Abstract
A minimal cell is one whose genome only encodes the minimal set of genes necessary for the cell to survive. Scientific reductionism postulates the best way to learn the first principles of cellular biology would be to use a minimal cell in which the functions of all genes and components are understood. The genes in a minimal cell are, by definition, essential. In 2016, synthesis of a genome comprised of only the set of essential and quasi-essential genes encoded by the bacterium Mycoplasma mycoides created a near-minimal bacterial cell. This organism performs the cellular functions common to all organisms. It replicates DNA, transcribes RNA, translates proteins, undergoes cell division, and little else. In this review, we examine this organism and contrast it with other bacteria that have been used as surrogates for a minimal cell.
The synthetic bacterial cell JCVI-Syn3.0, reported in 2016, is the first real minimal cell. Because its genome encodes essential genes and little else, it will help us to understand basic principles of cellular life.
In 2016, the construction of a bacterial cell that encoded only the genes necessary for cell life and rapid cell growth in rich laboratory media was announced. The creation of this “minimal cell” was widely heralded as a scientific milestone. The organism was called JCVI-Syn3.0. It has a smaller genome (531,490 bp) than that of any known organism that can be grown in axenic culture. There are only 438 protein-coding genes and 35 RNA-coding genes (Hutchison et al. 2016). Although there have been more than 2000 scientific papers published that mentioned minimal bacterial cells, and creating one has been an ambition of the scientific community since the 1930s, the bacterium JCVI-Syn3.0 is the first real minimal cell. This review discusses the concept of a minimal cell, why they are important, the evolution of that concept, and how the creation of a living minimal cell has changed that concept.
THE MINIMAL CELL CONCEPT
In the 19th and 20th centuries, physicists used the hydrogen atom to gain deep understanding about atomic structure and the nature of matter. Their reasoning was what is true for hydrogen, the simplest of all atoms, is also true for the other more complex elements. Even today, the hydrogen atom is still a tool for the investigation matter because some calculations about atomic structure are just too complex for the other atoms. By analogy, the minimal cell is the hydrogen atom of biology.
Even though cells are the simplest units of life, a huge number of different interacting parts comprise even the simplest cells. Motivated by the same reductionist view that led physicists to study the hydrogen atom, in the 1930s, physicist Max Delbrück founded the American Phage Group. This was a group of physicists, chemists, and biologists who reasoned that an understanding of the first principles of cellular life would come through study of the simplest biological systems (Morange 2000). They adopted a reductionist approach to understand how life works by identifying and determining how each essential element in a living cell functions (obviously, the technology of the day was not up to the task): hence, the minimal cell concept. A minimal cell would contain only the essential genes for independent growth under ideal laboratory conditions. It would live in a stress-free environment where all necessary nutrients would be provided in the growth media. No single gene can be removed without loss of viability. A minimal cell has all of the machinery for independent cellular life. There would be no unnecessary redundancy. From there, one can imagine that if the function(s) of every gene is determined, then it may be possible to achieve a complete understanding of what it takes to be alive. Additionally, it may be possible to model the minimal cell’s behavior on a computer and eventually predict the effects of environmental variations or the effects of added metabolic pathways, etc. From that understanding of the first principles of cellular life, one may be able to build cells that are more complex by addition of genes and metabolic pathways to the minimal cell.
ESSENTIAL, QUASI-ESSENTIAL, AND NONESSENTIAL GENES
In discussion of minimal cells, the term “essential genes” is often used. Genes code for proteins or RNAs with precise functions that may be enzymatic, regulatory, or structural. In the case of essential genes, they code for essential functions. Part of establishing what genes to include in a minimal cell involves classifying them as to whether their functions are essential or not. In a given bacterium, a gene is essential (E) if the cell in which that gene is inactivated cannot be indefinitely propagated. The cell may continue to divide for a time, but once the E gene product is sufficiently diluted among the progeny, no more growth occurs. In this case, the gene specifies an essential function that is not supplied by any other gene in the cell. However, if this same function is supplied by two different either homologous or nonhomologous genes, then in the usual single-gene knockout studies, these two genes are both classed as nonessential, even though the genetic function they specify is an essential function. The pair of genes comprises what is called a “synthetic lethal.” Essentiality is a measure of genetic redundancy. If a genome is completely redundant, there are no E genes. On the other hand, if there is no genetic functional redundancy, then E genes correspond one-to-one with essential genetic functions.
A gene is nonessential (NE) if it can be inactivated without affecting the viability or growth rate of the cell for a specific environment. It can specify either a nonessential genetic function or an essential genetic function. In the latter case, it is dispensable because another gene in the cell supplies the same essential function.
A third class of genes, quasi-essential (QE) genes, which when disrupted result in impaired growth, has been observed in several studies when pools of transposon mutagenized cells are grown competitively (Smith et al. 1996; Badarinarayana et al. 2001; Lluch-Senar et al. 2015; Hutchison et al. 2016). The degree of growth impairment can vary from modest to severe. QE genes specify genetic functions that are important for robust growth, but are not strictly essential. As an example, several QE genes could be collectively involved in supplying a critical essential function such as transport of an amino acid. Deletion of any one gene might make the transport less efficient, but two or more deletions might eliminate the transport and produce a lethal effect. In general, if several QE genes are involved in a similar function, some combinations may act as synthetic lethals when simultaneously disrupted.
