulated genes in control versus TET3 knockdown neurons. (B) Heatmap showing the relative expression of all voltage and Ca2+-activated K+ channels in TET3 knockdown neurons. (C) Heatmap showing the relative expression of all voltage-gated Na+ channels in TET3 knockdown neurons. (D) Heat-map showing the relative expression of all voltage-and ligand-gated Ca2+ channels in TET3 knockdown neurons. (A to D) Data are based on RNA-seq analyses of hippocampal neurons expressing an shRNA against Tet3 (sh-Tet3) compared to those expressing a control shRNA (n = 3 samples each; GSE67245). Asterisks indicate genes that exhibited significant differential expression in TET3 knockdown neurons [false discovery rate (FDR) < 0.05].
DNMTi reduces the voltage threshold and decreases the mAHP of single APs
In the rat neocortex, neurons that have overlapping expression patterns of genes encoding ion channels have similar electrophysiological properties (76, 77). However, within large families of ion channels, there is considerable overlap in biophysical properties and electrophysiological function (78, 79), a feature known as degeneracy (80–82). The concept of degeneracy is demonstrated by experimental (83–85) and computational studies (81, 84, 86), showing that multiple conductance distributions can give rise to membrane properties that are biologically indistinguishable. Because cytosine methylation seems to broadly regulate the expression of genes encoding ion channels (Fig. 4), it is difficult to predict the precise transcriptional changes occurring in response to DNMTi or the combinatorial influence on neuronal membrane properties.
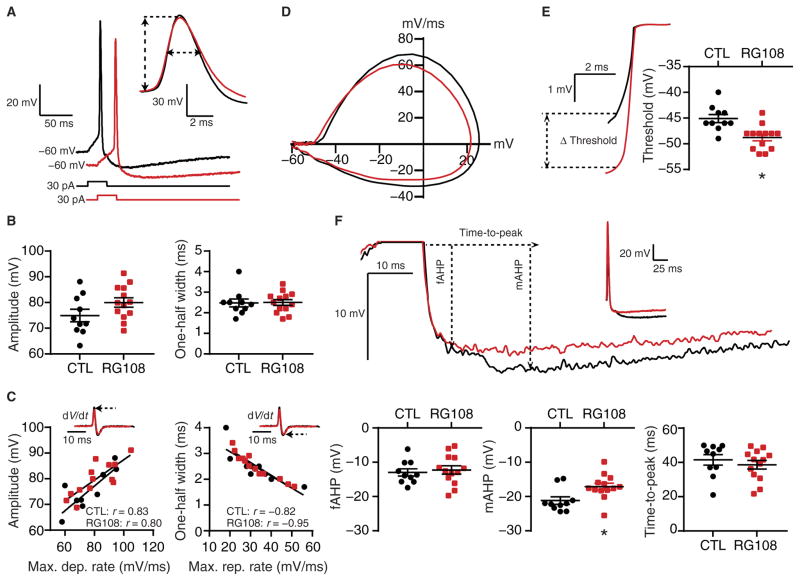
Therefore, we examined how DNMTi affects single APs, which represent the functional output unit of the active neuronal membrane. By measuring waveform characteristics of single APs, we sought to identify potential families of voltage-gated or Ca2+-activated ion channels that may drive DNMTi-enhanced IME. After 24-hour exposure of cultured cortical neurons to RG108, we performed patch-clamp recordings of pyramidal cells. RG108 did not affect passive membrane properties (Table 1). We then elicited single APs using a 30-ms-long suprathreshold current injection from −60 mV (Fig. 5A), measured AP waveform characteristics, and constructed phase plane plots of single APs by calculating the first derivative of the change in membrane potential with respect to time (that is, the rate of change in mV/ms) and plotting those values at specific points in time against the membrane voltage (87). RG108 had no effect on maximal depolarization or repolarization rates, amplitude, or one-half width (Fig. 5B and Table 2), indicating that DNMTi did not change the overall distribution or properties of the voltage-dependent ion channel families that underlie these components of the AP waveform. Additionally, in both the control and RG108-exposed groups, we found correlations between the maximal rate of depolarization and the AP amplitude and the maximal rate of repolarization and the AP one-half width, likely due to the similar contributions of inward Na+ conductances activated during the AP rising phase and outward K+ conductances activated during the AP repolarization phase (Fig. 5C).
Fig. 5. DNMTi alters specific components of the AP waveform.
(A) Representative recordings of single APs elicited with a 30-ms current pulse after 24-hour CTL or RG108 (red) treatment. Inset: Overlay of recordings at an expanded time scale demonstrates the measurement of amplitude and one-half width. (B) AP amplitudes (left) and one-half widths (right) from cortical pyramidal neurons treated with RG108. (C) Examples of rates of change (insets) calculated from recordings in (A) and associations of amplitude versus maximal depolarization rate and one-half width versus maximal repolarization rate. (D) Phase plane plots (dV/dt versus membrane voltage) constructed from example records from (A). (E) Representative recordings at an expanded scale and voltage thresholds (left) and graph of all threshold measurements (right). (F) Representative recordings demonstrate the AHP components measured in pyramidal cells [upper trace at two scales; lower graphs show fast AHP (fAHP) amplitudes, mAHP amplitudes, and time-to-peak AHP measured from CTL and RG108-treated cortical neurons]. Graphs of grouped data show individual cells as well as means ± SEM from cells pooled from at least three experiments for each condition (CTL, n = 10 cells; RG108, n = 13 cells). *P < 0.05, Student’s unpaired t test.
Table 2.
Single AP waveform characteristics of control and RG108-treated neurons.
| Treatment (n) | CTL (10) | RG108 (13) |
|---|---|---|
| AP waveform | ||
| Vthr (mV) | −45.1 ± 0.8 | −48.8 ± 0.7* |
| Maximal depolarization rate (mV/ms) | 75.4 ± 4.2 | 80.9 ± 3.7 |
| Amplitude (mV) | 75.0 ± 2.4 | 80.0 ± 1.8 |
| Maximal repolarization rate (mV/ms) | 32.5 ± 3.4 | 32.5 ± 2.7 |
| One-half width (ms) | 2.5 ± 0.2 | 2.5 ± 0.1 |
| fAHP (mV) | −13.0 ± 1.1 | −12.3 ± 1.2 |
| mAHP (mV) | −21.2 ± 1.1 | −17.2 ± 1.0* |
| Time-to-peak AHP (ms) | 41.5 ± 3.0 | 38.6 ± 2.5 |
These values represent the neurons used for Fig. 5. All values are means ± SEM.
P < 0.05, Student’s t test. n, number of cells; Vthr, voltage threshold.
However, RG108 decreased the voltage threshold (measured as the abrupt increase in dV/dt; Fig. 5, D and E, and Table 2), such that an AP occurred at more negative potentials, indicating a change in the voltage-dependent currents that underlie AP generation. RG108 also altered specific components of the after-hyperpolarization (AHP) potential that follows the repolarization phase. RG108 did not affect the amplitude of the fast AHP but decreased the amplitude of the medium AHP (mAHP) without affecting the time-to-peak AHP amplitude (Fig. 5F and Table 2). These results indicated that pharmacological DNMTi with RG108 induces specific changes in the expression, properties, or distribution of the outward K+ conductances that drive the AHP in cortical pyramidal neurons (88–90). Together, our results suggested that DNMTi increases IME through specific changes in the AP waveform, namely, the voltage threshold and the mAHP, indicating that the transcriptional changes induced by the inhibition of DNA methylation have specific functional consequences at the neuronal membrane.
