Key Points
Question
What is the genomic profile of small-bowel adenocarcinoma (SBA) and how is it unique from other cancers of the gastrointestinal tract?
Findings
In this cohort study of 7559 patients, small-bowel adenocarcinoma demonstrated distinct genomic differences compared with colorectal adenocarcinoma and gastric adenocarcinoma, including variations in the frequency and types of alterations of KRAS, APC, BRAF, ERBB2/HER2, and a number of other genes. Microsatellite instability and high tumor mutational burden were enriched in small-bowel adenocarcinoma, and 91% of small-bowel adenocarcinomas harbored potentially actionable genomic alterations.
Meaning
Small-bowel adenocarcinoma is a molecularly unique intestinal cancer with a relatively high incidence of genomic alterations predictive of response to targeted and immunotherapies.
Abstract
Importance
Small-bowel adenocarcinomas (SBAs) are rare cancers with a significantly lower incidence, later stage at diagnosis, and worse overall survival than other intestinal-derived cancers. To date, comprehensive genomic analysis of SBA is lacking.
Objective
To perform in-depth genomic characterization of a large series of SBAs and other gastrointestinal tumors to draw comparisons and identify potentially clinically actionable alterations.
Design, Setting, and Participants
Prospective analysis was performed of clinical samples from patients with SBA (n = 317), colorectal cancer (n = 6353), and gastric carcinoma (n = 889) collected between August 24, 2012, and February 3, 2016, using hybrid-capture–based genomic profiling, at the request of the individual treating physicians in the course of clinical care for the purpose of making therapy decisions.
Results
Of the 7559 patients included in analysis, 4138 (54.7%) were male; the median age was 56 (range, 12-101) years. The frequency of genomic alterations seen in SBA demonstrated distinct differences in comparison with either colorectal cancer (APC: 26.8% [85 of 317] vs 75.9% [4823 of 6353], P < .001; and CDKN2A: 14.5% [46 of 317] vs 2.6% [165 of 6353], P < .001) or gastric carcinoma (KRAS: 53.6% [170 of 317] vs 14.2% [126 of 889], P < .001; APC: 26.8% [85 of 317] vs 7.8% [69 of 889], P < .001; and SMAD4: 17.4% [55 of 317] vs 5.2% [46 of 889], P < .001). BRAF was mutated in 7.6% (484 of 6353) of colorectal cancer and 9.1% (29 of 317) of SBA samples, but V600E mutations were much less common in SBA, representing only 10.3% (3 of 29) of BRAF-mutated cases. The ERBB2/HER2 point mutations (8.2% [26 of 317]), microsatellite instability (7.6% [13 of 170]), and high tumor mutational burden (9.5% [30 of 317]) were all enriched in SBA. Significant differences were noted in the molecular profile of unspecified SBA compared with duodenal adenocarcinoma, as well as in inflammatory bowel disease–associated SBAs. Targetable alterations in several additional genes, including PIK3CA and MEK1, and receptor tyrosine kinase fusions, were also identified in all 3 series.
Conclusions and Relevance
This study presents to our knowledge the first large-scale genomic comparison of SBA with colorectal cancer and gastric carcinoma. The distinct genomic differences establish SBA as a molecularly unique intestinal cancer. In addition, genomic profiling can identify potentially targetable genomic alterations in the majority of SBA cases (91%), and the higher incidence of microsatellite instability and tumor mutational burden in SBA suggests a potential role for immunotherapy.
This cohort study evaluates the genomic profile of patients with small-bowel adenocarcinoma compared with colorectal cancer and gastric carcinoma.
Introduction
Small-bowel adenocarcinoma (SBA) is a rare cancer, and current understanding of its genomic alterations is limited. This lack of molecular knowledge results from not only the rarity of this disease, but also the limited availability of high-quality tissue samples. Frequently, SBA is not diagnosed preoperatively; instead, it is found incidentally during surgical exploration for a small-bowel obstruction. Thus, there is a great need for a robust genomic analysis of paraffin-based tissue.
The surface area of the small intestine is much larger than that of the large intestine, yet the rate of cancer is approximately 50- to 100-fold less. Proposed theories for this difference include the rapid transit time in the small bowel, lower relative bacterial load, more alkaline environment, and greater lymphoid infiltrate in the small-intestinal mucosa. However, the cause of the lower incidence is currently unknown, and without a fuller genomic understanding of SBA, it is unlikely that the apparent protective environment of the small intestine will begin to be understood.
At the present time, SBA is clinically approached in a similar manner to colorectal cancer (CRC) since both are intestinal-derived cancers. However, aside from the disparity in incidence, SBA has a significantly worse outcome compared with CRC when stratified stage for stage. In addition, SBA is more likely to be poorly differentiated and stage IV at presentation. Multiple small studies have consistently demonstrated that the frequency of adenomatous polyposis coli (APC [NM_000038]) gene mutations in sporadic SBA ranges from 7% to 13%, which is significantly lower than reported in CRC, where rates of APC gene mutations exceed 80%.
To our knowledge, this project provides the largest genomic characterization of SBA, including duodenal adenocarcinoma (DA), to date and provides an improved biological insight into this cancer in relation to cancers of neighboring sites of the stomach and large intestine. In-depth genomic profiling (GP) also reveals many targetable alterations that guide the potential use of targeted therapeutics in clinical care.
