Abstract
Abnormal intracellular accumulation of the fused in sarcoma (FUS) protein is the characteristic pathological feature of cases of familial amyotrophic lateral sclerosis (ALS) caused by FUS mutations (ALS-FUS) and several uncommon disorders that may present with sporadic frontotemporal dementia (FTLD-FUS). Although these findings provide further support for the concept that ALS and FTD are closely related clinical syndromes with an overlapping molecular basis, important differences in the pathological features and results from experimental models indicate that ALS-FUS and FTLD-FUS have distinct pathogenic mechanisms.
Some cases of ALS and frontotemporal dementia exhibit abnormal accumulation of the FUS protein. Although these are closely related clinical syndromes with an overlapping molecular basis, they have distinct pathogenic mechanisms.
Over the past few decades, there has been increasing evidence that amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are closely related conditions with an overlapping molecular basis. Approximately 10% of patients who initially present with ALS develop dementia, which often has features of FTD (Murphy et al. 2007), and a significant proportion of patients with FTD eventually develop pyramidal motor-system dysfunction (Lomen-Hoerth et al. 2002). In addition, a number of families have been reported with autosomal dominant disease in which affected members develop ALS, FTD, or both (Vance et al. 2006; Boxer et al. 2011). In 2006, the transactive response DNA-binding protein with Mr of 43 kDa (TDP-43) was identified as the pathological protein that accumulates in most cases of ALS and in the most common pathological subtype of FTD (previously known as frontotemporal lobar degeneration [FTLD] with ubiquitin-positive inclusions, FTLD-U, and subsequently renamed FTLD-TDP) (Arai et al. 2006; Neumann et al. 2006; Davidson et al. 2007). More recently, abnormal expansion of a hexanucleotide repeat in a noncoding region of the chromosome 9 open reading frame 72 gene (C9orf72) was discovered to be the most common genetic cause of both ALS and FTD and the basis on which both ALS and FTD occur in most families (Dejesus-Hernandez et al. 2011; Renton et al. 2011).
In early 2009, mutations in the fused in sarcoma (FUS) gene (also known as the translocated in liposarcoma [TLS] gene) were reported to cause a small proportion of familial ALS (fALS) cases (ALS-FUS), in which the associated pathology is characterized by inclusions that are FUS-immunoreactive (FUS-ir) but TDP-43-negative (Kwiatkowski et al. 2009; Vance et al. 2009). Subsequently, the role of FUS was investigated in the remaining subtypes of FTLD for which the molecular basis was still uncertain. The discovery that most of these tau/TDP-negative conditions are characterized by FUS-ir pathology (Munoz et al. 2009; Neumann et al. 2009a,b) led to the formation of a new molecular class of FTLD (FTLD-FUS) (Mackenzie et al. 2010a) and provided further evidence that FTD and ALS are closely related conditions. However, despite some overlap in the phenotype and pathological features of ALS-FUS and FTLD-FUS, significant differences in their molecular pathology have recently been identified that have helped to elucidate their distinct underlying pathogenic mechanisms (Dormann et al. 2010, 2012; Neumann et al. 2011, 2012; Mackenzie and Neumann 2012).
NORMAL FUNCTION OF FUS
The biology and genetics of FUS are covered in detail in Guo and Shorter (2016) and have recently been reviewed by Sama et al. (2014). The FUS gene, located on chromosome 16, consists of 15 exons that encode a 526-amino-acid protein (Aman et al. 1996). The N-terminus functions in transcriptional activation, whereas the C-terminus region contains multiple nucleic acid–binding domains and a nonclassical nuclear localization signal (Prasad et al. 1994; Sama et al. 2014). FUS is a ubiquitously expressed protein (Aman et al. 1996; Andersson et al. 2008) that binds to RNA (Crozat et al. 1993; Zinszner et al. 1997) and DNA (Baechtold et al. 1999) and is involved in diverse cellular processes including cell proliferation (Bertrand et al. 1999), DNA repair (Baechtold et al. 1999; Bertrand et al. 1999), transcription regulation, RNA splicing (Yang et al. 1998), and RNA transport between intracellular compartments (Zinszner et al. 1997). In most cell types, FUS is present in both the nucleus and cytoplasm; however, in neurons, there is proportionally more in the nucleus, and its expression in glia is exclusively nuclear (Andersson et al. 2008). In the central nervous system, FUS has been reported to bind to RNA from more than 5500 genes, primarily through a GUGGU-binding motif (Lagier-Tourenne et al. 2012), and it may be involved in neuronal plasticity and the maintenance of dendritic integrity by transporting mRNA (including those encoding actin-related proteins) to dendritic spines for local translation in response to synaptic stimulation (Fujii and Takumi 2005; Fujii et al. 2005).
As the name indicates, the FUS gene was first identified because chromosomal translocation of the 5′ portion of FUS results in several fusion oncogenes that are each associated with specific types of human cancer, including myxoid liposarcoma, Ewing's sarcoma, and acute myeloid leukemia (Law et al. 2006). The discovery that other types of FUS mutations cause fALS was the first association between this protein and a neurodegenerative condition (Kwiatkowski et al. 2009; Vance et al. 2009).
ALS WITH FUS PATHOLOGY
ALS is a common neurodegenerative disorder in which the relentless destruction of upper and lower motor neurons causes progressive weakness, typically leading to death within a few years. Approximately 10% of ALS is inherited, and fALS has been genetically linked to a number of chromosomal loci with causal mutations identified in several genes (http://alsod.iop.kcl.ac.uk/index.aspx) (Chen et al. 2013).
In 2009, mutations in the FUS gene were identified as the cause of fALS type 6 (Kwiatkowski et al. 2009; Vance et al. 2009). Subsequently, genetic screening of ALS cohorts from many centers in North America, Europe, and Asia have confirmed that FUS mutations are responsible for ∼3% of fALS cases and <1% of sporadic ALS (sALS) cases (collectively referred to here as ALS-FUS) (reviewed in Mackenzie et al. 2010b). The associated clinical phenotype is usually classical ALS, with early features of both upper and lower motor neuron involvement but without any associated cognitive dysfunction.
Most of the FUS mutations that cause ALS affect highly conserved regions in exons 12–15 that encode the C-terminus of the protein and disrupt or delete a region characterized as a nonclassical nuclear localization signal. These changes result in impaired transportin (TRN)-mediated nuclear import with redistribution of FUS to the cytoplasm (Bosco et al. 2010; Dormann et al. 2010; Ito et al. 2011). Several studies have shown a correlation between disease severity (age of onset and disease duration) and the degree to which the causal FUS mutation impairs FUS nuclear transport (Dormann et al. 2010; Kino et al. 2011; Niu et al. 2012). Mutations in other regions of the FUS gene (mainly exons 3–6) have mostly been reported in cases of sALS or fail to segregate with disease in fALS and are of uncertain pathogenicity. Furthermore, there have been no autopsy reports describing FUS-ir pathology in an ALS patient carrying one of these mutations.
Neuropathology of ALS-FUS
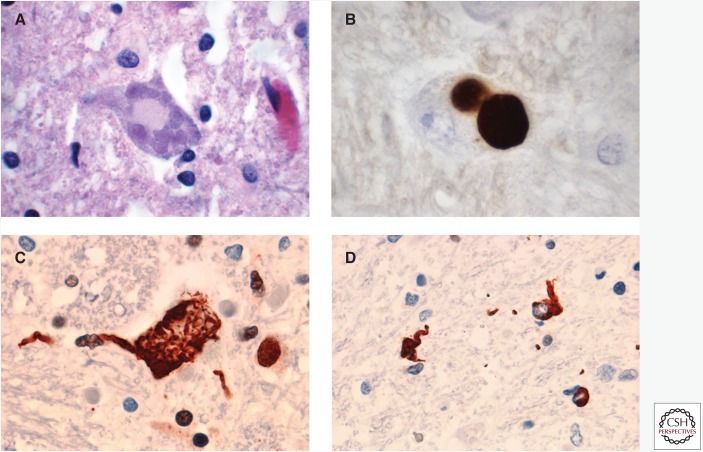
Cases with FUS mutations show typical features of ALS, including loss of primary upper and lower motor neurons, chronic reactive changes, and degeneration of the corticospinal tracts and motor nerves. Bunina bodies, which are a characteristic finding in ALS with TDP-43 pathology, are absent. In most cases, at least some motor neurons contain sharply defined, single or multiple, round, oval, or multilobulated cytoplasmic inclusions with a pale blue-gray color on hematoxylin and eosin (H&E) staining, referred to as basophilic inclusions (BIs) (Fig. 1A) (Mackenzie et al. 2011a). In addition to classical BIs, some cases also have smaller, more darkly staining basophilic deposits in motor neurons. A consistent feature is the presence of neuronal cytoplasmic inclusions (NCIs), which are immunoreactive for FUS (Fig. 1B,C) and variably reactive for components of the ubiquitin–proteasome system (such as ubiquitin and p62) but negative for TDP-43 (Kwiatkowski et al. 2009; Vance et al. 2009; Blair et al. 2010; Groen et al. 2010; Hewitt et al. 2010; Huang et al. 2010; Kobayashi et al. 2010; Rademakers et al. 2010; Yamamoto-Watanabe et al. 2010; Robertson et al. 2011; Suzuki et al. 2012). FUS immunohistochemistry (IHC) also shows variable numbers of glial cytoplasmic inclusions (Fig. 1D), primarily in oligodendrocytes (Hewitt et al. 2010; Kobayashi et al. 2010; Yamamoto-Watanabe et al. 2010; Mackenzie et al. 2011a; Suzuki et al. 2012). Although most inclusion-bearing cells show an absence of normal nuclear FUS immunoreactivity, there may be some retention (Rademakers et al. 2010; Robertson et al. 2011).
