In this study, Inoue et al. investigated the mechanism underlying Xist imprinting in female mammals. They demonstrate that the Xist locus is coated with a broad H3K27me3 domain that is established during oocyte growth and persists through preimplantation development in mice, thus identifying maternal H3K27me3 as the imprinting mark of Xist.
Keywords: H3K27me3, X-chromosome inactivation, genomic imprinting, mouse early development
Abstract
Maternal imprinting at the Xist gene is essential to achieve paternal allele-specific imprinted X-chromosome inactivation (XCI) in female mammals. However, the mechanism underlying Xist imprinting is unclear. Here we show that the Xist locus is coated with a broad H3K27me3 domain that is established during oocyte growth and persists through preimplantation development in mice. Loss of maternal H3K27me3 induces maternal Xist expression and maternal XCI in preimplantation embryos. Our study thus identifies maternal H3K27me3 as the imprinting mark of Xist.
In females of certain therian mammals, including rodents, one of the two X chromosomes is inactivated to achieve gene dosage compensation (Augui et al. 2011; Lee and Bartolomei 2013). During development, X-chromosome inactivation (XCI) can take place in either an imprinted or a random manner (Augui et al. 2011; Lee and Bartolomei 2013). For imprinted XCI, the paternal X chromosome (Xp) is selectively inactivated during preimplantation development (Okamoto et al. 2005). Although imprinted XCI is maintained in the extraembryonic cell lineage, it is lost in the pre-epiblast cell lineage of late blastocysts (Takagi and Sasaki 1975). At the peri-implantation stage, epiblast cells undergo random XCI, resulting in the silencing of either the Xp or maternal X chromosome (Xm) (Lyon 1961). Previous studies have demonstrated a critical role of Xist, an X-linked long noncoding RNA, in both imprinted and random XCI. The Xist RNA participates in XCI by coating and inactivating the X chromosome in cis (Penny et al. 1996; Marahrens et al. 1997, 1998; Borensztein et al. 2017).
To selectively silence the Xp during preimplantation development, Xist is imprinted in the Xm with a long-sought-after but yet-to-be-identified mechanism. Previous studies using nuclear transfer approaches have suggested that genomic imprinting of Xist is established during oogenesis (Tada et al. 2000; Oikawa et al. 2014). However, analyses of DNA methyltransferase maternal knockout embryos revealed that oocyte DNA methylation is dispensable for Xist imprinting (Chiba et al. 2008). A recent study demonstrated that overexpression of an H3K9me3 demethylase, Kdm4b, in parthenogenetic (PG) embryos partially derepresses Xist (Fukuda et al. 2014), suggesting the involvement of H3K9me3 in imprinted Xist silencing. However, the fact that PG embryos undergo nonphysiological Xist derepression of a single maternal allele (Nesterova et al. 2001) raises the question of whether the derepression effect observed in the H3K9me3-depleted PG embryos is physiologically relevant. Our recent discovery that maternal H3K27me3 serves as an imprinting mark for DNA methylation-independent autosomal gene imprinting (Inoue et al. 2017) prompted us to ask whether it is also responsible for Xist imprinting.
Results and Discussion
H3K9me3 is dispensable for Xist silencing in biparental embryos
To determine whether Kdm4b-mediated loss of H3K9me3 can induce Xist derepression in biparental embryos, we injected Kdm4b mRNA into in vitro fertilization-derived embryos. Immunostaining analysis confirmed that Kdm4b mRNA injection effectively depleted H3K9me3 in zygotes in a concentration-dependent manner (Fig. 1A). To assess Xist RNA expression, we performed RNA fluorescent in situ hybridization (FISH) analysis in four-cell embryos. To distinguish between male and female embryos, we simultaneously labeled X chromosomes by DNA FISH using a probe specific for the Rnf12 locus (Fukuda et al. 2015). As such, each blastomere of the male or female embryos should have one or two DNA FISH signals, respectively. The “no injection” control male embryos showed no Xist RNA signal, and the majority of female embryos showed one RNA cloud or spot signal (Fig. 1B–D). Similarly, Kdm4b-injected embryos did not induce maternal Xist expression in either male or female embryos (Fig. 1B–D), indicating that H3K9me3 does not play a major role in maternal Xist silencing under physiological biparental conditions.
Figure 1.
