Abstract
As more clinically relevant cancer genes are identified, comprehensive diagnostic approaches are needed to match patients to therapies, raising the challenge of optimization and analytical validation of assays that interrogate millions of bases of cancer genomes altered by multiple mechanisms. Here we describe a test based on massively parallel DNA sequencing to characterize base substitutions, short insertions and deletions (indels), copy number alterations and selected fusions across 287 cancer-related genes from routine formalin-fixed and paraffin-embedded (FFPE) clinical specimens. We implemented a practical validation strategy with reference samples of pooled cell lines that model key determinants of accuracy, including mutant allele frequency, indel length and amplitude of copy change. Test sensitivity achieved was 95–99% across alteration types, with high specificity (positive predictive value >99%). We confirmed accuracy using 249 FFPE cancer specimens characterized by established assays. Application of the test to 2,221 clinical cases revealed clinically actionable alterations in 76% of tumors, three times the number of actionable alterations detected by current diagnostic tests.
Systemic cancer treatment is undergoing a fundamental change, moving away from a paradigm in which histopathologically defined disease is treated primarily with cytotoxic chemotherapy, toward the use of molecularly targeted drugs prescribed to selected subsets of patients across multiple tumor types. Targeted therapies promise to be both safer and more efficacious as demonstrated by the success of trastuzumab (Herceptin) in ERBB2 (also known as HER2)-amplified breast cancer, imatinib (Gleevec) in Philadelphia chromosome– positive chronic myelogenous leukemia, erlotinib (Tarceva) in EGFR-mutated non-small cell lung cancer and vemurafenib (Zelboraf) in BRAF-mutant melanoma1. Informed by rapid genomic discovery2–5, there are now hundreds of compounds in clinical development targeting more than 100 genomic alterations in cancer-related genes representing multiple cellular pathways6,7. More personalized cancer therapy will be achieved by addressing the specific molecular drivers of a patient’s disease8.
Essential to the successful delivery of personalized cancer therapy are diagnostic tests that comprehensively characterize the genomic alterations occurring within individual tumors. These tests enable patients to be matched with available targeted therapies, either approved or in clinical trials. Several technologies, including PCR, Sanger sequencing, mass spectrometric genotyping, fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC)9–13, are currently in use for the clinical assessment of a limited number of oncogenic markers. Owing to technical limitations and the small amount of material obtained from biopsies, none of these methodologies can be scaled to address the increasing number and variety of therapeutically relevant genomic alterations that occur across hundreds of cancer-related genes, both in primary disease and in cases of acquired resistance to therapy14–18.
Massively parallel (or ‘next-generation’) DNA sequencing, successfully applied in the research setting to elucidate the complexity of the cancer genome, is thus becoming an attractive clinical diagnostic technology because it can accurately detect most genomic alterations in all therapeutically relevant cancer genes in a single assay19–21. However, the adoption of this technology in a clinical context as a routine test to support the selection of therapy for cancer patients faces multiple challenges. First, the majority of cancer specimens are FFPE, a process that may damage DNA22, such that robust DNA extraction and sequencing library construction protocols must be applied. Second, many samples available for testing are small-core needle biopsies, fine-needle aspirations or cell blocks prepared from malignant pleural, pericardial or peritoneal effusions, necessitating protocols that accommodate limited amounts of tissue and extracted DNA23. Third, whereas selecting specimens with high tumor content is feasible in the research setting, accurate profiling in clinical specimens must be achieved when the relative proportion of tumor nuclei is low24. Thus, uniformly high sequence coverage across all test regions and appropriately designed analysis algorithms are required. Finally, because millions of bases within the tumor genome are assayed, rigorous analytical approaches for validation are needed to demonstrate the readiness of the technology for clinical use.
We have developed and validated a next-generation sequencing (NGS)-based cancer genome profiling test that interrogates 4,557 exons of 287 cancer-related genes and have established performance benchmarks supporting direct clinical use. We assessed the analytical sensitivity, specificity, accuracy and precision across the reportable range of the assay, in line with guidelines established by the Next Generation Sequencing: Standardization of Clinical Testing work-group25. Relevant sample types were represented, including FFPE. Base substitutions, indels, focal gene amplifications and homozygous gene deletions were tested. We report our experience with the first 2,221 patient tumor FFPE specimens submitted to our Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited laboratory and provide insights for the successful incorporation of NGS into routine oncologic practice, including clinical trials.
RESULTS
NGS-based clinical cancer gene test
We have developed a cancer genome profiling test based on massively parallel DNA sequencing (Fig. 1)26,27. Briefly, DNA was extracted from routine FFPE biopsy or surgical specimens, 50–200 ng of which underwent whole-genome shotgun library construction and hybridization-based capture of 4,557 exons from 287 cancer-related genes and 47 introns from 19 genes frequently rearranged in solid tumors (Supplementary Table 1). Using the Illumina HiSeq2000 platform, hybrid-capture–selected libraries were sequenced to high uniform depth (targeting >500× coverage by non-PCR duplicate read pairs, with >99% of exons at coverage >100×). Protocols and reagents were optimized to assure even coverage and robust performance for a wide variety of specimens28–30. Sequence data were processed using a customized analysis pipeline designed to accurately detect multiple classes of genomic alterations: base substitutions, indels, focal gene amplifications, homozygous gene deletions and selected gene fusions in routine clinical specimens. All testing was done in a CLIA-certified, CAP-accredited laboratory.
Figure 1.
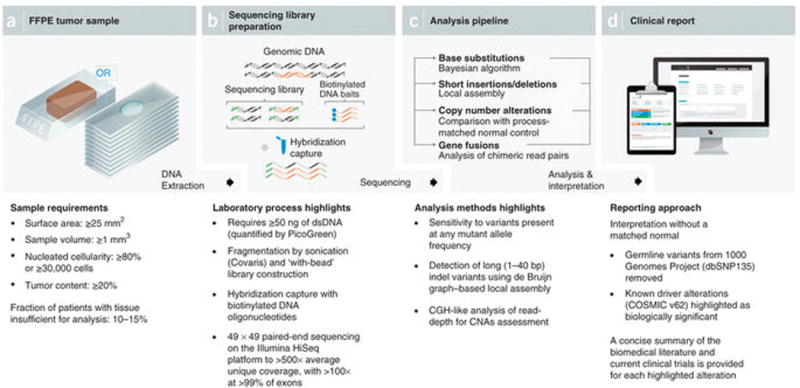
NGS-based cancer genomic profiling test workflow. (a) DNA is extracted from routine FFPE biopsy or surgical specimens. (b) 50–200 ng of DNA undergoes whole-genome shotgun library construction and hybridization-based capture of 4,557 exons of 287 cancer-related genes and 47 introns of 19 genes frequently rearranged in solid tumors. Hybrid-capture libraries are sequenced to high depth using the Illumina HiSeq2000 platform. (c) Sequence data are processed using a customized analysis pipeline designed to accurately detect multiple classes of genomic alterations: base substitutions, short insertions/deletions, copy-number alterations and selected gene fusions. (d) Detected mutations are annotated according to clinical significance and reported.
Validation approach
In contrast to diagnostic assays for a limited number of genomic sites, analytical validation of an NGS-based genomic profiling test assaying ~1.5 Mb of target sequence is a complex challenge. A single tumor specimen can harbor multiple types of genomic alterations, at any position in the tested region, at a wide range of mutant allele frequencies (MAF) or copy number levels. Reference specimens containing all possible somatic alterations in all cancer-related genes do not exist. We therefore developed a representative validation approach with reference samples created to span the key axes of mutation and specimen variability affecting detection performance, allowing the accuracy of the test to be robustly estimated.