Figure 1.
Neuropathology of ALS-FUS. (A) Basophilic inclusions (BIs) in the cytoplasm of motor neurons are a common feature of most cases of ALS caused by FUS mutations. In early-onset cases, the predominant finding with fused in sarcoma (FUS) immunohistochemistry (IHC) is (B) large, round neuronal cytoplasmic inclusions (NCIs) that correspond to BI. In late-onset cases, (C) FUS-positive, tangle-like neuronal inclusions predominate and (D) glial cytoplasmic inclusions are more numerous. Neuropathology in A is shown by hematoxylin and eosin staining; in B–D, by FUS IHC.
There is significant variation in the morphology, abundance, and anatomical distribution of BIs and FUS-ir inclusions among ALS-FUS cases. Most of the published descriptions are individual case reports or very small series that vary in methodology and the amount of detail provided, making it difficult to compare cases directly and to determine genetic, clinical, and pathological correlations. The largest series published to date provides a comparative analysis of six cases of ALS-FUS, which include sporadic and familial cases with both juvenile and adult onset and with four different FUS mutations (Mackenzie et al. 2011a). There was significant pathological heterogeneity among these cases, with two distinct patterns identified that correlated with disease severity and the specific mutation (Table 1). Three cases with early onset and rapid disease progression, including two with the p.P525L mutation, were found to have moderate numbers of BIs in lower motor neurons and the primary motor cortex. These BIs were most often large, round, and pale bodies (classical); however, all of these early-onset cases also had some basophilic inclusions that were smaller, darker-staining, and sometimes multiple in number. Basophilic inclusions were also occasionally found in other regions, such as the substantia nigra and the nuclei of the basis pontis. The predominant FUS-ir pathology shown in early-onset cases was large, round, compact NCIs that likely corresponded to BIs (Fig. 1B). In contrast, cases with later disease onset and slower progression, including two cases with the p.R521C mutation, had few BIs (always with classical morphology) that were restricted to the lower motor neurons. In these cases, FUS IHC primarily showed filamentous-tangle-like NCIs (Fig. 1C) and numerous glial cytoplasmic inclusions (Fig. 1D). There were also differences noted in the anatomic distribution of FUS-ir pathology between the two groups of cases. All cases had many NCIs in the lower motor neurons; however, the early-onset cases had significant pathology in the primary motor cortex, substantia nigra, basis pontis, and cerebellar dentate nucleus, whereas the late-onset cases had more consistent involvement of the basal ganglia and thalamus. It is important to note that although many of the other published case reports of ALS-FUS have described clinical, genetic, and pathological correlations consistent with one of these two patterns (Kwiatkowski et al. 2009; Vance et al. 2009; Blair et al. 2010; Groen et al. 2010; Hewitt et al. 2010; Huang et al. 2010; Yamamoto-Watanabe et al. 2010), other combinations have also been reported (Hewitt et al. 2010; Kobayashi et al. 2010; Tateishi et al. 2010; Suzuki et al. 2012), indicating a wider range of variability.
Table 1.
Frequency and morphology of cellular inclusions in ALS-FUS
| BI | BI | BI | FUS-ir | FUS-ir | FUS-ir | FUS-ir | FUS-ir | |
|---|---|---|---|---|---|---|---|---|
| NCI | NCI | NCI | NCI | GCI | ||||
| Motor cortex | Subcort | Lower motor neurons | Motor cortex | Subcort | Lower motor neurons | Morph | ||
| Early onset, short duration | ++ | + | ++ | +++ | ++a | +++ | Round | + |
| Late onset, long duration | − | − | + | + | ++b | +++ | Tangles | +++ |
Based on data in study by Mackenzie et al. 2011a.
Grading: +, few; ++, moderate; +++, numerous.
ALS-FUS, Amyotrophic lateral sclerosis caused by FUS mutations; BI, basophilic inclusion; ir, immunoreactive; NCI, neuronal cytoplasmic inclusion; GCI, glial cytoplasmic inclusion.
aNucleus of the basis pontis, cerebellar dentate nucleus, and substantia nigra.
bBasal ganglia, thalamus, and substantia nigra.
Following the discovery of FUS mutations, a number of previously reported cases of ALS with BIs were reevaluated (Baumer et al. 2010; Huang et al. 2010; Kobayashi et al. 2010; Suzuki et al. 2010; Tateishi et al. 2010; Yamamoto-Watanabe et al. 2010; Fujita et al. 2011; Matsuoka et al. 2011). These studies showed that the BIs in cases of ALS are consistently FUS-ir and that most are associated with a FUS mutation (Baumer et al. 2010; Huang et al. 2010; Kobayashi et al. 2010; Suzuki et al. 2010; Tateishi et al. 2010; Yamamoto-Watanabe et al. 2010). Cases without a mutation (Huang et al. 2010; Fujita et al. 2011; Matsuoka et al. 2011) are much less common and may represent part of the sporadic BI body disease (BIBD) spectrum (see below).
FTD WITH FUS PATHOLOGY
FTD is a clinical syndrome characterized by abnormalities of behavior, personality, and language (Neary et al. 1998, McKhann et al. 2001) that accounts for 10%–15% of all dementia cases and is the second most common cause of dementia in individuals under the age of 65 years (Bird et al. 2003; Feldman et al. 2003). The neuropathology that underlies clinical FTD is heterogeneous, with a consistent feature being the relatively selective degeneration of the frontal and temporal cerebral lobes (Cairns et al. 2007; Neumann et al. 2009c). FTLD is often used as the general term for those pathological conditions that commonly present with clinical FTD, and there is a growing trend to classify FTLD based on the molecular defect that is presumed to be most closely related to the underlying pathogenic process (Mackenzie et al. 2009, 2010a). The majority of FTD cases are associated with abnormal, intracellular accumulation of either tau or the TDP-43 protein (FTLD-tau and FTLD-TDP, respectively) (Neumann et al. 2009c). However, there remain ∼10% of FTLD cases composed of a heterogeneous collection of uncommon disorders that do not have significant tau or TDP-43 pathology (Cairns et al. 2007; Mackenzie et al. 2008; Roeber et al. 2008). Following the discovery of FUS mutations in ALS (Kwiatkowski et al. 2009; Vance et al. 2009), the role of FUS was investigated in these tau/TDP-negative forms of FTLD (Munoz et al. 2009; Neumann et al. 2009a,b; Holm et al. 2009). In most of the FTLD subtypes where the major molecular defect was not yet known, the characteristic pathological inclusions were found to be FUS-ir. These included cases previously designated as atypical FTLD with ubiquitinated inclusions (aFTLD-U) (Mackenzie et al. 2008; Roeber et al. 2008; Neumann et al. 2009a), neuronal intermediate filament inclusion disease (NIFID) (Cairns et al. 2003, 2004a; Neumann et al. 2009b), and BIBD (Munoz-Garcia and Ludwin 1984; Munoz et al. 2009). As a result, these three conditions have now been grouped together within the broad molecular category of FTLD-FUS (Mackenzie et al. 2010a). Studies of several pathological series have confirmed FTLD-FUS in 5%–10% of clinical FTD cases (Mackenzie et al. 2008; Roeber et al. 2008; Neumann et al. 2009a; Seelaar et al. 2010), and a large international study found 92% of tau/TDP-negative FTLD to have FUS-ir pathology (Urwin et al. 2010).
