Abstract
Over the past decade, since epigenetic mechanisms were first implicated in memory formation and synaptic plasticity, dynamic DNA methylation reactions have been identified as integral to long-term memory formation, maintenance, and recall. This review incorporates various new findings that DNA methylation mechanisms are important regulators of non-Hebbian plasticity mechanisms, suggesting that these epigenetic mechanisms are a fundamental link between synaptic plasticity and metaplasticity. Because the field of Neuroepigenetics is so young and the biochemical tools necessary to probe gene specific questions are just now being developed and used, this review also speculates about the direction and potential of therapeutics that target epigenetic mechanisms in the CNS and the unique pharmacokinetic and pharmacodynamic properties that epigenetic therapies may possess. Mapping the dynamics of the epigenome in response to experiential learning, even a single epigenetic mark in isolation, remains a significant technical and bioinformatic hurdle facing the field, but will be necessary to identify changes to the methylome that govern memory-associated gene expression and effectively drug the epigenome.
Keywords: Epigenetics, neuroepigenetics, psychopharmacology, neuropharmacology, pharmacokinetics, DNMT, TET Oxidase, Tcf4, Ube3a, cytosine methylation, DNA methylation, demethylation, learning, memory, cognition, cognitive disorders, Pitt-Hopkins Syndrome, Angelman Syndrome
Introduction
A methyl group is a fairly simple biochemical moiety, yet if bonded to DNA at certain genomic pressure points, can completely silence a gene in perpetuity. If the relevant methyl group is removed, genes inactivated through imprinting or differentiation can be unlocked and rendered fully functional. In the brain, changes in DNA methylation states are necessary for neurodevelopment, plasticity, and even the long-term stability of memory, and accordingly their dysregulation has been linked to neurodevelopmental, neurodegenerative, and psychiatric disease. Neuroepigenetic tools that target DNA methylation mechanisms or alter the methylation patterns of specific gene locus are currently being developed and present a potential paradigm shift in how diseases of the mind can be studied and approached therapeutically. This review seeks to put into context the seemingly strange rules that apply to epigenetic therapies, the advances in the area of therapeutic development, and the neurological disease states that are most likely to benefit from their introduction.
Imagine a drug that could be administered once and, after being cleared from the body, remain efficacious and fully functional, indefinitely. One would probably be hesitant to take this kind of drug, especially if they were looking for relief from a temporary illness. But what if such a drug existed for depression, schizophrenia, or Alzheimer’s disease? In pharmacology, small molecules that covalently and irreversibly bond to an enzyme’s active site are known as suicide inhibitors, because they eliminate enzyme activity for the lifetime of the target protein with stoichiometric efficiency. However, even suicide inhibitors have a limited effect, as they are metabolized and target proteins resynthesized. Epigenetic therapies, and specifically those that induce changes in DNA methylation, may function on an entirely different plane by irreversibly affecting not simply a copy of a protein but the very gene that encodes the protein.
The field of neuroepigenetics is only just now entering its second decade, and the dynamics of epigenetic regulation in the functioning adult CNS remains an active area of research. However, it has become apparent that the epigenome in general and DNA methylation and histone acetylation states specifically are altered by experiential learning and memory and can be modulated to tune underlying neuronal plasticity. This review also hopes to frame the problems facing the field of neuroepigenetics conceptually, experimentally, and therapeutically, and to remark upon ways to potentially not only address them but also capitalize upon them.
DNA Methylation as a Mnemogenic Reaction
Learned information, even episodic memory, can persist for a lifetime, as compared to proteins and mRNA within the activated neurons that subserve plasticity and memory formation, which are usually degraded within hours (Crick, 1984). This leads to the necessity of self-perpetuating biochemical mechanisms by which the memory of a neuron’s activation can be maintained, so-called memory-forming or “mnemogenic” reactions (Roberson and Sweatt, 2001). On the protein level, mnemogenic reactions are known to occur at a minimum at the post-synaptic terminal. For example, the auto-phosphorylation of the calcium-activated protein kinase CaMKII, induced by glutamate signaling at AMPA and NMDA receptors across the synapse (Figure 1), is a well-studied example of local enzymatic signal propagation to mediate synaptic plasticity (Lisman, 1985; Roberson and Sweatt, 2001; Sweatt, 2009). According to the synaptic tagging hypothesis, proposed by Frey and Morris near the end of the 20th century in a landmark publication, these local mnemogenic reactions might explain how long-term potentiation, dependent on transcription occurring in the nucleus, could function at a small subset of synapses among the ten thousand or so throughout the dendritic tree of a single neuron (Frey and Morris, 1998). Indeed, more recent work using two-photon microscopy has elucidated that many cortical synaptic connections can persist on the order of years, despite dynamic synaptic structural changes occurring at neighboring synapses (Attardo et al., 2015). Fascinatingly, synaptic connections appear unstable in the hippocampus after only a few weeks, which potentially explains a great deal about the role of the hippocampus in memory formation and consolidation, but is perplexingly hard to explain with the synaptic tagging hypothesis in isolation, since many of the mnemogenic reactions engaged at the synapse are thought to occur in both cortical and hippocampal neurons.
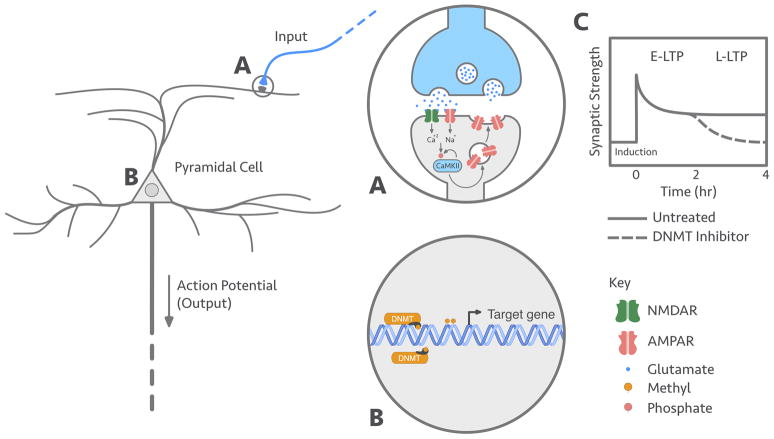
Figure 1.
Molecular mechanisms of memory. A. The induction and early-phase stabilization of LTP (E-LTP) from a glutamatergic presynaptic input requires dual AMPAR and NMDAR activation, flooding the postsynaptic compartment with calcium and sodium. This burst of intracellular calcium leads to the phosphorylation of CaMKII, which increases AMPAR sensitivity and postsynaptic density. CaMKII activation causes CaMKII auto-phosphorylation, a local and short-term mnemogenic reaction that facilitates E-LTP. B. The late-phase stabilization of LTP (L-LTP) requires active DNA methylation of and gene expression of plasticity-related genes. C. Small molecule inhibition of DNMT activity inhibits L-LTP (Miller et al., 2008), in a similar fashion to inhibitors of gene transcription like Actinomycin D (Frey et al., 1996), without affecting induction and E-LTP.