To validate the accuracy of base substitution detection, we created pools of normal cell lines from the 1000 Genomes Project31, leveraging the abundance and population distribution of known germline base- substitution variation to create test specimens that include thousands of variants across the targeted exons and span a broad range of MAF (5–100%). Similarly, to validate the accuracy of indel detection, we mixed tumor cell lines with known somatic indel alterations into variably sized pools to represent both a range of MAF (10–100% and indel length (1–40 bp). Because unique sequence coverage, a function of sample quality, is a key driver of accuracy for substitutions and indels, in silico down-sampling (the random selection of subsets of reads) was carried out to examine performance over a wide coverage range (150–500×). For copy number alterations (CNAs), pairs of matched tumor and normal cell lines bearing diverse somatic focal gene amplifications and homozygous gene deletions were mixed in ratios ranging from low to high tumor content (20–75%). The test was applied blinded to all pools and the results compared to expectation based on the constituents in each pool. Initial fusion gene validation was described previously30,32, and extended in manuscripts in preparation or under review.
To confirm that performance criteria established by cell line validation are representative of routine clinical samples, we examined the concordance between NGS test results and the results obtained from a variety of current clinical technologies, including Sequenom mass-spectrometry analysis, gel sizing, FISH and IHC, for a large series of FFPE specimens. Finally, we validated test reproducibility by examining mutation calls in replicates of clinical FFPE specimens processed independently as components of several process batches and control specimens tested repeatedly by inclusion in every process batch.
Taken together, these experiments assessed test performance across a broad range of genomic alteration types and tumor specimen properties to support clinical use.
Base substitution detection performance
Base substitution detection was done using a Bayesian method allowing the detection of somatic mutations at low MAF across the 1.5 Mb assayed with increased sensitivity for mutations at hotspot sites. To validate base substitution detection, we created two pools of ten normal cell lines each, containing a total of 2,057 known base substitutions, representing a broad range of allele frequencies (Fig. 2a and Supplementary Tables 2 and 3). We compared the alterations detected to those expected from base substitutions present in the individual cell line constituents. Median exon sequencing coverage of 738× and 580× was obtained for the two pools, with >99% of the exons in each pool covered at >250×. Overall substitution detection performance was high; >99% of base substitutions expected to be present at MAF ≥ 10% were successfully detected (1,036/1,036), as were 99% of substitutions at MAF < 10% (1,013/1,021) (Supplementary Table 4). In addition, high specificity was maintained with a positive predictive value (PPV, the fraction of substitution calls in the pools traceable to a substitution in a constituent cell line) >99% (2,577/2,579, with two false-positive calls at MAF < 5%).
Figure 2.
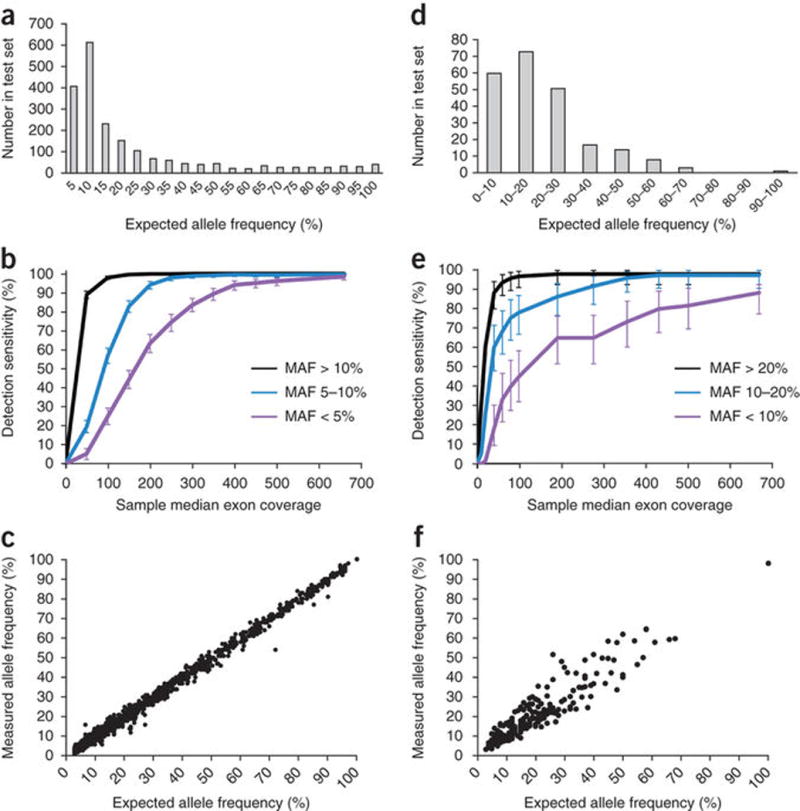
Base substitution and indel detection performance. (Base substitutions given in panels a–c, indels in panels d–f.) (a) Expected allele frequencies of base substitution alterations within the test set. (b) Detection sensitivity as a function of sample median exon coverage. Error bars, s.e.m. (c) Allele frequencies measured in pooled samples (y axis) match the frequencies expected based on the genotypes and mixing ratios of constituent cell lines (x axis). (d) Expected allele frequencies of indel alterations within the test set. (e) Detection sensitivity as a function of sample median exon coverage. Error bars, s.e.m. (f) Allele frequencies measured in pooled samples (y axis) match the frequencies expected based on the genotypes, ploidy and mixing ratios of constituent cell lines (x axis).
We next assessed the effect of reduced sequence coverage by down-sampling data from each of the two pools, targeting median exon coverage from 500× to 150× in intervals of 50× (Fig. 2b). As expected, detection sensitivity dropped as coverage decreased, especially for substitutions occurring at low MAF (<10%), although high sensitivity was obtained down to 250× median exon coverage. At 250× median coverage, >99% of base substitutions present at MAF ≥ 10% were successfully detected (1,035/1,036), as were 98% of substitutions at 5% ≤ MAF ≤ 10% (601/614). Furthermore, PPV remained high (>99%) across the full coverage range. We observed a high correlation between expected and observed MAFs at tested sites (Fig. 2c), highlighting the quantitative nature of an optimized NGS-based test.
To further quantify the impact of sequencing coverage and MAF on base substitution detection sensitivity, we generated a smoothed representation of the relationship between these variables in our down-sampled data at a given mutated site (in contrast to aggregate performance in Fig. 2b) and examined how performance degrades under reduced coverage depth (Supplementary Fig. 1). As expected, a marked reduction in sensitivity was observed for alterations with MAF < 10% when local coverage falls below 100×, emphasizing the importance of high, uniform sequencing coverage across the reportable exon range of the assay.
Indel detection performance
To allow discovery of longer events, we detected indels by de novo local assembly using the de Bruijn approach. To validate indel detection, we used 28 tumor cell lines containing a total of 47 known somatic indel alterations in 22 genes to generate 41 pools of two to ten cell lines each (Supplementary Tables 5 and 6), thereby creating a test set of 227 indels spanning a wide range of MAF and indel lengths (1–40 bp) (Fig. 2d). An average sequence coverage of 667× was obtained across all pools, with >99% of the exons covered at >250×. Overall, indel detection was high: 98% (92/94) of indels at MAF ≥ 20% were successfully detected, as well as 97% (71/73) of indels at 10% ≤ MAF ≤ 20% and 88% (53/60) of indels at 5% ≤ MAF ≤ 10%. Very few false-positive calls were observed, with PPV > 99% (875/878, all false-positive calls occurred at MAF < 20%).