Atypical FTLD-U
Most cases of tau-negative FTLD were previously defined by the presence of neuronal inclusions that could only be shown with IHC for ubiquitin (FTLD-U) (Mackenzie et al. 2006). In the majority of these cases, the ubiquitinated pathological protein was discovered to be TDP-43 (FTLD-TDP) (Arai et al. 2006; Neumann et al. 2006; Davidson et al. 2007). However, it was subsequently recognized that 10%–20% of FTLD-U cases do not show evidence of abnormal TDP-43 metabolism (Mackenzie et al. 2008; Roeber et al. 2008). These cases of aFTLD-U have an unusual and remarkably consistent clinical phenotype characterized by sporadic, early-onset, severe, and rapidly progressive psychobehavioral abnormalities in the absence of significant language or motor deficits (Mackenzie et al. 2008; Roeber et al. 2008; Urwin et al. 2010).
Neuropathology of aFTLD-U
In cases of aFTLD-U, there is symmetric atrophy of the frontal and temporal lobes as well as the caudate nucleus (Mackenzie et al. 2008; Roeber et al. 2008; Neumann et al. 2009a). Histological evidence of chronic degeneration is present in the frontal and temporal neocortex, hippocampal CA1 sector and subiculum (hippocampal sclerosis), striatum, globus pallidus, and substantia nigra. Characteristic cellular inclusions are not seen with histochemical staining (including those for amyloid, such as thioflavin) and can only be detected with IHC.
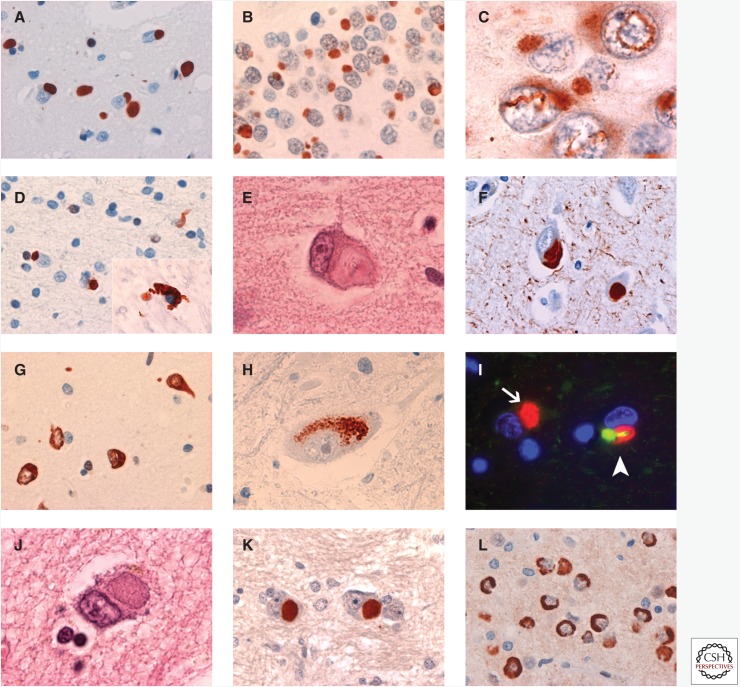
FUS IHC labels variable numbers of well-defined, round, oval, or kidney-shaped NCIs in the middle and deeper layers of the neocortex (Fig. 2A) (Neumann et al. 2009a; Lashley et al. 2011; Mackenzie et al. 2011b). Similar NCIs are moderate to numerous in the hippocampal dentate fascia (Fig. 2B), striatum, and periaqueductal gray mater, whereas other cortical and subcortical regions are less consistently involved and to a milder degree. Neuronal cytoplasmic inclusions with other morphologies, such as crescentic and granular forms, are much less frequent. In addition to cytoplasmic inclusions, neuronal intranuclear inclusions are a consistent feature and appear as a single straight, curved, or twisted (vermiform) thick filament (Fig. 2C). Neuronal intranuclear inclusions are most numerous in the dentate granule cells, but they are also found in pyramidal neurons of the neocortex and hippocampus and occasionally in subcortical regions. Although most neurons harboring inclusions (either neuronal cytoplasmic or neuronal intranuclear inclusions) show the absence of normal physiological nuclear staining, this finding is inconsistent. FUS IHC also shows occasional short dystrophic neurites and a moderate number of oval or flame-shaped glial cytoplasmic inclusions in white-matter oligodendrocytes (Fig. 2D). The neuronal inclusions are also demonstrated with IHC for ubiquitin and p62; however, FUS IHC tends to be more sensitive, and some types of inclusions, such as glial cytoplasmic inclusions, are only FUS-ir.
Figure 2.
Neuropathology of FTLD-FUS. In cases of atypical FTLD with ubiquitinated inclusions (aFTLD-U), moderate numbers of compact, round, or oval FUS immunoreactive (FUS-ir) NCIs are found in the (A) cerebral neocortex, (B) dentate granule cells of the hippocampus, and some subcortical regions. These cases also have long, twisted, filamentous neuronal intranuclear inclusions that are most numerous in the (C) dentate fascia and neocortex. (D) FUS-ir glial cytoplasmic inclusions are a common feature of all types of FTLD-FUS. Neuronal intermediate filament inclusion disease (NIFID) is characterized by NCI of varying morphologies, including (E) hyaline conglomerate inclusions, which are sometimes immunoreactive for (F) neuronal intermediate filaments (IFs). In NIFID, FUS IHC shows an even wider range of inclusion types, which include (G) annular, crescentic, and tangle-like NCI, as well as collections of (H) cytoplasmic granules. Many of the NCIs in NIFID are immunoreactive for FUS but not IFs (arrow, I), and all neurons that contain an IF-ir inclusion also show a discrete focal accumulation of FUS (arrowhead, I). Basophilic inclusion body disease (BIBD) is defined by the presence of (J) BI in the cytoplasm of neurons in a wide range of anatomical regions. FUS IHC consistently labels (K) BI and shows additional types of (L) NCI in BIBD. Neuropathology in A–D, G, H, K, and L is shown by FUS IHC; in E and J, by hematoxylin and eosin; and in I, by double-label immunofluorescence for FUS (red) and α-internexin (green).
Neuronal Intermediate Filament Inclusion Disease
NIFID is an uncommon neurological disorder that typically presents as early-onset, sporadic FTD associated with a pyramidal and/or extrapyramidal movement disorder (Cairns et al. 2003, 2004a). The defining feature is the presence of NCIs that are immunoreactive for all class IV intermediate filaments (light, medium, and heavy neurofilament subunits and α-internexin) (Cairns et al. 2004b).
Neuropathology of NIFID
In cases of NIFID, the brain is usually small with symmetric atrophy of the frontal lobes (Cairns et al. 2004a; Neumann et al. 2009b). Chronic degenerative changes are prominent in the frontal and temporal neocortex as well as many subcortical regions including the basal ganglia, thalamus, substantia nigra, periaqueductal gray matter, inferior olivary nucleus, and cerebellar dentate nucleus. Decreased myelin staining of the corticospinal tracts and appreciable loss of lower motor neurons may also be present.
Many different types of neuronal inclusions may be seen with histochemical staining and IHC (Cairns et al. 2004a; Neumann et al. 2009b,c). Although immunoreactivity for intermediate filaments is the defining feature of NIFID, only a small number of the NCIs are intermediate filament immunoreactive (IF-ir) (Cairns et al. 2004b; Neumann et al. 2009c; Armstrong et al. 2011; Lashley et al. 2011). Small, round, Pick-body-like inclusions; hyaline conglomerate inclusions (Fig. 2E); and crescentic, annular, and tangle-like inclusions are all variably immunoreactive for ubiquitin, p62, and intermediate filaments (Fig. 2F). Large, round, brightly eosinophilic neuronal intranuclear inclusions have been described in about half of the reported cases of NIFID and are intensely ubiquitin/p62-immunoreactive but negative for intermediate filaments. Finally, ubiquitin IHC shows filamentous neuronal intranuclear inclusions in dentate granule cells and cortical pyramidal neurons that are similar to those found in aFTLD-U.
All of the morphological types of inclusions are labeled with antibodies against FUS but in greater numbers and staining intensity than with other methods (Fig. 2G) (Neumann et al. 2009b; Armstrong et al. 2011; Mackenzie et al. 2011b). FUS IHC also shows additional types of inclusions that are not seen with other techniques, including aggregates of coarse cytoplasmic granules in some neuronal populations (Fig. 2H) and glial cytoplasmic inclusions in oligodendrocytes. FUS-ir pathology is abundant in virtually all neocortical, limbic, and subcortical gray-matter regions, with the exception of the cerebellar cortex. Inclusion-bearing cells often show an absence or a reduction of the normal diffuse nuclear FUS immunoreactivity. Double labeling confirms that all ubiquitin/p62-ir inclusions are also FUS positive (Neumann et al. 2009b; Lashley et al. 2011). Double labeling with antibodies against FUS and intermediate filaments shows that many neurons contain NCIs or neuronal intranuclear inclusions that are only immunoreactive for FUS (Fig. 2I) (Neumann et al. 2009b; Lashley et al. 2011). Moreover, all cells with IF-ir NCIs also have focal accumulation of FUS; however, in these cells, the two antibodies label discrete inclusions or different regions of an inclusion with minimal overlap (Fig. 2I) (Neumann et al. 2009b; Lashley et al. 2011). These results suggest that FUS plays a more central role in the pathogenesis of NIFID and that the abnormal accumulation of intermediate filaments is likely a secondary phenomenon.
