This cohort study assesses what impact mitochondrial DNA haplogroup variation has on risk of autism spectrum disorders.
Key Points
Question
Is there an association between mitochondrial DNA haplogroup-linked functional variants and risk of autism spectrum disorders (ASD)?
Findings
In this study, based on 1624 patients with ASD and 2417 healthy parents and siblings from 933 Autism Genetic Resource Exchange families, individuals with European haplogroups I, J, K, O-X, T, and U and Asian and Native American haplogroups A and M are at significantly increased risk of ASD compared with the most common European haplogroup HHV.
Meaning
Functional mitochondrial DNA variants associated with haplogroups may contribute to the risk for ASD, supporting the role of mitochondrial dysfunction in ASD.
Abstract
Importance
Autism spectrum disorders (ASD) are characterized by impairments in social interaction, communication, and repetitive or restrictive behavior. Although multiple physiologic and biochemical studies have reported defects in mitochondrial oxidative phosphorylation in patients with ASD, the role of mitochondrial DNA (mtDNA) variation has remained relatively unexplored.
Objective
To assess what impact mitochondrial lineages encompassing ancient mtDNA functional polymorphisms, termed haplogroups, have on ASD risk.
Design, Setting, and Participants
In this cohort study, individuals with autism and their families were studied using the Autism Genetic Resource Exchange cohort genome-wide association studies data previously generated at the Children’s Hospital of Philadelphia. From October 2010 to January 2017, we analyzed the data and used the mtDNA single-nucleotide polymorphisms interrogated by the Illumina HumanHap 550 chip to determine the mtDNA haplogroups of the individuals. Taking into account the familial structure of the Autism Genetic Resource Exchange data, we then determined whether the mtDNA haplogroups correlate with ASD risk.
Main Outcomes and Measures
Odds ratios of mitochondrial haplogroup as predictors of ASD risk.
Results
Of 1624 patients with autism included in this study, 1299 were boys (80%) and 325 were girls (20%). Families in the Autism Genetic Resource Exchange collection (933 families, encompassing 4041 individuals: 1624 patients with ASD and 2417 healthy parents and siblings) had been previously recruited in the United States with no restrictions on age, sex, race/ethnicity, or socioeconomic status. Relative to the most common European haplogroup HHV, European haplogroups I, J, K, O-X, T, and U were associated with increased risk of ASD, as were Asian and Native American haplogroups A and M, with odds ratios ranging from 1.55 (95% CI, 1.16-2.06) to 2.18 (95% CI, 1.59-3) (adjusted P < .04). Hence, mtDNA haplogroup variation is an important risk factor for ASD.
Conclusions and Relevance
Because haplogroups I, J, K, O-X, T, and U encompass 55% of the European population, mtDNA lineages must make a significant contribution to overall ASD risk.
Introduction
Autism spectrum disorders (ASD) are characterized by impairments in social interaction, communication, and repetitive or restrictive behaviors but can also present with a broad range of associated symptoms,1,2 including general weakness, gastrointestinal dysfunction, and epilepsy. The incidence of ASD is rising in the United States and as of 2010, the prevalence of ASD among 8-year-old children was 1 in 42 in boys and 1 in 189 in girls (male to female ratio is 4:1).3 Analysis of nuclear DNA (nDNA) genetic variation has revealed large numbers of associated heterozygous copy number variants (CNVs)4,5 and loss-of-function mutations,6,7,8 most of which account for only a few cases,1,6,7,9,10 suggesting a polygenic origin of ASD.11 Metabolic profiles of patients with ASD are consistent with mild mitochondrial defects,12,13 with oxidative phosphorylation (OXPHOS) defects having been reported in ASD lymphocytes,14 muscle,15 and brain.16,17 Autism spectrum disorder CNVs frequently delete nDNA genes that could perturb mitochondrial function,18 and ASD loss-of-function gene mutations can affect mitochondrially related functions such as variants in the Wnt β-catenin signaling pathway and associated chromodomain helicase DNA-binding protein 8 genes,6,19 as well as calcium regulation and fatty acid oxidation genes.7 Although arising heteroplasmic mitochondrial DNA (mtDNA) base substitution mutations have been observed in patients with ASD,20,21,22 the role of ancient mtDNA polymorphisms has not been rigorously evaluated, to our knowledge.