As was the case for base substitutions, indel detection sensitivity decreased with reduced coverage depth, especially at low MAF, but remained high above ~250× unique median coverage. Ninety eight percent (92/94) of indels at MAF > 20% and 92% (67/73) of indels at 10% ≤ MAF ≤ 20% were successfully detected (Fig. 2e and Supplementary Table 7). Similarly to base substitutions, the correlation between measured and expected MAF for indels was high (Fig. 2f). Likewise, the relationships between sensitivity, MAF and local sequence coverage depth were maintained (Supplementary Fig. 1b).
Comparison with other variant-calling approaches
We evaluated the contribution of our customized variant-calling pipeline to the performance observed in the cell line models above, focusing on two key distinguishing characteristics of our approach. First, we apply statistical models that allow for the identification of a mutation at low MAF. Second, we incorporate local assembly to allow for the reconstruction and detection of longer indel events. We therefore selected publicly available algorithms that were similar overall to our approach, with the key exception of the two features above. To assess the benefit of modeling low-frequency variants, we compared the substitution detection performance of our pipeline on the two normal cell line pools to calls produced by SAMtools33, a robust and widely used software package for sequence data manipulation and germline genotype analysis. To measure the effect of local assembly, we compared our indel results on both individual cancer cell lines and cell line pools to those produced by Dindel34, an advanced indel detection algorithm based on candidate alternate haplotype identification and read re-alignment. Both packages include call filtering on widely accepted metrics (e.g., base quality, mapping quality) and were run using recommended parameters (Supplementary Fig. 2).
SAMtools was able to detect nearly all (97.8%, 636/650) substitution variants present at MAF ≥ 20% (Supplementary Fig. 2a). Performance was substantially reduced at MAF below these levels, with only 25.1% (97/386) of variants present at 10% ≤ MAF ≤ 20% detected and no variants (0/1,021) at MAF < 10% being called. Dindel, on the other hand, detected 80% (217/272) of all indel events (Supplementary Fig. 2b). Of the variants missed, 73% (40/55) were ≥23 bp. Indeed, no indels of >23 bp were called in any sample. All other missed variants had MAF <20% or occurred in regions of low mapping quality (Supplementary Table 8). Nearly all SAMtools calls (1,039/1,048) were also seen in our pipeline results, suggesting that SAMtools maintained tight specificity control, whereas Dindel calls outnumbered ours more than sevenfold (8,923 versus 1,154), highlighting that additional filtering is likely required.
Although these comparisons are necessarily imperfect, and additional differences in variant calling likely affect the results, these findings are consistent with the importance of somatic-variant modeling to allow the detection of low-frequency alterations and the deep leveraging of information across multiple short reads by means of local assembly to allow detection of larger indel events.
CNA detection performance
We detected CNAs by fitting a statistical copy-number model to normalized coverage and allele frequencies at all exons and ~3,500 genome-wide, single-nucleotide polymorphisms (SNPs), accounting for stromal admixture35–37. To validate CNA detection, we pooled seven tumor cell lines bearing 19 focal gene amplifications (6–15 copies, 15 genes) and 9 homozygous gene deletions (6 genes) with their matched normal cell lines (thereby maintaining consistent genotypes) in five ratios ranging from low to high tumor content (20–75%), creating a total test set of 210 CNAs (Supplementary Table 9).
High performance was achieved for both high-level amplifications (copy number ≥ 8) and homozygous deletions when tumor purity was as low as 30%: sensitivity was 99% (91/92) with PPV > 99% (127/127) (Fig. 3 and Supplementary Table 10). Performance was reduced for lower CNAs (6–7 copies) and at lower sample purities (20–30%), with overall sensitivity >80%. Our results demonstrate that an optimized NGS-based test can accurately detect most clinically actionable CNAs in a broad spectrum of patient specimens. These results also highlight the scope for further improvements in this methodology, including the robust detection of heterozygous loss.
Figure 3.
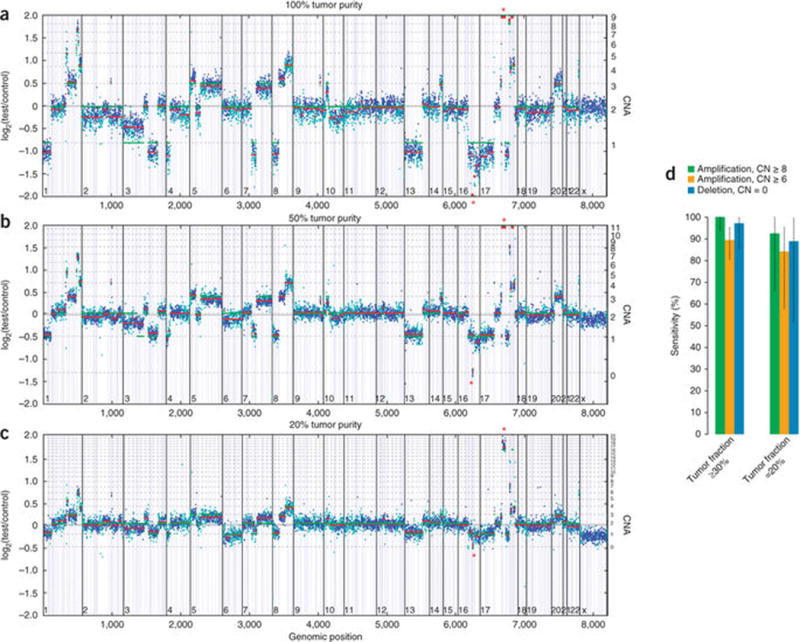
CNA detection performance. (a) Example of CNA data. HCC2218 cell line mixed with matched normal sample at 100% (a), 50% (b) and 20% tumor content (c). Y axes denote log-ratio measurements of coverage obtained in test samples versus a normal reference sample, with assessed copy numbers marked by dashed lines. Each point denotes a genomic region measured by the assay (blue exon, cyan SNP), and these are ordered by genomic position. Red lines indicate average log-ratio in a segment, whereas green lines illustrate the model prediction. Asterisks denote the detected CDH1 homozygous deletion (chr16) and ERBB2 amplification (chr17). (d) Summary of sensitivity results of CNA calling validation study for focal amplifications and homozygous deletions in samples with tumor fractions ≥30% and 20–30%. Error bars, s.e.m.
Concordance between NGS and other test platforms
The above studies demonstrate that the NGS-based test has the performance characteristics necessary to accurately detect base substitutions, indels and CNAs. We further validated test accuracy by blinded comparisons to four alternative clinical diagnostic technologies for 249 FFPE cancer specimens.
To assess base substitution and indel detection in routine clinical cancer samples, we selected 118 FFPE resection specimens (67 lung cancers, 31 colorectal cancers and 20 melanoma) previously tested for 91 mutations in eight oncogenes using Sequenom mass spectrometry genotyping or a PCR fragment gel-sizing approach (Fig. 4b). For this study, DNA for NGS was extracted from 4 × 10 μm, new, unstained sections from the originally tested FFPE block. High concordance was noted across platforms; of the 101 mutations identified by Sequenom or gel sizing, 97 were also called by NGS (using an earlier version of the bait set targeting 182 genes; Supplementary Table 1b), and NGS found an additional 7 mutations at mutually tested sites (Fig. 4a). Discordant samples did not have lower average coverage (857× versus 922× for concordant samples; t-test, P = 0.51, n =118), and did not otherwise differ in processing. Several discordances showed weak or equivocal evidence of mutation by Sequenom, indicating that high-uniform-coverage NGS may have even greater sensitivity, although tumor heterogeneity may have also contributed to the discordances observed (Supplementary Table 11). More than 25% of concordant calls had an MAF ≤ 10%, emphasizing the high sensitivity needed in a clinical cancer genomic test (Fig. 4c).