Basophilic Inclusion Body Disease
BIBD is a term that has been used for a small number of clinically and pathologically heterogeneous cases, in which the common finding is NCIs that appear blue with H&E staining (BIs) (Fig. 2J). Clinical phenotypes include sALS (Kusaka et al. 1993), fALS (Tsuchiya et al. 2001), ALS with dementia (Ishihara et al. 2006), and pure FTD (Munoz-Garcia and Ludwin 1984).
Neuropathology of BIBD
In cases of BIBD, there is chronic degeneration of the frontal and temporal neocortex, the striatum, and the substantia nigra. Even in cases with clinical FTD, BIs tend to be most numerous in subcortical regions (Munoz-Garcia and Ludwin 1984). Basophilic inclusions are round, oval, or crescentic; weakly argyrophilic; and may be detected with histochemical staining for RNA. They are variably immunoreactive for ubiquitin and p62 but negative for tau, α-synuclein, intermediate filaments, tubulin, actin, and TDP-43.
A recent study found that BIs in BIBD are consistently FUS-ir (Fig. 2K) and that FUS IHC shows additional neuronal and glial pathology that had not previously been recognized in these cases (Munoz et al. 2009). FUS-ir NCIs are moderate to numerous in the cerebral neocortex, hippocampal pyramidal layer, globus pallidus, thalamus, midbrain, pons, inferior olivary nucleus, and the lower motor neurons of the spinal cord (Munoz et al. 2009; Mackenzie et al. 2011b). The hippocampal dentate granule cells, striatum, hypoglossal nucleus, and cerebellar dentate nucleus are also consistently affected but to a more variable degree. In addition to large round NCIs that correspond to BIs, other morphologies include crescentic, annular, and tangle-like inclusions in the cerebral neocortex, hippocampus, and basal ganglia, and noncompact collections of coarse granules in the thalamus, many brainstem nuclei, and the cerebellar dentate nucleus (Fig. 2L). Affected areas also show variable numbers of small round or crescentic FUS-ir glial cytoplasmic inclusions and short cell processes. Neuronal intranuclear inclusions are exceptionally rare in BIBD. Absent or reduced levels of nuclear FUS in both inclusion-bearing and normal-appearing neurons in affected anatomical regions is a more consistent feature than in other types of FTLD-FUS (Munoz et al. 2009).
Comparison of FUS Pathology among FTLD-FUS Subtypes
BIBD, NIFID, and aFTLD-U were each originally defined by the presence of specific neuropathological features (BIs, IF-ir NCIs, and ubiquitin-ir NCIs that are only apparent with IHC, respectively) (Munoz-Garcia and Ludwin 1984; Cairns et al. 2003; Mackenzie et al. 2008). The common finding of abundant FUS-ir pathology indicates that these conditions are more closely related than previously understood and has resulted in them being subsumed within the broad molecular class of FTLD-FUS (Mackenzie et al. 2010a). However, despite there being significant overlap, there are also sufficient differences in the pattern of FUS-ir pathology to allow the FTLD-FUS subtypes to be distinguished from one another (Table 2) (Mackenzie et al. 2011b; Lashley et al. 2011; Lee et al. 2013). Cases of aFTLD-U have the most distinct pattern of FUS pathology, characterized by relatively modest involvement of subcortical regions, less variation in the morphology of NCIs, and the most numerous and anatomically widespread neuronal intranuclear inclusions. Cases of NIFID and BIBD both have more numerous and morphologically diverse NCIs in a broad anatomic distribution; however, the absence of neuronal intranuclear inclusions in the hippocampus and neocortex is unique to BIBD cases. These findings validate the specificity of the classical neuropathological diagnostic criteria and support these as representing closely related but distinct entities that share a common molecular pathogenesis.
Table 2.
Frequency and morphology of cellular inclusions in FTLD-FUS
| ub-ir | IF-ir | BI | FUS-ir | FUS-ir | FUS-ir | FUS-ir | |
|---|---|---|---|---|---|---|---|
| NCI | NCI | NCI | NCI | NCI | NII | ||
| Cortex | Cortex | Subcort | Cortex | Subcort | Morph | HC | |
| aFTLD-U | +++ | +/− | +/− | ++ | ++ | Round | ++ |
| NIFID | +++ | ++ | +/− | +++ | +++ | Variable | ++ |
| BIBD | ++ | +/− | +++ | +++ | +++ | Variable | − |
Based on data in study by Mackenzie et al. 2011b.
Grading: +/−, rare and not present in every case; +, few; ++, moderate; +++, numerous.
FTLD, Frontotemporal lobar degeneration; FUS, fused in sarcoma; aFTLD-U, atypical FTLD with ubiquitinated inclusions; NIFID, neuronal intermediate filament inclusion disease; BIBD, basophilic inclusion body disease; ub, ubiquitin; ir, immunoreactive; IF, intermediate filament; FUS, fused in sarcoma; NCI, neuronal cytoplasmic inclusions; NII, neuronal intranuclear inclusion.
Biochemical and Genetic Analysis of FTLD-FUS
In all FTLD-FUS subtypes, the pathological changes are immunoreactive with FUS antibodies that recognize different epitopes across the entire protein (Neumann et al. 2009a,b; Lashley et al. 2011). Immunoblot analysis of postmortem brain tissue shows a relative shift of FUS protein to the insoluble fraction; however, there are no additional protein bands of higher or lower molecular mass (Neumann et al. 2009a,b, 2011).
Genetic analysis has only been reported for a small number of pathologically confirmed FTLD-FUS cases; none of which has been found to have any abnormality of the FUS gene (Neumann et al. 2009a,b; Urwin et al. 2010; Lashley et al. 2011; Snowden et al. 2011). A few patients have been reported with fALS due to FUS mutations who also developed clinical FTD (Ticozzi et al. 2009; Blair et al. 2010; Broustal et al. 2010; Yan et al. 2010), and one study identified a novel FUS mutation (M254V) in a patient with sporadic, adult-onset FTD without evidence of ALS (Van Langenhove et al. 2010); however, none of these reports included neuropathological evaluation. A family with autosomal dominant dementia and autopsy-proven FTLD-FUS (aFTLD-U subtype) in both the proband and their mother was reported in a study from the United Kingdom; however, no FUS mutation was identified (Lashley et al. 2011).
FUS PATHOLOGY IN OTHER NEURODEGENERATIVE DISORDERS
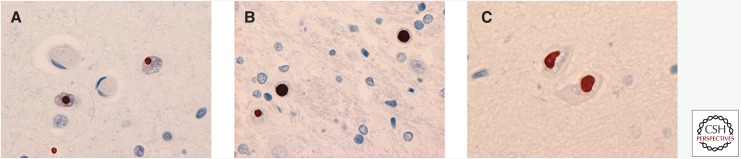
FUS IHC does not label the characteristic pathological changes of most other common neurodegenerative disorders, including Alzheimer’s disease, Lewy body disease, Pick's disease, corticobasal degeneration, progressive supranuclear palsy, multiple system atrophy, FTLD-TDP, ALS-TDP, or ALS-SOD (Neumann et al. 2009a; Rademakers et al. 2010). Important exceptions are the neuronal intranuclear inclusions that characterize several of the polyglutamine (polyQ) repeat disorders and neuronal intranuclear inclusion body disease (NIIBD) (Woulfe et al. 2010; Mori et al. 2012, 2014). Prior to the discovery of FUS mutations in ALS and FUS-ir pathology in ALS and FTD, FUS was identified as a component of the neuronal intranuclear inclusions of Huntington’s disease (Fig. 3A) (Doi et al. 2008). A more comprehensive study of neurodegenerative disease with neuronal intranuclear inclusions found that FUS IHC labeled the neuronal intranuclear inclusions of other polyQ diseases, including spinocerebellar ataxia types 1 and 3 (SCA1 and SCA3, respectively) (Fig. 3B) (Woulfe et al. 2010). This study also showed FUS labeling of neuronal intranuclear inclusions in an experimental mouse model of SCA1 and in cultured cells transfected with mutant human huntingtin. No FUS reactivity was seen in the neuronal intranuclear inclusions of disorders caused by other repeat expansions (oculopharyngeal muscular dystrophy or fragile X tremor ataxia syndrome), inherited forms of FTLD-TDP with mutations in either the granulin or valosin-containing protein genes, or in multiple system atrophy. The only other condition with FUS-ir neuronal intranuclear inclusions is NIIBD (Fig. 3C) (Woulfe et al. 2010; Mori et al. 2012, 2014). NIIBD is a heterogeneous neurodegenerative condition of unknown etiology that is defined by the presence of eosinophilic intranuclear inclusions in neurons and sometimes glia, which are also ubiquitin-ir and show variable labeling with an antibody that recognizes expanded polyQ (1C2). Although these findings suggest that the recruitment of FUS may be a relatively specific feature of polyQ containing neuronal inclusions (Nukina 2010), these inclusions are known to also contain a variety of other proteins, and the role of FUS in pathogenesis remains uncertain.