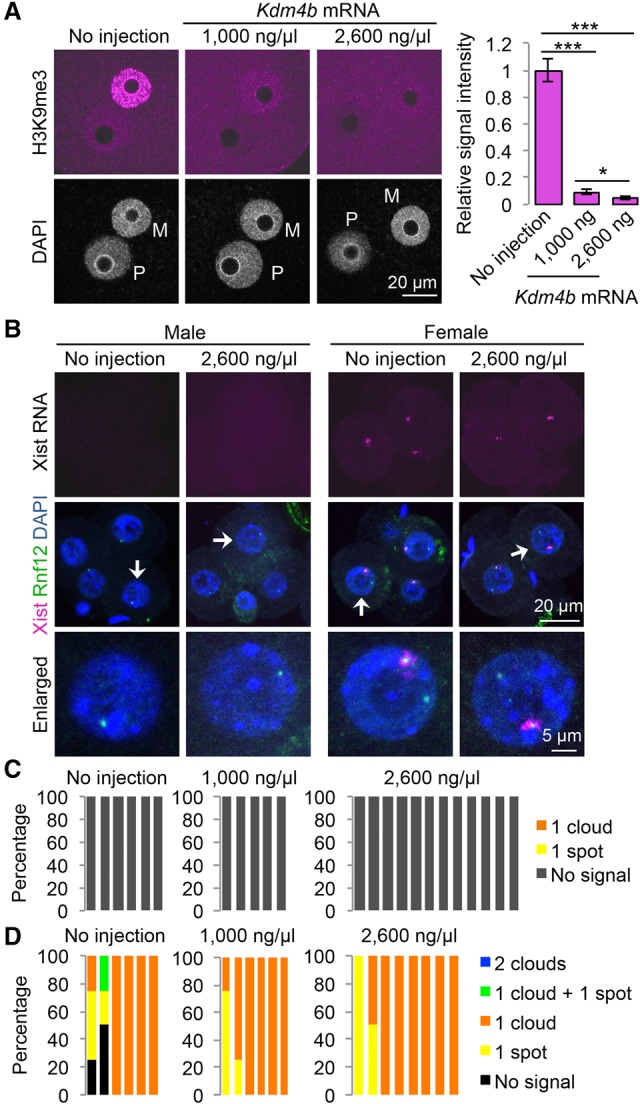
Ectopic removal of H3K9me3 does not induce maternal Xist expression. (A) Representative images of zygotes stained with anti-H3K9me3 antibody. (M) Maternal pronucleus; (P) paternal pronucleus. The bar graph shows relative H3K9me3 signal intensity of the maternal pronuclei. The average signal intensity of noninjected zygotes was set as 1.0. The total numbers of embryos examined were eight (no injection), nine (1000 ng/µL Kdm4b), and nine (2600 ng/µL Kdm4b). Error bars indicate SE. (***) P < 0.001; (*) P < 0.05, two-tailed Student's t-test. (B) Representative images of Xist RNA FISH (magenta) in Kdm4b-injected four-cell embryos. The gender of each embryo was assessed by simultaneous DNA FISH against the Rnf12 locus (green). Arrows indicate the blastomeres enlarged in the bottom panels. (C,D) The ratio of blastomeres that show the indicated number of Xist RNA clouds and spots in male (C) and female (D) embryos. Each bar represents an individual embryo. The numbers of embryos examined were six (no injection), five (1000 ng/µL Kdm4b), and 12 (2600 ng/µL Kdm4b) males and six (no injection), six (1000 ng/µL Kdm4b), and eight (2600 ng/µL Kdm4b) females.
Maternal H3K27me3 coats Xist in oocytes and preimplantation embryos
Since maternal H3K27me3 can function as an imprinting mark (Inoue et al. 2017), we examined its potential involvement in Xist imprinting. Analysis of H3K27me3 ChIP-seq (chromatin immunoprecipitation [ChIP] combined with high-throughput sequencing) data sets (Zheng et al. 2016) revealed that Xist is coated with a broad H3K27me3 domain, which spans ∼450 kb in mature oocytes and is established during oocyte growth (Fig. 2A). Analyses of the oocyte DNase I sequencing (DNase I-seq) (Inoue et al. 2017) and DNA methylome (Kobayashi et al. 2012) data sets revealed that this entire H3K27me3 domain exhibits low chromatin accessibility and low DNA methylation (Fig. 2A), suggesting formation of a heterochromatin domain independent of DNA methylation. Analyses of the ChIP-seq data sets of post-fertilization embryos (Zheng et al. 2016) revealed that the maternal H3K27me3 domain is maintained throughout preimplantation development but lost in the embryonic day 6.5 (E6.5) epiblast (Fig. 2B). Notably, the upstream ∼200-kb region, which spans Xist to Zcchc13 but does not include the Tsix promoter, maintains the maternal allele bias of H3K27me3 enrichment in blastocyst embryos (Fig. 2C). These data support a potential role of maternal H3K27me3 in maternal Xist silencing.