Figure 4.
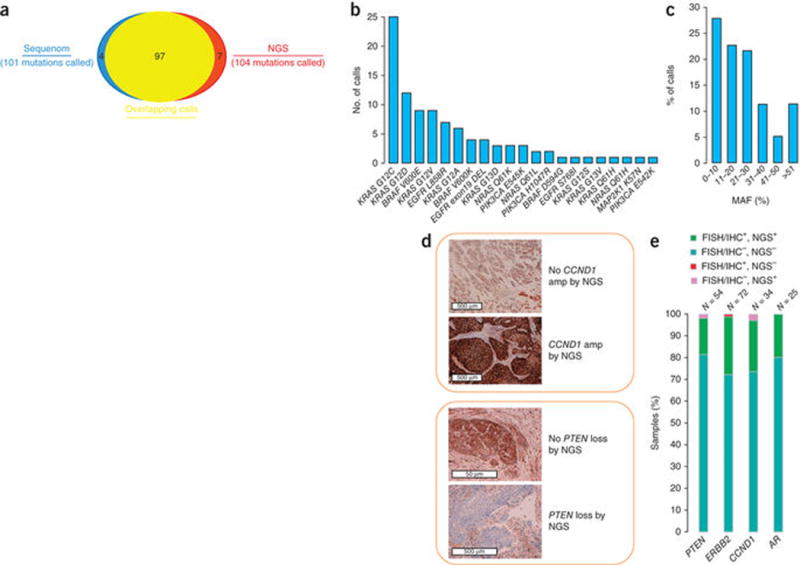
Concordance with clinical testing on FFPE specimens. Tumor specimens (N = 249) were assayed using the NGS-based test and by several other methods. (a) Overlap between positive alteration calls by NGS and Sequenom or gel sizing at 91 mutually tested sites in 118 FFPE clinical cancer specimens. (b) Specific alterations comprising the 97 concordant calls. (c) Histogram of NGS MAF in the 97 concordant calls. The high prevalence of low MAF in clinical cancer specimens is highlighted. (d) Examples of confirmation of NGS CNAs by IHC. (e) Summary of concordance between NGS, FISH and IHC findings across four genes (Supplementary Table 12).
Additional patient sample cohorts were assembled to assess the accuracy of CNA detection: 72 breast cancer specimens tested clinically for ERRB2 amplification by FISH, 54 combined prostate and head & neck cancer specimens tested for PTEN loss by FISH or IHC, 34 head & neck cancer specimens tested for CCND1 amplification by IHC, and 25 prostate samples tested for amplification of AR by FISH. High concordance was again observed, with the accuracy of NGS ranging from 95–100% relative to other tests (Fig. 4d,e and Supplementary Table 12).
In total, these results demonstrate that the optimized NGS test can be as accurate as clinical assays currently in use; it also comprehensively assays a much larger series of clinically relevant cancer genes.
Mutation detection reproducibility in FFPE samples
In addition to establishing test analytical sensitivity, specificity and accuracy with respect to alternative technologies, we validated test reproducibility by comparing the equivalence of test results on separate aliquots of the same tumor DNA25.
We first evaluated reproducibility by testing six clinical FFPE specimens in five replicates, each on three different assay batches, including three replicates in a single batch. Taken together, these samples contained 35 known genomic alterations of all types, providing a diverse set of test variants for comprehensive assessment of both inter- and intrabatch reproducibility. Concordance between replicates was 97% (Supplementary Table 13), with no significant differences between inter- and intrabatch replicates observed. Most discordant calls could be ascribed to inconsistency in copy number assessment for a single low-purity specimen.
We further assessed long-term test reproducibility by examining mutation calls for two FFPE colon cancer resections that were repeatedly tested as regular process controls. Each of the two samples contained two or three known alterations (substitutions or indels) and each was sequenced repeatedly over 4–8 months (79 and 71 replicates, respectively). All alterations were successfully detected in all replicates, including one alteration occurring at a low (~4%) MAF. The quantitative nature of deep NGS was again highlighted, with MAFs of detected mutations showing stability even over this extended time period (Fig. 5).
Figure 5.
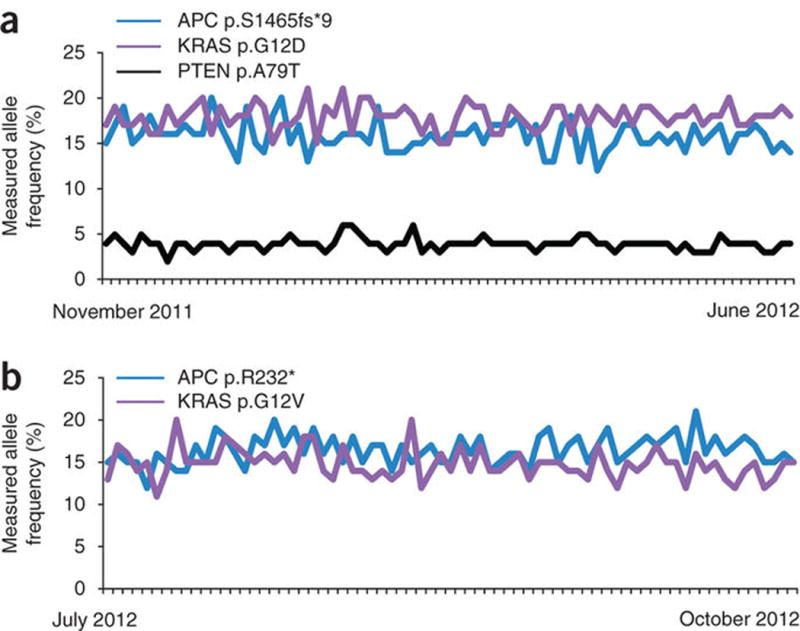
Reproducibility of mutation detection in FFPE specimens. Two large colon cancer resections were repeatedly tested in separate process batches. DNA was first extracted from all of the tissue in each FFPE block in several batches, pooled, and then used to make 200 ng aliquots. The reproducibility of mutation detection and measured MAF were examined. The MAF measured for the known somatic alterations in these samples is shown, with samples ordered left to right from earliest to latest. (a) One specimen was tested 79 times between November 9, 2011 and June 17, 2012. Three known somatic mutations (COSMIC) were detected in this sample: APC c.4394-4395insAG p.S1465fs*9, KRAS c.35G>A p.G12D, and PTEN c.235G>A p.A79T. Each of these mutations was successfully detected in 79/79 tests. (b) A second specimen was tested 71 times between July 12, 2012 and October 21, 2012. Two known somatic mutations were detected in this sample: APC c.694C>T p.R232* and KRAS c.35G>T p.G12V. Each of these mutations was successfully detected in 71/71 tests.
Finally, we explored the potential impact of FFPE-related artifacts by comparing mutation calls in five pairs of matched normal FFPE and blood specimens derived from the same patient. If FFPE-related artifacts significantly affected the spectrum of called alterations, we would expect alterations called in FFPE normal specimens to be absent from the matching blood controls. Indeed, of the 13 alterations called in FFPE specimens, all were present in the corresponding blood sample (Supplementary Fig. 3), indicating that these were likely real (although rare, i.e., not in NCBI dbSNP) germline variation. In contrast, the number of sites with more than three observations of an alternate base (substitution call ‘candidates’, in our method) approximately doubled in FFPE samples to an average of 935, versus 485 (t-test, P < 0.02), with most excess sites present at MAF < 5%. Although this sample set is too small to draw definitive conclusions, we surmise that any FFPE-related artifacts exist below the call thresholds of our test, an observation reinforced by the high concordance with the results of mass spectrometric genotyping reported above.