DNMT knockdown decreases the mAHP, decreases SK channel mRNA, and increases IME
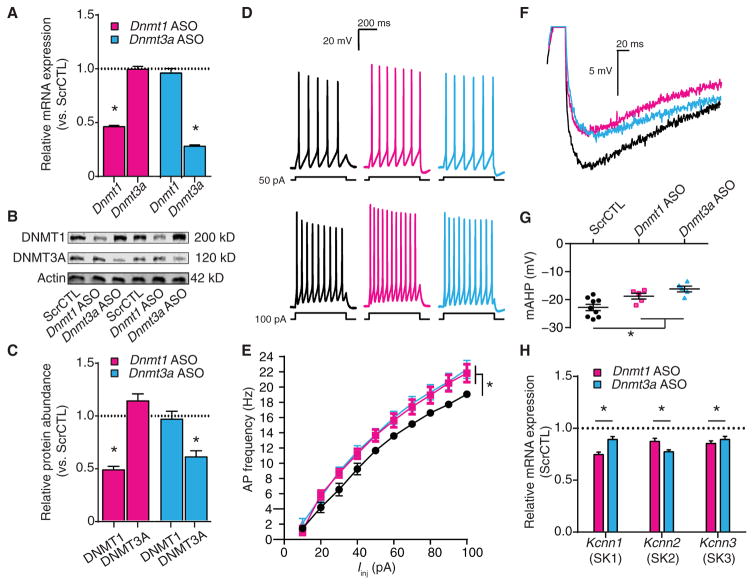
In postmitotic neurons, DNA methylation occurs predominantly through the activity of DNMT1 and DNMT3A (16, 17). Because RG108 is a nonselective inhibitor of DNMT activity (91), we used an ASO-mediated knockdown approach to evaluate the contribution of each methyltransferase on target gene expression and IME. We designed ASOs to specifically reduce the abundance of DNMT1 and DNMT3A (Fig. 6, A to C) (34). After a 6- to 7-day ASO treatment, we recorded passive membrane properties (Table 1) and measured evoked firing rates and single AP waveform characteristics (Table 3). DNMT3A knockdown hyperpolarized the resting membrane potential, whereas DNMT1 knockdown had no effect on resting membrane potential (Table 1). Moreover, knocking down neither DNMT1 nor DNMT3A affected other passive properties (Table 1). However, reducing the abundance of either DNMT1 or DNMT3A increased AP firing rates evoked by current injection (Fig. 6, D and E), suggesting that the decreased abundance of either methyltransferase alone is sufficient to increase IME. Last, individual knockdown of DNMT1 or DNMT3A produced distinct effects on AP waveform characteristics. In contrast to pharmacological DNMTi (Fig. 5), a reduction in either methyltransferase did not affect the voltage threshold (Table 3), suggesting that inhibition of both DNMT1 and DNMT3A is necessary to change the abundance, properties, or distribution of the voltage-dependent channels that underlie AP generation. Similar to pharmacological DNMTi, DNMT1 or DNMT3A knockdown did not affect depolarization or repolarization rates, AP amplitude, or AP one-half widths (Table 3). Last, knocking down only DNMT3A decreased the fast AHP (Table 3), whereas knocking down either DNMT1 or DNMT3A decreased the mAHP (Fig. 6, F and G, and Table 3).
Fig. 6. DNMT1 or DNMT3A knockdown increases excitability, decreases the mAHP, and decreases expression of SK channel–encoding genes.
Neurons were exposed to scrambled control, Dnmt1 ASO, or Dnmt3a ASO. (A) Relative Dnmt1 and Dnmt3a mRNA expression after ASO exposure (scrambled control, n = 9 biological replicates; Dnmt1 ASO, n = 9 biological replicates; Dnmt3a ASO, n = 9 biological replicates). (B and C) Representative immunoblot (B) and relative DNMT1 and DNMT3A protein abundance (C) after ASO exposure (scrambled control, n = 9 biological replicates; Dnmt1 ASO, n = 9 biological replicates; Dnmt3a ASO, n = 8 biological replicates). (D and E) Representative recordings of evoked firing responses (D) and mean firing rates (E) of ASO-treated neurons (scrambled control, n = 12 cells; Dnmt1 ASO, n = 9 cells; Dnmt3a ASO, n = 7 cells). (F) Representative recordings of single APs from ASO-treated pyramidal neurons overlaid to show the mAHP. (G) Graph of mAHP amplitudes after ASO exposure (scrambled control, n = 9 cells; Dnmt1 ASO, n = 5 cells; Dnmt3a ASO, n = 5 cells). (H) Relative transcript expression of the SK channel genes Kcnn1, Kcnn2, and Kcnn3 after ASO exposure (scrambled control, n = 9 biological replicates; Dnmt1 ASO, n = 9 biological replicates; Dnmt3a ASO, n = 9 biological replicates). (A, C, and H) Data are normalized to scrambled control and are means ± SEM from three experiments. (E and G) Graphs show means ± SEM from cells pooled from at least three experiments for each condition. (A, C, G, and H) *P < 0.05, one-way ANOVA followed by Dunnett’s test. (E) *P < 0.05, RM-ANOVA.
Table 3.
Single AP waveform characteristics of scrambled CTL, Dnmt1 ASO–treated, and Dnmt3a ASO–treated neurons in Fig. 6.
| Treatment (n) | Scrambled CTL (9) | Dnmt1 ASO (5) | Dnmt3a ASO (5) |
|---|---|---|---|
| AP waveform | |||
| Vthr (mV) | −45.2 ± 1.1 | −48.4 ± 1.6 | −49.6 ± 1.6 |
| Maximal depolarization rate (mV/ms) | 78.6 ± 4.7 | 81.4 ± 6.5 | 82.0 ± 2.5 |
| Amplitude (mV) | 75.8 ± 3.3 | 79.5 ± 3.1 | 82.1 ± 1.8 |
| Maximal repolarization rate (mV/ms) | 28.8 ± 7.6 | 37.8 ± 5.4 | 31.4 ± 4.5 |
| One-half width (ms) | 2.2 ± 0.1 | 2.2 ± 0.2 | 2.6 ± 1.8 |
| fAHP (mV) | −14.9 ± 0.7 | −12.4 ± 1.0 | −12.1 ± 0.8* |
| mAHP (mV) | −22.8 ± 1.1 | −18.8 ± 1.0* | −16.2 ± 1.0* |
| Time-to-peak AHP (ms) | 37.9 ± 2.2 | 38.6 ± 3.3 | 41.8 ± 1.3 |
All values are means ± SEM.
P < 0.05, one-way ANOVA followed by Dunnett’s test. n, number of cells.
The reduced mAHP observed after DNMT1 or DNMT3A knockdown suggested that the decreased activity of these methyltransferases results in decreased abundance of the outward K+ current–mediating ion channels that are active during AP repolarization. In cortical pyramidal cells, the activity of small-conductance Ca2+-activated K+ (SK) channels partially shapes the mAHP (92, 93). Notably, decreased SK channel abundance, such as that resulting from behavioral experience or genetic or pharmacological manipulation, decreases the mAHP and increases IME (89, 94, 95). We therefore examined the effect of DNMT1 or DNMT3A knockdown on the mRNA abundance of the three subtypes of SK channels that are present in the rat brain (96). Knocking down either DNMT1 or DNMT3A decreased the abundance of all three SK channel mRNAs relative to that in the neurons receiving the scrambled control ASO (Fig. 6H). These results implied that DNA cytosine methylation directly or indirectly regulates the expression or transcript abundance of the entire family of genes encoding SK channels, and effect that could contribute to a decreased mAHP and increased IME resulting from reduced DNMT activity.
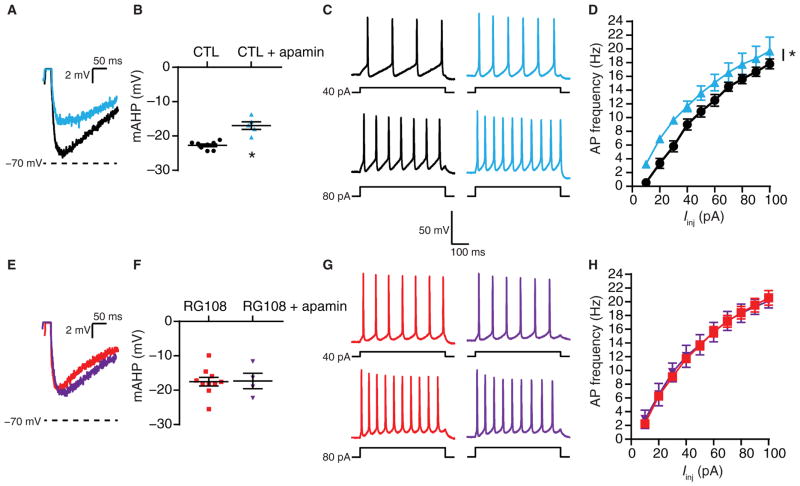
The SK channel–specific blocker apamin is ineffective after DNMTi
If reduced SK channel abundance or activity underlies the increased IME caused by the chronic inhibition of DNA cytosine methylation, then subsequent pharmacological blockade of these channels should not trigger any additional effect changes in AP firing rates and mAHP amplitude above that already produced by DNMTi. Thus, we used apamin, a bee venom toxin that is a selective SK channel blocker (97), to examine the contribution of SK channels to evoked AP firing rates and mAHP amplitudes after DNMTi in cultured pyramidal neurons. Apamin, in the presence or absence of RG108, had no effect on the passive properties of the neurons (Table 1). In control cells, apamin decreased the mAHP amplitude of single APs (Fig. 7, A and B, and Table 4) and increased the evoked AP firing rates compared to control cells recorded in the absence of extracellular apamin (Fig. 7, C and D). In contrast, apamin did not change the mAHP amplitudes of single APs or evoked firing rates of neurons exposed to RG108 (Fig. 7, E to H, and Table 4). These results indicated that reduced SK channel conductance underlies the observed DNMTi-mediated reductions in mAHP amplitude and increased AP firing rates. Collectively, our results suggested that the regulation of SK channel transcript abundance by DNA cytosine methylation contributes to AP waveform characteristics and IME of individual pyramidal neurons.