Figure 2.
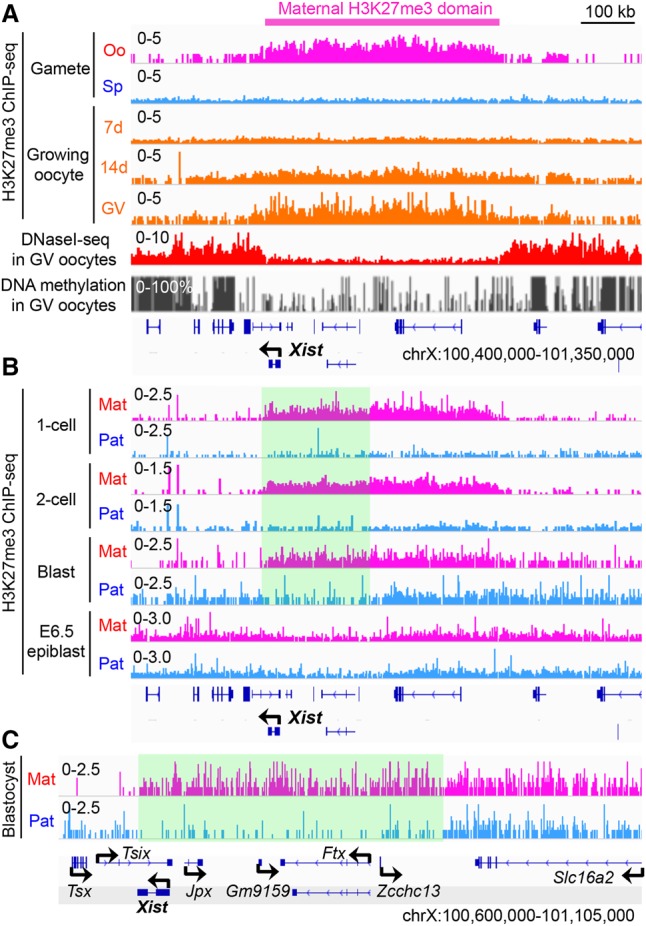
Maternal H3K27me3 coats Xist in oocytes and preimplantation embryos. (A) Genome browser view of H3K27me3 ChIP-seq (Zheng et al. 2016), DNase I-seq (Inoue et al. 2017), and DNA methylation levels (Kobayashi et al. 2012) at the Xist locus. (Oo) MII oocyte; (Sp) sperm; (7 d) growing oocytes collected from 7-d-old females; (14 d) growing oocytes collected from 14-d-old females; (GV) fully grown GV stage oocytes. (B) Genome browser view of allelic H3K27me3 in one-cell, two-cell, and blastocyst embryos and E6.5 epiblast at the Xist locus. The highlighted square indicates a computationally determined region where the maternal allele bias of H3K27me3 enrichment is retained in blastocyst embryos. (Mat) Maternal allele; (Pat) paternal allele. The H3K27me3 ChIP-seq data sets were from Zheng et al. (2016). (C) A higher-resolution view of the Xist locus in blastocyst embryos. The maternal allele-biased H3K27me3 domain is highlighted.
Maternal H3K27me3 is responsible for maternal Xist silencing
To examine whether H3K27me3 is responsible for maternal Xist silencing, we depleted H3K27me3 in zygotes by injecting mRNA coding an H3K27me3-specific demethylase, Kdm6b (Fig. 3A). As a negative control, we prepared zygotes injected with its catalytic mutant, Kdm6bMUT, harboring a point mutation at the catalytic domain (Fig. 3A; Inoue et al. 2017). Despite the transient expression of the exogenous Kdm6b, the H3K27me3 level in Kdm6bWT embryos was significantly lower than that of Kdm6bMUT embryos at the four-cell and morula stages (Supplemental Fig. S1A,B).
Figure 3.