Methods
A series of 317 SBA, 6353 CRC, and 889 gastric adenocarcinomas were prospectively assayed with a validated GP platform (August 24, 2012, to February 3, 2016). DNA was extracted from 40 µm of formalin-fixed, paraffin-embedded sections, which underwent hematoxylin-eosin review to ensure a minimum of 20% DNA derived from tumor cells. Secondary review by a board-certified pathologist (J.S.R.) was performed on a subset of cases noted in the provided pathology reports as periampullary carcinomas to include those with intestinal histology and exclude those with pancreaticobiliary or mixed histology. Genomic profiling was performed on hybridization-captured, adaptor ligation–based libraries to a mean coverage depth of ×651 for 236 or 315 cancer-related genes plus select introns from 19 or 28 genes frequently rearranged in cancer. All classes of genomic alterations (GAs) were identified, including base substitutions, insertions and deletions, copy number alterations, and rearrangements. Tumors were classified as microsatellite unstable-high (MSI-H) or microsatellite stable (MSS) using a principal component 1 cutoff value of less than −8.5 or greater than −4, respectively. Tumor mutational burden (TMB) was calculated as the number of somatic base substitutions or indels per megabase (Mb) of the coding region target territory of the test (currently, 1.11 Mb) after filtering to remove known somatic and deleterious mutations and extrapolating that value to the exome or genome as a whole. Categorical relationships were examined using Pearson χ2 test with Yates' continuity correction.
Approval for this study, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board. The data were recorded for the purposes of this study in deidentified format, consistent with the institutional review board–approved protocol.
Results
Genomic profiling of 317 SBAs (130 unspecified SBA and 187 DA) was performed prospectively at the request of treating physicians. Molecular profiles were compared with those of 6353 CRC and 889 gastric carcinomas (GCs) also assayed by GP during the course of clinical care. The overall median age was 56 years (range, 12-101 years), with 60 years (range, 18-86 years) for patients with SBA, 56 years (range, 13-88 years) for patients with CRC, and 58 years (range, 12-88 years) for those with GC. Of patients with SBA, 161 (50.8%) were male compared with 3491 (55.0%) of those with CRC and 486 (54.7%) of those with GC (Table). Of patients typically undergoing GP, the majority had stage IV disease, including 78.1% (207 of 265) of SBA cases for which stage information was available. For SBA, 60.3% (191 of 317) of the samples were from the primary small bowel, and 39.7% (126 of 317) were from metastatic sites, most commonly the liver (n = 34). We observed similar rates of GA per gene in primary (6.3% [12 of 191]) vs metastatic (13.5% [17 of 126]) biopsies tested, with BRAF (NM_004333) GA more common in metastases (P = .047) (eFigure in the Supplement). Means (SDs) of 6.23 (4.77), 5.94 (4.35), and 4.65 (3.81) GAs per sample were identified among SBAs, CRCs, and GCs, respectively (Table).
Table. Comparison of Available Clinical and Molecular Characteristics.
| Characteristic | SBA | CRC | GC |
|---|---|---|---|
| No. of patients | 317 | 6353 | 889 |
| Age, median (range), y | 60 (18-86) | 56 (13-101) | 58 (12-88) |
| Sex, No. (%) | |||
| Male | 161 (50.8) | 3491 (55.0) | 486 (54.7) |
| Female | 156 (49.2) | 2862 (45.0) | 403 (45.3) |
| Stage, No. (%)a | NA | NA | |
| I | 2 (0.8) | ||
| II | 11 (4.2) | ||
| III | 45 (17.0) | ||
| IV | 207 (78.1) | ||
| GA | |||
| Mean (SD) | 6.23 (4.77) | 5.94 (4.35) | 4.65 (3.81) |
| Median (range) | 5 (0-47) | 5 (0-92) | 3 (0-33) |
| MSI status, No. (%)a | |||
| MSS | 156 (91.8) | 3050 (95.3) | 361 (94.5) |
| MSI-ambiguous | 1 (0.6) | 23 (0.7) | 6 (1.6) |
| MSI-H | 13 (7.6) | 129 (4.0) | 15 (3.9) |
| TMB status, No. (%) | |||
| <10 mut/Mb (low) | 278 (87.7) | 5868 (92.4) | 799 (89.9) |
| 10-20 mut/Mb (intermediate) | 9 (2.8) | 209 (3.3) | 40 (4.5) |
| >20 mut/Mb (high) | 30 (9.5) | 276 (4.3) | 50 (5.6) |
Abbreviations: CRC, colorectal carcinoma; GA, genomic alteration; GC, gastric carcinoma; MSI, microsatellite instability; MSI-H, MSI-high; MSS, microsatellite stable; mut/Mb, mutations per megabase; NA, not assessed; SBA, small-bowel adenocarcinoma; TMB, tumor mutational burden.
Values shown for the subset of cases with available data. Comparison of cases classified as MSS vs MSI-H was statistically significant when comparing SBA and CRC (SBA vs CRC, P = .04; SBA vs GC, P = .11). Comparison of the cases classified as TMB-high was statistically significant when comparing SBA with CRC (P < .001) and GC (P = .03).