Memory biochemists, aware of the limitations presented by bi-stable self-regenerating mnemogenic biochemical reactions occurring at the synapse, then turned their attention to the nucleus, specifically gene transcription, to explain how the plasticity of a neuron could be altered for the long-term. The synthesis of new proteins had been known to be necessary for the formation of memory for several decades. Indeed, the finding by Flexner, Flexner, and Stellar in 1963 in many ways initiated the field of biochemistry of learning and memory because for the first time memory was shown to be dependent on biochemical processes (Flexner et al., 1963). In the many years since, active transcription and translation have been shown to be necessary for the maintenance of long-term potentiation, memory consolidation, memory reconsolidation after recall, and metaplasticity, such as homeostatic plasticity and intrinsic excitability (Alberini, 2009; Alberini and Kandel, 2015; Guzman-Karlsson et al., 2014; Kozyrev and Nikitin, 2013; Nader et al., 2000). Today it is hypothesized that epigenetic mechanisms, known to integrate environmental influences into the functional genome of a cell, can help explain long-term changes in transcriptional tone orchestrated by the nucleus of a neuron long after learning.
DNA methylation is an important inhibitory epigenetic mark utilized during differentiation to silence genes deemed unnecessary for the future of the cell and its progeny (Smith and Meissner, 2013; Suzuki and Bird, 2008). In the nucleus, DNA is wound tightly around histone proteins forming a nucleosome, the smallest subunit of chromatin. Epigenetic modifications can occur on the histone tails or directly on the DNA at the 5 position of cytosine (Figure 2). DNA methylation is mediated by de novo DNA methyltransferases (DMNTs) that catalyze the methyl transfer from S-adenosyl methionine (SAM) to the cytosine of a cytidine – guanosine (CpG) dinucleoside site. This epigenetic mark can then self-perpetuate through the activity of maintenance DNMTs, which recognize monomethylation and methylate the complementary strand of the CpG site, producing a dimethylated mark (Feng et al., 2010). The persistence of the CpG dimethylated mark is self-evident: mature neurons do not divide, and if a 5-methylcytosine is damaged and needs replacement, the new cytosine can be methylated through maintenance to reconstitute the epigenetic mark. Thus, one current view is that epigenetic molecular mechanisms such as DNA methylation can be both dynamic but also self-perpetuating, by virtue of being a manifestation of a bi-stable biochemical positive feedback system involving self-reinforcing DNMT activity.
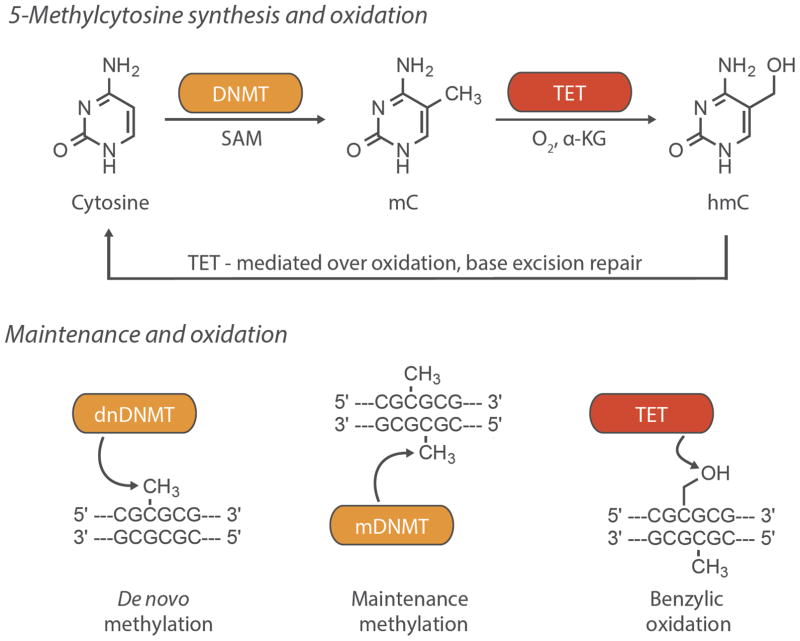
Figure 2.
DNA methylation and maintenance. Top. Cytosine can be methylated at the 5-position by DNMTs, using SAM (S-adenosyl methionine) as an activated methyl source, to form 5-methylcytosine (mC). The methyl group is stable in vivo until oxidized by Tet dioxygenases to generate hydroxymethylcytosine (hmC), which is the first step in the de-methylation pathway. Bottom. De novo DNMTs methylate unmodified DNA to form a hemi-methylated CpG. Hemi-methylation recruits maintenance DNMTs that methylate and encode the mark on the complementary strand. Maintenance DNMT activity provides the mnemogenic nature of the DNA methylation epigenetic mark, which are stable until oxidized by Tet enzymes.
That is not to say de-methylation cannot occur. Active CpG de-methylation mechanisms have been an active area of research over the last several years (Ito et al., 2011). The growing consensus is that Tet proteins, which are a family of dioxygenases, bind double-stranded DNA, detect 5-methylcytosine nucleotides, and oxidize the methyl group by an Fe (II) / alpha-ketoglutarate-dependent mechanism to produce 5-hydroxymethylcytosine (Tahiliani et al., 2009). The hydroxymethyl mark itself is stable and exists in elevated levels in the brain; however, it can be deaminated to 5-hydroxymethyluracil and repaired by base or nucleotide excision mechanisms, erasing cytosine substitution (Williams et al., 2011). Thus the cytosine methylation cycle and regulation of the methylome, insulated from DNA damage by duplicity, is perpetuated by maintenance mechanisms, yet dynamic and potentially plastic when desired.
Over the past decade, since the first discoveries that active DNA methylation was involved in hippocampal LTP and memory formation (Miller and Sweatt, 2007; Lubin et al., 2008; Levenson et al., 2006; Miller et al., 2008), the field of Neuroepigenetics, including DNA methylation in long-term memory, has grown considerably. A host of experiments has been published that catalog the negative effects small molecule inhibitors of the DNMT family have on plasticity and memory. Neuronal activation is sufficient to induce changes in DNA methylation (Day et al., 2013; Guo et al., 2011). Within memory circuits, experiential learning initiates changes in DNA methylation at memory-associated genes (Figure 1) (Day et al., 2013; Malleret et al., 2001; Miller et al., 2010; Miller and Sweatt, 2007; Roth et al., 2009; Tognini et al., 2015). Altering DNA methylation within these circuits is necessary for memory formation (Day et al., 2013; Feng et al., 2010; Kaas et al., 2013; Maddox and Schafe, 2011; Miller et al., 2010; Miller and Sweatt, 2007; Monsey et al., 2011; Rudenko et al., 2013).