Loss of H3K27me3 induces maternal Xist expression. (A) Representative images of zygotes stained with anti-H3K27me3 antibody. (M) Maternal pronucleus, (P) paternal pronucleus. The bar graph indicates the relative H3K27me3 signal intensity of maternal pronuclei. The average signal of Kdm6bMUT-injected zygotes was set as 1.0. The total numbers of embryos examined were 15 (Kdm6bMUT) and 13 (Kdm6bWT). Error bars indicate SE. (***) P < 0.001, two-tailed Student's t-test. (B) Genome browser view of the Xist locus showing loss of maternal H3K27me3 ChIP-seq signal in Kdm6bWT-injected morula embryos. (Mat) Maternal allele; (Pat) paternal allele. (C) Representative images of Xist RNA FISH (magenta) in Kdm6b-injected four-cell embryos. The gender of each embryo was assessed by simultaneous DNA FISH against the Rnf12 locus (green). (D,E) The ratio of blastomeres showing the indicated number of Xist RNA clouds and spots in male (D) and female (E) four-cell embryos. Each bar represents an individual embryo. The numbers of embryos examined were eight (Kdm6bMUT) and 15 (Kdm6bWT) for males and eight (Kdm6bMUT) and 12 (Kdm6WT) for females.
To confirm that H3K27me3 is lost at the Xist locus in Kdm6bWT-injected embryos, we performed ultralow input native ChIP-seq (ULI-NChIP) analysis (Brind'Amour et al. 2015), which worked efficiently using 500–2000 mouse embryonic stem cells (Supplemental Fig. S2A). We then performed H3K27me3 ULI-NChIP using ∼2000 blastomeres from Kdm6bWT- or Kdm6bMUT-injected morula embryos and validated the data quality by comparing Kdm6bMUT-injected embryos with a public morula embryo data set (Supplemental Fig. S2B–D; Liu et al. 2016). The number of H3K27me3 peaks in Kdm6bWT-injected embryos was much smaller than that of Kdm6bMUT-injected embryos (Supplemental Fig. S2E). Importantly, the Xist locus exhibits a marked decrease of H3K27me3 enrichment throughout the entire domain in Kdm6bWT embryos (Supplemental Fig. S2F). Furthermore, analysis of single-nucleotide polymorphism (SNP) information revealed that the maternal H3K27me3 domain is lost in Kdm6bWT embryos (Fig. 3B).
We then asked whether maternal Xist is depressed in Kdm6bWT-injected four-cell embryos. RNA/DNA FISH analysis revealed that the majority of Kdm6bWT-injected males showed one Xist RNA cloud or spot, while all of the Kdm6bMUT-injected males showed no signal (Fig. 3C,D). Furthermore, the majority of Kdm6bWT-injected females showed two Xist RNA clouds or/and spots, while most of the Kdm6bMUT-injected females showed one cloud (Fig. 3C,E). These results demonstrate that loss of maternal H3K27me3 at the Xist locus induces maternal Xist derepression at the four-cell stage.
Loss of H3K27me3 induces maternal XCI
To examine whether maternal Xist expression continues until the morula stage (Fig. 4A), we performed RNA/DNA FISH analysis. Strikingly, the majority of Kdm6bWT-injected male and female embryos showed one and two RNA clouds, respectively, while most of the Kdm6bMUT-injected male and female embryos showed none and one RNA cloud, respectively (Fig. 4B–D), indicating that the reactivated Xist is persistent. Although Xist can be up-regulated by Rnf12 overexpression (Tan et al. 2016), RT-qPCR analysis found no evidence of Rnf12 up-regulation in Kdm6bWT-injected embryos (Supplemental Fig. S3), excluding the possibility that Kdm6bWT-mediated Xist depression is due to Rnf12 overexpression.
Figure 4.