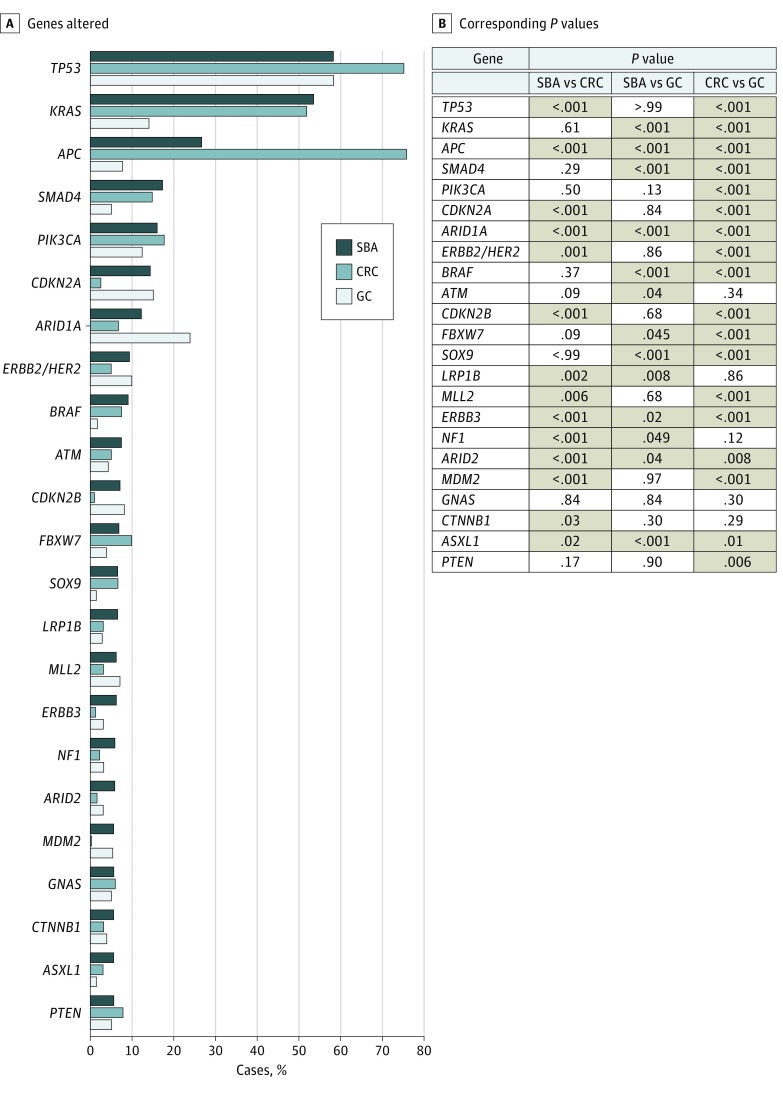
The most common GAs in our series of SBA affected TP53 (NM_000546) (58.4% [185 of 317]), KRAS (NM_004985) (53.6% [170 of 317]), APC (26.8% [85 of 317]), SMAD4 (NM_005359) (17.4% [55 of 317]), PIK3CA (NM_006218) (16.1% [51 of 317]), CDKN2A (NM_000077) (14.5% [46 of 317]), and ARID1A (NM_006015) (12.3% [39 of 317]) (Figure 1). Distinct differences were observed in the rates of GAs in these top 7 genes between series. Only PIK3CA GAs occurred at a similar rate in both CRC and GC (16.1% [51 of 317] vs 17.7% [1127 of 6353] vs 12.5% [111 of 889]). APC and ARID1A GA rates were significantly different from both CRC and GC, while KRAS and SMAD4 GAs occurred at similar rates to CRC, and TP53 (NM_000546) and CDKN2A GAs occurred at similar rates to GC. Compared with CRC, the greatest differences were in APC (26.8% [85 of 317] vs 75.9% [4823 of 6353], P < .001), TP53 (58.4% [185 of 317] vs 75.0% [4766 of 6353], P < .001), and CDKN2A (14.5% [46 of 317] vs 2.6% [165 of 6353], P < .001). In comparison with GC, the greatest differences were in KRAS (53.6% [170 of 317] vs 14.2% [126 of 889], P < .001), APC (26.8% [85 of 317] vs 7.8% [69 of 889], P < .001), and SMAD4 (17.4% [55 of 317] vs 5.2% [46 of 889], P < .001) (Figure 1). Genomic alterations in other wingless integration site family member (WNT) pathway genes were largely mutually exclusive from APC and were altered in 11.9% (19 of 160) of APC wild-type SBA (CTNNB1 [NM_001904], 6.3% [10 of 160]; RNF43 [NM_017763], 5.0% [8 of 160]; and AXIN1 [NM_003502], 0.6% [1 of 160]).
Figure 1. Frequency of Genomic Alterations in Small-Bowel Adenocarcinoma (SBA), Colorectal Cancer (CRC), and Gastric Carcinoma (GC) Cases.
A, Genes altered in more than 5% of SBA cases in this series. B, Corresponding P values comparing frequency of alteration of a gene across tumor types. Cases for which the difference was statistically significant (P < .05) are highlighted in tan.