Fig. 7. Bath application of the SK channel blocker apamin is ineffective after DNMTi.
(A) Representative recordings of single APs from pyramidal neurons exposed to 24-hour CTL or 24-hour CTL + bath application of apa-min, overlaid to show the mAHP amplitude relative to −70 mV. (B) Graph of mAHP amplitudes after CTL or CTL + apamin (CTL, n = 8 cells; CTL + apamin, n = 5 cells). *P < 0.05, Student’s t test. (C and D) Representative evoked voltage responses (C) and mean firing rates (D) of neurons after CTL or CTL + apamin (CTL, n = 18 cells; CTL + apamin, n = 5 cells). *P < 0.05, RM-ANOVA. (E) Representative recordings of single APs from neurons exposed to 24-hour RG108 or 24-hour RG108 + bath application of apamin. Scale is the same as in (A). (F) Graph of mAHP amplitudes after RG108 or RG108 + apamin (RG108, n = 10 cells; RG108 + apamin, n = 4 cells). (G and H) Representative evoked voltage responses (G) and mean firing rates (H) of neurons after RG108 or RG108 + apamin (RG108, n = 19 cells; RG108 + apamin, n = 4 cells). (B, D, F, and H) Graphs show means ± SEM from cells pooled from at least three experiments for each condition.
Table 4.
Single AP waveform characteristics of neurons exposed to CTL versus CTL + apamin and RG108 versus RG108 + apamin in Fig. 7.
| Treatment (n) | CTL (8) | CTL + apamin (5) | RG108 (10) | RG108 + apamin (4) |
|---|---|---|---|---|
| AP waveform | ||||
| Vthr (mV) | −45.1 ± 1.0 | 49.0 ± 0.9 | −49.1 ± 0.6 | −47.3 ± 2.1 |
| Maximal depolarization rate (mV/ms) | 75.0 ± 4.2 | 78.2 ± 3.7 | 80.7 ± 4.5 | 73.0 ± 8.8 |
| Amplitude (mV) | 73.0 ± 2.4 | 78.3 ± 2.5 | 79.0 ± 2.0 | 72.8 ± 4.7 |
| Maximal repolarization rate (mV/ms) | 33.8 ± 3.8 | 28.4 ± 4.5 | 34.0 ± 3.4 | 28.5 ± 4.5 |
| One-half width (ms) | 2.3 ± 0.1 | 2.8 ± 0.4 | 2.4 ± 0.2 | 2.5 ± 0.2 |
| fAHP (mV) | −13.5 ± 0.9 | −10.6 ± 1.2 | −9.4 ± 3.2 | −11.3 ± 2.4 |
| mAHP (mV) | −22.7 ± 0.4 | −17.5 ± 1.3* | 17.0 ± 1.1 | 17.3 ± 2.2 |
| Time-to-peak (ms) | 40.3 ± 3.7 | 37.6 ± 3.0 | 41.1 ± 3.0 | 43.0 ± 2.8 |
All values are means ± SEM.
P < 0.05, Student’s t test. n, number of cells.
DISCUSSION
Dynamic epigenomic modulation regulates the coordinated gene expression profiles necessary for the formation and maintenance of long-term memories resulting from behavioral training (6, 9). Although the modulation of behavior by an experientially altered nuclear readout is thought to be due to the induction of neuronal plasticity, the full spectrum of necessary cellular adaptations and their underlying mechanisms are areas of intense investigation (1, 2, 4). Epigenetic modifications, such as DNA cytosine methylation, regulate forms of synaptic plasticity that involve local modifications in postsynaptic strength, including long-term potentiation (18, 20, 30, 31, 98–100) and long-term depression (20, 26), and forms of homeostatic plasticity, such as synaptic scaling, that involve cell-wide modifications in postsynaptic strength (33, 34). In addition to plasticity at the synapse, transcription-dependent changes in the intrinsic excitability of the neuronal membrane contribute to the cellular adaptations underlying learning and memory (4, 56). Although growing evidence implicates epigenetic mechanisms in intrinsic plasticity (3), a mechanistic understanding of how DNA methylation influences neuronal IME is lacking. This study established activity-dependent DNA cytosine methylation as a critical regulator of neuronal IME. Collectively, our results showed that epigenetic regulation by DNA methylation affects transcript abundance of a family of K+ channels to ultimately shape the AP waveform and repetitive AP firing behavior of individual cortical pyramidal neurons.
Here, we observed that inhibition of DNMT activity by pharmacological antagonism or by ASO-mediated DNMT knockdown increased neuronal IME, evidenced by an increased evoked AP frequency. Notably, DNMT1 or DNMT3A knockdown phenocopied the increased evoked AP frequency observed after exposure to the pharmacological DNMTi RG108 (Figs. 1, B and C, and 6, B and C). This effect contrasts our previous findings that both DNMT1 and DNMT3A knockdown was required to produce glutamatergic synaptic upscaling of equal magnitude to the up-scaling produced by RG108 (34). Together, these studies highlight the important, and perhaps overlapping, roles of DNMT1 and DNMT3A in regulating neuronal adaptations and also suggest that key distinctions may exist in the induction of intrinsic plasticity and synaptic scaling by dynamic DNA cytosine methylation. Future studies are needed to examine whether DNMT1 and DNMT3A converge on the same genes that encode individual or families of ion channels involved in neuronal excitability, yet diverge on the genes encoding proteins involved in glutamatergic signaling and AMPA receptor trafficking.
In further support of our interpretation that distinct yet overlapping mechanisms exist between intrinsic plasticity and synaptic scaling, we showed here that increased IME caused by DNMT inhibition depended on synaptic transmission through both AMPA and NMDA receptors (Fig. 2), whereas previously we found that synaptic upscaling caused by DNMT inhibition was not dependent on NMDA receptor signaling (34). These observations are consistent with other studies showing that homeostatic intrinsic plasticity relies on NMDA receptor signaling, whereas homeostatic synaptic upscaling does not (54, 55, 101, 102). However, an additional layer of complexity is that the molecular mechanisms that regulate homeostatic synaptic downscaling are not inverse of those that regulate homeostatic synaptic upscaling; in particular, downscaling depends on NMDA receptor signaling (103). Thus, future investigations are needed to examine the mechanisms of receptor signaling involved in the regulation of IME and cell-wide synaptic scaling by DNA cytosine methylation.
Our experiments implicated transcriptional repression or mRNA destabilization of SK channels (Fig. 6F) and subsequent reduction in the mAHP amplitude (Figs. 5 to 7) as a probable mechanism underlying the DNMTi-mediated enhancement of IME. Our post hoc analysis of previously published results (33) also revealed a TET3 knockdown–mediated alteration in SK channel transcripts (Fig. 4B), a finding that further reinforces a link between DNA cytosine methylation and the expression of genes encoding SK channels. However, our studies do not exclude the possibility that the activity or abundance of additional families of voltage-or ligand-activated ion channels, or a combination thereof, may be altered by DNMTi. For example, we interpret our finding that DNMTi hyperpolarized the resting membrane potential and decreased the AP threshold as indicative of demethylation-associated secondary transcriptional changes resulting in altered activity or abundance of other ion channel families, such as inwardly rectifying K+ channels (104) and voltage-gated Na+ channels (105). This interpretation is supported by the widespread alterations in ion channel transcripts revealed by our post hoc analyses of neurons in which TET3 was knocked down, which also identified alterations in transcripts for inwardly rectifying K+ channels (Fig. 4B) and voltage-gated Na+ channels (Fig. 4C). Additionally, homeostatic intrinsic plasticity caused by prolonged exposure to TTX is associated with decreased expression of genes encoding voltage-gated K+ channels, Ca2+-activated K+ channels, and tandempore K+ channels (55). Collectively, these observations suggest that dynamic alterations in DNA cytosine methylation broadly affect the expression of genes encoding ion channels.