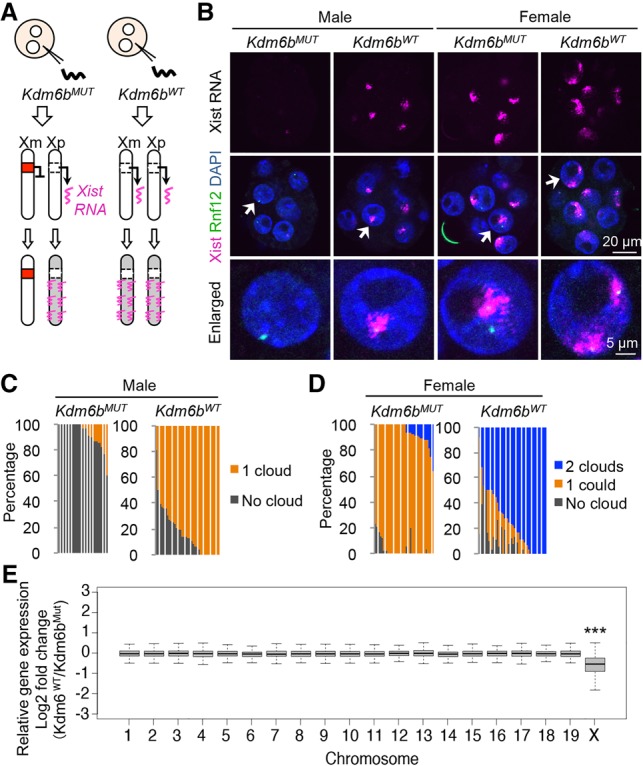
Loss of H3K27me3 induces maternal XCI. (A) Illustration depicting maternal XCI caused by Kdm6WT-mediated maternal Xist expression. (B) Representative images of Xist RNA FISH (magenta) in Kdm6b-injected morula embryos. The gender of each embryo was assessed by simultaneous DNA FISH against the Rnf12 locus (green). (C,D) The ratio of blastomeres showing the indicated number of Xist RNA clouds in male (C) and female (D) morula embryos. Each bar represents an individual embryo. The numbers of embryos examined were 19 (Kdm6bMUT) and 35 (Kdm6bWT) for males and 34 (Kdm6bMUT) and 35 (Kdm6bWT) for females. (E) Box plot showing the relative expression of genes on individual maternal chromosomes between Kdm6bMUT- and Kdm6bWT-injected blastocysts. Genes with enough SNP reads (SNP reads > 10, RPM [reads per million] > 0.5) were analyzed. The middle lines in the boxes represent the medians. Box edges and whiskers indicate the 25th/75th and 2.5th/97.5th percentiles, respectively. (***) P < 0.001, Mann-Whitney-Wilcoxon test.
To determine whether maternal Xist expression leads to maternal XCI (Fig. 4A), we preformed RNA sequencing (RNA-seq) analysis on early blastocyst stage hybrid mouse embryos with biological duplicates (Supplemental Fig. S4A; Supplemental Table S1). Analysis of SNP information allowed us to examine Xm-linked genes, which revealed that the expression level of Xm-linked genes, but not those of autosomal genes, was significantly down-regulated in Kdm6bWT-injected embryos (Fig. 4E). Consistently, the maternal allele expression bias [Mat/(Mat + Pat)] of X-linked genes was significantly compromised in Kdm6bWT-injected embryos, although it was still >50% (Supplemental Fig. S4B). These data demonstrate that maternal XCI occurs at a level milder than paternal XCI in Kdm6bWT-injected embryos. A closer examination of individual X-linked genes confirmed that most genes were down-regulated in Kdm6bWT-injected embryos (Supplemental Fig. S4C), while genes known to escape imprinted XCI (Borensztein et al. 2017) were unchanged (Supplemental Fig. S4D), suggesting that Kdm6b-mediated maternal XCI could recapitulate physiological XCI.
Taken together, our data provide evidence that H3K27me3 serves as the imprinting mark of Xist. The domain-based regulation of maternal H3K27me3 at the Xist locus is an attractive observation and raises important questions. For example, how are the boundaries of the H3K27me3 domain defined during oogenesis? How does the first half (∼200 kb) of the domain, spanning Xist and Zcchc13, maintain the maternal allele-specific H3K27me3 enrichment in blastocyst embryos? Notably, this ∼200-kb region appears to form a topological association domain (Giorgetti et al. 2014), implicating potential involvement of a protein such as CTCF in regulating chromatin boundaries. Interestingly, this domain highly overlaps transgenes whose insertion into an autosome recapitulated imprinted XCI (Okamoto et al. 2005). This raises a possibility that a core element attracting Polycomb group complexes might exist in the region and contribute to imprinting establishment during oogenesis. Further studies are needed to answer these questions. Our study thus not only identifies Xist as a new member of H3K27me3-dependent imprinted genes (Inoue et al. 2017) but also demonstrates the biological significance of H3K27me3-dependent genomic imprinting in safeguarding the Xm from XCI.
Materials and methods
Collection of mouse oocytes
All animal studies were performed in accordance with guidelines of the Institutional Animal Care and Use Committee at Harvard Medical School. The procedures of oocyte collection and in vitro fertilization were described previously (Inoue et al. 2017). The hybrid embryos used for SNP analysis in this study were obtained by in vitro fertilization of B6D2F1/J (BDF1) oocytes and PWK sperm (Jackson Laboratory, 003715).
mRNA preparation and injection
The construction and preparation of Kdm6b mRNA and microinjection into fertilized oocytes were described previously (Inoue et al. 2017). The Kdm4b construct was generated by cloning its cDNA amplicon into the pcDNA3.1-Flag-poly(A)83 plasmid. mRNA was synthesized with mMESSAGE mMACHINE T7 Ultra kit (Life technologies), purified by lithium chloride precipitation, and dissolved with nuclease-free water. The concentrations of injected mRNA of Kdm6bWT and Kdm6bMUT were 1800 ng/µL, and those of Kdm4b were 1000 or 2600 ng/µL.