After the discovery that DNMT inhibitors could block memory formation and stabilization (Miller and Sweatt, 2007; Lubin et al. 2008) it was not clear which, if any, specific roles the various DNMT isoforms might play in memory formation. This was not a trivial issue, because the canonical role of the DNMT1 “maintenance” isoform was to recognize hemi-methylated DNA and methylate the second strand, whereas the DNMT3a and 3b “de novo” isoforms were assumed to uniquely methylate previously unmethylated sites in the genome. Thus one of the original hypotheses, proposed early on after the first indications that active DNA methylation was critical to memory formation, was that de novo methylation was responsible for encoding new information, while maintenance methylation contributed to maintenance of the memory (reviewed in (Day and Sweatt, 2010). The basic experimental observations were consistent with this view: broad non-isoform-selective small molecule DNMT inhibitors functioned to inhibit memory formation (Day et al., 2015). These broad inhibitors were administered into the hippocampus during the consolidation window shortly after learning and in addition were able to effectively wipe long-term memory recall months after training when infused into the anterior cingulate cortex (Lubin et al. 2008; Miller et al., 2010; Miller and Sweatt, 2007). Therefore, these early studies using small molecule DNMT inhibitors seems at odds with prior results showing that gene knockouts for either DNMT1 or DNMT3a yielded no such observable memory or plasticity phenotypes (Feng et al., 2010). Subsequent work has clarified this apparent discrepancy (Feng et al., 2010; Arand et al.,, 2012). It is now known that altogether there are three DNMTs (DNMT1, DNMT3a, and DNMT3b), and each can function in some capacity as both a de novo and a maintenance enzyme (Arand et al., 2012). There also exists the homologous DNMT3L that is not truly a methyltransferase, since it is incapable of performing that chemistry due to an active site mutation, but does dimerize with DNMT3a to dramatically increase its activity (Suetake et al., 2004). Therefore, due to this functional redundancy, double knockout of both DNMT1 and DNMT3a in excitatory hippocampal and forebrain neurons was necessary block L-LTP and cause long-term memory deficits, validating the original experiments performed with broad small molecule inhibitors (Feng et al., 2010).
It is important to also note that this early debate was ongoing before the mechanistic discovery that Tet enzymes metabolize methylated cytosine, enabling active demethylation (Tahiliani et al., 2009). The early findings of effects of DNMT inhibitors on cognitive function led to considerable debate concerning whether DNMT inhibition could have any effect at all in a non-dividing cell such as a neuron. Central epigenetics dogma at the time was that DNA cytosine methylation was irreversible once established. (Ooi and Bestor, 2008; Klose and Bird, 2006; Suzuki and Bird, 2008; Pastor et al., 2013). Indeed, for some time the idea that active cytosine de-methylation occurred in fully differentiated neurons was thought to be unlikely. However, recent work has clarified this issue, revealing that Tet enzymes mediate active demethylation (Tahiliaini et al., 2009; Ito et al., 2011; Guo et al., 2011; Pastor et all, 2013) and that altering Tet enzyme activity has pronounced effects on memory formation and stabilization. Thus, oxidation of methylated cytosine to the hydroxymethyl mark followed by over oxidation to formyl- and carboxy- cytosine, leads to erasure of the covalent mark through base excision repair, and it is now clear that this reaction occurs extensively in neurons and responds to neuronal activation (Kangaspeska et al., 2008; Métivier et al., 2008). Moreover, Tet1 knockout mice exhibit impaired memory extinction (reviewed in Tsai, 2013), and viral overexpression of Tet1 in the CA1 region of the hippocampus induces deficits in contextual fear conditioning (Kaas et al., 2013; Rudenko et al., 2013). Gadd45b, a gene involved in aiding de-methylation via base excision repair, when knockout produced an enhanced memory phenotype (Sultan and Sweatt, 2013; Sultan et al., 2012). Taken together, these data suggest a negative regulatory role for de-methylation mechanisms, and that inhibiting the removal of DNA methylation marks laid down after learning enhances memory maintenance.
We also note that the discovery of the dynamic, activity-regulated neuronal methylome also raises another apparent conundrum that has not yet been resolved. These discoveries imply that there must exist some as-yet-unidentified mechanism that serves to compartmentalize the plasticity-associated malleable methylome from the stable methylome associated with perpetuation of cell fate and cellular phenotype. The nature of and mechanism underlying this implied compartmentalization, or indeed even the existence of such a mechanism, remains an important area for future resolution (Sweatt, 2013a).
Tuning Plasticity
Mnemogenic reactions at the synapse are thought to tag that synapse after stimulation and self-propagate biochemically utilizing local resources, to in effect maintain an acquired modification sustained over time by the arrival of gene products originating from the nucleus and the attendant transcriptional machinery. In this model mnemogenic reactions in the nucleus should impact and regulate the entire neuronal system of synapses (on the order of tens of thousands per neuron), or at least those that are tagged and waiting for mRNA to arrive. But as noted previously, synaptic tagging only lasts on the timescale of hours, while nuclear changes to the methylome and transcriptome persist much longer, maybe indefinitely. Conceptually speaking, the long-term consequences of altered epigenetic mechanisms of memory should therefore be systemic, and biochemical processes that regulate long-term changes in electrophysiology ought to be primary targets for changes in DNA methylation.
Homeostatic plasticity is a type of non-Hebbian plasticity that functions to modulate synaptic sensitivity up or down across all synapses in a concerted manner (Nelson and Turrigiano, 2008). The easiest way to think about homeostatic plasticity is to imagine a room with many light bulbs of varying intensity all controlled by the same dimmer switch. Adjusting the dimmer switch regulates the relative brightness of each of the light bulbs in concert, but in the homeostatic model, the brighter light bulbs become brighter or dimmer at a steeper slope than bulbs of a lower wattage. In this metaphor the light bulbs are synapses and the dimmer switch is the nucleus, which controls the weight of each synapse in unison and multiplicatively. The synaptic mechanism underlying homeostatic plasticity is synaptic scaling, where there is an increase in postsynaptic AMPA receptor density in response to reduced neuronal firing (upscaling) and a decrease in AMPA receptor density after heightened periods of neuronal activity (downscaling) (Kilman et al., 2002; Rannals and Kapur, 2011; Shin et al., 2012; Turrigiano et al., 1998; Wierenga et al., 2005). Notably, the direction of AMPA receptor trafficking in response to activity is opposite to that of traditional mechanisms of Hebbian plasticity, in which the strength of individual synapses can be altered to encode the memory trace. Thus, via scaling, the excitability of all synapses in aggregate can be tuned without compromising the relative synaptic weights established during learning, allowing neurons a mechanism to maintain relatively stable overall firing rates (Turrigiano, 2008).
Understanding how synaptic scaling is mediated at the synapse can tell us quite a lot about how neurons control their long-term excitability, but very little about how the nucleus orchestrates the change. It seems at least likely that epigenetic mechanisms play a part is steadying the transcriptional output of the nucleus, and very recently two simultaneous studies probed the question of whether changes in DNA methylation might be behind homeostatic plasticity. Applying the sodium channel blocker tetrodotoxin (TTX), the neurotoxin found in pufferfish, to primary cultured neurons decreased global synaptic activity resulting in upscaling, an effect that was blocked by silencing Tet1 expression (Yu et al., 2015). Conversely, silencing Tet3 expression blocked downscaling induced by the GABA receptor antagonist bicuculline, which increased global synaptic activity. Moreover, inhibiting DNMT activity with the competitive inhibitor RG-108 blocked TTX-induced scaling, suggesting that active DNA methylation was necessary for upscaling to occur (Meadows et al., 2015). This effect was also observed by knocking down both DNMT1 and DNMT3a with targeted antisense oligonucleotides. All of these data taken together suggest that the dynamic regulation of DNA methylation states is required to alter synaptic scaling, a nice concept convoluted by the fact that each of these treatments that block DNMT or Tet activity themselves cause changes in synaptic scaling. For example, RG108 blocks TTX-induced upscaling, suggesting active DNA methylation is required for upscaling, but if TTX is removed and just RG108 applied, the neuron scales up. We therefore can only conclude that DNA methylation is important for tuning homeostatic plasticity and that new methods of altering local DNA methylation states at particular genes of interest rather than global blocking DNMT or Tet activity will be necessary to resolve these findings.