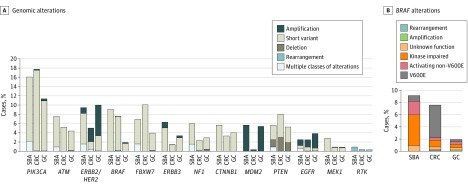
The most common potentially targetable alterations in SBA were identified in PIK3CA (NM_006218) (16.1% [51 of 317]), ERBB2/HER2 (NM_004448) (9.5% [30 of 317]), BRAF (9.1% [29 of 317]), ATM (NM_000051) (7.6% [24 of 317]), FBXW7 (NM_033632) (6.9% [22 of 317]), ERBB3 (NM_001982) (6.3% [20 of 317]), NF1 (NM_001042492) (6.0% [19 of 317]), CTNNB1 (5.7% [18 of 317]), MDM2 (NM_002392) (5.7% [18 of 213]), and PTEN (NM_000314) (5.7% [18 of 317]) (Figure 2A). ERBB2/HER2 was amplified in 7 (2.2% [7 of 317]) SBA cases (median copy number, 8), and ERBB2/HER2 point mutations were observed in 26 (8.2% [26 of 317]) cases including known activating mutations: S310F/Y (11 cases), V777L (5 cases), V842I, (4 cases), D769Y (3 cases), and L755S (2 cases). Three SBA cases harbored both ERBB2/HER2 amplification and point mutation. Alterations in ERBB2/HER2 were also observed in 9.5% (89 of 889) of GCs and 5.1% (324 of 6353) of CRCs, including ERBB2/HER2 amplification in 61 (6.9%) GC cases and 215 (3.4%) CRC cases. Alterations in EGFR (NM_005228) were observed in 2.5% (8 of 317) of SBAs, 2.5% (158 of 6353) of CRCs, and 4.0% (36 of 889) GCs. Amplification of EGFR was less frequent in SBA and CRC than in GC (1.6%, 1.6%, and 3.4% of cases, respectively). Activating receptor tyrosine kinase rearrangements were identified in 3 (0.9%) SBA cases including EML4-ALK, GOPC-ROS1, and FGFR2-CCAR1; 3 (0.3%) GC cases, all involving truncation of FGFR2/3 after exon 17; and 24 (0.4%) CRC cases involving fusions of ALK, ROS1, RET, and NTRK1/3. Overall, clinically relevant GAs, defined as those associated with an approved therapy or active clinical trial, were identified in 91.5% (290 of 317) of SBA, 88.7% (5638 of 6353) of CRC, and 71.1% (632 of 889) of GC cases.
Figure 2. Frequency of Targetable Genomic Alterations in Small-Bowel Adenocarcinoma (SBA), Colorectal Cancer (CRC), and Gastric Carcinoma (GC) Cases .
A, Targetable classes of genomic alterations found in more than 5% of SBA cases in this series as well as potentially targetable EGFR and MEK1 alterations, and RTK rearrangements. B, Frequency of classes of BRAF alterations.
BRAF alterations occurred at a similar frequency (7.6% [484 of 6353] and 9.1% [29 of 317]) in SBA and CRC, respectively, but only 10.3% (3 of 29) of BRAF-mutated SBAs harbored V600E mutations. Instead, BRAF mutations in these tumors were more likely to be associated with impaired kinase function (55.2% [16 of 29]). In contrast, V600E mutations were found in 73.2% (342 of 467) BRAF-mutated CRCs. BRAF mutations were less common in GC (1.8% [16 of 889]), but were also less likely to occur at V600E (3 of 17 cases [17.6%]) (Figure 2B). Alterations in PIK3CA (16.1% [51 of 317] of SBA cases) included activating mutations in exon 9 (21 cases) and exon 20 (15 cases). Mutations of MAP2K1 (MEK1) (2.8% [9 of 317] of SBA) were primarily Q56P, K57E/N/T, D67N, C121S, and E203K.
The MSI status was available for a subset of tumors in these series, and 3.9% and 4.0% of GC and CRC were MSI-H, respectively, whereas 7.6% (13 of 170) of SBA were MSI-H (Table). Alterations in mismatch repair genes, including MSH2 (NM_000251), MSH6 (NM_000179), and MLH1 (NM_000249), were observed in SBA at 1.9%, 3.8%, and 2.2%, respectively. In all cases, at least 1 mismatch repair gene harbored a deletion or truncation predicted to cause loss of function. Of 13 MSI-H SBA cases, 10 (76.9%) had 1 or more inactivating alterations in MSH2, MSH6, or MLH1. In CRC and GC MSH2, MSH6, and MLH1, alterations were observed in 1.0%, 1.7%, and 0.9% of cases, and 0.8%, 1.8%, and 1.6% of cases. Although limited by the small sample size and not statistically significant, alterations in MSH6 were 2-fold higher in SBA when compared with either CRC or GC. POLE (NM_006231) alterations were rare, observed in 0.3% of both SBA and CRC cases, and 0.2% of GC cases. Of SBA, 9.5% (30 of 317) had high TMB (>20 mutations/Mb) compared with 4.3% (276 of 6353) of CRC (P = .001) and 5.6% (50 of 889) of GC (P = .03). The majority (92.3%-100% of 13) of MSI-H cases in each series had high TMB, and all had a TMB of more than 10 mutations/Mb. However, 3.7% (114 of 3050) of MSS CRC and 4.4% (16 of 361) of MSS GC cases also had high or intermediate TMB (Table).