Probe for FISH
A probe for Xist RNA was prepared by using Nick translation reagent kit (Abbott Molecular, 07J00-001) with Cy3-dCTP (GE Healthcare, PA53021). The template DNA was a plasmid coding the full-length mouse Xist gene (Addgene, 26760) (Wutz and Jaenisch 2000). A probe for DNA FISH was prepared using the same kit with Green-dUTP (Abbott Molecular, 02N32-050). The template DNA was a BAC clone containing the Rnf12 locus (RP23-36C20) (Fukuda et al. 2015). The fluorescent probes were ethanol-precipitated with 5 µg of Cot-1 DNA (Life technologies), 5 µg of herring sperm DNA (Thermo Fisher Scientific), and 2.5 µg of yeast tRNA (Thermo Fisher Scientific, AM7119) and then dissolved with 20 µL of formamide (Thermo Fisher Scientific, 17899). The probes were stored at 4°C. Before being used, the probes (0.75 µL each) were mixed with 0.75 µL of Cot-1 DNA/formamide and 2.25 µL of 4× SSC/20% dextran (Millipore S4030). The probe mixtures were heated for 30 min at 80°C and then transferred to a 37°C incubator (“preannealed probes”).
Whole-mount RNA/DNA FISH
Four-cell or morula embryos were fixed at 46 or 78 h post-fertilization (hpf) in 2% paraformaldehyde (PFA) in PBS containing 0.5% Triton X-100 for 20 min at room temperature. After three washes with 0.1% BSA/PBS, embryos were treated with 0.1 N HCl containing 0.02% Triton X-100 for 15 min at 4°C. After three washes with 0.1% BSA/2× SSC, embryos were incubated in a series of 10%, 20%, and 50% formamide/2× SSC in a glass dish (Electron Microscopy Science, 705430-30) and incubated for 30 min. The samples were covered with mineral oil, heated for 30 min at 80°C, and then incubated for >30 min at 37°C. The embryos were then transferred into 4.5 µL of “preannealed probes,” covered with mineral oil on another glass dish, and incubated for >24 h at 37°C. Embryos were washed with 42°C prewarmed 0.1% BSA/2× SSC, left in the last drop for 30 min, and mounted on a glass slide in VectaShield with DAPI (Vector Laboratories). Fluorescence was detected under a laser-scanning confocal microscope (Zeiss, LSM800).
Whole-mount immunostaining
The procedure of immunostaining and quantification was described previously (Inoue et al. 2017).
Identification of the maternal allele-biased H3K27me3 domain
The BED files, including RPKM (reads per kilobase per million mapped reads) values in 100-base-pair (bp) bins for H3K27me3 ChIP-seq in the inner cell mass (ICM), were from GSE76687 (Zheng et al. 2016). BED files labeled maternal- or paternal-containing RPKM values for two parental alleles, and allelic reads were normalized to total read number. “bedtools makewindows” was used to generate 1-kb bins for the entire mm9 genome, and the RPKM value for each bin was calculated by “bedtools map.” All of the bins were classified to three categories of “no signal,” “biallelic,” and “maternal-biased” using a signal cutoff of 1 and a fold change cutoff of 4. A sliding window approach was used to identify domains that were enriched for “maternal-biased” H3K27me3 bins. The criteria used were as follows: Within a window of 20 kb, the minimum number of “maternal-biased” bins was three, and the percentage of “maternal-biased” bins was larger than “biallelic” bins. Overlapped windows were merged by “bedtools merge.” A total of 5986 domains was identified in the genome.
ULI-NChIP
At 78 hpf, ∼110 morula embryos per group were briefly treated with acid Tyrode's solution (Sigma-Aldrich) to remove zona pellucida, washed with 0.2% BSA/PBS, and transferred to 1.5-mL tubes. The ULI-NChIP was performed using H3K27me3 antibody (Diagenode, C15410069) as described previously (Brind'Amour et al. 2015) with the following modifications. First, we used Beckman SPRIselect beads (Beckman Coulter) instead of Agencourt Ampure XP beads. Second, the sequencing library was prepared using NEBNext Ultra II DNA library preparation kit for Illumina (New England Biolabs). Third, PCR amplification was performed using Kapa Hifi hot start ready mix (Kapa Biosystems). Last, no size selection was performed. For input samples, 10% volume of the chromatin lysate was taken and used for library construction and sequencing. The quantification and sequencing of the libraries were described previously (Inoue et al. 2017).