Another form of non-Hebbian plasticity that is functionally distinct from synaptic scaling, yet modifies global excitability, is intrinsic plasticity (reviewed in (Guzman-Karlsson et al., 2014). Intrinsic plasticity involves the regulation of the population of sodium and potassium ion channels that determine the biophysical properties of neuronal firing. Shortly after training with classical conditioning, pyramidal cells become hyperexcitable, eliciting a larger number of spikes for a given depolarizing applied voltage (Coulter et al., 1989; Disterhoft and Oh, 2006; Thompson et al., 1996). The effects of intrinsic plasticity associated with learning tend to be short-lived relative to the behavioral memory, especially in the case of hippocampus-dependent tasks (Kaczorowski and Disterhoft, 2009; McKay et al., 2009; Ohno et al., 2006). Therefore, it is possible that these alterations in the expression of voltage-gated sodium and potassium channels do not require a long-lived mnemogenic reaction in the nucleus and instead rely on second messengers (cAMP) and their transcriptional targets (CREB-mediated) to elicit short-term changes in membrane machinery (McClung and Nestler, 2003). However, outside the hippocampus there are examples of truly extended changes in channel expression, such as the downregulation of sodium channel Nav1.8 and potassium channel Kv4.3 in the case of animal modes of neuropathic pain following nerve injury (Uchida et al., 2010a; Uchida et al., 2010b). This decrease in expression is correlated with decreased histone acetylation at the respective gene promoters, so it is entirely possible that epigenetic mechanisms, and even DNA methylation, may serve to regulate changes in intrinsic plasticity.
Transcription
The question that resides at the heart of designing therapeutics to target DNA methylation mechanisms is: how does DNA methylation regulate transcription? And for that matter, how does DNA methylation communicate with or even dictate other epigenetic marks that regulate chromatin structure? The simplest mechanism by which DNA methylation regulates transcription is through the direct steric inhibition of transcription factor binding. A pair of methyl groups tucked into the major groove of DNA hardly seems like a major steric force to be reckoned with, yet that is precisely where many transcription factors scan to recognize their preferred binding sequences (Miranda and Jones, 2007). Methylation of a CTCF binding site within the H19 imprinting control region proximal to insulin-like growth factor 2 (Igf2) blocks CTCF binding on the paternal allele (Felsenfeld and Bell, 2000). Since CTCF binding inhibits transcription, the paternal allele is freely transcribed. The maternal allele, which lacks methylation through this region, binds CTCF and is silenced. Igf2 is an important growth factor in neuronal plasticity, and targeting insulin-like growth factors is currently being investigated as therapies applicable for a range of learning and memory disorders and diseases, such as Rett Syndrome to Alzheimer’s disease (Pascual-Lucas et al., 2014; Pini et al., 2012). One can therefore imagine a therapeutic tool that selectively methylates this insulator region, freeing the maternal copy of Igf2, and boosting growth factor production in vivo utilizing naturally occurring mechanisms.
DNA methylation also increases nucleosome compaction by inhibiting or attracting histone-remodeling enzymes (Figure 3) (Cedar and Bergman, 2009). Proteins that contain CXXC domains detect unmethylated CpG sites and recruit modifiers of histone methylation (Allen et al., 2006; Blackledge et al., 2010; Lee and Skalnik, 2005), an interaction blocked by DNA methylation (Lee et al., 2001). However, it appears most histone-remolding proteins are actually attracted to methylated DNA by methyl CpG-binding domain (MBD) containing proteins. MBD proteins are relatively small factors that scan the major groove of DNA and preferentially detect dimethylated or hemi-methylated CpG sites. Once bound, MBD proteins can recruit histone deacetylases (HDACs), which increase the cationic nature of the associated nucleosomes and inhibit transcription by compacting local chromatic structure, and can attract repressive H3K9 methyltransferases (Fujita et al., 2003; Kass et al., 1997; Ng et al., 1999). MBD proteins bind methylated CpG sites with high fidelity, but only weakly bind hemi-methylated sites (Hashimoto et al., 2012). Notably, mutations in the MBD genes MeCP2 and MBD5 are the genetic causes for the developmental disabilities Rett Syndrome and 2q23.1 microdeletion syndrome, also known as Pseudo-Angelman Syndrome (Lyst and Bird, 2015; Williams et al., 2010). Hemi-methylated CpG sites are recognized by the ubiquitin ligase UHRF1, which recruits DNMT1 to facilitate maintenance activity and further recruitment of HDACs (Klose and Bird, 2006; Liu et al., 2013). Interestingly, hydroxymethylation through Tet activity of the CpG site inhibits both MBD and UHRF1 binding. This suggests that the stable hydroxymethyl epigenetic mark may have selective inhibitory functions, where it continues to inhibit transcription factor binding sterically without recruiting repressive histone-modifiers to contract local chromatin structure (Pastor et al., 2013).
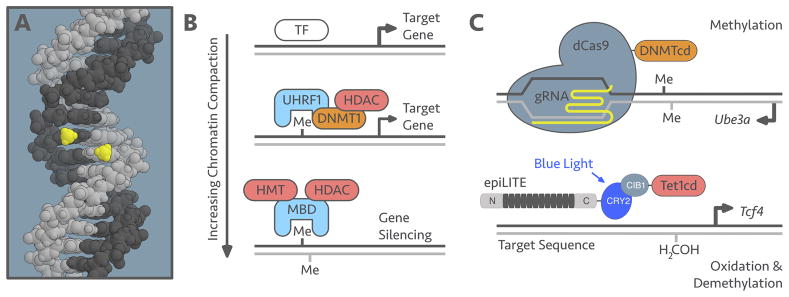
Figure 3.
Structure and Function. A. Despite being on opposite strands, CpG methylation is present in a cis relationship within the major groove of DNA. Methyl groups on the CpG site shown in yellow. B. DNA methylation can blocks transcription factor binding. Hemi-methylation recruits UHRF1 that localizes DNMT1 and HDACs. CpG methylation recruits MBD proteins that bind HDACs and histone methyltransferases (HMTs). These processes lead to an increasing chromatin compaction and gene silencing. C. dCas9 and TALE epigenetic modifiers targeted therapies could be utilized to increase the expression of Ube3a and Tcf4 for the treatment of Angelman Syndrome and Pitt-Hopkins Syndrome, respectively. TF, transcription factor; HMT, histone methyltransferase, gRNA, guide RNA; DNMTcd, DNMT catalytic domain; TETcd, TET catalytic domain.