Taken together, these experiments demonstrate the robustness of the platform and offer confidence that the high performance characteristics of the test can be sustained over a long period.
Clinically actionable alterations in patients
Full clinical validation of comprehensive cancer genomic profiling in oncologic care, through demonstration of broadly improved outcomes, is an active area of investigation that will engage the research community for some time38. Nevertheless, our deployment of an analytically validated, comprehensive, NGS-based cancer gene test allows for several initial insights to be drawn, and points the way toward successfully incorporating NGS in clinical oncology and clinical trials.
We report our experience with 2,221 solid-tumor FFPE specimens (Fig. 6a) submitted to our CLIA-certified, CAP-accredited laboratory for comprehensive genomic profiling to inform clinical decision making for cancer patients worldwide. In total, 95.1% (2,112/2,221) of specimens were successfully tested (mean coverage 1,134×, Supplementary Table 14). Despite the diversity of tumor types assayed and concerns that the quality of sample DNA from tissues embedded in paraffin may degrade over time, the site of origin and specimen age had only a modest impact on assay performance (Supplementary Fig. 4). As expected, larger specimens arising from tumor resections perform somewhat better than needle biopsies, and samples yielding abundant double-stranded DNA (dsDNA) outperformed those with more marginal DNA yields (Supplementary Fig. 4). Overall, clinical cancer specimens obtained in the course of routine care appear amenable to an optimized NGS-based diagnostic assay, highlighting the broad applicability of the approach.
Figure 6.
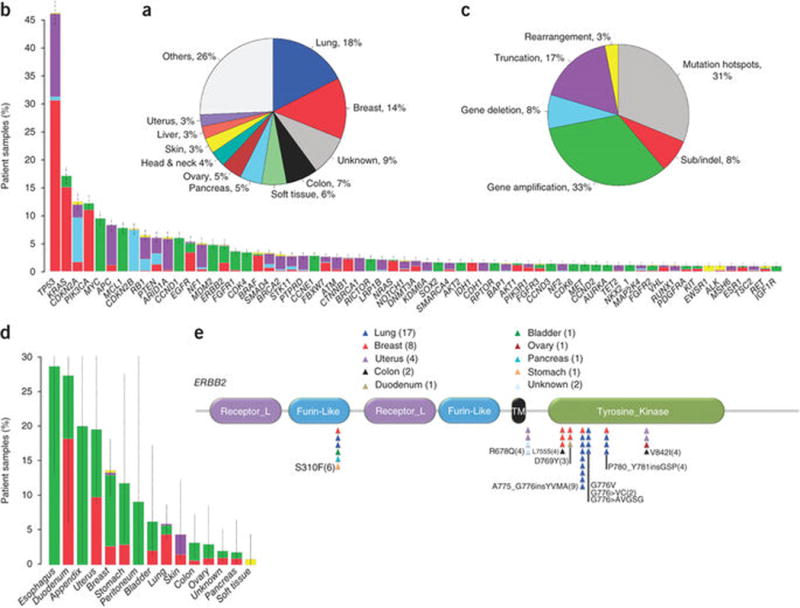
Clinically actionable alterations in patient samples. (a) Distribution of tumor tissue of origin (type) for profiled specimens. (b) Frequency of all reported alterations in most commonly altered genes among the specimens. Alterations are colored according to alteration class, as depicted in panel c. Error bars, s.e.m. (c) Distribution of clinically actionable alteration classes detected. (d) Frequency of ERBB2 alterations detected among specimens of various tumor types. Alterations are colored according to class, as in panel c. Error bars, s.e.m. (e) Distribution of substitution and indel mutations, across the domain structure of the ERBB2 protein. Individual mutations are represented as triangles colored according to the tumor type.
Given that matched normal specimens are not routinely collected in clinical practice, reporting focused on known sites of somatic mutation39, truncations or homozygous deletions of known tumor suppressor genes40, as well as known amplifications of oncogenes and gene fusions in genes known to be rearranged in solid tumors. Alterations were reported in 174/189 (92%) of tested genes, with an average of 3.06 alterations per sample (range, 0–23). When the focus was limited to alterations associated with a clinically available targeted treatment option or a mechanism-driven clinical trial (i.e., clinically actionable)26,41,42, the average number of alterations per sample was 1.57 (range, 0–16), with 76% of samples containing at least one such alteration, suggesting that clinical application of individual patient results is feasible.
Although the number of actionable alterations in any individual cancer patient’s sample was low (average, 1.57), a wide variety of alterations was observed across all samples, with 1,579 unique alterations reported. Their frequency displayed a long-tail distribution, with a handful of common alterations in cancer complemented by many rarer events (Fig. 6b and Supplementary Table 15). It was thus not surprising to observe that current clinical testing paradigms comprising only mutation hotspots10,11,43 capture less than one-third of total actionable results (Fig. 6c).
The therapeutic implications of the long tail were particularly notable for proven targets of therapy, as exemplified by ERBB2. Although ERBB2 is currently clinically validated only as an amplified or overexpressed drug target in breast and gastro-esophageal cancer, we observed ERBB2 alterations in 12 additional solid tumor types (Fig. 6d), spanning most known sites of activation throughout the gene (Fig. 6e), and comprising 5% of total cases. Importantly, more than 40% of all ERBB2 alterations were point mutations or indels in nonamplified specimens that would have been negative by limited biomarker tests. Although robust clinical evidence for targeting these alterations must still be generated, recent preclinical work on their functional importance44 highlights the promise of improving cancer care through comprehensive genomic profiling using an optimized NGS diagnostic test.
DISCUSSION
Cancer treatment has entered a new age in which sophisticated diagnostics reveal the molecular drivers of individual patient tumors, offering the opportunity to select appropriate targeted therapy. However, as more clinically relevant cancer genes are identified, matching patients with the optimum targeted therapy becomes more challenging, particularly as routine clinical practice presents hurdles not typically encountered in the research setting. Of note, the majority of clinical specimens are FFPE, which can damage DNA and requires optimized methodologies to assay accurately. Biopsies of advanced disease, which is associated with the greatest risk of morbidity, are becoming smaller, with needle biopsies common, necessitating the development of methodologies that can detect all classes of genomic alteration in a single, tissue-sparing test. Furthermore, the selection of tumors with the highest percentage of tumor nuclei, common practice in the research setting, is not a clinical option. In cancer specimens with a low percentage of tumor tissue (high levels of normal cell contamination), high test sensitivity is required. High specificity is also required as false-positive reports may lead to suboptimal therapy choice. Finally, the long tail of genomic alterations that are individually rare, but which cumulatively form a substantial fraction of clinically and biologically relevant genomic alterations, requires the interrogation of thousands of exons from hundreds of cancer-related genes to maximize targeted treatment options.
To overcome these challenges, we have developed an NGS-based diagnostic test to accurately detect all clinically relevant genomic alterations across all coding exons of 287 cancer genes in routine FFPE clinical specimens, including needle biopsies. We validated this test by creating pooled cell-line models spanning key determinants of detection accuracy for somatic alterations, including MAF, indel length, degree of stromal admixture and amplitude of copy number change. We verified accuracy in FFPE specimens by examining concordance in tumors clinically characterized for selected mutations by current validated tests.