Data analysis of ULI-NChIP
The procedures of alignment and peak calling and the summary of SNP-tracked data analyses are described in Supplemental Table S2. For normalization between Kdm6bMUT and Kdm6bWT samples, we used a strategy similar to MAnorm (Shao et al. 2012). First, we identified the common peaks between the Kdm6bMUT and Kdm6bWT samples. Next, all samples were normalized to the highest coverage samples based on the RPKM value for each sample within the common peaks. SNP-specific reads were normalized to total uniquely mapped reads per library and further normalized to the highest-coverage allele based on the sum of SNP-specific reads per allele. The procedures of reanalysis of public data sets are described in Supplemental Table S2.
Reverse transcription and real-time PCR analysis
Kdm6b-injected embryos were collected at 46 hpf (four-cell), 60 hpf (eight-cell), and 78 hpf (morula). The procedures of reverse transcription and real-time PCR analysis were described previously (Inoue and Zhang 2014) except for the use of random primers for reverse transcription in this study. Primer sequences used for real-time PCR were as follows: 18S-F (5′-TTGACGGAAGGGCACCACCAG-3′), 18S-R (5′-GCACCACCACCCACGGAATCG-3′), Rnf12-F (5′-TTTGTCGCAGGGCAGTCTTA-3′), and Rnf12-R (5′-GTTTGCCCATCACTATTCCAGC-3′) (Tan et al. 2016).
RNA-seq and data analysis
Blastocyst embryos at 96 hpf were treated briefly with acid Tyrode's solution, washed by 0.2% BSA/PBS, and stored in PCR tubes at −80°C. Forty to 46 embryos per group were pooled and used for RNA-seq. RNA-seq libraries were prepared as described previously (Inoue et al. 2017) with the exception of using Nextera XT DNA library preparation kit (Illumina) following cDNA amplification. The procedure of data analysis is described in Supplemental Table S1.
Statistical analyses
Statistical analyses were implemented with R (http://www.r-project.org). Pearson's R coefficient was calculated using the “cor” function with default parameters. For Supplemental Figure S4B, Mann-Whitney U-Test was performed with the R function “wilcox.test.”
Code availability
A customized pipeline was used to split the alignment of sequencing data from hybrid embryos to their parental origin based on SNP information. The code is available at https://github.com/lanjiangboston/UniversalSNPsplit.
Data availability
ChIP-seq and RNA-seq data sets generated in this study are summarized in Supplemental Table S2 and were deposited at the Gene Expression Omnibus database under accession number GSE103714. The WGBS data set for GV oocytes was downloaded from http://www.nodai-genome.org/mouse.html?lang=en (Kobayashi et al. 2012). H3K27me3 ChIP-seq data sets were downloaded from GSE76687 (Zheng et al. 2016). The oocyte DNase I-seq data set was from GSE92605 (Inoue et al. 2017). The BAM file and peak file of ENCODE data were downloaded from https://www.encodeproject.org/files/ENCFF001KDT. Morula embryo H3K27me3 ChIP-seq data sets were from GSE73952 (Liu et al. 2016).
Supplementary Material
Acknowledgments
We are grateful to Michihiko Sugimoto (Kumamoto University) and Hirosuke Shiura (Tokyo Medical and Dental University) for technical advice about RNA FISH. We thank Luis Tuesta for critical reading of the manuscript, and Shogo Matoba (RIKEN BioResource Center) and Tatsuya Ohhata (Hamamatsu University School of Medicine) for helpful discussion. This project was supported by the Howard Hughes Medical Institute. L.F. was supported by a Charles A. King Trust Post-doctoral Research Fellowship. Y.Z. is an Investigator of the Howard Hughes Medical Institute. A.I. and Y.Z. conceived the project, A.I. designed and performed the experiments, L.J. analyzed sequencing data sets, F.L. performed RNA-seq, and A.I., L.J., and Y.Z. interpreted the data and wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.304113.117.