The mitochondrial genome consists of 1000 to 2000 nDNA genes plus hundreds to thousands of copies of the mtDNA per cell. The mtDNA codes for 13 of the most important polypeptides of OXPHOS plus the ribosomal RNA and transfer RNA for their translation in the mitochondrion. Hence, mtDNA variants directly affect cellular energy metabolism.23
There are 3 clinically relevant types of mtDNA variation: ancient adaptive haplogroup-associated polymorphisms that are homoplasmic (single pure mtDNA allele), recent deleterious mutations that have arisen in the female germline within the past 10 generations and can be either homoplasmic or heteroplasmic (mixture of 2 mtDNA alleles), and developmental arising somatic mutations that are invariably heteroplasmic.23 Ancient haplogroup-associated mtDNA variants have accumulated along radiating maternal lineages throughout human history, initially in Africa giving rise to macrohaplogroup L. About 65 000 years ago, women carrying 2 mtDNAs (M and N) left Africa for Eurasia, giving rise to macrohaplogroups M and N. Macrohaplogroup N settled in Europe and generated haplogroups HHV, J, T, U, K (Uk), I, W, and R. Macrohaplogroups M and N settled in Asia and generated 49% and 51% of the Asian mtDNAs, respectively. Asian macrohaplogroup M gave rise to multiple additional M sublineages (M1-M80), which include haplogroups C, D, G, and Z, while Asian macrohaplogroup N gave rise to Asian lineages A, B, F, O, and others. Eurasian haplogroups A, B, C, D, and X mtDNAs then came to the Americas.24 Because the mtDNA does not recombine, all single-nucleotide variants (SNVs) of an mtDNA lineage are in total disequilibrium and function as a unit. Hence, defining the haplogroup also defines its linked functional variants.
The various mtDNA haplogroup lineages arose and radiated within regional indigenous populations and are functionally different. Therefore, their proliferation within specific environments was due to adaptive selection. Although these variants are adapted to 1 regional environment, changes in the environment, such as migration, changes in diet, or new infection agents, can be maladaptive for a particular mtDNA lineage resulting in clinical phenotypes.24
Given the genetic complexity of ASD and the association of autism with mitochondrial bioenergetic defects, mtDNA variation should contribute to ASD risk. Because our objective was to determine whether inherited mtDNA variation contributes to ASD risk, we analyzed the Autism Genetic Resource Exchange (AGRE) family cohort.25 This cohort is particularly appropriate for detecting inherited subclinical risk factors, such as mtDNA variation, because it has been found that in 21 of 30 AGRE multiplex families affected by known risk CNVs (70%), a de novo or inherited event was not shared by all affected children.5
Methods
Participants
The mtDNA SNVs were determined by the Children’s Hospital of Philadelphia Center for Applied Genomics using the Illumina HumanHap 550 (Illumina Inc) array during the genome-wide association study analysis of AGRE families.26,27 Individuals were recruited in the United States with no restrictions on age, sex, race/ethnicity, or socioeconomic status, and they completed informed consent through the genome-wide association studies separately. Acquisition and analysis of the data for this study were approved by the Children’s Hospital of Philadelphia institutional review board and began in October 2010, and analysis ended in January 2017. The AGRE database was downloaded as PLINK files along with the family relationships and phenotypes. Among the samples in the PLINK files, 27 individuals (7 families) were missing from the downloaded AGRE phenotype file and were excluded from further analysis. The resulting genotyped AGRE data set included 4300 individuals, comprising 934 families with 1 to 12 members. Based on the Autism Diagnosis Interview score, 1624 individuals were diagnosed as having autism, 1 as having ASD, 156 as being on a broad spectrum, 74 as not quite having autism, 1 as having paranoia personality disorder–not otherwise specified, and 27 as not meeting the criteria for autism. The last 259 individuals without autism were excluded from analysis. The final total sample encompassed 933 families, including 1624 patients with ASD and 2417 healthy parents and siblings (N = 4041). Most of the AGRE samples in this study had mtDNA lineages of European origin, with some Asian, Native American, and African contributions.