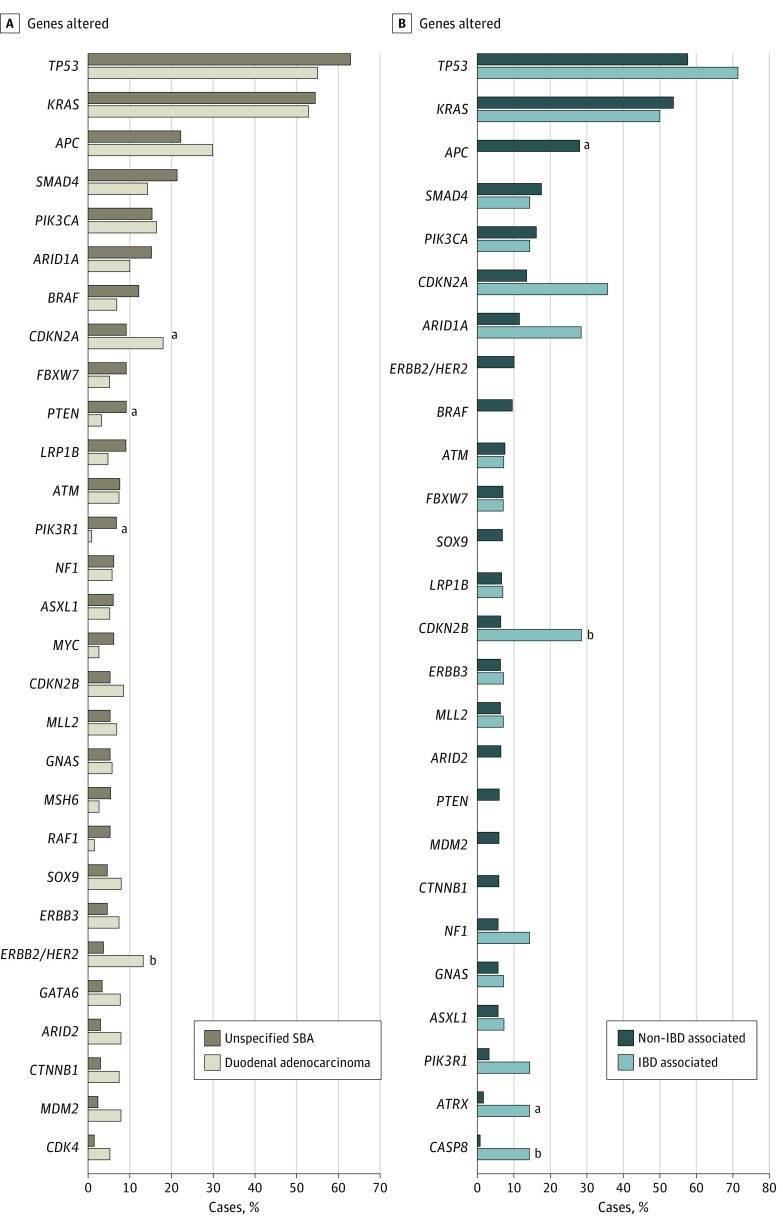
For tumors specified as DA, significant enrichment in the alteration frequency of ERBB2/HER2 and CDKN2A was observed compared with unspecified SBA, and conversely, alterations in BRAF, PTEN, and PIK3R1 were more frequent in unspecified SBA vs DA (Figure 3A). The rate of MSI-H was higher in unspecified SBA (9.0% [6 of 67]) compared with DA (7.3% [7 of 96]), but the difference was not statistically significant (P = .80). The TMB was significantly higher (P = .046) in unspecified SBA (median, 4.4; mean, 11.3 mutations/Mb) compared with DA (median, 4.0; mean, 8.8 mutations/Mb).
Figure 3. Genomic Comparison of Small-Bowel Adenocarcinoma (SBA) Subtypes by Location and Inflammatory Bowel Disease (IBD) Association.
A, Frequency of genomic alterations in duodenal adenocarcinoma (n = 187) compared with unspecified SBA (n = 130) cases. Graph includes genes altered in more than 5% of cases of duodenal adenocarcinoma or unspecified SBA. B, Frequency of genomic alterations in IBD-associated SBA (n = 14, including 11 cases with Crohn disease noted) compared with non–IBD-associated SBA (n = 303), as noted in the pathology report provided by the treating physician. Graph includes genes altered in more than 5% of cases of non–IBD-associated SBA or in more than 1 case of IBD-associated SBA.
aP < .05.
bP < .01.
Finally, through review of pathology reports provided by the treating physician, inflammatory bowel disease–associated SBA (IBD-SBA) was noted in 14 of 317 (4.4%) SBA cases, including 11 with concurrent Crohn disease, and alterations in CDKN2A/B, CASP8, and ATRX were significantly more frequent in IBD-SBA (Figure 3B). However, APC alterations were exclusive to non–IBD-SBA (85 of 303 [28.1%] vs 0 of 14 cases, P = .045).