Figure 3.
FUS pathology in other neurodegenerative conditions. FUS IHC labels the neuronal intranuclear inclusions that are characteristic of (A) Huntington's disease, (B) other polyglutamine disorders such as spinocerebellar ataxia type 1, and (C) cases of neuronal intranuclear inclusion body disease. Pathology in A–C is shown by FUS IHC.
OTHER FET PROTEINS IN FTLD-FUS AND ALS-FUS
FUS belongs to a group of DNA/RNA binding proteins known as the FET protein family that includes FUS, Ewing’s sarcoma protein (EWS), and TATA-binding protein-associated factor 15 (TAF15) (Law et al. 2006; Schwartz et al. 2015). All of the FET proteins were initially discovered as components of fusion oncogenes that cause specific types of human cancer. They are all involved in various aspects of RNA metabolism and are predominantly nuclear in localization but shuttle between the nucleus and cytoplasm via a common TRN-mediated mechanism. Protein-interaction studies suggest that FET proteins are able to interact with each other and form protein complexes (Kovar 2011; Thomsen et al. 2013), suggesting that TAF15 and EWS might also be altered in FUS-opathies.
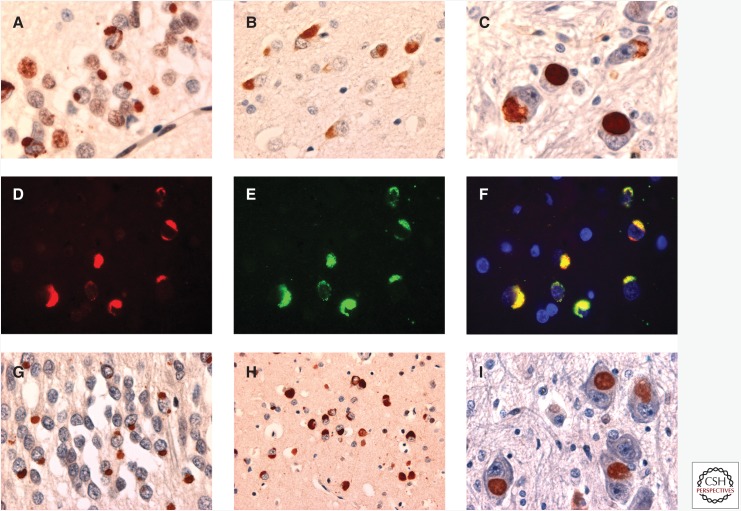
Studies have shown that in cases of ALS-FUS with a range of different mutations, there is no co-accumulation of other FET proteins into FUS-positive inclusions, and cells retain the normal physiological nuclear staining of TAF15 and EWS (Neumann et al. 2011). In striking contrast, in all FTLD-FUS subtypes, TAF15 and EWS are also found to co-accumulate in FUS-positive inclusions, and inclusion-bearing cells show a reduction in the normal nuclear staining of all three FET proteins, particularly TAF15 (Fig. 4A–F) (Neumann et al. 2011; Davidson et al. 2013). Moreover, inclusions in FTLD-FUS also show consistent labeling for the FET carrier protein TRN (Fig. 4G–I), whereas the inclusions in cases of ALS-FUS are TRN-negative (Brelstaff et al. 2011; Neumann et al. 2012; Davidson et al. 2013; Troakes et al. 2013).
Figure 4.
Other FET proteins in FTLD-FUS. All of the different types of FUS immunoreactive cellular inclusions, in all subtypes of FTLD-FUS, including (A) aFTLD-U, (B) NIFID, and (C) BIBD, can also be labeled with antibodies against the other FET proteins, TAF-15 (A and C) and Ewing’s sarcoma protein (EWS) (B). Double-label immunofluorescence confirms the co-localization of FET proteins in the inclusions (D–F). The inclusions of FTLD-FUS also contain the FET carrier protein transportin (TRN) (G, aFTLD-U; H, NIFID; I, BIBD). TAF-15 IHC is shown in A and C. EWS IHC is shown in B. Double-label immunofluorescence is shown in D–F for FUS (red); in E for TAF-15 (green); and in F for a merged image. TRN IHC is shown in G–I.
These findings indicate that the FUS-ir pathology in FTLD-FUS and ALS-FUS result from two distinct pathogenic processes. In ALS-FUS, the causal mutations in FUS affect the nuclear localization signal and specifically interfere with the normal binding of FUS to TRN, which results in the accumulation of FUS in the cytoplasm where it forms inclusions (Dormann et al. 2010; Ito et al. 2011). In FTLD-FUS, a more general defect of TRN-mediated nuclear import seems to be present that affects the intracellular distribution of all FET proteins. Although this could be the result of some primary abnormality of TRN itself (such as genetic variation in the TRN gene [TNPO1] or altered expression levels), the fact that a number of other protein cargoes of TRN show normal intracellular distribution and are not incorporated into the FET-ir inclusions in FTLD-FUS suggests that this is not the case (Neumann et al. 2012). Alternatively, the proper nuclear import of FET proteins might be affected by some abnormal posttranslational modification that affects all of the FET proteins. One recent study provided evidence that in FTLD-FUS there is reduced methylation of the arginine-glycine rich region, immediately adjacent to the nuclear localization signal, which results in abnormally avid binding of FET proteins to TRN, producing protein complexes that cannot dissociate and that aggregate as inclusion bodies (Dormann et al. 2012). The cause of the arginine hypomethylation in FTLD-FUS is not known, but it does not appear to be due to variation in the genes encoding the responsible protein N-arginine methyltransferases (Ravenscroft et al. 2013).
EXPERIMENTAL MODELS AND PATHOGENIC MECHANISMS OF FUS-OPATHIES
Although the mechanisms of FUS cellular redistribution and inclusion-body formation in ALS-FUS and FTLD-FUS are now fairly well understood, it is still uncertain whether neurodegeneration is the result of loss of some essential normal function of FUS or gain of a novel toxic function. Loss of function is suggested by reduced nuclear FUS immunoreactivity in cases of both ALS-FUS and FTLD-FUS and by in vitro models that show a correlation between the degree of protein loss from the nucleus and the severity of human disease associated with specific FUS mutations (Dormann et al. 2010; Kino et al. 2011; Niu et al. 2012). The wide range and essential nature of the many functions performed by nuclear FUS makes it easy to speculate that reduced levels would be detrimental. FUS also plays an important role in transporting mRNAs within the cytoplasmic compartment, which could be compromised if insoluble protein aggregates sequester FUS and make it unavailable. Alternatively, a gain of toxic function is commonly implicated in neurodegenerative disorders characterized by the aggregation of misfolded proteins. Redistribution of FUS to the cytoplasm could result in aberrant binding to cytosolic RNA targets, and abnormal FUS aggregates might overwhelm important cellular functions such as stress-granule dynamics and proteasomal degradation. Results from in vivo models have been somewhat inconsistent but provide evidence for both types of mechanism, possibly acting in combination (reviewed by Da Cruz and Cleveland 2011; Lanson and Pandey 2012; Dormann and Haass 2013; Deng et al. 2014). Briefly, loss-of-function pathogenicity is supported by studies showing that complete knockout of FUS in mice has devastating effects (including perinatal lethality), whereas depletion of FUS in Drosophila and zebra fish results in abnormal motor-neuron morphology and a motor phenotype that can be rescued by wild-type but not mutant FUS. In contrast, many studies have shown that overexpression of wild-type or mutant FUS leads to cytoplasmic accumulation and aggregation of the protein, is associated with neurotoxicity, and recapitulates various features of disease including degeneration of cortical, hippocampal, and motor neurons, impaired neuromuscular transmission, and development of weakness and learning and memory deficits.