Nevertheless, our electrophysiological recordings indicated a high level of functional degeneracy at the neuronal membrane, because much of the passive and active membrane properties remained unaffected by DNMTi. Presumably, a portion of the underlying transcriptional changes could be subtractive or antagonistic in nature, resulting in limited functional consequence on AP waveform and repetitive firing behavior. Future studies will be necessary to identify the precise transcriptome-wide changes in the expression of ion channel–encoding genes mediated by reduced DNMT activity and to illuminate the physiological consequences produced by discrete and combinatorial patterns of altered channel expression.
Furthermore, although our studies indicated a link between DNA cytosine methylation, the expression of SK channel–encoding genes, and SK channel activity, we do not exclude the possibility that DNMTi may affect the expression or activity of SK channel modifiers involved in fine-tuning the Ca2+ sensitivity and gating of these channels (106–108). Another possibility is that DNMTi affects SK channel activity through alterations in the expression of genes encoding proteins involved in the accumulation of intracellular Ca2+ by entry through voltage-gated Ca2+ channels or by release from intracellular Ca2+ stores (89, 94, 95). In our post hoc analyses of TET3 knockdown neurons, we found alterations in transcripts for various voltage- and ligand-gated Ca2+ channels (Fig. 4D), suggesting that the expression of these genes falls within the transcription-regulating potential of DNA cytosine methylation. However, future studies will be needed to directly examine the role of cytosine methylation in the transcriptional regulation of genes that encode proteins involved in the modulation of SK channel activity.
We believe that the discovery that DNMT inhibition can increase neuronal IME has three important implications related to neuroepigenetic research. First, from a pathoetiological perspective, our findings suggest new mechanistic hypotheses for neuropsychiatric disorders associated with alterations in intrinsic plasticity. Disorders characterized by alterations in IME might have underlying dysregulation in the expression of ion channel–encoding genes driven by altered cytosine methylation. This is certainly possible for epilepsy (109) and neuropathic pain (110) but may extend to other disorders, such as anxiety (111), depression (112), Alzheimer’s disease (113), and drug addiction (114). Second, interventions or manipulations that rely on broad-spectrum or even isoform-specific modulation of DNMT function will affect neuronal excitability. For example, if alterations in intrinsic excitability are critical to the pathogenesis of a specific disorder, then a genome-wide epigenetic therapeutic approach would be a possible avenue for therapeutics. If not, approaches that use zinc finger nucleases (ZFNs), transcriptional activator–like effectors (TALEs), or the clustered regularly interspaced short palindromic repeats (CRISPR)/ Cas9 system (91, 115) to target DNA methylation enzymes to specific genomic sequences would be preferred to limit unintentional consequences on intrinsic excitability.
Finally, an expanded repertoire of neuronal adaptations regulated by DNA cytosine methylation requires a renewed examination of previous studies in which DNMTi impaired memory formation (18, 19, 22, 69, 116) and maintenance (21). At the time, these findings were attributed to DNMTi-mediated dysregulation of the necessary gene expression profiles required for synaptic plasticity. Although this effect is likely contributory, DNMTi might also produce memory deficits by disrupting the regulation of other neuronal adaptations, such as homeostatic synaptic scaling and intrinsic plasticity. Normally, synaptic strength and IME are carefully tuned, such that neurons within memory circuits act as “high-pass” saliency filters, selectively responding with APs to robust and behaviorally relevant synaptic activity, thus generating an appropriate behavior response through targeted local synaptic plasticity. In contrast, under a highly excitable and upscaled neuronal state, any and all synaptic inputs, including nonsalient and behaviorally irrelevant ones, could potentially activate memory circuits, resulting in saturated and nonselective synaptic plasticity and degraded memory fidelity. Simply put, such an interpretation suggests that DNMTi-mediated IME and glutamatergic upscaling decrease information storage capacity by decreasing a given neuron’s overall signal-to-noise ratio of synaptic activity to AP generation in response to learning-related stimuli (114). Undoubtedly, future investigation will be required to explore these topics among others to obtain a comprehensive understanding of the role of DNA methylation in neuronal information storage.
MATERIALS AND METHODS
Cell culture and drug treatments
Embryonic rat cortical cultures were prepared as previously described (34). Briefly, 12-well tissue culture plates (Corning) were coated overnight at 24°C with poly-L-lysine (50 μg/ml) (Sigma-Aldrich) and rinsed three times with diH2O. For electrophysiological experiments, poly-L-lysine was added to nitric acid–treated glass coverslips (Menzel-Gläser). Embryonic day 18 cortical tissues were dissected in Hanks’ balanced salt solution (Thermo Scientific), digested in papain (Worthington), and dissociated with fire-polished glass pipettes in Neurobasal medium (Thermo Scientific) supplemented with L-glutamine and B-27 (Thermo Scientific). Dissociated cells were filtered with a 70-μm cell strainer and centrifuged. Cells were resuspended in the supplemented Neurobasal medium at a concentration of 250,000/ml and plated. Cells were grown in a humidified CO2 (5%) incubator at 37°C. One-half of the medium was changed at 2 days in vitro (DIV) and every 4 DIV thereafter. Patch-clamp recordings were made after 24-hour drug treatments or 6 to 7 days after a single ASO dose. RNA was collected at 21 DIV after a 6- to 7-day ASO treatment. Cultured neurons were treated for 24 hours with pharmacological agents at the following concentrations: 0.1% DMSO (control), 100 μM RG108 (Calbiochem), 1 μM TTX, 10 μM NBQX, 50 μM APV, and 1.25 μM actinomycin D (Sigma-Aldrich).
ASO design and neuronal treatment
The design, preparation, and use of Dnmt1-, Dnmt3a-, and Tet1-targeting ASOs were conducted as previously described (34). ASOs were 20-mers constructed on a phosphorothioate-modified backbone, containing a central region of 10 2'-deoxynucleotides flanked 5' and 3' by 5 2'-O-methyl (OMe)–modified nucleotide “wings” (“5-10-5 2'-OMe gapmers”). ASO sequences were as follows: scrambled control, 5'-mA-mC-mU-mC-mG-C-A-A-C-T-G-C-C-A-C-mU-mC-mU-mC-mC-3'; Dnmt1, 5'-mC-mC-mC-mU-mU-C-C-C-T-T-T-C-C-C-T-mU-mU-mC-mC-mC-3'; Dnmt3a, 5'-mC-mC-mU-mC-mC-G-G-C-A-T-T-T-C-C-T-mC-mC-mU-mC-mU-3'; and Tet1, mU-mG-mC-mC-mG-T-T-C-A-T-C-T-T-C-C-mA-mC-mC-mU-mG. ASOs were resuspended in phosphate-buffered saline (Life Technologies) to 150 μM, and the stock was added to the culture medium to a final concentration of 1.5 μM.