MYCOPLASMAS ARE NEAR-MINIMAL CELLS
Mycoplasmas are a group of bacteria characterized their lack of a cell wall, obligate parasitic lifestyle, metabolic simplicity, and most importantly, for this review, small genomes. Long before the advent of the genomic era, it was clear to many biologists that the mycoplasmas were already near-minimal cells. In 1984, physicist Harold Morowitz, recognizing that the mycoplasmas were the simplest cells capable of autonomous growth, proposed that these bacteria be used as models for understanding the basic principles of life (Morowitz 1984). Although the mycoplasmas are often called atypical bacteria because of their salient characteristics, for minimal cell purposes, the exact opposite is true. Mycoplasmas are excellent embodiments of what is constant in all bacteria and indeed in all cellular life. Although mycoplasmas are the simplest cells capable of independent growth in laboratory media, they did not evolve as simpler forms of life. They are not ancient bacteria found at the base of the tree of life. Mycoplasmas are great models for minimal cells because of how they evolved. They did not originally evolve with small genomes and the other unusual features. Rather, they descended from conventional bacteria of the Firmicutes class (e.g., Bacillus subtilis or Staphylococcus aureus) through a process of massive gene loss, which was presumably allowed because they adopted parasitic lifestyles constraining them to live in highly nutrient-rich, stable environments (Woese et al. 1980). Indeed, the three bacteria with the smallest genomes that can be grown in axenic culture are all parasites of the human urogenital tract (this suggests that the mammalian urogenital system may be the most stress-free, hospitable-to-bacteria environment on earth). One of those three species, Mycoplasma genitalium, has the smallest genome at 580,076 (Fraser et al. 1995). It has long been referred to as a minimal or near-minimal cell and used as a surrogate for a minimal cell in many studies. The genes that remain in modern mycoplasmas likely consist of only the minimum number of genes needed for life in their natural habitat. This is because mycoplasmas are under some evolutionary pressure to discard NE genes. Thus, the mycoplasmas have very few redundant genes. Furthermore, their obligate parasitic lifestyle enables easy access to almost all their needed nutrients. These are imported rather than synthesized, which can be done with fewer genes. Essentially, all that mycoplasmas do is synthesize DNA, RNA, and proteins from imported precursors and replicate themselves. Those functions are done the same way in all living cells. Although the idea of using mycoplasmas as starting points for minimal cell studies predates the first genome sequences, it was the availability of whole-genome sequences for mycoplasma species (as well as other bacteria) that really jump-started minimal cell studies.
COMPUTATIONAL DETERMINATION OF MINIMAL GENE SETS
In 1995, the era of genome sequencing began with the shotgun sequencing and computational assembly of the M. genitalium and Haemophilus influenza Rd bacterial genomes. This ushered in a revolution in microbial genome sequencing. Not long after there were two genomes to compare, comparative genomics became a new field. In 1996, Mushegian and Koonin compared orthologs between the two genomes, one Gram- negative and one Gram-positive, and determined a set of 256 genes thought to specify the core functions of a minimal cell. To supply a few apparently missing essential functions not represented by orthologs, they proposed the concept of “nonorthologous gene displacements,” representing recruitment of unrelated or distantly related proteins for the same function (Mushegian and Koonin 1996). Nonorthologous gene displacements may be independently evolved proteins or simply proteins diverged too far to have recognizable relatedness.
Koonin proposed one possible mechanism for establishing nonorthologous gene displacements (Koonin et al. 1996). Suppose a cell has an essential genetic function that is specified by two separate nonhomologous genes. These form an essential function redundancy. As rounds of cell division take place, an occasional daughter cell may lose one member of the pair. Eventually, two lineages will be established, each with a single gene providing the essential function. When the genomes of the two lineages are compared, one gene will be identified as supplying the essential function, but in the other lineage, no orthologous gene can be identified as supplying that essential function. It is necessary to postulate that a nonorthologous displacement has taken place. The apparently missing essential genetic function is actually present, but is unrecognized because it is provided by a gene that is not orthologous to the known gene. Over evolutionary time, this happens many times and gradually the overlap of recognizable orthologs between different bacterial species decreases. Koonin (2003) found that the intersection of orthologs common to a collection of organisms decreases to a low of 65 when comparing all sequenced organisms available in 2003. Charlebois and Doolittle (2004) compared 147 prokaryotic genomes available in 2004 and found less than 50 orthologous genes in common, most of which were involved in translation. Even in the relatively narrow mycoplasma family, comparison of 20 sequenced strains yields a core of only 196 orthologs (Liu et al. 2012). Nonorthologous gene displacements seriously impact the usefulness of comparative genomics for determining minimal gene sets.