CONCLUDING REMARKS
Abnormal intracellular accumulation of FUS is a characteristic pathological feature of fALS caused by FUS mutations and a newly recognized subset of sporadic FTD (FTLD-FUS). Although the mechanisms that lead to FUS pathology in these two classes of disease appear to be distinct, the finding of a common molecular defect provides further support for the concept that ALS and FTD are closely related clinical syndromes with an overlapping pathogenesis, and adds FUS to the growing list of RNA-binding proteins involved in neurodegenerative disease.
ACKNOWLEDGMENTS
This work was supported by grants from the Canadian Institutes of Health Research (I.R.A.M.), the Pacific Alzheimer Research Foundation (I.R.A.M.), the German Helmholtz Association (M.N.), and the International Foundation for Research on Alzheimer Disease (IFRAD) (M.N.).
Footnotes
Editor: Stanley B. Prusiner
Additional Perspectives on Prion Diseases available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this subject collection.
- Aman P, Panagopoulos I, Lassen C, Fioretos T, Mencinger M, Toresson H, Höglund M, Forster A, Rabbitts TH, Ron D, et al. 1996. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics 37: 1–8. [DOI] [PubMed] [Google Scholar]
- Andersson MK, Stahlberg A, Arvidsson Y, Olofsson A, Semb H, Stenman G, Nilsson O, Aman P. 2008. The multifunctional FUS, EWS, and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell Biol 9: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, et al. 2006. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun 351: 602–611. [DOI] [PubMed] [Google Scholar]
- Armstrong RA, Gearing M, Bigio EH, Cruz-Sanchez FF, Duyckaerts C, Mackenzie IR, Perry RH, Skullerud K, Yokoo H, Cairns NJ. 2011. The spectrum and severity of FUS-immunoreactive inclusions in the frontal and temporal lobes of ten cases of neuronal intermediate filament inclusion disease. Acta Neuropathol 121: 219–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baechtold H, Kuroda M, Sok J, Ron D, Lopez BS, Akhmedov AT. 1999. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J Biol Chem 274: 34337–34342. [DOI] [PubMed] [Google Scholar]
- Baumer D, Hilton D, Paine AML, Turner MR, Lowe J, Talbot K, Ansorge O. 2010. Juvenile ALS with basophilic inclusions is a FUS proteinopathy with FUS mutations. Neurology 75: 611–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand P, Akhmedov AT, Delacote F, Durrbach A, Lopez BS. 1999. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: Both POMp75 and POMp100 DNA homologous pairing activities are associated with cell proliferation. Oncogene 18: 4515–4521. [DOI] [PubMed] [Google Scholar]
- Bird T, Knopman D, Van Swieten J, Rosso S, Feldman H, Tanabe H, Graff-Raford N, Geschwind D, Verpillat P, Hutton M. 2003. Epidemiology and genetics of frontotemporal dementia/Pick's disease. Ann Neurol 54: S29–S31. [DOI] [PubMed] [Google Scholar]
- Blair IP, Williams KL, Warrich ST, Durnall JC, Thoeng AD, Manavis J, Blumbergs PC, Vucic S, Kiernan MC, Nicholson GA. 2010. FUS mutations in amyotrophic lateral sclerosis: Clinical, pathological, neurophysiological and genetic analysis. J Neurol Neurosurg Psychiatry 81: 639–645. [DOI] [PubMed] [Google Scholar]
- Bosco DA, Lemay N, Ko HK, Zhou H, Burke C, Kwiatkowski TJ, Sapp P, McKenna-Yasek D, Brown RH, Hayward LJ. 2010. Mutant FUS proteins that cause amyotrophic lateral sclerosis incorporate into stress granules. Hum Mol Genet 19: 4160–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer AL, Mackenzie IR, Boeve BF, Baker M, Seeley WW, Crook R, Feldman H, Hsiung GYR, Rutherford N, Laluz V, et al. 2011. Clinical, neuroimaging and neuropathological features of a chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatr 82: 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brelstaff J, Lashley T, Holton JL, Lees AJ, Rossor MN, Bandopadhyay R, Revesz T. 2011. Transportin1: A marker of FTLD-FUS. Acta Neuropathol 122: 591–600. [DOI] [PubMed] [Google Scholar]
- Broustal O, Camuzat A, Guillot-Noël L, Guy N, Millecamps S, Deffond D, Lacomblez L, Golfier V, Hannequin D, Salachas F, et al. 2010. FUS mutations in frontotemporal lobar degeneration with amyotrophic lateral sclerosis. J Alzheimers Dis 22: 765–769. [PubMed] [Google Scholar]
- Cairns NJ, Bigio EH, Mackenzie IRA, Neumann M, Lee VMY, Hatanpaa KJ, White CL, Schneider JA, Grinberg LT, Halliday G, et al. 2007. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: Consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol 114: 2–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Grossman M, Arnold SE, Burn DJ, Jaros E, Perry RH, Duyckaerts C, Stankoff B, Pillon B, Skullerud K, et al. 2004a. Clinical and neuropathological variation in neuronal intermediate filament inclusion disease. Neurology 26: 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Perry RH, Jaros E, Burn D, McKeith IG, Lowe JS, Holton J, Rossor MN, Skullerud K, Duyckaerts C, et al. 2003. Patients with a novel neurofilamentopathy: Dementia with neurofilament inclusions. Neurosci Lett 341: 177–180. [DOI] [PubMed] [Google Scholar]
- Cairns NJ, Zhukareva V, Uryu K, Zhang B, Bigio E, Mackenzie IR, Gearing M, Duyckaerts C, Yokoo H, Nakazato Y, et al. 2004b. α-Internexin is present in the pathological inclusions of neuronal intermediate filament inclusion disease. Am J Path 164: 2153–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sayana P, Zhang X, Le W. 2013. Genetics of amyotrophic lateral sclerosis: An update. Mol Neurodegener 8: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat A, Aman P, Mandahl N, Ron D. 1993. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature 363: 640–644. [DOI] [PubMed] [Google Scholar]
- Da Cruz S, Cleveland DW. 2011. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol 21: 904–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson Y, Kelley T, Mackenzie IRA, Pickering Brown S, Du Plessis D, Neary D, Snowden JS, Mann DMA. 2007. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol 113: 521–533. [DOI] [PubMed] [Google Scholar]
- Davidson YS, Robinson AC, Hu Q, Mishra M, Baborie A, Jaros E, Perry RH, Cairns NJ, Richardson A, Gerhard A, et al. 2013. Nuclear carrier and RNA binding proteins in frontotemporal lobar degeneration associated with fused in sarcoma (FUS) pathological changes. Neuropathol Appl Neurobiol 39: 157–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Gao K, Jankovic J. 2014. The role of FUS gene variants in neurodegenerative disease. Nat Rev Neurol 10: 337–348. [DOI] [PubMed] [Google Scholar]
- Doi H, Okamura K, Bauer PO, Furukawa Y, Shimizu H, Kurosawa M, Machida Y, Miyazaki H, Mitsui K, Kuroiwa Y, et al. 2008. RNA-binding protein TLS is a major nuclear aggregate-interacting protein in Huntingtin exon 1 with expanded polyglutamine-expressing cells. J Biol Chem 283: 6489–6500. [DOI] [PubMed] [Google Scholar]
- Dormann D, Haass C. 2013. Fused in sarcoma (FUS): An oncogene goes awry in neurodegeneration. Mol Cell Neurosci 56: 475–486. [DOI] [PubMed] [Google Scholar]
- Dormann D, Madl T, Valori CF, Bentmann E, Tahirovic S, Abou-Ajram C, Kremmer E, Ansorge O, Mackenzie IRA, Neumann M, et al. 2012. Arginine methylation next to the PY-NLS modulates transportin binding and nuclear import of FUS. EMBO J 31: 4258–4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. 2010. ALS-associated fused in sarcoma (FUS) mutations disrupt transportin-mediated nuclear import. EMBO J 29: 2841–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman H, Levy AR, Hsiung GY, Peters KR, Donald A, Black SE, Bouchard RW, Gauthier SG, Guzman DA, Hogan DB, et al. 2003. A Canadian cohort study of cognitive impairment and related dementias (ACCORD): Study methods and baseline results. Neuroepidemiology 22: 265–274. [DOI] [PubMed] [Google Scholar]
- Fujii R, Okabe S, Urushido T, Inoue K, Yoshimura A, Tachibana T, Nishikawa T, Hicks GG, Takumi T. 2005. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr Biol 15: 587–593. [DOI] [PubMed] [Google Scholar]