Whole-cell recordings
Because the density of voltage-dependent currents increases with post-natal age, plateauing about 3 to 5 weeks in neocortical pyramidal cells (117), cultured cortical cells were allowed to grow for 20 to 21 DIV. After drug or ASO treatment, whole-cell patch-clamp recordings were made from visually identified pyramidal neurons using a Zeiss Axiovert 25 inverted microscope. The extracellular solution contained the following: 120 mM NaCl, 3 mM KCl, 15 mM Hepes, 20 mM glucose, 2 mM CaCl2, and 1 mM MgCl2 (pH 7.3). Patch pipettes had resistances of 3 to 5 megohms when filled with the recording solution containing 125 mM K-gluconate, 10 mM KCl, 10 mM Hepes, 10 mM creatine phosphate, 2 mM Mg–adenosine triphosphate, 0.2 mM Na–guanosine triphosphate, and 0.5 mM EGTA (pH 7.3). All recordings were performed at room temperature (21° to 23°C) in the presence of 20 μM 6-cyano-7-nitroquinoxaline-2,3-dione and 50 μM APV to block fast excitatory transmission and 20 μM bicuculline to block fast inhibitory transmission. Bath application of 100 nM apamin (Abcam) was used to block SK channels. Recorded signals were amplified with an Axopatch 200B amplifier (Molecular Devices), filtered at 2 kHz, and digitized at 10 kHz with Digidata 1200 Series (Molecular Devices). Data were compiled and analyzed using Clampfit (Molecular Devices), Microsoft Excel, and Prism (GraphPad Software Inc.). Interspike intervals were measured using Mini Analysis (Synaptosoft) software.
In all experiments, tip potentials were zeroed before membrane-pipette seals were formed. Capacitance and series resistance were determined in voltage-clamp mode upon entering the whole-cell configuration and estimated using a voltage step of −10 mV from a holding potential of −70 mV. Series resistances ranged between 12 and 30 megohms and did not differ among any of the groups. Current-clamp recordings were obtained from cells with stable resting membrane potentials more negative or equal to −50 mV. All recorded potentials were corrected for a −15-mV liquid junction potential. Cell input resistance was determined by injecting a small (20 pA) hyperpolarizing current pulse (500 ms) and calculated by dividing the steady-state voltage response by the current pulse amplitude. The membrane time constant was measured by fitting the membrane potential trajectory during a small hyperpolarizing current injection with a single exponential equation. To evaluate active IME, we somatically injected a series of 500-ms depolarizing current pulses (10-pA steps, 10 to 100 pA) to evoke trains of APs. Because of potential differences in resting membrane potential among treatment groups, all APs were evoked from an adjusted membrane potential of −60 mV. AP frequency was determined by averaging the reciprocal of each interspike interval at a given current pulse for each cell. Single APs were elicited by a 30-ms suprathreshold step from −60 mV, and waveform characteristics were determined by differentiating the spike voltage with respect to time (dV/dt). Plane phase plots were constructed by plotting the first derivative in mV/ms against the membrane voltage (87). Threshold was defined as the membrane voltage corresponding to the abrupt increase in dV/dt. Maximum depolarization and repolarization rates corresponded to the maximum rate of change during the AP upstroke and downstroke, respectively. AP amplitude was measured from the voltage threshold to the peak positive deflection, and one-half width was measured as the duration at the halfway point between the threshold and peak amplitude. The amplitudes of the fast and medium AHPs of single APs were determined, respectively, by measuring the maximal negative deflections in potential 3.5 to 5 ms and 10 to 50 ms after the membrane potential reached the equipotential point of the AP threshold during repolarization.
RNA extraction and qRT-PCR
mRNAwas extracted with the RNeasy Mini Kit (Qiagen), deoxyribonuclease-treated (Qiagen), eluted in 30 μl of EB buffer (Qiagen), and quantified spectrophotometrically using the NanoDrop 200c (Thermo Scientific). Complementary DNA (cDNA) synthesis was performed on 300 ng of input RNA using the iScript cDNA Synthesis Kit (Bio-Rad). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was carried out using a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with either SSO Advanced Universal SYBR Green Supermix (Bio-Rad) and 500 nM of intron-spanning primers (Table 5) or TaqMan Universal Master Mix II, no UNG, and TaqMan gene expression assays (Thermo Scientific) with hydrolysis probes for Gapdh (glyceraldehyde-3-phosphate dehydrogenase) (catalog no. Rn01775763_g1) and Kcnn2 (catalog no. Rn00570910_m1). PCR amplifications were performed in triplicate under the following cycling conditions: 95°C for 30 s, followed by 40 cycles of 95°C for 10 s and 60°C (or 58°C) for 30 s, followed by real-time melt analysis to verify product specificity (SYBR), or 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s (TaqMan). Differential gene expression between samples was determined by the comparative Ct (ΔΔCt) method using Gapdh as an internal control.
Table 5.
Primers used in this study.
| Gene | Sense primer (5′ ≥ 3′) | Antisense primer (5′ ≥ 3′) | Annealing temp (°C) |
|---|---|---|---|
| Gapdh | ACCTTTGATGCTGGGGCTGGC | GGGCTGAGTTGGGATGGGGACT | 58 |
| Dnmt1 | GTGTGCGGGAATGTGCTCGCT | CAGTGGTGGTGGCACAGCGT | 58 |
| Dnmt3A | AGCAAAGTGAGGACCATTACCACCA | TGTGTAGTGGACAGGGAAGCCA | 58 |
| Tet1 | GCCAACCAGGAAGAGGCGACTG | GAGGAAGCCTGCAGGGGACAG | 58 |
| Kcnn1 | AGGTGAATGATCAGGCCAACA | GCTATGAGACTTGGTAGGGCC | 60 |
| Kcnn3 | CACCAACTGAGGGGTGTCAA | GGCTGCCAATCTGCTTTTCC | 60 |
Immunoblots
Cortical neuron cultures were directly homogenized in 2× Laemmli buffer. Protein samples were prepared with 2-mercaptoethanol, heated for 10 min at 95°C, then separated on 7.5% Mini-Protean TGX Gels (Bio-Rad), and transferred to Immobilon-FL PVDF membranes (Millipore) using the Trans-Blot Turbo semidry transfer system (Bio-Rad). The membrane was blocked in LI-COR Odyssey blocking buffer for 1 hour at room temperature and incubated with the appropriate primary antibody. The specific primary antibody treatment parameters were as follows: DNMT1 (Abcam, ab87656; 1:1000, overnight at 4°C), DNMT3A (Abcam, ab2850; 1:200, overnight at 4°C), and actin (Abcam, ab3280; 1:1000, 1 hour at room temperature). After primary antibody treatment, membranes were washed three times in tris-buffered saline with 0.1% Tween (TBS-T), followed by incubation for 1 hour with Alexa Fluor 800–conjugated goat antibodies specific for mouse immunoglobulin G (1:20,000; LI-COR). Membranes were then washed three times in TBS-T, followed by a single wash in TBS, imaged on the LI-COR Odyssey fluorescence imaging system, and quantified using LI-COR Image Studio.
RNA-seq data analysis
To identify voltage- and calcium-gated ion channels homeostatically regulated in response to alterations in DNA methylation, we used publicly available RNA-seq data sets from embryonic hippocampal neurons treated with either control- or Tet3-targeting shRNAs [see (33); GSE67245]. Raw paired-end sequenced reads were downloaded, quality-controlled (FASTQC v0.63, Galaxy), filtered for read quality (FASTX toolkit, Galaxy), and aligned to the mouse genome (mm10 assembly) in Galaxy (118) using TopHat v2.0.14 (with custom settings –read-realign-edit-dist 0 –r 131 –mate-std-dev 64 –no-coverage-search –microexon-search –no-novel-juncs –G iGenomesmm10UCSCRefSeq.GTF –b2-very-sensitive) (119). Gene expression differences between groups were calculated in Galaxy using Cuffdiff v2.2.1 (with custom settings –library-norm-method quartile –dispersion-method per-condition –u –b –c 10 –FDR 0.05) (120). Gene ontology overrepresentation of significantly altered genes was conducted with PANTHER (v10.0) (72, 73), using the Gene Ontology database (06 August 2015 release) (74, 75) and a Bonferroni multiple testing correction.
Statistics
Results are means ± SEM. An unpaired Student’s t or Mann-Whitney test was used when comparing two groups. A one-way ANOVA was used when comparing more than two groups. Differences in AP frequency versus current injection were assessed with two-way RM-ANOVAs. The threshold for significance was P < 0.05 for all analyses.
Footnotes
Author contributions: J.P.M., M.C.G.-K., J.D.S., and J.J.H. contributed to designing the experiments and interpreting the results. S.P. designed the ASOs used in the study. J.P.M., M.C.G.-K., J.A.B., and S.K.S. executed the experiments and analyzed data. J.P.M., M.G.-K., J.J.H., and J.D.S. wrote the manuscript.
Competing interests: The authors declare that they have no competing financial interests.
Data and materials availability: The publicly available RNA-seq data set used in this work can be accessed at the Gene Expression Omnibus (GSE67245).