Still, through rigorous curation, multi-bacteria comparative genomics studies can be useful. In one of the most often cited comparative genomics studies, Gil and collaborators present a set of 206 protein-coding genes that are highly conserved in both near-minimal organisms such as M. genitalium and endosymbiotic bacteria such as Buchnera aphidicola, as well as in the biotechnology workhorse species Escherichia coli and B. subtilis. The genes are described as the core elements of a possible minimum gene set necessary to sustain life (Gil et al. 2004). Numerous instances of nonorthologous gene displacement are explained showing how some E bacterial gene sets only appear to differ. These genes are almost all present in both the minimal cell JCVI-Syn3.0 and in the near-minimal species M. genitalium. Members of the E gene set in Gil et al. (2004) study not present in the mycoplasmas generally have the necessary cellular tasks performed by different genes.
EXPERIMENTAL DETERMINATION OF ESSENTIAL GENE SETS BY SINGLE-GENE INACTIVATION STUDIES
Hutchison and collaborators introduced global transposon mutagenesis for identifying NE genes in bacterial genomes (Hutchison et al. 1999). Their work was done in two closely related Mycoplasma species. The Tn4001 transposon carried on an E. coli plasmid, which did not replicate in mycoplasma, was introduced by electroporation into an M. genitalium culture. Then, cells with transposon insertions were selected on gentamycin. The principle here is that the transposon, which contains an antibiotic resistance marker, randomly inserts in the M. genitalium genome. Insertions in NE genes result in antibiotic resistant mutants. Insertions in E genes result in nonviable mutants. After more than 30 generations of growth, the locations of insertions in the genome were determined by inverse polymerase chain reaction (PCR) and sequencing from the transposon ends to determine junction points between Tn4001 and the mycoplasma genome. In parallel experiments, Mycoplasma pneumoniae, which contains orthologs of all but one of the 482 M. genitalium genes, plus an additional 197 genes, was mutagenized. For the combined data, 1354 unique insertions were obtained. Insertions were found in 140 M. genitalium genes and 179 M. pneumoniae genes. Combined analysis based on shared orthologs between the two species led to an estimate of between 265 and 351 E genes in M. genitalium (Hutchison et al. 1999). In a follow-up study, Glass and collaborators reanalyzed M. genitalium, this time actually isolating individual clones with insertions in different genes. The number of clones with inserts in NE genes reached a level of 101 insertions asymptotically (Table 1) (Glass et al. 2006). Likely, many of the NE genes identified only in the initial study were not found in 2006 because they were actually QE mutants, and grew too slowly to be recovered.
Table 1.
Essential genes in Mycoplasma species as determined by transposon bombardment
| Mycoplasma species | Genome size (kb) | Total genes | NE genes | Total–NE |
|---|---|---|---|---|
| M. mycoides JCVI-Syn1.0a | 1080 | 901 | 432 | 469 |
| M. pulmonisb | 963 | 789 | 321 | 468 |
| M. pneumoniaec | 816 | 739 | 259 | 480 |
| M. genitaliumd | 580 | 507 | 101 | 406 |
| M. mycoides JCVI-Syn2.0e | 576 | 516 | 90 | 426 |
| M. mycoides JCVI-Syn3.0e | 531 | 473 | 48 | 425 |
NE, Nonessential.
cLluch-Senar et al. 2015. Note that the small previously unannotated noncoding RNAs found by these researchers were not included in these calculations.
In 2008, a Tn4001 transposon study was done in Mycoplasma pulmonis, which identified insertion sites in 1700 clones (French et al. 2008). The 963-kb genome has 789 annotated protein-coding and RNA-coding genes, of which 321 were inactivated (NE) and thus 468 could be classed as E and QE. Because of the small number of insertions analyzed, 321 might be somewhat lower than the true NE gene value.
As a follow-up to the Hutchison et al. study, preliminary determination of NE genes in M. pneumoniae used as a control in to judge when they had achieved saturation of their M. genitalium transposon bombardment (Hutchison et al. 1999), Lluch-Senar and collaborators performed a much more extensive and insightful transposon mutagenesis study of M. pneumoniae (Lluch-Senar et al. 2015). Cells with mini-transposon inserts were grown in culture for 12 days to identify NE genes. The only cells that persisted were those with transposon insertions in NE genes with little to no growth impairment. They found 259 NE genes and 332 E genes. They also identified 93 fitness (QE) genes using a statistical criterion to identify slow growing mutants that persisted but decreased relative to the overall population with increasing passages.
In the most recent mycoplasma study by Hutchison et al. (2016), the synthetic cell Mycoplasma mycoides JCVI-Syn1.0 (Gibson et al. 2010), whose genome was an almost exact copy of the wild-type M. mycoides, subspecies capri genome was subjected to global Tn5 mini-transposon mutagenesis. Mutagenized cells were plated and selected for puromycin resistance conferred by Tn5 insertions. Colonies were counted and pooled. Inserts were mapped to the genome using inverse PCR (Hutchison et al. 1999, 2016), thus providing a zero passage, P0 data set. A sample of the pool was subjected to four serial passages of approximately 40 generations and Tn5 inserts were again mapped to the genome giving a P4 data set. Leaving out RNA-coding genes, the P4 data set showed disrupting Tn5 insertions in 432 genes. These were classified as NE by the criteria used above. There were 205 E genes without Tn5 inserts in P0 and 229 growth-impaired QE genes with P0 inserts, but few if any P4 inserts.