- Fujii R, Takumi T. 2005. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J Cell Sci 118: 5755–5765. [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita S, Takatama M, Ikeda M, Okamoto K. 2011. Numerous FUS-positive inclusions in an elderly woman with motor neuron disease. Neuropathology 31: 170–176. [DOI] [PubMed] [Google Scholar]
- Groen EJN, van Es MA, van Vught PWJ, Spliet WG, van Engelen-Lee J, de Visser M, Wokke JH, Schelhaas HJ, Ophoff RA, Fumoto K, et al. 2010. FUS mutations in familial amyotrophic lateral sclerosis in the Netherlands. Arch Neurol 67: 224–230. [DOI] [PubMed] [Google Scholar]
- *.Guo L, Shorter J. 2016. Biology and pathobiology of TDP-43 and emergent therapeutic strategies. Cold Spring Harb Perspect Med 10.1101/cshperspect.a024554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt C, Kirby J, Highley R, Hartley JA, Hibberd R, Hollinger HC, Williams TL, Ince PG, McDermott CJ, Shaw PJ. 2010. Novel FUS/TLS mutations and pathology in familial and sporadic amyotrphic lateral sclerosis. Arch Neurol 67: 455–461. [DOI] [PubMed] [Google Scholar]
- Holm IE, Isaacs A, Mackenzie IRA. 2009. Absence of FUS-immunoreactive pathology in frontotemporal dementia linked to chromosome 3 (FTD-3) caused by mutation in the CHMP2B gene. Acta Neuropathol 118: 719–720. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Zhang J, Geser F, Trojanowski JQ, Strober JB, Dickson DW, Brown RH, Shapiro BE, Lomen-Hoerth C. 2010. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol 20: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K, Araki S, Ihori N, Shiota J, Kawamura M, Nakano I. 2006. An autopsy case of frontotemporal dementia with severe dysarthria and motor neuron disease showing numerous basophilic inclusions. Neuopathology 26: 447–454. [DOI] [PubMed] [Google Scholar]
- Ito D, Seki M, Tsunoda Y, Uchiyama H, Suzuki N. 2011. Nuclear transport impairment of amyotrophic lateral sclerosis-linked mutations in FUS/TLS. Ann Neurol 69: 152–162. [DOI] [PubMed] [Google Scholar]
- Kino Y, Washizu C, Aquilanti E, Okuno M, Kurosawa M, Yamada M, Doi H, Nukina N. 2011. Intracellular localization and splicing regulation of FUS/TLS are variably affected by amyotrophic lateral sclerosis-linked mutations. Nucleic Acids Res 39: 2781–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Z, Tsuchiya K, Arai T, Aoki M, Hasegawa M, Ishizu H, Akiyama H, Mizusawa H. 2010. Occurrence of basophilic inclusions and FUS-immunoreactive neuronal and glial inclusions in a case of familial amyotrophic lateral sclerosis. J Neurol Sci 293: 6–11. [DOI] [PubMed] [Google Scholar]
- Kovar H. 2011. Dr. Jekyll and Mr. Hyde: The two faces of the FUS/EWS/TAF15 protein family. Sarcoma 2011: 837474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusaka H, Matsumoto S, Imai T. 1993. Adult-onset motor neuron disease with basophilic intraneuronal inclusion bodies. Clin Neuopathol 12: 215–218. [PubMed] [Google Scholar]
- Kwiatkowski TJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, Davis A, Gilchrist J, Kasarskis EJ, Munsat T, et al. 2009. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 323: 1205–1208. [DOI] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Polymenidou M, Hutt KR, Vu AQ, Baughn M, Huelga SC, Clutario KM, Ling SC, Liang TY, Mazur C, et al. 2012. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci 15: 1488–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanson NA, Pandey UB. 2012. FUS-related proteinopathies: Lessons from animal models. Brain Res 1462: 44–60. [DOI] [PubMed] [Google Scholar]
- Lashley T, Rohrer JD, Bandopadhyay R, Fry C, Ahmed Z, Isaacs AM, Brelstaff JH, Borroni B, Warren JD, Troakes C, et al. 2011. A comparative clinical, pathological, biochemical and genetic study of fused in sarcoma proteinopathies. Brain 134: 2548–2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law WJ, Cann KL, Hicks GG. 2006. TLS, EWS, and TAF15: A model for transcriptional integration of gene expression. Brief Funct Genomic Proteomic 5: 8–14. [DOI] [PubMed] [Google Scholar]
- Lee EB, Russ J, Jung H, Elman LB, Chahine LM, Kremens D, Miller BL, Branch Coslett H, Trojanowski JQ, Van Deerlin VM, et al. 2013. Topography of FUS pathology distinguishes late-onset BIBD from aFTLD-U. Acta Neuropathol Commun 1: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomen-Hoerth C, Anderson T, Miller B. 2002. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology 59: 1077–1079. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Ansorge O, Strong M, Bilbao J, Zinman L, Ang LC, Baker M, Stewart H, Eisen A, Rademakers R, et al. 2011a. Pathological heterogeneity in amyotrophic lateral sclerosis with FUS mutations: Two distinct patterns correlating with disease severity and mutation. Acta Neuropathol 122: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IRA, Foti D, Woulfe J, Hurwitz TA. 2008. Atypical frontotemporal lobar degeneration with ubiquitin-positive, TDP-43-negative neuronal inclusions. Brain 131: 1282–1293. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Munoz DG, Kusaka H, Yokota O, Ishihara K, Roeber S, Kretzschmar HA, Cairns NJ, Neumann M. 2011b. Distinct pathological subtypes of FTLD-FUS. Acta Neuropathol 121: 207–218. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Neumann N. 2012. FET proteins in frontotemporal dementia and amyotrophic lateral sclerosis. Brain Res 1462: 40–43. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, et al. 2009. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: Consensus recommendations. Acta Neuropathol 117: 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IRA, Neumann M, Bigio EH, Cairns NJ, Alafuzoff I, Kril J, Kovacs GG, Ghetti B, Halliday G, Holm IE, et al. 2010a. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: An update. Acta Neuropathol 119: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie IRA, Rademakers R, Neumann M. 2010b. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol 9: 995–1007. [DOI] [PubMed] [Google Scholar]
- Mackenzie IRA, Shi J, Shaw CL, DuPlessis D, Neary D, Snowden JS, Mann DMA. 2006. Dementia lacking distinctive histology (DLDH) revisited. Acta Neuropathol 112: 551–559. [DOI] [PubMed] [Google Scholar]
- Matsuoka T, Fujii N, Kondo A, Iwaki A, Hokonohara T, Honda H, Sasaki K, Suzuki SO, Iwaki T. 2011. An autopsied case of sporadic adult-onset amyotrophic lateral sclerosis with FUS-positive basophilic inclusions. Neuropathology 31: 71–76. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Albert MS, Grossman M, Miller B, Dickson D, Trojanowski JQ. 2001. Clinical and pathological criteria for fronto-temporal dementia. Arch Neurol 58: 1803–1909. [DOI] [PubMed] [Google Scholar]
- Mori F, Tanji K, Kon T, Odagiri S, Hattori M, Hoshikawa Y, Kono C, Yasui K, Yokoi S, Hasegawa Y, et al. 2012. FUS immunoreactivity of neuronal and glial intranuclear inclusions in intranuclear inclusion body disease. Neuropathol Appl Neurobiol 38: 322–328. [DOI] [PubMed] [Google Scholar]
- Mori F, Toyoshima Y, Tanji K, Kakita A, Takahashi H, Wakabayashi K. 2014. FUS colocalizes with polyglutamine, but not with TDP-43 in neuronal intranuclear inclusions in spinocerebellar ataxia type 2. Neuropathol Appl Neurobiol 40: 351–355. [DOI] [PubMed] [Google Scholar]
- Munoz-Garcia D, Ludwin SK. 1984. Classic and generalized variants of Pick's disease: A clinicopathological, ultrastructural, and immunocytochemical comparative study. Ann Neurol 16: 467–480. [DOI] [PubMed] [Google Scholar]
- Munoz DG, Neumann M, Kusaka H, Yokota O, Ishihara K, Terada S, Kuroda S, Mackenzie IR. 2009. FUS pathology in basophilic inclusion body disease. Acta Neuropathol 118: 617–627. [DOI] [PubMed] [Google Scholar]
- Murphy JM, Henry RG, Langmore S, Kramer JH, Miller BL, Lomen-Hoerth C. 2007. Continuum of frontal lobe impairment in amyotrophic lateral sclerosis. Arch Neurol 64: 530–534. [DOI] [PubMed] [Google Scholar]
- Neary D, Snowden JS, Gustafson L, Passant U, Stuss D, Black S, Freedman M, Kertesz A, Robert PH, Albert M, et al. 1998. Frontotemporal lobar degeneration: A consensus on clinical diagnostic criteria. Neurology 51: 1546–1554. [DOI] [PubMed] [Google Scholar]