The overall performance of our test was high. In cell line models, sensitivity was >99% for base substitutions, 98% for indels and >95% for CNAs. False-negative calls were predominantly low MAF (<10%) substitutions and indels or low-magnitude (6,7) CNAs. Because it is impractical to independently confirm all alterations arising from a clinical NGS test, it was equally important that high specificity was maintained, with the PPV of alteration calls exceeding 99%. The raw read support for all variants is additionally subject to expert review in our clinical practice, to further guard against the potential for false-positive reports. Robust performance translated to FFPE specimens: concordance on mutually tested markers exceeded 95% for all alteration types compared to current clinical tests. Comprehensive NGS-based genomic profiling was successful for most (95.1%) of >2,200 consecutive clinical cases, identifying more actionable alterations (in 76% of patients) than otherwise practical today.
We present a validation study and performance metrics for a comprehensive NGS-based diagnostic test for use in oncologic care. Given the capability of optimized NGS to detect a broader range of genomic alterations than current clinical assays, particularly when the amount of tissue is limited, we advocate that NGS-based genomic profiling be employed to maximize the appropriate use of targeted therapy, allow populations of patients with uncommon or rare alterations to be routinely identified for clinical trials and thus expand treatment choices for cancer patients.
METHODS
Methods and any associated references are available in the online version of the paper.
ONLINE METHODS
NGS-based clinical cancer gene test
DNA extraction and library construction
A 4-μm section of a hematoxylin and eosin–stained slide was reviewed for pathology (J.R.) to ensure a sample volume of ≥1 mm3, nucleated cellularity ≥80% or ≥30,000 cells and that ≥20% of the nuclei in the sample were derived from the tumor. A macro-dissection to enrich specimens of ≤20% tumor content was performed when warranted. DNA was extracted from 40 μm of unstained FFPE sections, typically 4 × 10 μm sections, by digestion in a proteinase K buffer for 12–24 h followed by purification with the Promega Maxwell 16 Tissue LEV DNA kit. Double-stranded DNA is quantified by a Picogreen fluorescence assay using the provided lambda DNA standards (Invitrogen); UV-based quantification was found unreliable for FFPE samples. 50–200 ng of dsDNA in 50–100 μl water in microTUBEs is fragmented to ~200 bp by sonication (3 min, 10% duty, intensity = 5, 200 cycles/burst; Covaris E210) before purification using a 1.8× volume of AMPure XP Beads (Agencourt). SPRI purification and subsequent library construction with the NEBNext kits (E6040S, NEB), containing mixes for end repair, dA addition and ligation, were performed in 96-well plates (Eppendorf) on a Bravo Benchbot (Agilent) using the “with-bead” protocol45 to maximize reproducibility and library yield. Indexed (6-bp barcodes) sequencing libraries are PCR amplified with HiFi (Kapa) for 10 cycles, 1.8× SPRI purified and quantified by qPCR (Kapa SYBR Fast) and sized on a LabChip GX (Caliper); size selection was not done. PCR yield was maximized by ensuring that no SPRI beads were transferred to the PCR tube. Samples yielding <50 ng of extracted DNA or 500 ng of sequencing library, or with a mean insert size >400 bp, were failed; the failure rate was 4.9% on the first 2,221 consecutive clinical samples.
Hybrid selection and sequencing
Solution hybridization was done using a >50-fold molar excess of a pool of 23,685 individually synthesized 5′-biotinylated DNA 120 bp oligonucleotides (Integrated DNA Technology). The baits targeted ~1.5 Mb of the human genome including 4,557 exons of 287 cancer-related genes, 47 introns of 19 genes frequently re-arranged in cancer, plus 3,549 polymorphisms located throughout the genome (Supplementary Table 1a). Baits were designed by taking overlapping 120 bp DNA sequence intervals covering target exons (60 bp overlap) and introns (20 bp overlap), with a minimum of three baits per target; SNP targets were allocated one bait each. Intronic baits were filtered for repetitive elements46 as defined by the UCSC Genome RepeatMasker track. Hybrid selection of targets demonstrating reproducibly low coverage was boosted by increasing the number of baits for these targets. In an earlier version of the test, hybridization capture was done using RNA-based baits (Agilent SureSelect), targeting 3,230 exons of 182 cancer genes and 37 introns of 14 genes frequently rearranged in cancer28–30 (Supplementary Table 1b). The long-term reproducibility and clinical analyses includes data using this method, as do the Sequenom and FISH/IHC concordance studies (Supplementary Table 12). As described previously47, 500–2,000 ng of sequencing library is lyophilized in a 96-well plate and suspended in water, heat denatured at 95 °C for 5 min and then incubated at 68 °C for 5 min before addition of the baitset reagent and Cot, salmon sperm and adaptor-specific blocker DNA in hybridization buffer. After a 24-h incubation, the library-bait duplexes are captured on paramagnetic MyOne streptavidin beads (Invitrogen) and off-target library is removed by washing once with 1× SSC at 25 °C and four times with 0.25× SSC at 55 °C. The PCR master mix is added to directly amplify (12 cycles) the captured library from the washed beads45. After amplification, the samples are 1.8× SPRI purified, quantified by qPCR (Kapa) and sized on a LabChip GX (Caliper). Libraries are normalized to 1.05 nM and pooled such that each Illumina HiSeq 2000 lane has up to four samples each (32 per flowcell), before 49 × 49 paired-end sequencing using manufacturer’s protocols.
Sequence data processing
Sequence data were mapped to the human genome (hg19) using BWA aligner v0.5.9 (ref. 48). PCR duplicate read removal and sequence metric collection was done using Picard 1.47 (http://picard.sourceforge.net/) and Samtools 0.1.12a33. Local alignment optimization was performed using GATK 1.0.4705 (ref. 49). Variant calling was done only in genomic regions targeted by the test.
Base substitution detection
We used a Bayesian methodology, which allows detection of novel somatic mutations at low MAF and increased sensitivity for mutations at hotspot sites through the incorporation of tissue-specific prior expectations39. Reads with mapping quality <25 are discarded, as are base calls with quality ≤2. The equations governing this evaluation are:
P(Mutation present| Read data “R”) = P(Frequency of mutation “F” > 0|R) = 1 – P(F = 0|R)
P(F = 0) = 1 – prior expectation “p” of the mutation in cancer type
P(R|F = i/n) is evaluated with a multinomial distribution of the observed allele counts at each candidate mutation site, using empirically observed error rates. Call candidates are issued if P(“F” >0|R) > 99%. Final calls are made at MAF ≥ 5% (MAF ≥ 1% at hotspots) after filtering for strand bias (Fisher’s test, P < 1e-6), read location bias (Kolmogorov–Smirnov test, P < 1e-6), and presence in two or more normal controls.
Indel detection
To detect indels, de novo local assembly in each targeted exon was performed using the de Bruijn approach50. Key steps are:
Collecting all read-pairs for which at least one read maps to the target region.
Decomposing each read into constituent k-mers and constructing an enumerable graph representation (de Bruijn) of all candidate nonreference haplotypes present.
Evaluating the support of each alternate haplotype with respect to the raw read data to generate mutational candidates. All reads are compared to each of the candidate haplotypes through ungapped alignment, and a ‘vote’ for each read is assigned to the candidate with best match. Ties between candidates are resolved by splitting the read vote, weighted by the number of reads already supporting each haplotype. This process is iterated until a ‘winning’ haplotype is selected.
Aligning candidates against the reference genome to report mutation calls. Indel candidates arising from direct read alignment were also considered.