In addition, designing and synthesizing the genome with most of the NE genes removed produced two reduced versions of Syn1.0. The first design resulted in JCVI-Syn2.0 (576 kb) with 516 genes. Transposon mutagenesis in this new genetic background yielded 90 apparent NE genes. Some of these had been classified as QE, but in the new slightly slower growing JCVI-Syn2.0, now appeared NE. The second reduced genome, JCVI-Syn3.0 (531 kb), had 508 total genes and 48 apparent NE genes by Tn5 mutagenesis. Again, many QE genes converted to apparent NE genes in this even slower growing cell (180 min doubling rate vs. 60 min for JVCI-Syn1.0 and 90 min for JCVI-Syn2.0).
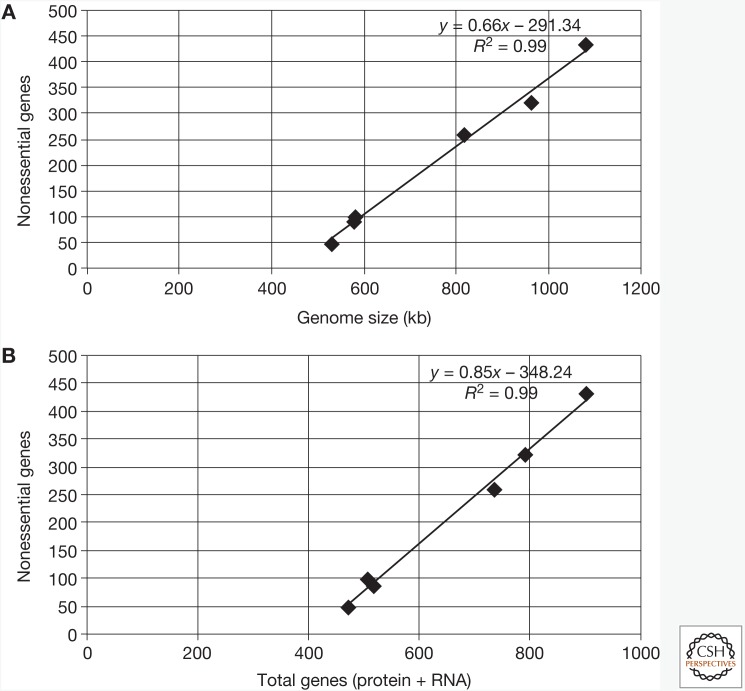
Data for the four mycoplasmas (above and in Fig. 1 and Table 1) show that the sum of E and QE genes is approximately constant at around 400 genes, whereas the number of NE genes varies directly with genome size. It is interesting that when NE genes are plotted against genome size, extrapolation to NE gene to zero yields a minimal genome size of ∼423 kb. Similarly, plotting NE genes versus total genes yields an extrapolated value of 413 genes when NE = 0.
Figure 1.
Plots of nonessential (NE) gene number versus genome size and total genes in Mycoplasma species and reduced strains. NE genes were determined by transposon mutagenesis in Mycoplasma genitalium (580 kb), Mycoplasma pneumoniae (816 kb), Mycoplasma pulmonis (963 kb), Mycoplasma mycoides JCVI-syn1.0 (1080 kb), M. mycoides JCVI-syn2.0 (576 kb), and M. mycoides JCVI-Syn3.0 (531 kb). (A) Plotting NE genes versus genome size yields an extrapolated value of 423 kb for the minimal genome size when NE = 0. (B) Plotting NE genes versus total genes yields an extrapolated value of 413 genes when NE = 0.
GENOME MINIMIZATION BY TOP-DOWN SERIAL DELETIONS
Two different approaches to genome minimization, bottom-up and top-down, have been used. The bottom-up approach involves design, synthesis, and installation of minimal or reduced genomes based on knowledge of gene sets that supply the essential functions for life. In the top-down approach, one starts with a viable “natural” cell and sequentially removes genes, testing for viability and appropriate phenotypic properties at each step. This process can be continued until the genome is substantially reduced, or until no more genes can be removed without severe growth impairment or loss of viability.
Top-down reduction of the M. mycoides JCVI-Syn1.0 genome (Gibson et al. 2010) by consecutive scar-less deletion of NE gene clusters has been recently reported (Hutchison et al. 2016). The 1078-kb JCVI-Syn1.0 genome that has 901 protein and RNA-coding genes was subjected to 22 consecutive deletions of multigene clusters resulting in removal of 255 genes (28%) and 357 kb of DNA (33%) based on knowledge of individual gene essentiality without significantly affecting the growth rate. As more and more genes were removed from the genome, the rate of reduction markedly slowed as NE gene cluster sizes decreased toward single genes. Removal of some genes, because one member of a synthetic lethal pair had already been removed, proved impossible. In other instances, gene removal resulted in an unacceptable slowing of growth rate.