- Neumann M, Bentmann M, Dormann D, Jawaid A, DeJesus-Hernandez M, Ansorge O, Roeber S, Kretzschmar HA, Munoz DG, Kusaka H, et al. 2011. FET proteins TAF15 and EWS are selective markers that distinguish FTLD-FUS from ALS with FUS mutations. Brain 134: 2595–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IRA. 2009a. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain 132: 2922–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Roeber S, Kretzschmar HA, Rademakers R, Baker M, Mackenzie IRA. 2009b. Abundant FUS pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol 118: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. 2006. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314: 130–133. [DOI] [PubMed] [Google Scholar]
- Neumann M, Tolnay M, Mackenzie IRA. 2009c. The molecular basis of frontotemporal dementia. Exp Rev Mol Med 11: e23. [DOI] [PubMed] [Google Scholar]
- Neumann M, Valori CF, Ansorge O, Kretzschmar HA, Munoz DG, Kusaka H, Yokota O, Ishihara K, Ang LC, Bilbao JM, et al. 2012. Transportin 1 accumulates specifically with FET proteins but no other transportin cargos in FTLD-FUS and is absent in FUS inclusions in ALS with FUS mutations. Acta Neuropathol 124: 705–716. [DOI] [PubMed] [Google Scholar]
- Niu C, Zhang J, Gao F, Yang L, Jia M, Zhu H, Gong W. 2012. FUS-NLS/Transportin 1 complex structure provides insights into the nuclear targeting mechanism of FUS and the implications in ALS. PLoS ONE 7: e47056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nukina N. 2010. FUS/TLS as a polyglutamine aggregate interacting protein. Rinsho Shinkeigaku 50: 945–947. [DOI] [PubMed] [Google Scholar]
- Prasad DD, Ouchida M, Lee L, Rao VN, Reddy ES. 1994. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene 9: 3717–3729. [PubMed] [Google Scholar]
- Rademakers R, Stewart H, DeJesus-Hernandez M, Krieger C, Graff-Radford N, Fabros M, Briemberg H, Cashman N, Eisen A, Mackenzie IRA. 2010. FUS gene mutations in familial and sporadic amyotrophic lateral sclerosis. Muscle Nerve 42: 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft TA, Baker MC, Rutherford N, Neumann M, Mackenzie IR, Josephs KA, Boeve BF, Petersen R, Halliday GM, Kril J, et al. 2013. Mutations in protein N-arginine methyltransferases are not the cause of FTLD-FUS. Neurobiol Aging 34: 2235.e11–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson J, Bilbao J, Zinman L, Hazrati L, Tokuhiro S, Sato C, Moreno D, Strome R, Mackenzie IR, Rogaeva E. 2011. A novel double mutation in FUS gene causing sporadic ALS. Neurobiol Aging 32: 553.e27–553.e30. [DOI] [PubMed] [Google Scholar]
- Roeber S, Mackenzie IR, Kretzschmar HA, Neumann M. 2008. TDP-43-negative FTLD-U is a significant new clinico-pathological subtype of FTLD. Acta Neuropathol 116: 147–157. [DOI] [PubMed] [Google Scholar]
- Sama RR, Ward CL, Bosco DA. 2014. Functions of FUS/TLS from DNA repair to stress response: Implications for ALS. ASN Neuro 1: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Cech TR, Parker RR. 2015. Biochemical properties and biological functions of FET proteins. Ann Rev Biochem 84: 355–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seelaar H, Klijnsma KY, de Koning I van der Lugt A, Chiu WZ, Azmani A, Rozemuller AJ, van Swieten JC. 2010. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J Neurol 257: 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden JS, Hu Q, Rollinson S, Halliwell N, Robinson A, Davidson YS, Momeni P, Baborie A, Griffiths TD, Jaros E, et al. 2011. The most common type of FTLD-FUS (aFTLD-U) is associated with a distinct clinical form of frontotemporal dementia but is not related to mutations in the FUS gene. Acta Neuropathol 122: 99–110. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Aoki M, Warita H, Kato M, Mizuno H, Shimakura N, Akiyama T, Furuya H, Hokonohara T, Iwaki A, et al. 2010. FALS with FUS mutation in Japan, with early onset, rapid progress and basophilic inclusions. J Hum Genet 55: 252–254. [DOI] [PubMed] [Google Scholar]
- Suzuki N, Kato S, Kato M, Warita H, Mizuno H, Kato M, Shimakura N, Akiyama H, Kobayashi Z, Konno H, et al. 2012. FUS/TLS-immunoreactive neuronal and glial cell inclusions increase with disease duration in familial amyotrophic lateral sclerosis with an R521C FUS/TLS mutation. J Neuropathol Exp Neurol 71: 779–788. [DOI] [PubMed] [Google Scholar]
- Tateishi T, Hokonohara T, Yamasaki R, Miura S, Kikuchi H, Iwaki A, Tashiro H, Furuya H, Nagara Y, Ohyagi Y, et al. 2010. Multiple system degeneration with basophilic inclusions in Japanese ALS patients with FUS mutation. Acta Neuropathol 119: 355–364. [DOI] [PubMed] [Google Scholar]
- Thomsen C, Grundevik P, Elias P, Stahlberg A, Aman P. 2013. A conserved N-terminal motif is required for complex formation between FUS, EWSR1, TAF15 and their oncogenic fusion proteins. FASEB J 27: 4965–4974. [DOI] [PubMed] [Google Scholar]
- Ticozzi N, Silani V, LeClerc AL, Keagle P, Gellera C, Ratti A, Taroni F, Kwiatkowski TJ, McKenna-Yasek DM, Sapp PC, et al. 2009. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology 73: 1180–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troakes C, Hortobagyi T, Vance C, Al-Sarraj S, Rogelj B, Shaw CE. 2013. Transportin 1 colocalization with fused in sarcoma (FUS) inclusions is not characteristic for amyotrophic lateral sclerosis-FUS confirming disrupted nuclear import of mutant FUS and distinguishing it from frontotemporal lobar degeneration with FUS inclusions. Neuropathol Appl Neurobiol 39: 553–561. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Matsunaga T, Aoki M, Haga C, Ooe K, Abe K, Ikeda K, Nakano I. 2001. Familial amyotrophic lateral sclerosis with posterior column degeneration and basophilic inclusion bodies: A clinical, genetic and pathological study. Clin Neuropathol 20: 53–59. [PubMed] [Google Scholar]
- Urwin H, Josephs KA, Rohrer JD, Mackenzie IR, Neumann M, Authier A, Seelaar H, van Swieten JC, Brown JM, Johannsen P, et al. 2010. FUS pathology defines the majority of tau- and TDP-43-negative frontotemporal lobar degeneration. Acta Neuropathol 120: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Langenhove T, van der Zee J, Sleegers K, Engelborghs S, Vandenberghe R, Gijselinck I, Van den Broeck M, Mattheijssens M, Peeters K, De Deyn PP, et al. 2010. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology 74: 366–371. [DOI] [PubMed] [Google Scholar]
- Vance C, Al-Chalabi A, Ruddy D, Smith BN, Hu X, Sreedharan J, Siddique T, Schelhaas HJ, Kusters B, Troost D, et al. 2006. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.p2–21.3. Brain 129: 868–876. [DOI] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. 2009. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science 323: 1208–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woulfe J, Gray DA, Mackenzie IRA. 2010. FUS-immunoreactive intranuclear inclusions in neurodegenerative disease. Brain Pathol 20: 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto-Watanabe Y, Watanabe M, Okamoto K, Fujita Y, Jackson M, Ikeda M, Nakazato Y, Ikeda Y, Matsubara E, Kawarabayashi T, et al. 2010. A Japanese ALS6 family with mutation R521C in the FUS/TLS gene: A clinical, pathological and genetic report. J Neurol Sci 296: 59–63. [DOI] [PubMed] [Google Scholar]
- Yan J, Deng HX, Siddique N, Fecto F, Chen W, Yang Y, Liu E, Donkervoort S, Zheng JG, Shi Y, et al. 2010. Frameshift and novel mutations in FUS in familial amyotrophic lateral sclerosis and ALS/dementia. Neurology 75: 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Embree LJ, Tsai S, Hickstein DD. 1998. Oncoprotein TLS interacts with serine-arginine proteins involved in RNA splicing. J Biol Chem 273: 27761–27764. [DOI] [PubMed] [Google Scholar]
- Zinszner H, Sok J, Immanuel D, Yin Y, Ron D. 1997. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J Cell Sci 110: 1741–1750. [DOI] [PubMed] [Google Scholar]