Data Acquisition
The mtDNA variants on the Illumina HumanHap 550 array were designed based on the African Yoruban mtDNA sequence (GenBank AF347015). Therefore, we converted the variant positions to the corresponding positions in the revised Cambridge Reference Sequence (GenBank NC012920).28 The corresponding SNVs are listed in eTable 1 in the Supplement. The 163 mtDNA SNVs interrogated by the Illumina HumanHap 550 single-nucleotide polymorphisms chip,26,27 which passed stringent quality control measures, were used in this analysis. Using the aggregate SNVs for each individual, the mtDNA haplogroup of each individual was deduced by identifying the best match of those variants to the variants in the mtDNAs encompassed within the global phylogeny of complete mtDNA sequences using HaploGrep.29
To determine the accuracy and specificity of mtDNA haplogroup interpolation using the 163 mtDNA SNVs, we obtained 30 589 mtDNA GenBank sequences and used HaploGrep29 to infer haplogroups at the 1-letter haplogroup level from the entire mtDNA sequence. We then extracted SNVs interrogated by the Illumina HumanHap 550 array and again used HaploGrep to infer haplogroups from the 163 SNVs (eTable 2 in the Supplement). Of the 28 haplogroups tested, haplogroups A, C, D, I, J, K, L, and M (haplogroups M1-M80, except M7 and M8, and G and D); N (all N haplogroups, except N1, N2, and N9, and A, X, and R); and T, U, V, and W were inferred correctly in essentially all cases.30 Minor ambiguities were found when deducing haplogroups B, E, F, G, H, HV, O, P, Q, and R (R1-R32, except R9, and P); R0; and X. The sensitivity ranged from 0.17 to 1.00 (mean, 0.89; median, 1.00), and specificity ranged from 0.99 to 1.0. To resolve misclassifications, we represented the haplogroup information as a graph, with each node being a haplogroup and an edge between 2 nodes if there was an ambiguity between the 2 haplogroups. We then computed connected components using the R software package with igraph package, version 1.0.0 (http://igraph.org) to cluster the ambiguous haplogroups into 3 computational macrohaplogroups, which we termed N+ (N [all N haplogroups, except N1, N2, and N9, and A, X, and R] and S), HHV+ (B, F, H, HV, P, and R [R1-R32, except R9, and P]; R0; and V), and O-X (O, X). Except for haplogroup C, all of the subhaplogroups in macrohaplogroup M were ambiguous and therefore clustered. Given that there was a relatively small number of individuals in both the M macrohaplogroup and C haplogroup in the AGRE population and that C falls under the M8 subtree, we treated macrohaplogroup M in totality including C. Although the N1 haplogroup includes haplogroup I, there were only 26 individuals who were classified as N1 but not part of I, so they were omitted from analysis. For completeness, we repeated our analyses with different combinations of classifications of individuals based on the phylogenetic tree30 including both collapsing and separating haplogroups C and macrohaplogroup M, I and N1, J and T, and U and K, and the corresponding results remained consistent.
Computational haplogroup HHV+ encompasses 44% of the AGRE mtDNAs consistent with European population studies that have revealed that haplogroup HHV encompasses between 40% and 42% of European mtDNAs.31 Although H, HV, R, and R0 are all European, closely related, and thus functionally similar, haplogroup B is Asian and Native American, and F is Southeast Asian. Because of its geographic origin, F is unlikely to be prevalent in the AGRE data set. Hence, in the HHV+ computational macrohaplogroup, only B is not closely related to European HHV. Assuming that in the AGRE sample, most North American B mtDNAs are Native American in origin, we can estimate the proportion of HHV+ that could be B. Among Native American individuals, N (A + B) = 59% and M (C + D) = 41%. Within N, A = 56% and B = 43%.32 Hence, B = 77% of A. In the AGRE sample, there are 128 individuals in haplogroup A. Therefore, the number of individuals in haplogroup B is approximately 98 or at most 6% of HHV+.32 Asian haplogroup O is also rare in North America, so O-X is essentially X. Last, the AGRE sample encompasses too few African haplogroup L mtDNAs to justify further subdivision. Hence, in our analysis, we aggregated all Ls under the single macrohaplogroup L.23
Statistical Analysis
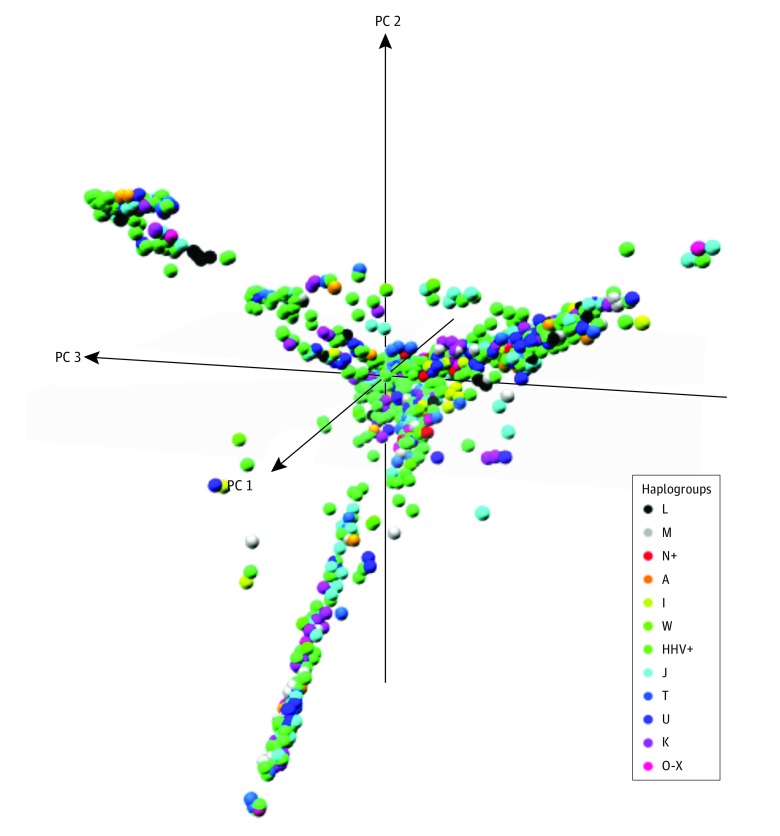
Because the sampling strategy for the AGRE sample set is not random, the degree of association between nDNA genetic variants and mtDNA variants could be a confounding factor. This prospect could be particularly problematic if there were high ethnic heterogeneity in the sample and strong assortative mating within the ethnic groups. To investigate the potential importance of nDNA population substructure, we analyzed the genome-wide association study nDNA SNV variation26,27 using multidimensional scaling (MDS) analyses performed using PLINK, version 2.0 (http://www.cog-genomics.org/plink/2.0/) and restricting the analysis to 5 clusters (Figure).