REFERENCES AND NOTES
- 1.Kim SJ, Linden DJ. Ubiquitous plasticity and memory storage. Neuron. 2007;56:582–592. doi: 10.1016/j.neuron.2007.10.030. [DOI] [PubMed] [Google Scholar]
- 2.Nelson SB, Turrigiano GG. Strength through diversity. Neuron. 2008;60:477–482. doi: 10.1016/j.neuron.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzman-Karlsson MC, Meadows JP, Gavin CF, Hablitz JJ, Sweatt JD. Transcriptional and epigenetic regulation of Hebbian and non-Hebbian plasticity. Neuropharmacology. 2014;80:3–17. doi: 10.1016/j.neuropharm.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W, Linden DJ. The other side of the engram: Experience-driven changes in neuronal intrinsic excitability. Nat Rev Neurosci. 2003;4:885–900. doi: 10.1038/nrn1248. [DOI] [PubMed] [Google Scholar]
- 5.McClung CA, Nestler EJ. Neuroplasticity mediated by altered gene expression. Neuropsychopharmacology. 2008;33:3–17. doi: 10.1038/sj.npp.1301544. [DOI] [PubMed] [Google Scholar]
- 6.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 9.Zovkic IB, Guzman-Karlsson MC, Sweatt JD. Epigenetic regulation of memory formation and maintenance. Learn Mem. 2013;20:61–74. doi: 10.1101/lm.026575.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graff J, Kim D, Dobbin MM, Tsai L-H. Epigenetic regulation of gene expression in physiological and pathological brain processes. Physiol Rev. 2011;91:603–649. doi: 10.1152/physrev.00012.2010. [DOI] [PubMed] [Google Scholar]
- 11.Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, Zhang K, Ming G-l, Gao Y, Song H. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Halder R, Hennion M, Vidal RO, Shomroni O, Rahman R-U, Rajput A, Centeno TP, van Bebber F, Capece V, Garcia Vizcaino JC, Schuetz A-L, Burkhardt S, Benito E, Navarro Sala M, Javan SB, Haass C, Schmid B, Fischer A, Bonn S. DNA methylation changes in plasticity genes accompany the formation and maintenance of memory. Nat Neurosci. 2016;19:102–110. doi: 10.1038/nn.4194. [DOI] [PubMed] [Google Scholar]
- 13.Guo JU, Su Y, Zhong C, Ming G, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, Verdine GL. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. doi: 10.1021/bi00110a002. [DOI] [PubMed] [Google Scholar]
- 15.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsycho-pharmacology. 2013;38:23–38. doi: 10.1038/npp.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goto K, Numata M, Komura J-I, Ono T, Bestor TH, Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 17.Inano K, Suetake I, Ueda T, Miyake Y, Nakamura M, Okada M, Tajima S. Maintenance-type DNA methyltransferase is highly expressed in post-mitotic neurons and localized in the cytoplasmic compartment. J Biochem. 2000;128:315–321. doi: 10.1093/oxfordjournals.jbchem.a022755. [DOI] [PubMed] [Google Scholar]
- 18.Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of bdnf gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nat Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, Tahir A, Sweatt JD. DNA methylation regulates associative reward learning. Nat Neurosci. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaPlant Q, Vialou V, Covington HE, III, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming G-l, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, Wei W, Zhao Q-Y, Widagdo J, Baker-Andresen D, Flavell CR, D’Alessio A, Zhang Y, Bredy TW. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proc Natl Acad Sci USA. 2014;111:7120–7125. doi: 10.1073/pnas.1318906111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai L-H. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 28.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, Fan G, Sun YE. DNA methylation-related chromatin remodeling in activity-dependent Bdnf gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 29.Nelson ED, Kavalali ET, Monteggia LM. Activity-dependent suppression of miniature neurotransmission through the regulation of DNA methylation. J Neurosci. 2008;28:395–406. doi: 10.1523/JNEUROSCI.3796-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levenson JM, Roth TL, Lubin FD, Miller CA, Huang I-C, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- 31.Sui L, Wang Y, Ju L-H, Chen M. Epigenetic regulation of reelin and brain-derived neurotrophic factor genes in long-term potentiation in rat medial prefrontal cortex. Neurobiol Learn Mem. 2012;97:425–440. doi: 10.1016/j.nlm.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Turrigiano GG. The self-tuning neuron: Synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu H, Su Y, Shin J, Zhong C, Guo JU, Weng Y-L, Gao F, Geschwind DH, Coppola G, Ming G-l, Song H. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat Neurosci. 2015;18:836–843. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meadows JP, Guzman-Karlsson MC, Phillips S, Holleman C, Posey JL, Day JJ, Hablitz JJ, Sweatt JD. DNA methylation regulates neuronal glutamatergic synaptic scaling. Sci Signal. 2015;8:ra61. doi: 10.1126/scisignal.aab0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coulter DA, Lo Turco JJ, Kubota M, Disterhoft JF, Moore JW, Alkon DL. Classical conditioning reduces amplitude and duration of calcium-dependent after-hyperpolarization in rabbit hippocampal pyramidal cells. J Neurophysiol. 1989;61:971–981. doi: 10.1152/jn.1989.61.5.971. [DOI] [PubMed] [Google Scholar]
- 36.Disterhoft JF, Coulter DA, Alkon DL. Conditioning-specific membrane changes of rabbit hippocampal neurons measured in vitro. Proc Natl Acad Sci USA. 1986;83:2733–2737. doi: 10.1073/pnas.83.8.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Disterhoft JF, Golden DT, Read HL, Coulter DA, Alkon DL. AHP reductions in rabbit hippocampal neurons during conditioning correlate with acquisition of the learned response. Brain Res. 1988;462:118–125. doi: 10.1016/0006-8993(88)90593-8. [DOI] [PubMed] [Google Scholar]
- 38.Farmer GE, Thompson LT. Learning-dependent plasticity of hippocampal CA1 pyramidal neuron postburst afterhyperpolarizations and increased excitability after inhibitory avoidance learning depend upon basolateral amygdala inputs. Hippocampus. 2012;22:1703–1719. doi: 10.1002/hipo.22005. [DOI] [PubMed] [Google Scholar]
- 39.Kaczorowski CC, Disterhoft JF. Memory deficits are associated with impaired ability to modulate neuronal excitability in middle-aged mice. Learn Mem. 2009;16:362–366. doi: 10.1101/lm.1365609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matthews EA, Linardakis JM, Disterhoft JF. The fast and slow afterhyperpolarizations are differentially modulated in hippocampal neurons by aging and learning. J Neurosci. 2009;29:4750–4755. doi: 10.1523/JNEUROSCI.0384-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKay BM, Matthews EA, Oliveira FA, Disterhoft JF. Intrinsic neuronal excitability is reversibly altered by a single experience in fear conditioning. J Neurophysiol. 2009;102:2763–2770. doi: 10.1152/jn.00347.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moyer JR, Jr, Thompson LT, Disterhoft JF. Trace eyeblink conditioning increases CA1 excitability in a transient and learning-specific manner. J Neurosci. 1996;16:5536–5546. doi: 10.1523/JNEUROSCI.16-17-05536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oh MM, Kuo AG, Wu WW, Sametsky EA, Disterhoft JF. Watermaze learning enhances excitability of CA1 pyramidal neurons. J Neurophysiol. 2003;90:2171–2179. doi: 10.1152/jn.01177.2002. [DOI] [PubMed] [Google Scholar]
- 44.Ohno M, Sametsky EA, Silva AJ, Disterhoft JF. Differential effects of αCaMKII mutation on hippocampal learning and changes in intrinsic neuronal excitability. Eur J Neurosci. 2006;23:2235–2240. doi: 10.1111/j.1460-9568.2006.04746.x. [DOI] [PubMed] [Google Scholar]