Filtering of indel candidates was carried out as described for base substitutions above (strand bias P < 1e-10, MAF ≥ 3% at hotspots), with an empirically increased MAF threshold at repeats and adjacent sequence quality metrics as implemented in GATK: percentage of neighboring base mismatches <25%, average neighboring base quality >25, average number of supporting read mismatches ≤2.
CNA detection
Using a comparative genomic hybridization (CGH)-like method, we obtained a log-ratio profile of the sample by normalizing the sequence coverage obtained at all exons and ~3,500 genome-wide SNPs against a process-matched normal control. This profile was corrected for GC-bias, segmented and interpreted using allele frequencies of sequenced SNPs to estimate tumor purity and copy number at each segment. Briefly, if Si is a genomic segment at constant copy number in the tumor, let li be the length of Si, rij be the coverage measurement of exon j within Si, and fik be the minor allele frequency of SNP k within Si. We estimate p, tumor purity, and Ci, the copy numbers of Si. We jointly model rij and fik, given p and Ci:
and,
where Mi is the copy number of minor alleles at Si, distributed as integer 0 ≤ Mi ≤ Ci. σri and σfi reflect noise observed in the CGH and SNP data, respectively. Fitting was performed using Gibbs sampling, assigning absolute copy number to all segments. Model quality was reviewed and alternative explanations considered36, and focal amplifications are called at segments with ≥6 copies (or ≥7 for triploid; ≥8 for tetraploid tumors) and homozygous deletions at 0 copies, in samples with purity >20%.
Gene fusion detection
Genomic rearrangements were identified by analyzing chimeric read pairs (read pairs for which reads map to separate chromosomes, or at a distance of over 10 Mbp). Pairs were clustered by genomic coordinate of the pairs, and clusters containing at least 10 chimeric pairs were identified as rearrangement candidates. Filtering of candidates was performed by mapping quality (MQ > 30) and distribution of alignment positions (s.d. >10). Rearrangements were annotated for predicted function (creation of fusion gene or tumor suppressor inactivation).
Analytical validation
Cell line specimens
For base substitution validation, purified DNA from 20 lymphoblastoid cell lines (Supplementary Table 2) from the 1000 Genomes Project were purchased from the Coriell Institute (http://ccr.coriell.org/). For indel validation, 28 immortalized tumor cell lines (Supplementary Table 5) were purchased from ATCC (http://www.atcc.org/). Purified gDNA was available for five tumor cell lines (A2058, DU-145, HCC1395, HCC1937, THP-1), whereas frozen cell pellets were used for the other 23. For copy number validation, seven pairs of matched tumor and normal cell lines were purchased from ATCC as either cell pellets or DNA (Supplementary Table 9). Cell pellets were thawed on ice and washed twice with PBS before cell lysis and DNA purification. Pool constituents were mixed in equals part using a Biomek NX (Beckman Coulter) to a total mass of 200 ng. Actual mixing ratios were calculated using a linear regression of SNP alternate allele frequencies. All sequence data generated for cell lines and pools have been deposited at NCBI Sequence Read Archive (SRA) under accession SRP028580.
FFPE tumor specimens
For concordance analyses, FFPE specimens and confirmatory assay results were obtained as detailed in Supplementary Table 12 and outlined below. Sequenom mass-spec genotyping: tumors were genotyped using the MassArray system (Sequenom). Samples were tested in duplicate using a series of multiplexed assays designed to interrogate AKT1 (codon 17), BRAF (codons 469, 594, 600), EGFR (codons 709, 719, 761, 768, 776, 790, 858, 861), ERBB2 (codons 755, 769, 777), KRAS (codons 12-13, 61, 117, 146), MAP2K1 (codons 56, 57, 67), NRAS (codons 12-13, 61) and PIK3CA (codons 88, 345, 420, 542, 545, 1043, 1047). Genomic DNA amplification and single base pair extension steps were conducted with primers designed using Assay Designer v3.1 (Sequenom). The allele-specific extension products were analyzed using MALDI-TOF/MS. Automated calls were confirmed by manual review of spectra.
EGFR exon 19 gel sizing: 200 ng of DNA was PCR amplified with FAM-labeled EGFR exon 19 specific primers (EGFR-Ex19-FWD1: 5′GCACCATCTCACAATTGCCAGTTA3′, EGFR-Ex19-REV1: 5′ Fam AAAAGGTGGGCCTGAGGTTCA3′). The resulting PCR product was checked by agarose gel electrophoresis, analyzed using a 3730XL instrument and Prism Software (Applied Biosystems). The size of the wild-type-labeled product is 207 bp, whereas mutated EGFR exon 19 appears as smaller sizes.
AR/PTEN FISH
These specimens have been described previously28. FFPE 0.4-μm tissue sections were hybridized with probes for PTEN+ reference probe (10q25.2) and separately with probes for Androgen Receptor (AR)+ and the centromeric region of the X chromosome (CEPX). For each case, at least 30 nonoverlapping nuclei were evaluated for PTEN deletion or AR amplification using a BX51 Olympus microscope based on the copy number of each probe, as well as the PTEN:10q25.2 and AR:CEPX ratios. Where NGS results were available for multiple specimens per patient, the highest purity specimen was used in concordance analysis (no within-patient discordances were noted).
ERBB2 FISH
5 μm tissue sections or core biopsies of invasive breast carcinomas were hybridized with probes for ERBB2 and the centromeric region of chromosome 17 (CEP17) that are components of the PathVysion HER2 FISH assay (Abbott-Vysis). Hybridized slides were digitally imaged and 20 nonoverlapping cells were evaluated for ERBB2 and CEP17 copy number using the Ikonscope HER2 Analysis Software. ERBB2 copy number, CEP17 copy number and ERBB2:CEP17 ratio were calculated and reported according to the package insert.
PTEN and CCND1 IHC
IHC was carried out using established methods51,52. Antibody 04-409 (Millipore-Merck KGaA) was used for PTEN staining, and antibody P2D11F11 (Novocastra) was used for CCND1 staining of 10-μm thick slides.
All confirmatory assays were performed on FFPE tissue. All NGS analyses were performed on FFPE tissue, except in study B, where tissue was preserved in RNAlater.
Assessment of base substitution detection performance
To determine the variants present in normal cell lines used for assessment of base substitution detection, all 20 HapMap DNA samples were sequenced individually. The 5,801 SNP sites from the dbSNP53 database (build 135) that overlap test gene coding regions (marked coding-synonymous, missense, or nonsense) were examined. SNP sites consistent with a homozygous (MAF > 90%) or heterozygous (40% ≤ MAF ≤ 60%) state were used in the test set. The expected MAF for each test base substitution in pooled samples was calculated based on the number of alternate alleles present in mix constituents and on mixing ratios (Supplementary Tables 2–4).
Assessment of indel detection performance
The test set of indels was generated from indels in cell lines in the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic/) occurring in the coding regions of test genes. The expected MAFs for indels in pooled samples were calculated based on the MAF of that indel in the individual tumor cell line, the ploidy of that locus in each of the pool constituents and on mixing ratios (Supplementary Tables 5 and 7).
Assessment of CNA detection performance
A total of 11 known CNAs occurring within 7 tumor cell lines were identified in the CGP database (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/cghviewer/CghHome.cgi) and confirmed to exist in the pure tumor cell lines. An additional 17 CNAs were discovered de novo in the pure tumor cell lines and added to the test set. Tumor and normal cell line pairs were combined to generate pools of 75%, 50%, 40%, 30%, 20% tumor fraction for each pair, and pools were sequenced and analyzed individually (Supplementary Tables 9 and 10).