The recruitment of HDACs by MDB proteins to sites of DNA methylation serves as a strong functional link between active DNA methylation post-experiential learning and chromatin reorganization during the formation of long-term memory (Cedar and Bergman, 2009; Miller et al., 2008). Histone acetylation is a powerful epigenetic mark that is strongly associated with increased gene expression, and HDACs are seductive targets for cognitive enhancement (Gräff and Tsai, 2013) and for treatment of neurodegenerative and psychiatric disorders (Abel and Zukin, 2008). Indeed, HDAC inhibition acts like a general cognitive boost across a diverse set of memory disease and disorder models. This is rather curious since DNMT inhibitors ablate memory so thoroughly, and HDACs and DNMTs co-localize to negatively regulate transcription in general. This phenomenon remains mostly unexplained and highlights the limitations of the tools at our disposal to probe neuroepigenetic mechanisms at present. Even if isoform-selective DNMT and HDAC inhibitors existed, their effects would be felt globally at both memory suppressing and enhancing genes. Knocking out or knocking down specific epigenetic modifiers is experimentally useful, but again act globally and almost certainly affect epigenetic modifying protein complexes, not just enzymatic activity. A new class of neuroepigenetic therapies is under development that holds the potential to selectively target individual epigenetic marks and harness their innate mnemogenic biochemistry to perpetually unlock genetic therapies already present in the genome.
Neuroepigenetic Therapies
Epigenetic marks by their nature are long-lived and function to determine the long-term individuality of a cell compared to other cells across the organism that carry the same genome. Therapeutics that add or remove methyl groups from DNA, therefore, have specific attributes that can in some instances apply to their pharmaco-kinetics and -dynamics that are unique relative to other classes of drugs, and that warrant the attention of medicinal chemists.
Attribute 1
The fidelity of DNA methylation patterns is clearly a high priority for the cell, since they are integral to maintaining cell-type identity. Any therapy that altered the methylation status of even a single CpG site, could be propagated by maintenance DNMT mechanisms perpetually over time or even trans-generationally. Traditional small molecule drugs are generally metabolized and have a limited half-life. Even suicide inhibitors are only effective on the timescale of protein turnover, but a change in methylation status at a CpG site could be perpetuated through maintenance DNMT activity and have transcriptional effects that last the lifetime of the cell. Therefore targeting cytosine methylation mechanisms in post-mitotic neurons could conceivably trigger lifelong effects – a potentially unique pharmacokinetic characteristic.
Attribute 2
We know from gene imprinting that, at least in a few dozen cases, the imprinting of genes can be canonical, where the expression of a gene is completely silenced through epigenetic mechanisms (Gregg et al., 2010a; Gregg et al., 2010b). Conceptually, the methylation of a single CpG site could completely silence a gene or selectively silence a single isoform or exon of that gene. Conversely, removing a methyl mark could render a gene previously silenced by imprinting fully functional, or cause an increase in the rate of transcription for a gene already being expressed. All-or-none pharmacodynamics of this sort is a rarity in traditional pharmacology. In principle, this holds the possibility of epigenetically targeted drugs being transformative for certain monogenic intellectual disabilities, such as Angelman Syndrome and Pitt-Hopkins Syndrome.
Angelman Syndrome is a rare developmental disorder on the autism spectrum that is caused by a loss of function mutation or deletion in the maternal copy of the ubiquitin ligase UBE3A, an imprinted gene in the CNS (Clayton-Smith and Laan, 2003). Methylation of the maternal allele silences a nuclear-localized long non-coding RNA, UBE3A antisense transcript (UBE3A-ATS), allowing for UBE3A expression. The paternal allele is hypomethylated, which leads to the expression of UBE3A-ATS and the subsequent silencing of UBE3A expression. Remarkably, the function of the inhibitory UBE3A-ATS transcript occurs in a very localized fashion without affecting the expression of the maternal allele, and anti-sense oligonucleotides that target UBE3A-ATS increase the expression of paternal UBE3A and rescue the memory phenotypes of Angelman Syndrome mouse model (Meng et al., 2015). Hypermethyalation of the UBE3A-ATS locus on the paternal allele is therefore a viable therapeutic strategy. If DNA methylation of the UBE3A-ATS is the underlying mechanism that perpetuates the imprinting of the UBE3A gene, then such an intervention would “turn on” the paternal allele and essentially take advantage of a gene therapy already present within Angelman Syndrome patients.
Pitt-Hopkins Syndrome (PTHS) presents very similarly to Angelman Syndrome. So much so in fact that until the genetic cause of PTHS was discovered to be the haploinsufficiency of transcription factor 4 (Tcf4), many PTHS patients were diagnosed with Angelman Syndrome (Sweatt, 2013). The role of Tcf4 in the CNS is poorly understood, but PTHS patients have profound learning and memory deficits, the most prominent of which is often a complete lack of language acquisition. PTHS patients have a loss of function mutation or deletion of the basic helix-loop-helix DNA binding domain common to E-protein transcription factors. Tcf4 does not appear to be an imprinted gene, but PTHS patients do have one functioning and one non-functioning Tcf4 copy. Theoretically, a neuroepigenetic therapy could be designed that de-methylates the Tcf4 gene causing an increase in expression to “normalize” the protein levels of functional Tcf4 protein (Figure 3). Beyond functioning solely as an inhibitory mark, DNA methylation can also facilitate an increase in gene expression; therefore, directed methylation at regions within the Tcf4 locus may also act to generally increase expression or to selectively increase the expression of select Tcf4 isoforms that may be more beneficial to treating PTHS.
Attribute 3
Of course, the same pharmaco-kinetic and dynamic principles would also apply to off-target and on-target side effects, potentially conferring unique toxicological properties on epigenetically targeted compounds. This non-trivial consequence should give the drug development community pause before advancing such therapies to the clinic. Since these therapies would target specific genomic locus, and patients vary in their genomes and epigenomes, the potential for idiosyncratic effects are particularly concerning.
Genomic Approaches
Genome editing is one of the fasting growing fields in the biological sciences, most notably the development of CRISPR-Cas9 systems, zinc fingers, and transcription activator-like effectors (TALEs) that can localize to selected DNA sequences (Hilton and Gersbach, 2015). Similarly, these tools have been adapted for editing the epigenome. In the case of the CRISPR-Cas9, the nuclease active site is deactivated (dCas9) creating not a genome editing protein, but a genome localizing protein that can be directed to a gene of interest using a guide RNA (Shalem et al., 2015). Tethering transcriptional activators, repressors, and epigenetic modifying enzymes to the dCas9 subunit facilitates highly localized biochemistry. The Gersbach lab has recently shown that tethering the histone acetyltransferase p300 and localizing them to promoter and enhancer regions significantly increased histone 3 lysine 17 acetylation and profoundly increased the expression of target genes, even more so than directly tethering transcriptional activators to the same promoter regions (Hilton et al., 2015). Tethering p300 to other DNA localizing constructs (both zinc fingers and TALEs) was also successful at upregulating target gene expression.