Figure. Multidimensional Scaling Analyses of Nuclear DNA Single-Nucleotide Polymorphisms From the Autism Genetic Resource Exchange Data Set.
Each point represents a single individual plotted against the first 3 dimensions from the multidimensional scaling analysis. Each haplogroup is depicted as a different color. PC indicates principle component.
To determine whether the nDNA MDS clusters correlated with the mtDNA haplogroups, we compared the distribution of mtDNA haplogroups associated with each MDS cluster. This analysis revealed that most mtDNA haplogroups were distributed across different clusters (Figure and Table 1) and that the mtDNA haplogroups did not significantly correlate with the clusters (weighted Cohen κ < 0.003). Hence, the haplogroups and MDS clusters represented different subgroups such that mtDNA haplogroup associations are unlikely to be the result of the spurious association between population-specific nDNA variants that are inherited along with mtDNA haplogroups due to assortative mating. Comparing the nDNA clusters with ASD revealed that only the first cluster was a significant predictor of ASD risk, consistent with nDNA genetic variation contributing to ASD risk, as expected.
Table 1. Distribution of mtDNA Haplogroups Across the nDNA MDS Clusters.
| MDS Cluster | Haplogroup, No. (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (n=143) |
HHV+ (n=1792) |
I (n=69) |
J (n=368) |
K (n=256) |
L (n=196) |
M (n=237) |
N+ (n=59) |
O-X (n=53) |
T (n=292) |
U (n=475) |
W (n=75) |
|
| 1 | 67 (46.9) | 1597 (89.1) | 68 (98.6) | 354 (96.2) | 251 (98.0) | 69 (35.2) | 67 (28.2) | 20 (33.9) | 50 (94.3) | 292 (100) | 467 (98.3) | 74 (98.7) |
| 2 | 70 (49.0) | 116 (6.5) | 1 (1.4) | 10 (2.7) | 5 (2.0) | 4 (2.0) | 73 (30.8) | 32 (54.2) | 3 (5.7) | 0 | 2 (0.4) | 0 |
| 3 | 2 (1.4) | 5 (0.3) | 0 | 4 (1.1) | 0 | 122 (62.2) | 7 (3.0) | 2 (3.3) | 0 | 0 | 2 (0.4) | 0 |
| 4 | 4 (2.8) | 62 (3.5) | 0 | 0 | 0 | 1 (0.5) | 77 (32.5) | 5 (8.5) | 0 | 0 | 2 (0.4) | 0 |
| 5 | 0 | 12 (0.7) | 0 | 0 | 0 | 0 | 13 (5.5) | 0 | 0 | 0 | 2 (0.4) | 1 (1.3) |
Abbreviations: MDS, multidimensional scaling; mtDNA, mitochondrial DNA; nDNA, nuclear DNA.