- 45.Song C, Detert JA, Sehgal M, Moyer JR., Jr Trace fear conditioning enhances synaptic and intrinsic plasticity in rat hippocampus. J Neurophysiol. 2012;107:3397–3408. doi: 10.1152/jn.00692.2011. [DOI] [PubMed] [Google Scholar]
- 46.Thompson LT, Moyer JR, Jr, Disterhoft JF. Transient changes in excitability of rabbit CA3 neurons with a time course appropriate to support memory consolidation. J Neurophysiol. 1996;76:1836–1849. doi: 10.1152/jn.1996.76.3.1836. [DOI] [PubMed] [Google Scholar]
- 47.Zelcer I, Cohen H, Richter-Levin G, Lebiosn T, Grossberger T, Barkai E. A cellular correlate of learning-induced metaplasticity in the hippocampus. Cereb Cortex. 2006;16:460–468. doi: 10.1093/cercor/bhi125. [DOI] [PubMed] [Google Scholar]
- 48.Brons JF, Woody CD. Long-term changes in excitability of cortical neurons after Pavlovian conditioning and extinction. J Neurophysiol. 1980;44:605–615. doi: 10.1152/jn.1980.44.3.605. [DOI] [PubMed] [Google Scholar]
- 49.Cohen-Matsliah SI, Rosenblum K, Barkai E. Olfactory-learning abilities are correlated with the rate by which intrinsic neuronal excitability is modulated in the piriform cortex. Eur J Neurosci. 2009;30:1339–1348. doi: 10.1111/j.1460-9568.2009.06894.x. [DOI] [PubMed] [Google Scholar]
- 50.Saar D, Grossman Y, Barkai E. Reduced after-hyperpolarization in rat piriform cortex pyramidal neurons is associated with increased learning capability during operant conditioning. Eur J Neurosci. 1998;10:1518–1523. doi: 10.1046/j.1460-9568.1998.00149.x. [DOI] [PubMed] [Google Scholar]
- 51.Santini E, Quirk GJ, Porter JT. Fear conditioning and extinction differentially modify the intrinsic excitability of infralimbic neurons. J Neurosci. 2008;28:4028–4036. doi: 10.1523/JNEUROSCI.2623-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Song C, Ehlers VL, Moyer JR., Jr Trace fear conditioning differentially modulates intrinsic excitability of medial prefrontal cortex–basolateral complex of amygdala projection neurons in infralimbic and prelimbic cortices. J Neurosci. 2015;35:13511–13524. doi: 10.1523/JNEUROSCI.2329-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desai NS, Rutherford LC, Turrigiano GG. Plasticity in the intrinsic excitability of cortical pyramidal neurons. Nat Neurosci. 1999;2:515–520. doi: 10.1038/9165. [DOI] [PubMed] [Google Scholar]
- 54.Lee KY, Chung HJ. NMDA receptors and L-type voltage-gated Ca2+ channels mediate the expression of bidirectional homeostatic intrinsic plasticity in cultured hippocampal neurons. Neuroscience. 2014;277:610–623. doi: 10.1016/j.neuroscience.2014.07.038. [DOI] [PubMed] [Google Scholar]
- 55.Lee KY, Royston SE, Vest MO, Ley DJ, Lee S, Bolton EC, Chung HJ. N-methyl-D-aspartate receptors mediate activity-dependent down-regulation of potassium channel genes during the expression of homeostatic intrinsic plasticity. Mol Brain. 2015;8:4. doi: 10.1186/s13041-015-0094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sehgal M, Song C, Ehlers VL, Moyer JR., Jr Learning to learn—Intrinsic plasticity as a metaplasticity mechanism for memory formation. Neurobiol Learn Mem. 2013;105:186–199. doi: 10.1016/j.nlm.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Abraham WC. Metaplasticity: Tuning synapses and networks for plasticity. Nat Rev Neurosci. 2008;9:387. doi: 10.1038/nrn2356. [DOI] [PubMed] [Google Scholar]
- 58.Benito E, Barco A. CREBs control of intrinsic and synaptic plasticity: Implications or CREB-dependent memory models. Trends Neurosci. 2010;33:230–240. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 59.Thoby-Brisson M, Simmers J. Transition to endogenous bursting after long-term decentralization requires de novo transcription in a critical time window. J Neurophysiol. 2000;84:596–599. doi: 10.1152/jn.2000.84.1.596. [DOI] [PubMed] [Google Scholar]
- 60.Ganguly K, Kiss L, Poo M-m. Enhancement of presynaptic neuronal excitability by correlated presynaptic and postsynaptic spiking. Nat Neurosci. 2000;3:1018–1026. doi: 10.1038/79838. [DOI] [PubMed] [Google Scholar]
- 61.Daoudal G, Hanada Y, Debanne D. Bidirectional plasticity of excitatory post-synaptic potential (EPSP)-spike coupling in CA1 hippocampal pyramidal neurons. Proc Natl Acad Sci USA. 2002;99:14512–14517. doi: 10.1073/pnas.222546399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quirk MC, Blum KI, Wilson MA. Experience-dependent changes in extracellular spike amplitude may reflect regulation of dendritic action potential back-propagation in rat hippocampal pyramidal cells. J Neurosci. 2001;21:240–248. doi: 10.1523/JNEUROSCI.21-01-00240.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frick A, Magee J, Johnston D. LTP is accompanied by an enhanced local excitability of pyramidal neuron dendrites. Nat Neurosci. 2004;7:126–135. doi: 10.1038/nn1178. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z, Xu N-l, Wu C-p, Duan S, Poo M-m. Bidirectional changes in spatial dendritic integration accompanying long-term synaptic modifications. Neuron. 2003;37:463–472. doi: 10.1016/s0896-6273(02)01189-3. [DOI] [PubMed] [Google Scholar]
- 65.Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: A Ca2+- and stimulus duration–dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- 66.Dash PK, Karl KA, Colicos MA, Prywes R, Kandel ER. cAMP response element-binding protein is activated by Ca2+/calmodulin- as well as cAMP-dependent protein kinase. Proc Natl Acad Sci USA. 1991;88:5061–5065. doi: 10.1073/pnas.88.11.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Faber ESL. Functional interplay between NMDA receptors, SK channels and voltage-gated Ca2+ channels regulates synaptic excitability in the medial prefrontal cortex. J Physiol. 2010;588:1281–1292. doi: 10.1113/jphysiol.2009.185645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J, Kang N, Jiang L, Nedergaard M, Kang J. Activity-dependent long-term potentiation of intrinsic excitability in hippocampal CA1 pyramidal neurons. J Neurosci. 2005;25:1750–1760. doi: 10.1523/JNEUROSCI.4217-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 70.Jones PA. Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 71.Wu H, Zhang Y. Reversing DNA methylation: Mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence–function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G Gene Ontology Consortium. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015;43:D1049–D1056. doi: 10.1093/nar/gku1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khazen G, Hill SL, Schürmann F, Markram H. Combinatorial expression rules of ion channel genes in juvenile rat (Rattus norvegicus) neocortical neurons. PLOS One. 2012;7:e34786. doi: 10.1371/journal.pone.0034786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Toledo-Rodriguez M, Blumenfeld B, Wu C, Luo J, Attali B, Goodman P, Markram H. Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb Cortex. 2004;14:1310–1327. doi: 10.1093/cercor/bhh092. [DOI] [PubMed] [Google Scholar]
- 78.Coetzee WA, Amarillo Y, Chiu J, Chow A, Lau D, McCormack T, Moreno H, Nadal MS, Ozaita A, Pountney D, Saganich M, Vega-Saenz de Miera E, Rudy B. Molecular diversity of K+ channels. Ann N Y Acad Sci. 1999;868:233–285. doi: 10.1111/j.1749-6632.1999.tb11293.x. [DOI] [PubMed] [Google Scholar]
- 79.Hille B, editor. Ion Channels of Excitable Membranes. Sinauer Associates; Sunderland, MA: 2001. p. 814. [Google Scholar]