Discussion
In this study, we report to our knowledge the largest genomic profiling of SBA and DA and also a comparative genomic analysis of SBA with neighboring intestinal tumors. These findings demonstrate that SBA represents a unique genomic entity with distinct alterations from both CRC and GC. Such molecular findings stress the need for further biological understanding of this rare cancer. In addition, this effort has uncovered a number of unique targetable alterations and a higher rate of MSI and increased TMB in SBA, representing direct clinical relevance.
This effort markedly furthers the current molecular characterization of SBA. Specifically, 317 SBA cases were tested with a large genomic platform covering 236 or 315 cancer-related genes and selected rearrangements. The largest prior effort examined 83 patients with a 46-gene panel and reported similar rates of GA (KRAS [43%], TP53 [41%], APC [13%], SMAD4 [10%], PIK3CA [8%], and ERBB2/HER2 [6%]) as seen in our study. In addition, a recent report utilizing an 18-gene panel in 24 SBA cases found similar rates of GA (TP53 [54%], KRAS [42%], and APC [11%]).
Two prior efforts examined larger exome-based analyses, but were limited to 14 and 18 cases and only studied duodenal adenocarcinomas. Recent profiling of GC and CRC, using the FoundationOne platform, has also been performed; however, these studies did not include SBA.
Herein, we demonstrate that the commonly altered genes in SBA are TP53, KRAS, APC, SMAD4, PIK3CA, CDKN2A, and ARID1A (>10% of cases). The relatively low rate of APC mutation (26.8% [85 of 317]) in SBA is consistent with the rate in prior studies and notably distinguishes the genomic profiles of SBA and CRC. However, in our analysis, alterations in other WNT pathway genes were relatively uncommon, and a substantial fraction (88.1%) of APC wild-type SBAs were also wild-type for CTNNB1, AX1N1, and RNF43. We also observed a paucity of APC alterations in IBD-associated SBA, consistent with recent findings in CRC.
We identified ERBB2/HER2 alterations in 9.5% (30 of 317) of SBAs, largely consistent with a prior study that identified ERBB2/HER2 alterations, primarily point mutations, in 10 of 83 cases (12.0%). We identified BRAF alterations in 9.1% (29 of 317) of SBA and 7.6% (484 of 6353) of CRC; however, the spectrum of variants was divergent. A total of 73.2% (342 of 467) of BRAF mutations in CRC were the canonical V600E, whereas V600E represented only 10.3% (3 of 29) of BRAF-mutated SBAs, which we believe is a novel and clinically relevant observation. Most BRAF mutations detected in SBA actually appeared to be inactivating, but may still be oncogenic through feedback activation on CRAF, and targetable using pan-RAF or MEK inhibitors. In CRC, tumors with non-V600E mutations have been associated with distinct clinical and pathologic characteristics and significantly greater overall survival.
The most notable finding from our study relates to the robust comparative analysis using a single genomic platform to assess nearly 7600 cases. Of 7 genes with GAs occurring in 10% or more of SBA cases, similarities to and marked differences from both CRC and GC were seen. The APC alteration frequencies were distinctly different between all 3 subtypes (CRC, 75.9%; SBA, 26.8%; and GC, 7.8%). Although the frequency of GAs for SMAD4 and KRAS were similar between SBA and CRC, the frequency of GAs for TP53 and CDKN2A were similar between SBA and GC. Such biological differences stress the need for further understanding and the clinical need to view SBA as an independent entity instead of a much rarer version of large-intestine cancer. At present, only 1 other report has attempted to compare SBA with its neighboring intestinal cancers, and this study was limited to copy number alterations via a comparative genomic hybridization approach. In this effort, SBA genomic-wide DNA copy number profiles appeared to be more similar to CRC than to GC. Although our data are not genome wide, copy number changes similar to both CRC and GC were evident for individual genes. In particular, amplification rates of ERBB2/HER2 and EGFR were more similar to CRC, and ERBB3 and MDM2 amplification rates were more similar to GC.
The large number of cases assessed herein also enabled what we believe to be the first robust attempt at comparing SBA genomics by subsite within the small intestine. As seen with prior reports, most cases within this study were from the duodenum, although determination of the subsite relied somewhat on individual clinician reporting. Nonspecified small-bowel site is reflective of either a jejunal or ileal location, as the distinction between these sites is often not easily known or reported, whereas duodenal lesions are easy to delineate by endoscopy or radiographic imaging. Our results demonstrate relatively similar genomics between DA and unspecified SBA; however, CDKN2A/B and ERBB2/HER2 alterations were enriched in duodenal compared with unspecified SBA, whereas DA tended overall to have a lower TMB. These findings suggest that the worse outcomes seen for patients with DA are likely more reflective of the anatomic complexity of the retroperitoneal-located duodenum. This complexity leads to a more challenging surgical approach for the duodenum, which is reflected in the markedly lower rate of assessed lymph nodes when comparing duodenum and jejunal or ileal resections.