References
- Augui S, Nora EP, Heard E. 2011. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Genet 12: 429–442. [DOI] [PubMed] [Google Scholar]
- Borensztein M, Syx L, Ancelin K, Diabangouaya P, Picard C, Liu T, Liang JB, Vassilev I, Galupa R, Servant N, et al. 2017. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol 24: 226–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brind'Amour J, Liu S, Hudson M, Chen C, Karimi MM, Lorincz MC. 2015. An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations. Nat Commun 6: 6033. [DOI] [PubMed] [Google Scholar]
- Chiba H, Hirasawa R, Kaneda M, Amakawa Y, Li E, Sado T, Sasaki H. 2008. De novoDNA methylation independent establishment of maternal imprint on X chromosome in mouse oocytes. Genesis 46: 768–774. [DOI] [PubMed] [Google Scholar]
- Fukuda A, Tomikawa J, Miura T, Hata K, Nakabayashi K, Eggan K, Akutsu H, Umezawa A. 2014. The role of maternal-specific H3K9me3 modification in establishing imprinted X-chromosome inactivation and embryogenesis in mice. Nat Commun 5: 5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A, Mitani A, Miyashita T, Umezawa A, Akutsu H. 2015. Chromatin condensation of Xist genomic loci during oogenesis in mice. Development 142: 4049–4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgetti L, Galupa R, Nora EP, Piolot T, Lam F, Dekker J, Tiana G, Heard E. 2014. Predictive polymer modeling reveals coupled fluctuations in chromosome conformation and transcription. Cell 157: 950–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. 2014. Nucleosome assembly is required for nuclear pore complex assembly in mouse zygotes. Nat Struct Mol Biol 21: 609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. 2017. Maternal H3K27me3 controls DNA methylation-independent genomic imprinting. Nature 547: 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. 2012. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8: e1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Bartolomei MS. 2013. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152: 1308–1323. [DOI] [PubMed] [Google Scholar]
- Liu X, Wang C, Liu W, Li J, Li C, Kou X, Chen J, Zhao Y, Gao H, Wang H, et al. 2016. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 537: 558–562. [DOI] [PubMed] [Google Scholar]
- Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Loring J, Jaenisch R. 1998. Role of the Xist gene in X chromosome choosing. Cell 92: 657–664. [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Barton SC, Surani MA, Brockdorff N. 2001. Loss of Xist imprinting in diploid parthenogenetic preimplantation embryos. Dev Biol 235: 343–350. [DOI] [PubMed] [Google Scholar]
- Oikawa M, Inoue K, Shiura H, Matoba S, Kamimura S, Hirose M, Mekada K, Yoshiki A, Tanaka S, Abe K, et al. 2014. Understanding the X chromosome inactivation cycle in mice. Epigenetics 9: 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto I, Arnaud D, Le Baccon P, Otte AP, Disteche CM, Avner P, Heard E. 2005. Evidence for de novo imprinted X-chromosome inactivation independent of meiotic inactivation in mice. Nature 438: 369–373. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. [DOI] [PubMed] [Google Scholar]
- Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. 2012. MAnorm: a robust model for quantitative comparison of ChIP-seq data sets. Genome Biol 13: R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T, Obata Y, Tada M, Goto Y, Nakatsuji N, Tan S, Kono T, Takagi N. 2000. Imprint switching for non-random X-chromosome inactivation during mouse oocyte growth. Development 127: 3101–3105. [DOI] [PubMed] [Google Scholar]
- Takagi N, Sasaki M. 1975. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature 256: 640–642. [DOI] [PubMed] [Google Scholar]
- Tan K, An L, Miao K, Ren L, Hou Z, Tao L, Zhang Z, Wang X, Xia W, Liu J, et al. 2016. Impaired imprinted X chromosome inactivation is responsible for the skewed sex ratio following in vitro fertilization. Proc Natl Acad Sci 113: 3197–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705. [DOI] [PubMed] [Google Scholar]
- Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, Li Y, Wang Q, Ma J, Peng X, et al. 2016. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell 63: 1066–1079. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-seq and RNA-seq data sets generated in this study are summarized in Supplemental Table S2 and were deposited at the Gene Expression Omnibus database under accession number GSE103714. The WGBS data set for GV oocytes was downloaded from http://www.nodai-genome.org/mouse.html?lang=en (Kobayashi et al. 2012). H3K27me3 ChIP-seq data sets were downloaded from GSE76687 (Zheng et al. 2016). The oocyte DNase I-seq data set was from GSE92605 (Inoue et al. 2017). The BAM file and peak file of ENCODE data were downloaded from https://www.encodeproject.org/files/ENCFF001KDT. Morula embryo H3K27me3 ChIP-seq data sets were from GSE73952 (Liu et al. 2016).