Although efforts to produce a minimal cell whose genome only encoded the minimal set of genes necessary for cell survival have focused on mycoplasmas, there are genome minimization projects that focus on other bacteria. The biotechnology workhorse species, E. coli and B. subtilis, are the subjects of several minimization efforts that seek to produce superior organisms for research and industry. Progress on reduction of E. coli and B. subtilis genomes was recently reviewed (Juhas et al. 2014).
In the first attempt at reduction of the E. coli genome in 2002 (Kolisnychenko et al. 2002), 12 strain-specific genomic islands (KI-islands) representing 8.1% of the genome were removed by scar-less deletion. Also, in 2002, Yu et al. (2002) introduced the Cre/loxP excision method for random removal of nonessential regions and reported a 6.7% reduction of E. coli. Hashimoto et al. (2005) constructed a series of deleted E. coli strains ranging up to 29% genome reduction. Significant cell morphological changes were observed in the 29% deleted strain, but this study did show that large parts of the genome could be removed without marked effects on cell growth. Posfai et al. (2006) used scar-less deletion to remove mobile elements, pathogenicity islands, and certain other nonessential regions totaling 15.3% of the E. coli genome. The resulting strain had good growth characteristics, improved electroporation efficiency, and decreased mutation rates. In 2008, Mizoguchi et al. (2008) and Kato and Hashimoto (2008) produced strains of E. coli lacking 22% and 30% of the genome, respectively. More recently, two E. coli strains lacking 29% and 35.2% of the genome were constructed (Hirokawa et al. 2013).
Several groups have achieved sizable reductions of the B. subtilis genome ranging from 20% to 35%. In 2008, Morimoto et al. (2008) constructed a reduced B. subtilis strain MGB874 genome lacking 874 kb (20.7%) of genomic DNA by consecutive deletion of 11 dispensable regions ranging from 11 kb to 195 kb in size with little effect on growth rate. Recently, Tanaka et al. (2013) and Reuss et al. (2017) produced B. subtilis strains lacking ∼35% of the genome by sequential deletions.
Although these strains of E. coli and B. subtilis with reduced genomes have some interesting properties, this is probably not a practical approach to achieving a minimal genome nor was this the intention of these studies. In the next section, we present work showing that the bottom-up synthetic path can yield an approximately minimal genome when performed in the low genetic redundancy mycoplasmas.
GENOME MINIMIZATION BY BOTTOM-UP DESIGN AND SYNTHESIS
In 1999, Hutchison and coworkers proposed building a cassette-based minimal cell based on a mycoplasma chassis, but this was not feasible at the time (Cho et al. 1999; Hutchison et al. 1999). In 2006, Forster and Church proposed a minimal set of 151 genes (113 proteins and 38 RNAs) encoded by a 113-kb genome that could constitute an in vitro system capable of replication and evolution. The system would be fed by needed small molecules through a membrane (Forster and Church 2006). Although this system was not cellular and was never successfully tested, it was a concrete proposal of a bottom-up synthetic genome design with some of the attributes of a living cell. Only very recently, Hutchison et al. (2016) successfully accomplished building a minimal cell using a bottom-up approach. A number of advances were required to make the synthetic approach feasible. Methods for synthesis of large DNA molecules and for installation and booting up of a naked synthetic DNA genome in recipient cell cytoplasm (genome transplantation) were developed (Lartigue et al. 2007; Gibson et al. 2008; Benders et al. 2010). Better methods for classification of genes as E, QE, or NE were developed (Hutchison et al. 2016). These new methods allowed minimal genome designs to be made, built, and tested. A design-build-test cycle was developed so that various designs could be tested for viability, modified as necessary, and then rebuilt and retested. For troubleshooting, genomes were built in eight overlapping segments, each representing about 1/8th of the genome. Individual synthetic reduced segments could then be tested for viability in the context of the other seven wild-type segments. Viable reduced segments could be put together in various combinations with wild-type segments to obtain partially reduced genomes for testing, and then finally viable genomes made only from reduced segments of various designs could be assembled and tested. Using these methods, Hutchison and coworkers started with M. mycoides JCVI-Syn1.0 (1078 kb, 863 protein, and 38 RNA-coding genes), the synthetic genome published in 2010 (Gibson et al. 2010), and in a series of three design, synthesis, and test cycles, constructed an ∼50% reduced version of the M. mycoides JCVI-Syn1.0 genome with >90% of the NE genes removed. This new “minimal” cell, JCVI-Syn3.0 (531 kb, 438 protein-coding, and 35 RNA-coding genes) grows with a doubling time of 180 min compared with 60 min for syn1.0. Syn3.0 and has a substantially smaller genome than that of any known natural cell capable of independent growth. How close to “minimal” is JCVI-Syn3.0? Hutchison and colleagues performed Tn5 mutagenesis on JCVI-Syn3.0 as reported in the supplementary materials and found 53 genes that did not affect growth rate in the slower growing JCVI-Syn3.0 cells. Most of these had been classified as QE genes in M. mycoides JCVI-Syn1.0 cells previously and had been retained in JCVI-Syn3.0. Adding the data for JCVI-Syn3.0 to the graph in Figure 1 suggests that additional genes could be removed to achieve a fully minimal cell. Total genes extrapolate to 413 when NE genes equals zero. Because JCVI-Syn3.0 has 473 genes (including the 35 RNA genes), as many as 60 genes might be removable while still retaining a viable cell. However, as the investigators point out, growth rate would likely be further impacted.