Because SBA is a rare cancer, there are currently no Food and Drug Administration–approved agents for treatment and no official guidelines regarding therapy for patients with metastatic disease. Thus, the finding of potentially targetable GAs in most (91%) SBAs is of great relevance, as it suggests further therapeutic options for patients for whom no well-established standards exist. At present, publications regarding use of targeted therapy in this disease are limited. Potentially targetable alterations presented in this series include ERBB2/HER2 amplification or mutation, EGFR amplification/mutation, MEK1 (NM_002755) mutation, receptor tyrosine kinase fusion, BRAF mutation, and PI3K pathway-activating alterations. Despite lower ERBB2/HER2 amplification frequency in SBA and CRC compared with GC, SBA had the highest ERBB2/HER2 mutation frequency, with most mutations confirmed as activating and some tumors harboring both mutation and amplification. These findings suggest the need for comprehensive next-generation sequencing in place of, or in addition to, immunohistochemistry or fluorescence in situ hybridization for optimal characterization. As broad GP becomes more common in SBA, there will likely be more opportunities for patients to receive benefit from matched targeted therapies, and future studies aimed at assessing clinical outcomes in these patients will be critical. Although no clinical trials of anti-EGFR therapies in SBA have been published, a small number of case reports have suggested some clinical benefit either alone or in combination with chemotherapy. It is possible that the detection of alterations in KRAS, NRAS, BRAF, EGFR, ERBB2/HER2, PIK3CA, PTEN, and MAP2K1 may predict resistance to EGFR antibodies and inform therapy decisions in SBA, as has been suggested in CRC.
Antiprogrammed death 1 checkpoint inhibitors have also demonstrated efficacy in patients with MSI-H cancers, including SBA. In this series, we found a higher incidence of MSI-H in SBA compared with CRC or GC, yet the frequency of MSI reported here is less than the frequency of mismatch repair deficiency reported in a prior study; however, the prior study included a significant fraction of early stage patients and found that high mismatch repair deficiency was associated with early tumor stage. We also found that all MSI-H tumors had an intermediate to high TMB, and a subset of MSS tumors, which would have been missed by standard MSI testing methods, also had a high TMB. Overall, the fraction of TMB-high SBA (9.5%) was significantly greater than the fraction of TMB-high CRC and GC, suggesting that further exploration of checkpoint inhibitors in SBA is warranted.
Limitations
Although this work represents what we believe to be the largest genomic characterization of SBA, it is limited by the genomic coverage of the assay utilized in which 236 or 315 genes were assessed. However, the genomic panel presented here includes full exon coverage and has undergone extensive clinical validation. The ability to perform such a broad characterization on formalin-fixed, paraffin-embedded tissue is critical in this disease because high-quality frozen tissues are rare since the diagnosis of SBA is frequently not known preoperatively. In addition, the initial categorization of DA was based on information in the provided pathology reports.
Conclusions
The characterization of SBA as a unique genomic entity, distinct from CRC and GC, should prompt further investigation into the optimal clinical management of this rare cancer. Identification of multiple clinically relevant genomic alterations and mutational profiles is encouraging in a population with limited treatment options and poor prognosis.
eFigure. Genomic Comparison of Primary Versus Metastatic SBA Tissue Biopsies
References
- 1.Raghav K, Overman MJ. Small bowel adenocarcinomas—existing evidence and evolving paradigms. Nat Rev Clin Oncol. 2013;10(9):534-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talamonti MS, Goetz LH, Rao S, Joehl RJ. Primary cancers of the small bowel: analysis of prognostic factors and results of surgical management. Arch Surg. 2002;137(5):564-570. [DOI] [PubMed] [Google Scholar]
- 3.Halfdanarson TR, McWilliams RR, Donohue JH, Quevedo JF. A single-institution experience with 491 cases of small bowel adenocarcinoma. Am J Surg. 2010;199(6):797-803. [DOI] [PubMed] [Google Scholar]
- 4.DeSesso JM, Jacobson CF. Anatomical and physiological parameters affecting gastrointestinal absorption in humans and rats. Food Chem Toxicol. 2001;39(3):209-228. [DOI] [PubMed] [Google Scholar]
- 5.Haselkorn T, Whittemore AS, Lilienfeld DE. Incidence of small bowel cancer in the United States and worldwide: geographic, temporal, and racial differences. Cancer Causes Control. 2005;16(7):781-787. [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7-30. [DOI] [PubMed] [Google Scholar]
- 7.Overman MJ, Hu C-Y, Kopetz S, Abbruzzese JL, Wolff RA, Chang GJ. A population-based comparison of adenocarcinoma of the large and small intestine: insights into a rare disease. Ann Surg Oncol. 2012;19(5):1439-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowenfels AB. Why are small-bowel tumours so rare? Lancet. 1973;1(7793):24-26. [DOI] [PubMed] [Google Scholar]
- 9.Calman KC. Why are small bowel tumours rare? an experimental model. Gut. 1974;15(7):552-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bläker H, Helmchen B, Bönisch A, et al. . Mutational activation of the RAS-RAF-MAPK and the WNT pathway in small intestinal adenocarcinomas. Scand J Gastroenterol. 2004;39(8):748-753. [DOI] [PubMed] [Google Scholar]
- 11.Miyaki M, Konishi M, Kikuchi-Yanoshita R, et al. . Characteristics of somatic mutation of the adenomatous polyposis coli gene in colorectal tumors. Cancer Res. 1994;54(11):3011-3020. [PubMed] [Google Scholar]
- 12.Alvi MA, McArt DG, Kelly P, et al. . Comprehensive molecular pathology analysis of small bowel adenocarcinoma reveals novel targets with potential for clinical utility. Oncotarget. 2015;6(25):20863-20874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laforest A, Aparicio T, Zaanan A, et al. . ERBB2 gene as a potential therapeutic target in small bowel adenocarcinoma. Eur J Cancer. 2014;50(10):1740-1746. [DOI] [PubMed] [Google Scholar]
- 14.Frampton GM, Fichtenholtz A, Otto GA, et al. . Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evaluation of microsatellite instability (MSI) status in gastrointestinal (GI) tumor samples tested with comprehensive genomic profiling (CGP) [abstract 528]. J Clin Oncol http://meetinglibrary.asco.org/content/159854-173. Published 2016. Accessed April 1, 2016.