Having defined the integrity of the haplogroup imputations and determined the potential effects of nDNA variation, we then analyzed the relationship between mtDNA haplogroup variation and ASD using MDS clusters 1-5 (C1-5) as a covariate along with haplogroup and sex. Our study comprises families of related individuals, and because the risk of autism increases if 1 family member has autism, family members cannot be analyzed as statistically independent. To take into account the family structure of the AGRE data set, we applied generalized linear modeling and adopted the generalized estimating equations approach to determine the significance of ASD-haplogroup associations (SAS PROC GENMOD; SAS Institute Inc).33,34 This generalized estimating equations approach involves computing the average response over the population and uses weighted combinations of observations for correlated data.35 When the number of classes is large as is the case here, the generalized estimating equations method provides a robust solution to modeling family-based, correlated samples.36 The data were modeled using a binary response variable of whether the individual was diagnosed as having ASD and a logistic link function and binomial distribution (SAS parameters: /DIST = binomial LINK = LOGIT). The exchangeable correlation matrix was used with the repeated individuals being family members to capture the family correlation structure.37,38 The stepdown Bonferroni and Benjamini-Hochberg false discovery rate correction procedures were used to adjust for multiple testing.39
Results
Using the AGRE data set consisting of 933 families, including 1624 individuals with known ASD status and 2417 healthy parents and siblings (N = 4041), we determined whether there was a difference between haplogroups, sex, and MDS C1-5. We chose as a reference the computational macrohaplogroup HHV+, which is composed primarily of European haplogroup HHV. Haplogroup HHV is the most prevalent European haplogroup and encompasses the revised Cambridge Reference Sequence for the mtDNA.28
The parameters of a logistic model for sex and mitochondrial haplogroup were determined (Table 2). As expected, sex was a significant predictor of increased risk of ASD in boys vs girls (odds ratio [OR], 3.93; 95% CI, 3.30-4.67; stepdown Bonferroni P < .001) (Table 2). This OR is similar to the reported ratio of affected boys vs girls, providing validation for the model.
Table 2. Generalized Linear Modeling Analysis Results With GEE Solution for the AGRE Family Data Seta.
| Covariate | Total, No. (%)
(N = 4015)b |
Patients With ASD, No. (%) (n = 1615)b | OR (95% CI) | P Value | Bonferroni | Benjamini-Hochberg |
|---|---|---|---|---|---|---|
| Intercept | NA | NA | 0.58 (0.48-0.71) | <.001 | <.001 | <.001 |
| Haplogroupc | ||||||
| A | 143 (3.6) | 64 (4.0) | 1.83 (1.32-2.53) | <.001 | .006 | .004 |
| I | 69 (1.7) | 27 (1.7) | 2.12 (1.26-3.55) | .004 | .08 | .04 |
| J | 368 (9.2) | 157 (9.7) | 2.18 (1.59-3) | <.001 | <.001 | <.001 |
| K | 256 (6.4) | 100 (6.2) | 1.76 (1.31-2.36) | <.001 | .003 | .002 |
| L | 196 (4.9) | 74 (4.6) | 1.01 (0.63-1.61) | .98 | >.99 | .98 |
| M | 237 (5.9) | 104 (6.4) | 1.55 (1.16-2.06) | .003 | .06 | .03 |
| N+ | 59 (1.5) | 28 (1.7) | 2.02 (1.19-3.42) | .009 | .16 | .06 |
| O-X | 53 (1.3) | 21 (1.3) | 2 (1.23-3.25) | .005 | .09 | .04 |
| T | 292 (7.3) | 108 (6.7) | 1.79 (1.3-2.46) | .003 | .006 | .004 |
| U | 475 (11.8) | 190 (11.8) | 1.98 (1.45-2.7) | <.001 | <.001 | <.001 |
| W | 75 (1.9) | 30 (1.9) | 1.41 (0.81-2.45) | .22 | >.99 | .89 |
| Male | 2431 (60.5) | 1290 (79.9) | 3.93 (3.3-4.67) | <.001 | <.001 | <.001 |
| MDS cluster | ||||||
| C1 | 3376 (84.1) | 1356 (84.0) | 156.44 (11.53-2122.39) | <.001 | .003 | .002 |
| C2 | 316 (7.9) | 133 (8.2) | 0.26 (0.01-4.49) | .35 | >.99 | .98 |
| C3 | 144 (3.6) | 56 (3.5) | 0.03 (0-0.94) | .05 | .83 | .23 |
| C4 | 151 (3.8) | 58 (3.6) | 669.14 (2.14-208 972.46) | .03 | .48 | .16 |
| C5 | 28 (0.7) | 12 (0.7) | 1.75 (0.30-10.17) | .53 | >.99 | .98 |
Abbreviations: AGRE, Autism Genetic Resource Exchange; ASD, autism spectrum disorders; GEE, generalized estimating equations; MDS, multidimensional scaling; NA, not applicable; OR, odds ratio.
Generalized estimating equations fit criteria, QIC = 5547.28; QICu = 5551.83.
In total, 931 families with 1 to 12 members were analyzed.
In the reference haplogroup (HHV+), there were 1792 total individuals and 712 patients with ASD.