THE GENES IN MINIMAL CELL JCVI-Syn3.0
The precise functions of 35 RNA genes and 240 protein-coding genes are known. Not unexpectedly, the largest category of genes in the minimal cell is tasked with the expression of genome information (195, 41%). Similarly, the cytosolic metabolism (81, 17%), cell membrane (79, 17%), and preservation of genome information (34, 7%) categories comprise significant fractions of the total gene content as would be expected. A more detailed breakdown of how many JCVI-Syn3.0 genes are involved with different cellular processes is shown in Table 2. What was unexpected was that 65 genes have unknown functions and 84 have only generic assignments such as hydrolase, esterase, ATPase, etc. That we have no clear idea of the functions performed by 149 of 473 genes in the minimal gene set makes it clear how incomplete our knowledge of cellular biology really is. To fully understand how life is specified by the genes of a minimal cell, one must know all of the functions of the cell components.
Table 2.
JCVI-Syn3.0 genes listed by functional category
| Functional category | Number of genes |
|---|---|
| Translation | 89 |
| Unassigned | 79 |
| RNA (rRNAs, tRNAs, small RNAs) | 35 |
| Membrane transport | 31 |
| Cofactor transport and salvage | 21 |
| Lipid salvage and biogenesis | 21 |
| Nucleotide salvage | 19 |
| tRNA modification | 17 |
| DNA replication | 16 |
| Glucose transport and glycolysis | 15 |
| Lipoprotein | 15 |
| Ribosome biogenesis | 14 |
| rRNA modification | 12 |
| Metabolic processes | 10 |
| Protein export | 10 |
| Proteolysis | 10 |
| Transcription | 9 |
| Regulation | 9 |
| Efflux | 7 |
| RNA metabolism | 7 |
| DNA repair | 6 |
| DNA topology | 5 |
| Redox homeostasis | 4 |
| Chromosome segregation | 3 |
| DNA metabolism | 3 |
| Protein folding | 3 |
| Transport and catabolism of nonglucose carbon sources | 2 |
| Cell division | 1 |
| TOTAL GENES | 373 |
COMPARISON OF MINIMAL CELL JCVI-Syn3.0 AND Mycoplasma genitalium GENE SETS BASED ON COMPARATIVE GENOMICS AND TRANSPOSON BOMBARDMENT
JCVI-Syn3.0 was designed to contain only the minimal set of genes needed in an environment where all needed nutrients were provided in the growth media and there were no environmental stressors. The genome encodes 438 protein-coding genes and 35 RNA-coding genes. Given that M. genitalium is often used as a surrogate for a minimal cell and given that extensive transposon bombardment studies have been reported for M. genitalium and JCVI-Syn3.0, we compared those two E gene sets. There are 29 disrupted M. genitalium genes that have homologs in JCVI-Syn3.0. For five of those homologous pairs, the JCVI-Syn3.0 gene was NE but just had not been deleted from the genome. For 21, the JCVI-Syn3.0 gene was QE. Because no growth rate data for the M. genitalium gene disruption mutants are available, we cannot tell if those mutations cause impaired growth, but it is likely. There were 3 JCVI-Syn3.0 E genes that could be disrupted in M. genitalium: chromosome segregation and condensation protein B (MG_214 and JCVISYN3_0328), DNA polymerase III, delta subunit (MG_315 and JCVISYN3_0826), and an ATP-binding cassette (ABC) transporter, ATP-binding protein (MG_290 and JCVISYN3_0707). The fact that JCVI-Syn3.0 divides every 3 hours and M. genitalium divides every 12–15 hours under the same growth conditions could be a reason these genes are essential in one species and not in the other. Of 438 protein-coding genes in JCVI-Syn3.0, 288 have reciprocal BLASTp homologs in M. genitalium, leaving 150 genes unpaired. Similarly, of the 383 protein coding M. genitalium genes that were not disrupted in the Glass et al. (2006) transposon bombardment study, 89 had no homolog in JCVI-Syn3.0. The vast majority of the genes in each organism lacking a homolog in the other encode hypothetical proteins and membrane associated proteins. Many more homologous pairs would likely be found if less stringent criteria for homology were applied. Plus, it is worth noting that M. mycoides (the organism minimized to produce JCVI-Syn3.0) and M. genitalium come from completely different branches of mycoplasma phylogeny. By the Koonin model for nonorthologous gene displacement (Koonin et al. 1996), it is likely that the two species have many genes performing the same functions but which have evolved so that there is no longer sequence similarity. Likely, as the functions of more genes of unknown function in these species are determined, the apparent difference in their gene content will diminish.