- 16.Chalmers ZR, Connelly CF, Fabrizio D, et al. . Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wan PTC, Garnett MJ, Roe SM, et al. ; Cancer Genome Project . Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855-867. [DOI] [PubMed] [Google Scholar]
- 18.Zheng G, Tseng L-H, Chen G, et al. . Clinical detection and categorization of uncommon and concomitant mutations involving BRAF. BMC Cancer. 2015;15:779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gingras M-C, Covington KR, Chang DK, et al. ; Australian Pancreatic Cancer Genome Initiative . Ampullary cancers harbor ELF3 tumor suppressor gene mutations and exhibit frequent WNT dysregulation. Cell Rep. 2016;14(4):907-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan W, Zhang Z, Dai B, et al. . Whole-exome sequencing of duodenal adenocarcinoma identifies recurrent Wnt/β-catenin signaling pathway mutations. Cancer. 2016;122(11):1689-1696. [DOI] [PubMed] [Google Scholar]
- 21.Ali SM, Sanford EM, Klempner SJ, et al. . Prospective comprehensive genomic profiling of advanced gastric carcinoma cases reveals frequent clinically relevant genomic alterations and new routes for targeted therapies. Oncologist. 2015;20(5):499-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rankin A, Klempner SJ, Erlich R, et al. . Broad detection of alterations predicted to confer lack of benefit from EGFR antibodies or sensitivity to targeted therapy in advanced colorectal cancer [published online September 28, 2016]. Oncologist. doi: 10.1634/theoncologist.2016-0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yaeger R, Shah MA, Miller VA, et al. . Genomic alterations observed in colitis-associated cancers are distinct from those found in sporadic colorectal cancers and vary by type of inflammatory bowel disease. Gastroenterology. 2016;151(2):278-287.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heidorn SJ, Milagre C, Whittaker S, et al. . Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cremolini C, Di Bartolomeo M, Amatu A, et al. . BRAF codons 594 and 596 mutations identify a new molecular subtype of metastatic colorectal cancer at favorable prognosis. Ann Oncol. 2015;26(10):2092-2097. [DOI] [PubMed] [Google Scholar]
- 26.Haan JC, Buffart TE, Eijk PP, et al. . Small bowel adenocarcinoma copy number profiles are more closely related to colorectal than to gastric cancers. Ann Oncol. 2012;23(2):367-374. [DOI] [PubMed] [Google Scholar]
- 27.Overman MJ, Hu CY, Wolff RA, Chang GJ. Prognostic value of lymph node evaluation in small bowel adenocarcinoma: analysis of the surveillance, epidemiology, and end results database. Cancer. 2010;116(23):5374-5382. [DOI] [PubMed] [Google Scholar]
- 28.Overman MJ, Hu C-Y, Wolff RA, Chang GJ. Prognostic value of lymph node evaluation in small bowel adenocarcinoma: analysis of the surveillance, epidemiology, and end results database. Cancer. 2010;116(23):5374-5382. [DOI] [PubMed] [Google Scholar]
- 29.Overman MJ, Wolff RA, Wang H. Cetuximab in small bowel adenocarcinoma: a new friend? Br J Cancer. 2010;103(8):1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poddar N, Raza S, Sharma B, Liu M, Gohari A, Kalavar M. Small bowel adenocarcinoma presenting with refractory iron deficiency anemia—case report and review of literature. Case Rep Oncol. 2011;4(3):458-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santini D, Fratto ME, Spoto C, et al. . Cetuximab in small bowel adenocarcinoma: a new friend? Br J Cancer. 2010;103(8):1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le DT, Uram JN, Wang H, et al. . PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.American Society of Clinical Oncology. Programmed death-1 blockade in mismatch repair deficient cancer independent of tumor histology [abstract 3003]. J Clin Oncol http://meetinglibrary.asco.org/content/170754-176. Published 2016. Accessed August 15, 2016.
- 34.Aparicio T, Svrcek M, Zaanan A, et al. . Small bowel adenocarcinoma phenotyping, a clinicobiological prognostic study. Br J Cancer. 2013;109(12):3057-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eFigure. Genomic Comparison of Primary Versus Metastatic SBA Tissue Biopsies