- 80.Edelman GM, Gally JA. Degeneracy and complexity in biological systems. Proc Natl Acad Sci USA. 2001;98:13763–13768. doi: 10.1073/pnas.231499798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Leary T, Williams AH, Caplan JS, Marder E. Correlations in ion channel expression emerge from homeostatic tuning rules. Proc Natl Acad Sci USA. 2013;110:E2645–E2654. doi: 10.1073/pnas.1309966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tononi G, Sporns O, Edelman GM. Measures of degeneracy and redundancy in biological networks. Proc Natl Acad Sci USA. 1999;96:3257–3262. doi: 10.1073/pnas.96.6.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goldman MS, Golowasch J, Marder E, Abbott LF. Global structure, robustness, and modulation of neuronal models. J Neurosci. 2001;21:5229–5238. doi: 10.1523/JNEUROSCI.21-14-05229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taylor AL, Hickey TJ, Prinz AA, Marder E. Structure and visualization of high-dimensional conductance spaces. J Neurophysiol. 2006;96:891–905. doi: 10.1152/jn.00367.2006. [DOI] [PubMed] [Google Scholar]
- 85.Schulz DJ, Goaillard J-M, Marder E. Variable channel expression in identified single and electrically coupled neurons in different animals. Nat Neurosci. 2006;9:356–362. doi: 10.1038/nn1639. [DOI] [PubMed] [Google Scholar]
- 86.Golowasch J, Goldman MS, Abbott LF, Marder E. Failure of averaging in the construction of a conductance-based neuron model. J Neurophysiol. 2002;87:1129–1131. doi: 10.1152/jn.00412.2001. [DOI] [PubMed] [Google Scholar]
- 87.Jenerick H. Phase plane trajectories of the muscle spike potential. Biophys J. 1963;3:363–377. doi: 10.1016/s0006-3495(63)86827-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sah P, Faber ESL. Channels underlying neuronal calcium-activated potassium currents. Prog Neurobiol. 2002;66:345–353. doi: 10.1016/s0301-0082(02)00004-7. [DOI] [PubMed] [Google Scholar]
- 89.Stocker M. Ca2+-activated K+ channels: Molecular determinants and function of the SK family. Nat Rev Neurosci. 2004;5:758–770. doi: 10.1038/nrn1516. [DOI] [PubMed] [Google Scholar]
- 90.Gulledge AT, Stuart GJ. Cholinergic inhibition of neocortical pyramidal neurons. J Neurosci. 2005;25:10308–10320. doi: 10.1523/JNEUROSCI.2697-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kennedy AJ, Sweatt JD. Drugging the methylome: DNA methylation and memory. Crit Rev Biochem Mol Biol. 2016;51:185–194. doi: 10.3109/10409238.2016.1150958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Abel HJ, Lee JCF, Callaway JC, Foehring RC. Relationships between in-tracellular calcium and afterhyperpolarizations in neocortical pyramidal neurons. J Neurophysiol. 2004;91:324–335. doi: 10.1152/jn.00583.2003. [DOI] [PubMed] [Google Scholar]
- 93.Lorenzon NM, Foehring RC. Relationship between repetitive firing and afterhyperpolarizations in human neocortical neurons. J Neurophysiol. 1992;67:350–363. doi: 10.1152/jn.1992.67.2.350. [DOI] [PubMed] [Google Scholar]
- 94.Adelman JP, Maylie J, Sah P. Small-conductance Ca2+-activated K+ channels: Form and function. Annu Rev Physiol. 2012;74:245–269. doi: 10.1146/annurev-physiol-020911-153336. [DOI] [PubMed] [Google Scholar]
- 95.Faber ESL. Functions and modulation of neuronal SK channels. Cell Biochem Biophys. 2009;55:127–139. doi: 10.1007/s12013-009-9062-7. [DOI] [PubMed] [Google Scholar]
- 96.Stocker M, Pedarzani P. Differential distribution of three Ca2+-activated K+ channel subunits, SK1, SK2, and SK3, in the adult rat central nervous system. Mol Cell Neurosci. 2000;15:476–493. doi: 10.1006/mcne.2000.0842. [DOI] [PubMed] [Google Scholar]
- 97.Blatz AL, Magleby KL. Single apamin-blocked Ca-activated K+ channels of small conductance in cultured rat skeletal muscle. Nature. 1986;323:718–720. doi: 10.1038/323718a0. [DOI] [PubMed] [Google Scholar]
- 98.Golshani P, Hutnick L, Schweizer F, Fan G. Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat Syst. 2005;3:227–233. doi: 10.1017/S1472928807000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morris MJ, Adachi M, Na ES, Monteggia LM. Selective role for DNMT3a in learning and memory. Neurobiol Learn Mem. 2014;115:30–37. doi: 10.1016/j.nlm.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Thiagarajan TC, Lindskog M, Malgaroli A, Tsien RW. LTP and adaptation to inactivity: Overlapping mechanisms and implications for metaplasticity. Neuropharmacology. 2007;52:156–175. doi: 10.1016/j.neuropharm.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 102.Thiagarajan TC, Lindskog M, Tsien RW. Adaptation to synaptic inactivity in hip-pocampal neurons. Neuron. 2005;47:725–737. doi: 10.1016/j.neuron.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 103.Siddoway B, Hou H, Xia H. Molecular mechanisms of homeostatic synaptic down-scaling. Neuropharmacology. 2014;78:38–44. doi: 10.1016/j.neuropharm.2013.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hibino H, Inanobe A, Furutani K, Murakami S, Findlay I, Kurachi Y. Inwardly rectifying potassium channels: Their structure, function, and physiological roles. Physiol Rev. 2010;90:291–366. doi: 10.1152/physrev.00021.2009. [DOI] [PubMed] [Google Scholar]
- 105.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bildl W, Strassmaier T, Thurm H, Andersen J, Eble S, Oliver D, Knipper M, Mann M, Schulte U, Adelman JP, Fakler B. Protein kinase CK2 is coassembled with small conductance Ca2+-activated K+ channels and regulates channel gating. Neuron. 2004;43:847–858. doi: 10.1016/j.neuron.2004.08.033. [DOI] [PubMed] [Google Scholar]
- 107.Allen D, Fakler B, Maylie J, Adelman JP. Organization and regulation of small conductance Ca2+-activated K+ channel multiprotein complexes. J Neurosci. 2007;27:2369–2376. doi: 10.1523/JNEUROSCI.3565-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schumacher MA, Rivard AF, Bächinger HP, Adelman JP. Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 109.Lubin FD. Epileptogenesis: Can the science of epigenetics give us answers? Epilepsy Curr. 2012;12:105–110. doi: 10.5698/1535-7511-12.3.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rahn EJ, Guzman-Karlsson MC, Sweatt JD. Cellular, molecular, and epigenetic mechanisms in non-associative conditioning: Implications for pain and memory. Neurobiol Learn Mem. 2013;105:133–150. doi: 10.1016/j.nlm.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rau AR, Chappell AM, Butler TR, Ariwodola OJ, Weiner JL. Increased basolateral amygdala pyramidal cell excitability may contribute to the anxiogenic phenotype induced by chronic early-life stress. J Neurosci. 2015;35:9730–9740. doi: 10.1523/JNEUROSCI.0384-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Friedman JJ, Walsh AK, Juarez B, Ku SM, Chaudhury D, Wang J, Li X, Dietz DM, Pan N, Vialou VF, Neve RL, Yue Z, Han M-H. Enhancing depression mechanisms in midbrain dopamine neurons achieves homeostatic resilience. Science. 2014;344:313–319. doi: 10.1126/science.1249240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Santos SF, Pierrot N, Octave JN. Network excitability dysfunction in Alzheimer’s disease: Insights from in vitro and in vivo models. Rev Neurosci. 2010;21:153–171. doi: 10.1515/revneuro.2010.21.3.153. [DOI] [PubMed] [Google Scholar]
- 114.Kourrich S, Calu DJ, Bonci A. Intrinsic plasticity: An emerging player in addiction. Nat Rev Neurosci. 2015;16:173–184. doi: 10.1038/nrn3877. [DOI] [PubMed] [Google Scholar]
- 115.Day JJ, Kennedy AJ, Sweatt JD. DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annu Rev Pharmacol Toxicol. 2015;55:591–611. doi: 10.1146/annurev-pharmtox-010814-124527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Han J, Li Y, Wang D, Wei C, Yang X, Sui N. Effect of 5-aza-2-deoxycytidine microinjecting into hippocampus and prelimbic cortex on acquisition and retrieval of cocaine-induced place preference in C57BL/6 mice. Eur J Pharmacol. 2010;642:93–98. doi: 10.1016/j.ejphar.2010.05.050. [DOI] [PubMed] [Google Scholar]
- 117.Guan D, Horton LR, Armstrong WE, Foehring RC. Postnatal development of A-type and Kv1- and Kv2-mediated potassium channel currents in neocortical pyramidal neurons. J Neurophysiol. 2011;105:2976–2988. doi: 10.1152/jn.00758.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goecks J, Nekrutenko A, Taylor J Galaxy Team. Galaxy: A comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]