We then analyzed the haplogroups relative to HHV+ and found that European haplogroups I, J, K, T, and U were all at significantly higher risk for ASD (range: OR, 1.76; 95% CI, 1.31-2.36 and OR, 2.18; 95% CI, 1.59-3) at the 0.05 Benjamini-Hochberg adjusted significance level. Computational macrohaplogroup O-X was also at increased ASD risk (OR, 2.00; 95% CI, 1.23-3.25), as were Asian and Native American haplogroup A and macrohaplogroup M (OR, 1.83; 95% CI, 1.32-2.53 and OR, 1.55; 95% CI, 1.16-2.06, respectively) (Table 2).
Because MDS C1 proved to be a strong covariate but with very high variance, we attempted to control for its effects by confining our mtDNA haplogroup analysis to only those samples encompassed by MDS C1. Multidimensional scaling C1 encompasses 3401 of 4041 individuals in the AGRE data set (84.2%), which includes 3153 of 3380 European mtDNAs (HHV+, I, J, K, T, U, and W) (93.3%) but 69 of 196 African L mtDNAs (35.2%) (Table 1). Hence, MDS C1 encompasses predominantly individuals of European ancestry.
The generalized estimating equations analysis of MDS C1 individuals revealed that European haplogroups I, J, K, T, and U had significantly higher risk of ASD than HHV+ (range: OR, 1.87; 95% CI, 1.34-2.61 to OR, 2.27; 95% CI, 1.35-3.81). Only European haplogroup W was not associated with increased ASD risk (Table 3). The Asian and Native American mtDNA haplogroups A and M were also at increased ASD risk (OR, 2.95; 95% CI, 1.92-4.52 and OR, 3.14; 95% CI, 2.08-4.75, respectively) as were computational O-X haplogroups and macrohaplogroups N+ (OR, 2.01; 95% CI, 1.21-3.34 and OR, 4.52; 95% CI, 1.96-10.41, respectively) (Table 3). Therefore, even if we focus only on MDS C1, the association between ASD risk and mitochondrial haplogroups remains intact and is in fact stronger.
Table 3. Generalized Linear Modeling Analysis Results With GEE Solution for the AGRE Family Data Set Using Samples from MDS Cluster 1a.
| Covariate | Total, No. (%) (N = 3376)b | Patients With ASD, No. (%) (n = 1356)b | OR (95% CI) | P Value | Bonferroni | Benjamini-Hochberg |
|---|---|---|---|---|---|---|
| Intercept | NA | NA | 0.51 (0.39-0.65) | <.001 | <.001 | <.001 |
| Haplogroupc | ||||||
| A | 67 (2.0) | 33 (2.4) | 2.95 (1.92-4.52) | <.001 | <.001 | <.001 |
| I | 68 (2.0) | 27 (2.0) | 2.27 (1.35-3.81) | .002 | .03 | .008 |
| J | 354 (10.5) | 149 (11.0) | 2.22 (1.59-3.11) | <.001 | <.001 | <.001 |
| K | 251 (7.4) | 97 (7.2) | 1.94 (1.39-2.69) | <.001 | .001 | <.001 |
| L | 69 (2.0) | 29 (2.1) | 1.81 (1.03-3.17) | .04 | .49 | .08 |
| M | 67 (2.0) | 36 (2.7) | 3.14 (2.08-4.75) | <.001 | <.001 | <.001 |
| N+ | 20 (0.6) | 11 (0.8) | 4.52 (1.96-10.41) | <.001 | .005 | .002 |
| O-X | 50 (1.5) | 19 (1.4) | 2.01 (1.21-3.34) | .007 | .09 | .02 |
| T | 292 (8.6) | 108 (8.0) | 1.87 (1.34-2.61) | <.001 | .003 | .001 |
| U | 467 (13.8) | 187 (13.8) | 2.07 (1.49-2.88) | <.001 | <.001 | <.001 |
| W | 74 (2.2) | 30 (2.2) | 1.4 (0.81-2.4) | .22 | >.99 | .22 |
| Male | 2054 (60.8) | 1083 (79.9) | 3.8 (3.13-4.62) | <.001 | <.001 | <.001 |
Abbreviations: AGRE, Autism Genetic Resource Exchange; ASD, autism spectrum disorders; GEE, generalized estimating equations; MDS, multidimensional scaling; NA, not applicable; OR, odds ratio.
Generalized estimating equations fit criteria, QIC = 4379.63; QICu = 4379.78.
In total, 820 families with 1 to 12 members were analyzed, which encompasses 3376 total individuals of which 1779 were patients with ASD.
In the reference haplogroup (HHV+), there were 1356 total individuals of which 630 were patients with ASD.