Zinc fingers and TALEs, which unlike the dCas9 system that can be used to target different loci simply by employing a different guide RNA, are multi-subunit proteins that target a unique nucleotide sequence, generally >12 nucleotides in length. Each has been used to target histone-modifying proteins including histone methyltransferase, acetyltransferase, and deacetylases. In a groundbreaking study, the Nestler lab utilized these tools to modify histone methylation and acetylation states at the known addition-related gene Fosb, which respectively repressed and enhanced sensitization to cocaine addiction in rats (Heller et al., 2014). The Zhang lab has introduced an additional layer of control by associating the histone modifier with the genomic anchoring system using the light-sensitive cryptochrome 2 protein (CRY2) and its partner CIB1 (Konermann et al., 2013). These epigenetic light-inducible transcriptional effectors (epiLITEs) and other optoepigenetic constructs provide the temporal control necessary to probe the dynamics of epigenetic regulation in memory formation at different time points along the formation and consolidation of the memory trace. An added benefit of using optoepigenetic constructs is the flexibility to created modular toolsets. A TALE fixed with CRY2 and designed to localize at a target gene that could be paired with many epigenetic modifying enzymes fused with a CIB1 binding partner.
Similar locus-selective tools can be designed to affect local DNA methylation, and TALEs tethered with DNMTs that target a CpG island at cyclin-dependent kinase inhibitor 2A increase local methylation and inhibition transcription in vitro (Bernstein et al., 2015). The progress in the development of these gene-selective targeted epigenetic modifiers has been truly remarkable over the past few years, and building on these tools to create temporally controlled in vivo DNA methylating and de-methylating systems is an obvious need to understand how DNA methylation functions to regulate learning and memory (Figure 3). To date, most of what is known about DNA and memory comes from correlative measures associated with memory and genome-wide ablation or enhancement of active DNA methylation mechanisms. The conceptual leap these locus-selective tools can provide in understanding how individual epigenetic marks interact to govern gene expression over the long-term cannot be overstated.
Summary
There is a set of important questions that currently confront the field of Neuroepigenetics. Very little is known about what type of changes to the methylome occur in neurons due to learning, most research to date being focused on methylation changes that occur at single genes. Evidence suggests that both active DNA methylation and de-methylation mechanisms occur is response to learning, yet it is mostly unknown what percentage of these overall changes are functionally relevant. How does the neuron know what genes or regions within and around genes to actively methylate in response to activation? Or de-methylate? One promising candidate is the inhibition or activation of epigenetic machinery around a single gene by locally concentrated RNAs. Extra coding RNAs (ecRNAs) are long non-coding RNAs that often include and span beyond the bounds of the gene body. These ecRNAs are potent inhibitors of DNMT1 and DNA methylation locally potentially without measurably affecting global DNA methylation levels or even nearby genes (Di Ruscio et al., 2013). If the expression of ecRNAs track with mRNA from the same gene locus, they may be a prominent force in altering methylation patterns at genes with the transcriptional changes underlying memory formation. How these ecRNAs can act as potent DNMT1 inhibitors in such a localized fashion along the genome is also a bit of a mystery.
Clearly active DNA methylation performs an important role in establishing threat recognition memory and addiction behavior. Spatial memory and representation are also regulated by DNA methylation, such as learning and memory in the Morris water maze and for the place field stability. Yet it remains unclear if these differing forms of learning require vastly different patterns of altered DNA methylation. Mapping these networks of genes with altered methylation statuses across the genome for multiple memory behavioral tasks in different memory circuits and at different time points after training is an important informatic set of barriers facing the field. Yet these data will be critical for designing epigenetic modifying tools to improve cognition or alter behavior across an array of heterogeneous diseases, disorders, and non-pathological causes of memory impairment, such as Alzheimer’s disease, addiction, post-traumatic stress disorder, and age-related cognitive decline to name a few.
If active DNA methylation is responsible for the formation of long-term memory and the regulation of metaplastic mechanisms that govern neuronal excitability, do DNA methylation mechanisms primarily function to stabilize the engram via altered excitability? DNMT inhibitors administered during the consolidation window disrupt memory formation (Miller and Sweatt, 2007), but they also disrupt long-term memory maintenance when infused into the anterior cingulate cortex (ACC) a month after fear conditioning (Miller et al., 2010), a region of the cortex shown to be a storage site for very long-lasting contextual memory (Frankland, 2004; Restivo et al., 2009; Wang et al., 2009). The erasure of the remote memory trace in the ACC by DNMT inhibition could be explained by an alteration in synaptic scaling that renders the trace destabilized, and the dynamics of DNA methylation in memory circuits may function as a rheostat for Hebbian and non-Hebbian forms of plasticity.
Understanding how DNA methylation mechanisms facilitate long-term potentiation and memory formation, function to stabilize circuits by altering homeostatic mechanisms of plasticity, and actively maintain long-term memory storage in the cortex is fundamental to the advancement of Neuroepigenetics. Moreover, such information is critical to designing a new generation of therapies that may potentially harness the unique chemical attributes that make epigenetic mechanisms so integral to memory function.
Acknowledgments
We thank Lynn A. Mandeltort, Mikael C Guzman Karlsson, Garrett Kaas, Kim Hawkins, and the other members of the Sweatt lab for stimulating discussion. This work is dedicated to the memory of Kindal Kivisto.
Footnotes
Declaration of Interest section
The authors report no declarations of competing interests. This work was supported by MH104158, MH57014, MH091122, Civitan International, the Evelyn F. McKnight Brain Research Foundation, and the Pitt-Hopkins Syndrome (PTHS) Foundation (JDS). AJK is supported by the PTHS Foundation.
Contributor Information
Andrew J. Kennedy, Department of Neurobiology, University of Alabama at Birmingham, SHEL 1074B1, 1825 University Blvd, Birmingham, AL 35294, 205-934-5298
J. David Sweatt, Department of Neurobiology, University of Alabama at Birmingham, SHEL 1010, 1825 University Blvd, Birmingham, AL 35294, 205-975-5196.