Calculation of performance statistics
For sensitivity analysis, all test variants were assigned either a true positive (TP) if detected in the pool or false negative (FN) if not detected. Sensitivity was calculated as TP/(TP+FN). For specificity analysis, each called variant was classified as a TP if a matching alteration was detected in the pure sample, or as a false positive (FP) if a matching alteration was not detected. PPV was calculated as TP/(TP + FP).
Supplementary Material
Acknowledgments
The authors would like to acknowledge L. Gay for advice and assistance with manuscript preparation. H.B. is the Damon Runyon-Gordon Family Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI-67-13). M.L. was supported by a Wellcome Trust Fellowship (WT093855MA) and by the Austrian Science Fund (J2856).
Footnotes
Accession codes. SRA: SRP028580.
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
G.M.F., D.L. and R.Y. designed the study, wrote the manuscript and developed and/or performed analyses. M.T.C. and P.J.S. designed the study and wrote the manuscript. V.A.M., J.S.R. and M.F.B. wrote the manuscript. G.A.O. designed the study. A.F., K.W., J.H., M.S.-L., J.W., E.M.S., P.A., J.S. and C.V. developed and/or performed analyses. G.A.O., S.R.D., K.B., F.J., V.B., S.B., J.B., A.D., L.G., K.I., A.M., K.M., T.R., S.T., E.W., M.Z., Z.Z., M.J., A.P., J.S.R. and J.C. planned and/or performed laboratory experiments. H.B., J.M.M., M.A.R., S.D., C.V.H., M.F.B., L.P., M.L. and C.B. planned and/or performed confirmatory experiments.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper.
References
- 1.Stegmeier F, Warmuth M, Sellers WR, Dorsch M. Targeted cancer therapies in the twenty-first century: lessons from imatinib. Clin Pharmacol Ther. 2010;87:543–552. doi: 10.1038/clpt.2009.297. [DOI] [PubMed] [Google Scholar]
- 2.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hudson TJ, et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mardis ER. Genome sequencing and cancer. Curr Opin Genet Dev. 2012;22:245–250. doi: 10.1016/j.gde.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pao W. New approaches to targeted therapy in lung cancer. Proc Am Thorac Soc. 2012;9:72–73. doi: 10.1513/pats.201112-054MS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomas RK, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet. 2007;39:347–351. doi: 10.1038/ng1975. [DOI] [PubMed] [Google Scholar]
- 10.MacConaill LE, et al. Profiling critical cancer gene mutations in clinical tumor samples. PLoS ONE. 2009;4:e7887. doi: 10.1371/journal.pone.0007887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dias-Santagata D, et al. Rapid targeted mutational analysis of human tumours: a clinical platform to guide personalized cancer medicine. EMBO Mol Med. 2010;2:146–158. doi: 10.1002/emmm.201000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ross JS. Update on HER2 testing for breast and upper gastrointestinal tract cancers. Biomark Med. 2011;5:307–318. doi: 10.2217/bmm.11.31. [DOI] [PubMed] [Google Scholar]
- 13.McCourt CM, Boyle D, James J, Salto-Tellez M. Immunohistochemistry in the era of personalised medicine. J Clin Pathol. 2013;66:58–61. doi: 10.1136/jclinpath-2012-201140. [DOI] [PubMed] [Google Scholar]
- 14.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding L, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens PJ, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerman PS, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roychowdhury S, et al. Personalized oncology through integrative high-throughput sequencing: a pilot study. Sci Transl Med. 2011;3:ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Craig DW, et al. Genome and transcriptome sequencing in prospective refractory metastatic triple negative breast cancer uncovers therapeutic vulnerabilities. Mol Cancer Ther. 2013;12:104–116. doi: 10.1158/1535-7163.MCT-12-0781. [DOI] [PubMed] [Google Scholar]
- 21.Liang WS, et al. Genome-wide characterization of pancreatic adenocarcinoma patients using next generation sequencing. PLoS ONE. 2012;7:e43192. doi: 10.1371/journal.pone.0043192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hadd AG, et al. Targeted, high-depth, next-generation sequencing of cancer genes in formalin-fixed, paraffin-embedded and fine-needle aspiration tumor specimens. J Mol Diagn. 2013;15:234–247. doi: 10.1016/j.jmoldx.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 23.Kerick M, et al. Targeted high throughput sequencing in clinical cancer settings: formaldehyde fixed-paraffin embedded (FFPE) tumor tissues, input amount and tumor heterogeneity. BMC Med Genomics. 2011;4:68. doi: 10.1186/1755-8794-4-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hiatt JB, Pritchard CC, Salipante SJ, O’Roak BJ, Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–854. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gargis AS, et al. Assuring the quality of next-generation sequencing in clinical laboratory practice. Nat Biotechnol. 2012;30:1033–1036. doi: 10.1038/nbt.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagle N, et al. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas RK, et al. Sensitive mutation detection in heterogeneous cancer specimens by massively parallel picoliter reactor sequencing. Nat Med. 2006;12:852–855. doi: 10.1038/nm1437. [DOI] [PubMed] [Google Scholar]
- 28.Beltran H, et al. Targeted next-generation sequencing of advanced prostate cancer identifies potential therapeutic targets and disease heterogeneity. Eur Urol. 2013;63:920–926. doi: 10.1016/j.eururo.2012.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giulino-Roth L, et al. Targeted genomic sequencing of pediatric Burkitt lymphoma identifies recurrent alterations in anti-apoptotic and chromatin-remodeling genes. Blood. 2012;120:5181–5184. doi: 10.1182/blood-2012-06-437624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipson D, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–384. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26:256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 32.Lovly CM, et al. Potentially actionable kinase fusions in inflammatory myofibroblastic tumors. J Clin Oncol. 2013;31(supplement, abstract):10513. [Google Scholar]
- 33.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Albers CA, et al. Dindel: accurate indel calls from short-read data. Genome Res. 2011;21:961–973. doi: 10.1101/gr.112326.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Loo P, et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci USA. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yau C, et al. A statistical approach for detecting genomic aberrations in heterogeneous tumor samples from single nucleotide polymorphism genotyping data. Genome Biol. 2010;11:R92. doi: 10.1186/gb-2010-11-9-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim ES, et al. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1:44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beroukhim R, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacConaill LE, Garraway LA. Clinical implications of the cancer genome. J Clin Oncol. 2010;28:5219–5228. doi: 10.1200/JCO.2009.27.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanton C. My Cancer Genome: a unified genomics and clinical trial portal. Lancet Oncol. 2012;13:668–669. [Google Scholar]
- 43.Beadling C, et al. Combining highly multiplexed PCR with semiconductor-based sequencing for rapid cancer genotyping. J Mol Diagn. 2013;15:171–176. doi: 10.1016/j.jmoldx.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Bose R, et al. Activating HER2 mutations in HER2 gene amplification negative breast cancer. Cancer Discov. 2013;3:224–237. doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Compeau PE, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nat Biotechnol. 2011;29:987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Djordjevic B, et al. Clinical assessment of PTEN loss in endometrial carcinoma: immunohistochemistry outperforms gene sequencing. Mod Pathol. 2012;25:699–708. doi: 10.1038/modpathol.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reis-Filho JS, et al. Cyclin D1 protein overexpression and CCND1 amplification in breast carcinomas: an immunohistochemical and chromogenic in situ hybridisation analysis. Mod Pathol. 2006;19:999–1009. doi: 10.1038/modpathol.3800621. [DOI] [PubMed] [Google Scholar]
- 53.Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