Although there is interplay between the nDNA and mtDNA in determining ASD susceptibility, our data demonstrate that mtDNA haplogroup variation significantly imparts differential risk for ASD. This result supports a mitochondrial component to the causes of ASD.
Discussion
Our linkage of mtDNA haplogroups to ASD supports the hypothesis that mitochondrial functional variation is important in the causes of ASD. A familial predisposition to ASD based on mtDNA background haplogroup may provide 1 explanation for why high-impact CNVs do not consistently correlate with the development of ASD among siblings.5 Because the mitochondrial genome encompasses both nDNA and mtDNA mitochondrial genes, in the face of predisposing mtDNA haplogroups, additional nDNA and mtDNA variants or environmental insults could lower mitochondrial function below the minimal bioenergetic threshold for normal brain function resulting in clinical manifestations and accounting for the polygenic nature of ASD.11
If mtDNA variation is an important component of ASD risk, then we would expect all 3 classes of clinically relevant mtDNA variants to correlate with ASD: ancient homoplasmic adaptive polymorphisms, recent homoplasmic or heteroplasmic deleterious mutations, and de novo heteroplasmic somatic mutations. In our study, we have found that homoplasmic functional mtDNA variants linked within mtDNA haplogroups24 contribute to ASD risk. Concurrent with our study, an analysis22 was performed of heteroplasmic mtDNA variation found in off-target mtDNA sequence data from 903 Simons Foundation Simplex mother-proband-sibling trios. This study revealed that ASD probands were more likely to harbor deleterious heteroplasmic mtDNA mutations than their unaffected siblings. The putative deleterious mutations were primarily present in 5% to 20% of the mtDNAs as heteroplasmic mutations. Although at least 1 high-confidence heteroplasmic mutation was found at an equal frequency in probands (21.2%) and siblings without autism (20.2%), the incidence of heteroplasmic mutations at nonpolymorphic sites was 53% higher in autistic probands compared with siblings. Moreover, probands harbored 52% more nonsynonymous mutations and 118% more predicted pathogenic mutations than siblings. Thus, nonsynonymous mutations were enriched approximately 1.5-fold, and potentially pathogenic mutations were enriched approximately 2.2-fold in probands. Hence, probands carrying nonsynonymous private heteroplasmic mutations had an ASD OR of 2.55 (95% CI, 1.26-5.51).22 Because these heteroplasmic mutations encompassed both mutations transmitted from heteroplasmic mothers as well as de novo mutations found only in the proband, these heteroplasmic mutations must encompass newly arising pathogenic germline mutations as well as deleterious somatic mutations arising during development.
Haplogroups I, J, K, O-X, T, and U represent approximately 55% of European mtDNAs. When combined with the more deleterious heteroplasmic mutations found in one-fifth of the newborns with ASD, mtDNA variation would appear to account for a significant proportion of ASD risk.22
The unique genetics of the mitochondrion can now account for many of the puzzling features of ASD genetics. The male bias of ASD is consistent with the male bias observed for the onset of blindness in patients with Leber hereditary optic neuropathy (male to female ratio is approximately 4:1), which is caused by mild homoplasmic mtDNA mutations.40 The female protection from blindness has been attributed to the presence of the estrogen receptor in the mitochondrion such that estradiol increases mitochondrion antioxidant defenses.41,42
Mitochondrial OXPHOS is also acutely sensitive to inhibition by agricultural and industrial toxins that are increasing in the environment and food supply.43 The added inhibition of OXPHOS by toxins could increase the penetrance of the milder haplogroup-associated variants, explaining the rising incidence of ASD.
The chance acquisition of a more deleterious de novo mtDNA mutation in 1 of the 20% of siblings who acquire a heteroplasmic mtDNA mutation or the chance inheritance of an nDNA CNV or loss-of-function mutation that partially impairs OXPHOS could all augment the haplogroup-based predisposition to ASD. This chance acquisition of additional mtDNA or nDNA mitochondrial variants or environmental insults could explain why in a maternal lineage with an ASD predisposing haplogroup, 1 sibling drops below the minimal bioenergetic threshold and manifests ASD while other siblings do not, giving rise to a bioenergetic polygenic cause.11
Finally, the neurobehavioral aspect of ASD can be related to the mitochondrial dysfunction through our mouse studies.44 We have found that mild mitochondrial OXPHOS inhibition perturbs the migration of the γ-aminobutyric acidergic inhibitory neurons during neuronal development without perturbing the migration of the excitatory glutamatergic neurons. This would cause reduced neuronal inhibition creating hyperexcitation that could account for the hyperexcitability and perseverative behaviors associated with ASD.44
Limitations
Because the mtDNA haplogroups in the present study were deduced from genome-wide association study single-nucleotide polymorphisms data, it was not possible to analyze the same samples for recent deleterious and somatic mtDNA mutations. This type of analysis will become possible when whole mtDNA sequence data become available.