References
- Abel T, Zukin RS. Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 2008;8:57–64. doi: 10.1016/j.coph.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM, Kandel ER. The regulation of transcription in memory consolidation. Cold Spring Harbor Perspectives in Biology. 2015;7:a021741–a021741. doi: 10.1101/cshperspect.a021741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen MD, Grummitt CG, Hilcenko C, Min SY, Tonkin LM, Johnson CM, Freund SM, Bycroft M, Warren AJ. Solution structure of the nonmethyl-CpG-binding CXXC domain of the leukaemia-associated MLL histone methyltransferase. The EMBO journal. 2006;25:4503–4512. doi: 10.1038/sj.emboj.7601340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arand J, Spieler D, Karius T, Branco MR, Meilinger D, Meissner A, Jenuwein T, Xu G, Leonhardt H, Wolf V, et al. In vivo control of CpG and non-CpG DNA methylation by DNA methyltransferases. PLoS genetics. 2012;8:e1002750. doi: 10.1371/journal.pgen.1002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo A, Fitzgerald JE, Schnitzer MJ. Impermanence of dendritic spines in live adult CA1 hippocampus. Nature. 2015;523:592–596. doi: 10.1038/nature14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein DL, Le Lay JE, Ruano EG, Kaestner KH. TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. The Journal of clinical investigation. 2015;125:1998–2006. doi: 10.1172/JCI77321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackledge NP, Zhou JC, Tolstorukov MY, Farcas AM, Park PJ, Klose RJ. CpG islands recruit a histone H3 lysine 36 demethylase. Molecular Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature Reviews: Genetics. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- Clayton-Smith J, Laan L. Angelman syndrome: a review of the clinical and genetic aspects. Journal of medical genetics. 2003;40:87–95. doi: 10.1136/jmg.40.2.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter DA, Lo Turco JJ, Kubota M, Disterhoft JF, Moore JW, Alkon DL. Classical conditioning reduces amplitude and duration of calcium-dependent afterhyperpolarization in rabbit hippocampal pyramidal cells. Journal of neurophysiology. 1989;61:971–981. doi: 10.1152/jn.1989.61.5.971. [DOI] [PubMed] [Google Scholar]
- Crick F. Memory and molecular turnover. Nature. 1984;312:101. doi: 10.1038/312101a0. [DOI] [PubMed] [Google Scholar]
- Day JJ, Childs D, Guzman-Karlsson MC, Kibe M, Moulden J, Song E, Tahir A, Sweatt JD. DNA methylation regulates associative reward learning. Nature neuroscience. 2013;16:1445–1452. doi: 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Kennedy AJ, Sweatt JD. DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annual review of pharmacology and toxicology. 2015;55:591–611. doi: 10.1146/annurev-pharmtox-010814-124527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Sweatt JD. DNA methylation and memory formation. Nature neuroscience. 2010;13:1319–1323. doi: 10.1038/nn.2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disterhoft JF, Oh MM. Learning, aging and intrinsic neuronal plasticity. Trends in neurosciences. 2006;29:587–599. doi: 10.1016/j.tins.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Bell AC. Methylation of a CTCF-dependent boundary controls imprinted expressionof the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature neuroscience. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flexner JB, Flexner LB, Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141:57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- Frankland PW. The Involvement of the Anterior Cingulate Cortex in Remote Contextual Fear Memory. Science. 2004;304:881–883. doi: 10.1126/science.1094804. [DOI] [PubMed] [Google Scholar]
- Frey U, Frey S, Schollmeier F, Krug M. Influence of actinomycin D, a RNA synthesis inhibitor, on long-term potentiation in rat hippocampal neurons in vivo and in vitro. J Physiol. 1996;490(Pt 3):703–711. doi: 10.1113/jphysiol.1996.sp021179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey U, Morris RG. Synaptic tagging: implications for late maintenance of hippocampal long-term potentiation. Trends Neurosci. 1998;21:181–188. doi: 10.1016/s0166-2236(97)01189-2. [DOI] [PubMed] [Google Scholar]
- Fujita N, Watanabe S, Ichimura T, Ohkuma Y, Chiba T, Saya H, Nakao M. MCAF Mediates MBD1-Dependent Transcriptional Repression. … and cellular biology. 2003 doi: 10.1128/MCB.23.8.2834-2843.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gräff J, Tsai LH. The potential of HDAC inhibitors as cognitive enhancers. Annual review of pharmacology and toxicology. 2013;53:311–330. doi: 10.1146/annurev-pharmtox-011112-140216. [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. Sex-specific parent-of-origin allelic expression in the mouse brain. Science. 2010a;329:682–685. doi: 10.1126/science.1190831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Weissbourd B, Luo S, Schroth GP, Haig D, Dulac C. High-resolution analysis of parent-of-origin allelic expression in the mouse brain. Science. 2010b;329:643–648. doi: 10.1126/science.1190830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Ma DK, Mo H, Ball MP, Jang MH, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Karlsson MC, Meadows JP, Gavin CF, Hablitz JJ, Sweatt JD. Transcriptional and epigenetic regulation of Hebbian and non-Hebbian plasticity. Neuropharmacology. 2014;80:3–17. doi: 10.1016/j.neuropharm.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic acids research. 2012;40:4841–4849. doi: 10.1093/nar/gks155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Cates HM, Peña CJ, Sun H, Shao N, Feng J, Golden SA, Herman JP, Walsh JJ, Mazei-Robison M, et al. Locus-specific epigenetic remodeling controls addiction- and depression-related behaviors. Nature neuroscience. 2014;17:1720–1727. doi: 10.1038/nn.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers : Nature Biotechnology : Nature Publishing Group. Nature. 2015 doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton IB, Gersbach CA. Enabling functional genomics with genome engineering. Genome research. 2015;25:1442–1455. doi: 10.1101/gr.190124.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito SS, Shen LL, Dai QQ, Wu SCS, Collins LBL, Swenberg JAJ, He CC, Zhang YY. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas GA, Zhong C, Eason DE, Ross DL, Vachhani RV, Ming G-l, King JR, Song H, Sweatt JD. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczorowski CC, Disterhoft JF. Memory deficits are associated with impaired ability to modulate neuronal excitability in middle-aged mice. Learning & Memory. 2009;16:362–366. doi: 10.1101/lm.1365609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kangaspeska S, Stride B, Métivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Current Biology. 1997 doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- Kilman V, van Rossum MCW, Turrigiano GG. Activity deprivation reduces miniature IPSC amplitude by decreasing the number of postsynaptic GABA(A) receptors clustered at neocortical synapses. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2002;22:1328–1337. doi: 10.1523/JNEUROSCI.22-04-01328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, Platt RJ, Scott DA, Church GM, Zhang F. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozyrev SA, Nikitin VP. Involvement of translation and transcription processes into neurophysiological mechanisms of long-term memory reconsolidation. Bulletin of Experimental Biology and Medicine. 2013;154:584–587. doi: 10.1007/s10517-013-2004-9. [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. The Journal of biological chemistry. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Lee JH, Voo KS, Skalnik DG. Identification and characterization of the DNA binding domain of CpG-binding protein. The Journal of biological chemistry. 2001;276:44669–44676. doi: 10.1074/jbc.M107179200. [DOI] [PubMed] [Google Scholar]
- Levenson JM, Roth TL, Lubin FD, Miller CA, Huang IC, Desai P, Malone LM, Sweatt JD. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:15763–15773. doi: 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Lisman JEJ. A mechanism for memory storage insensitive to molecular turnover: a bistable autophosphorylating kinase. PNAS. 1985;82:3055–3057. doi: 10.1073/pnas.82.9.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nature communications. 2013;4:1563. doi: 10.1038/ncomms2562. [DOI] [PubMed] [Google Scholar]