MINIMAL CELL COMPUTATIONAL MODELS
As stated at the beginning of this review, one of the aspirations for the minimal cell is to learn the functions of all the gene products and cell components. Those data would be used to create a computational model of the cell that would mimic cell behavior and accurately predict how the cell would respond to genome changes or environmental stress. Richard Feynman’s statement “What I cannot create, I do not understand” (quoted in Gleick 1992), which has become a mantra for the synthetic biology community, is applicable here. Until we understand all aspects of the minimal cell well enough to build computational models that can replicate minimal cell biology, we do not understand the cell. Thus, computational models that consider all of the minimal cell systems will become the yardstick for measuring how well we understand the cell. Already, computational models have been constructed based on the annotated genome of M. genitalium that showed promise in embodying several aspects of the biology of the organism (Tomita et al. 1999; Karr et al. 2012); however, they are far from perfect. Efforts are now underway by several groups to develop computational models for JCVI-Syn3.0.
EPILOGUE
Recently, Bruce Alberts, former president of the United States National Academy of Sciences, wrote about the astonishing finding that 149 genes in the JCVI-Syn3.0 minimal cell were of unknown function. “Hundreds of talented young scientists should be leaping to fill this huge gap in understanding of fundamental biological mechanisms, perhaps earning several Nobel Prizes along the way. Over the long term, such results are certain to lead to powerful new approaches for improving human health and welfare” (Alberts 2016). Having a minimal bacterial cell has been a long-standing goal of cell biologists. No longer are we limited to working with imaginary minimal cells or naturally occurring or naturally occurring organisms with small genomes as surrogates. A minimal cell has now been constructed. Clearly, there is much about its biology that biologists do not understand. First principles of cellular life are waiting to be discovered.
ACKNOWLEDGMENTS
We thank Synthetic Genomics (SGI), the Defense Advanced Research Projects Agency’s Living Foundries program (contract HR0011-12-C-0063), and the J. Craig Venter Institute for funding this work. We also thank the members of the J. Craig Venter Institute and SGI who contributed to the creation of the minimal cell, JCVI-Syn3.0.
Footnotes
Editors: Daniel G. Gibson, Clyde A. Hutchison III, Hamilton O. Smith, and J. Craig Venter
Additional Perspectives on Synthetic Biology available at www.cshperspectives.org
REFERENCES
- Alberts B. 2016. Ensuring an innovative and productive future for the next generation of scientists: The 2016 Lasker-Koshland special achievement award in medical science. JAMA 316: 1256–1257. [DOI] [PubMed] [Google Scholar]
- Badarinarayana V, Estep PW III, Shendure J, Edwards J, Tavazoie S, Lam F, Church GM. 2001. Selection analyses of insertional mutants using subgenic-resolution arrays. Nat Biotechnol 19: 1060–1065. [DOI] [PubMed] [Google Scholar]
- Benders GA, Noskov VN, Denisova EA, Lartigue C, Gibson DG, Assad-Garcia N, Chuang RY, Carrera W, Moodie M, Algire MA, et al. 2010. Cloning whole bacterial genomes in yeast. Nucleic Acids Res 38: 2558–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois RL, Doolittle WF. 2004. Computing prokaryotic gene ubiquity: Rescuing the core from extinction. Genome Res 14: 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho MK, Magnus D, Caplan AL, McGee D. 1999. Policy forum: Genetics. Ethical considerations in synthesizing a minimal genome. Science 286: 2087, 2089–2090. [DOI] [PubMed] [Google Scholar]
- Forster AC, Church GM. 2006. Towards synthesis of a minimal cell. Mol Syst Biol 2: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270: 397–403. [DOI] [PubMed] [Google Scholar]
- French CT, Lao P, Loraine AE, Matthews BT, Yu H, Dybvig K. 2008. Large-scale transposon mutagenesis of Mycoplasma pulmonis. Mol Microbiol 69: 67–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, et al. 2008. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science 319: 1215–1220. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Glass JI, Lartigue C, Noskov VN, Chuang RY, Algire MA, Benders GA, Montague MG, Ma L, Moodie MM, et al. 2010. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 329: 52–56. [DOI] [PubMed] [Google Scholar]
- Gil R, Silva FJ, Pereto J, Moya A. 2004. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev 68: 518–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, Hutchison CA III, Smith HO, Venter JC. 2006. Essential genes of a minimal bacterium. Proc Natl Acad Sci 103: 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleick J. 1992. Genius: The life and science of Richard Feynman. Random House, New York. [Google Scholar]