- Lubin FD, Roth TL, Sweatt JD. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J Neurosci. 2008;28:10576–10586. doi: 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Bird A. Rett syndrome: a complex disorder with simple roots. Nature Reviews: Genetics. 2015;16:261–275. doi: 10.1038/nrg3897. [DOI] [PubMed] [Google Scholar]
- Maddox SA, Schafe GE. Epigenetic alterations in the lateral amygdala are required for reconsolidation of a Pavlovian fear memory. Learn Mem. 2011;18:579–593. doi: 10.1101/lm.2243411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, Mansuy IM. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- McClung CA, Nestler EJ. Regulation of gene expression and cocaine reward by CREB and DeltaFosB. Nat Neurosci. 2003;6:1208–1215. doi: 10.1038/nn1143. [DOI] [PubMed] [Google Scholar]
- McKay BM, Matthews EA, Oliveira FA, Disterhoft JF. Intrinsic neuronal excitability is reversibly altered by a single experience in fear conditioning. Journal of neurophysiology. 2009;102:2763–2770. doi: 10.1152/jn.00347.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JP, Guzman-Karlsson MC, Phillips S, Holleman C, Posey JL, Day JJ, Hablitz JJ, Sweatt JD. DNA methylation regulates neuronal glutamatergic synaptic scaling. Science signaling. 2015;8:ra61. doi: 10.1126/scisignal.aab0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Métivier R, Gallais R, Tiffoche C, Le Péron C, Jurkowska RZ, Carmouche RP, Ibberson D, Barath P, Demay F, Reid G, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- Miller CA, Campbell SL, Sweatt JD. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol Learn Mem. 2008;89:599–603. doi: 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Gavin CF, White JA, Parrish RR, Honasoge A, Yancey CR, Rivera IM, Rubio MD, Rumbaugh G, Sweatt JD. Cortical DNA methylation maintains remote memory. Nature neuroscience. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miranda TB, Jones PA. DNA methylation: The nuts and bolts of repression. Journal of cellular physiology. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nader K, Schafe GE, Le Doux JE. Fear memories require protein synthesis in the amygdala for reconsolidation after retrieval. Nature. 2000;406:722–726. doi: 10.1038/35021052. [DOI] [PubMed] [Google Scholar]
- Nelson SB, Turrigiano GG. Strength through diversity. Neuron. 2008;60:477–482. doi: 10.1016/j.neuron.2008.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nature genetics. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Ohno M, Sametsky EA, Silva AJ, Disterhoft JF. Differential effects of alphaCaMKII mutation on hippocampal learning and changes in intrinsic neuronal excitability. The European journal of neuroscience. 2006;23:2235–2240. doi: 10.1111/j.1460-9568.2006.04746.x. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Pascual-Lucas M, Viana da Silva S, Di Scala M, Garcia-Barroso C, González-Aseguinolaza G, Mulle C, Alberini CM, Cuadrado-Tejedor M, Garcia-Osta A. Insulin-like growth factor 2 reverses memory and synaptic deficits in APP transgenic mice. EMBO molecular medicine. 2014;6:1246–1262. doi: 10.15252/emmm.201404228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pini G, Scusa MF, Congiu L, Benincasa A, Morescalchi P, Bottiglioni I, Di Marco P, Borelli P, Bonuccelli U, Della-Chiesa A, et al. IGF1 as a Potential Treatment for Rett Syndrome: Safety Assessment in Six Rett Patients. Autism research and treatment. 2012;2012:679801. doi: 10.1155/2012/679801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rannals MD, Kapur J. Homeostatic strengthening of inhibitory synapses is mediated by the accumulation of GABA(A) receptors. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011;31:17701–17712. doi: 10.1523/JNEUROSCI.4476-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restivo L, Vetere G, Bontempi B, Ammassari-Teule M. The formation of recent and remote memory is associated with time-dependent formation of dendritic spines in the hippocampus and anterior cingulate cortex. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:8206–8214. doi: 10.1523/JNEUROSCI.0966-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberson EDE, Sweatt JDJ. Memory-forming chemical reactions. Reviews in the Neurosciences. 2001;12:41–50. doi: 10.1515/revneuro.2001.12.1.41. [DOI] [PubMed] [Google Scholar]
- Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko A, Dawlaty MM, Seo J, Cheng AW, Meng J, Le T, Faull KF, Jaenisch R, Tsai LH. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nature Reviews: Genetics. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin SM, Zhang N, Hansen J, Gerges NZ, Pak DTS, Sheng M, Lee SH. GKAP orchestrates activity-dependent postsynaptic protein remodeling and homeostatic scaling. Nature neuroscience. 2012;15:1655–1666. doi: 10.1038/nn.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZDZ, Meissner AA. DNA methylation: roles in mammalian development. Nature Reviews: Genetics. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. The Journal of biological chemistry. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- Sultan FA, Sweatt JD. The role of the gadd45 family in the nervous system: a focus on neurodevelopment, neuronal injury, and cognitive neuroepigenetics. Advances in experimental medicine and biology. 2013;793:81–119. doi: 10.1007/978-1-4614-8289-5_6. [DOI] [PubMed] [Google Scholar]
- Sultan FA, Wang J, Tront J, Liebermann DA, Sweatt JD. Genetic deletion of Gadd45b, a regulator of active DNA demethylation, enhances long-term memory and synaptic plasticity. J Neurosci. 2012;32:17059–17066. doi: 10.1523/JNEUROSCI.1747-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Sweatt JD. Mechanisms of Memory. Academic Press; 2009. [Google Scholar]
- Sweatt JD. The emerging field of neuroepigenetics. Neuron. 2013;80:624–32. doi: 10.1016/j.neuron.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweatt JD. Pitt-Hopkins Syndrome: intellectual disability due to loss of TCF4-regulated gene transcription. Experimental & molecular medicine. 2013;45:e21–e21. doi: 10.1038/emm.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LT, Moyer JR, Disterhoft JF. Transient changes in excitability of rabbit CA3 neurons with a time course appropriate to support memory consolidation. Journal of neurophysiology. 1996;76:1836–1849. doi: 10.1152/jn.1996.76.3.1836. [DOI] [PubMed] [Google Scholar]
- Tognini P, Napoli D, Tola J, Silingardi D, Della Ragione F, D'Esposito M, Pizzorusso T. Experience-dependent DNA methylation regulates plasticity in the developing visual cortex. Nature neuroscience. 2015;18:956–958. doi: 10.1038/nn.4026. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Uchida H, Ma L, Ueda H. Epigenetic gene silencing underlies C-fiber dysfunctions in neuropathic pain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010a;30:4806–4814. doi: 10.1523/JNEUROSCI.5541-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida H, Sasaki K, Ma L, Ueda H. Neuron-restrictive silencer factor causes epigenetic silencing of Kv4.3 gene after peripheral nerve injury. Neuroscience. 2010b;166:1–4. doi: 10.1016/j.neuroscience.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Wang SH, Teixeira CM, Wheeler AL, Frankland PW. The precision of remote context memories does not require the hippocampus. Nature neuroscience. 2009;12:253–255. doi: 10.1038/nn.2263. [DOI] [PubMed] [Google Scholar]
- Wierenga CJ, Ibata K, Turrigiano GG. Postsynaptic expression of homeostatic plasticity at neocortical synapses. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2005;25:2895–2905. doi: 10.1523/JNEUROSCI.5217-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KK, Christensen JJ, Pedersen MTM, Johansen JVJ, Cloos PACP, Rappsilber JJ, Helin KK. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SR, Mullegama SV, Rosenfeld JA, Dagli AI, Hatchwell E, Allen WP, Williams CA, Elsea SH. Haploinsufficiency of MBD5 associated with a syndrome involving microcephaly, intellectual disabilities, severe speech impairment, and seizures. European Journal of Human Genetics. 2010;18:436–441. doi: 10.1038/ejhg.2009.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Su Y, Shin J, Zhong C, Guo JU, Weng YL, Gao F, Geschwind DH, Coppola G, Ming G-l, et al. Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nature neuroscience. 2015;18:836–843. doi: 10.1038/nn.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]