- Hashimoto M, Ichimura T, Mizoguchi H, Tanaka K, Fujimitsu K, Keyamura K, Ote T, Yamakawa T, Yamazaki Y, Mori H, et al. 2005. Cell size and nucleoid organization of engineered Escherichia coli cells with a reduced genome. Mol Microbiol 55: 137–149. [DOI] [PubMed] [Google Scholar]
- Hirokawa Y, Kawano H, Tanaka-Masuda K, Nakamura N, Nakagawa A, Ito M, Mori H, Oshima T, Ogasawara N. 2013. Genetic manipulations restored the growth fitness of reduced-genome Escherichia coli. J Biosci Bioeng 116: 52–58. [DOI] [PubMed] [Google Scholar]
- Hutchison CA, Peterson SN, Gill SR, Cline RT, White O, Fraser CM, Smith HO, Venter JC. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286: 2165–2169. [DOI] [PubMed] [Google Scholar]
- Hutchison CA 3rd, Chuang RY, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, et al. 2016. Design and synthesis of a minimal bacterial genome. Science 351: aad6253. [DOI] [PubMed] [Google Scholar]
- Juhas M, Reuss DR, Zhu B, Commichau FM. 2014. Bacillus subtilis and Escherichia coli essential genes and minimal cell factories after one decade of genome engineering. Microbiology 160: 2341–2351. [DOI] [PubMed] [Google Scholar]
- Karr JR, Sanghvi JC, Macklin DN, Gutschow MV, Jacobs JM, Bolival B Jr, Assad-Garcia N, Glass JI, Covert MW. 2012. A whole-cell computational model predicts phenotype from genotype. Cell 150: 389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Hashimoto M. 2008. Construction of long chromosomal deletion mutants of Escherichia coli and minimization of the genome. Methods Mol Biol 416: 279–293. [DOI] [PubMed] [Google Scholar]
- Kolisnychenko V, Plunkett G III, Herring CD, Feher T, Posfai J, Blattner FR, Posfai G. 2002. Engineering a reduced Escherichia coli genome. Genome Res 12: 640–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2003. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol 1: 127–136. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Mushegian AR, Bork P. 1996. Non-orthologous gene displacement. Trends Genet 12: 334–336. [PubMed] [Google Scholar]
- Lartigue C, Glass JI, Alperovich N, Pieper R, Parmar PP, Hutchison CA III, Smith HO, Venter JC. 2007. Genome transplantation in bacteria: Changing one species to another. Science 317: 632–638. [DOI] [PubMed] [Google Scholar]
- Liu W, Fang L, Li M, Li S, Guo S, Luo R, Feng Z, Li B, Zhou Z, Shao G, et al. 2012. Comparative genomics of Mycoplasma: Analysis of conserved essential genes and diversity of the pan-genome. PLoS ONE 7: e35698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluch-Senar M, Delgado J, Chen WH, Llorens-Rico V, O’Reilly FJ, Wodke JA, Unal EB, Yus E, Martinez S, Nichols RJ, et al. 2015. Defining a minimal cell: Essentiality of small ORFs and ncRNAs in a genome-reduced bacterium. Mol Syst Biol 11: 780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi H, Sawano Y, Kato J, Mori H. 2008. Superpositioning of deletions promotes growth of Escherichia coli with a reduced genome. DNA Res 15: 277–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morange M. 2000. A history of molecular biology. Harvard University Press, Cambridge. [Google Scholar]
- Morimoto T, Kadoya R, Endo K, Tohata M, Sawada K, Liu S, Ozawa T, Kodama T, Kakeshita H, Kageyama Y, et al. 2008. Enhanced recombinant protein productivity by genome reduction in Bacillus subtilis. DNA Res 15: 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morowitz HJ. 1984. The completeness of molecular biology. Isr J Med Sci 20: 750–753. [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci 93: 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai G, Plunkett G III, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, et al. 2006. Emergent properties of reduced-genome Escherichia coli. Science 312: 1044–1046. [DOI] [PubMed] [Google Scholar]
- Reuss DR, Altenbuchner J, Mader U, Rath H, Ischebeck T, Sappa PK, Thurmer A, Guerin C, Nicolas P, Steil L, et al. 2017. Large-scale reduction of the Bacillus subtilis genome: Consequences for the transcriptional network, resource allocation, and metabolism. Genome Res 27: 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V, Chou KN, Lashkari D, Botstein D, Brown PO. 1996. Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science 274: 2069–2074. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Henry CS, Zinner JF, Jolivet E, Cohoon MP, Xia F, Bidnenko V, Ehrlich SD, Stevens RL, Noirot P. 2013. Building the repertoire of dispensable chromosome regions in Bacillus subtilis entails major refinement of cognate large-scale metabolic model. Nucleic Acids Res 41: 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita M, Hashimoto K, Takahashi K, Shimizu TS, Matsuzaki Y, Miyoshi F, Saito K, Tanida S, Yugi K, Venter JC, et al. 1999. E-CELL: Software environment for whole-cell simulation. Bioinformatics 15: 72–84. [DOI] [PubMed] [Google Scholar]
- Woese CR, Maniloff J, Zablen LB. 1980. Phylogenetic analysis of the mycoplasmas. Proc Natl Acad Sci 77: 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BJ, Sung BH, Koob MD, Lee CH, Lee JH, Lee WS, Kim MS, Kim SC. 2002. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat Biotechnol 20: 1018–1023. [DOI] [PubMed] [Google Scholar]