Conclusions
We have shown that mitochondrial haplogroups, with their associated functional variants, contribute a significant proportion of ASD risk, thus confirming that mitochondrial dysfunction is a significant factor in the cause of ASD. The interaction between the ancient haplogroup functional variants, recent heteroplasmic mtDNA mutations, mutation or deletion of 1 or more nDNA genes, and environmental insults that modulate bioenergetics may all combine to explain many of the unusual features of ASD genetics.
eTable 1. List of 163 Illumina 550 Chip SNPs
eTable 2. Correspondence Between Haplogroup Calls Using 30,893 Whole mtDNA Sequences
References
- 1.Robinson EB, St Pourcain B, Anttila V, et al. ; iPSYCH-SSI-Broad Autism Group . Genetic risk for autism spectrum disorders and neuropsychiatric variation in the general population. Nat Genet. 2016;48(5):552-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bresnahan M, Hornig M, Schultz AF, et al. Association of maternal report of infant and toddler gastrointestinal symptoms with autism: evidence from a prospective birth cohort. JAMA Psychiatry. 2015;72(5):466-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Developmental Disabilities Monitoring Network Surveillance Year 2010 Principal Investigators; Centers for Disease Control and Prevention (CDC) . Prevalence of autism spectrum disorder among children aged 8 years: autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014;63(2):1-21. [PubMed] [Google Scholar]
- 4.Zhao X, Leotta A, Kustanovich V, et al. . A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104(31):12831-12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leppa VM, Kravitz SN, Martin CL, et al. Rare inherited and de novo CNVs reveal complex contributions to ASD risk in multiplex families. Am J Hum Genet. 2016;99(3):540-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37(2):95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Rubeis S, He X, Goldberg AP, et al. ; DDD Study; Homozygosity Mapping Collaborative for Autism; UK10K Consortium . Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosmicki JA, Samocha KE, Howrigan DP, et al. Refining the role of de novo protein-truncating variants in neurodevelopmental disorders by using population reference samples. Nat Genet. 2017;49(4):504-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop SL, Farmer C, Bal V, et al. Identification of developmental and behavioral markers associated with genetic abnormalities in autism spectrum disorder. Am J Psychiatry. 2017;174(6):576-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brandler WM, Antaki D, Gujral M, et al. Frequency and complexity of de novo structural mutation in autism. Am J Hum Genet. 2016;98(4):667-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiner DJ, Wigdor EM, Ripke S, et al. ; iPSYCH-Broad Autism Group; Psychiatric Genomics Consortium Autism Group . Polygenic transmission disequilibrium confirms that common and rare variation act additively to create risk for autism spectrum disorders. Nat Genet. 2017;49(7):978-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filipek PA. Medical aspects of autism In: Volkmar FR, Klin A, Paul R, Cohen DJ, eds. Handbook of Autism and Pervasive Developmental Disorders. New York, NY: John Wiley and Sons; 2005:534-578. [Google Scholar]
- 13.Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17(3):290-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giulivi C, Zhang YF, Omanska-Klusek A, et al. Mitochondrial dysfunction in autism. JAMA. 2010;304(21):2389-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guevara-Campos J, González-Guevara L, Cauli O. Autism and intellectual disability associated with mitochondrial disease and hyperlactacidemia. Int J Mol Sci. 2015;16(2):3870-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang G, Gutierrez Rios P, Kuo SH, et al. Mitochondrial abnormalities in temporal lobe of autistic brain. Neurobiol Dis. 2013;54:349-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goh S, Dong Z, Zhang Y, DiMauro S, Peterson BS. Mitochondrial dysfunction as a neurobiological subtype of autism spectrum disorder: evidence from brain imaging. JAMA Psychiatry. 2014;71(6):665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith M, Flodman PL, Gargus JJ, et al. Mitochondrial and ion channel gene alterations in autism. Biochim Biophys Acta. 2012;1817(10):1796-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010;24(14):1507-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pons R, Andreu AL, Checcarelli N, et al. Mitochondrial DNA abnormalities and autistic spectrum disorders. J Pediatr. 2004;144(1):81-85. [DOI] [PubMed] [Google Scholar]
- 21.Picard M, Zhang J, Hancock S, et al. Progressive increase in mtDNA 3243A>G heteroplasmy causes abrupt transcriptional reprogramming. Proc Natl Acad Sci U S A. 2014;111(38):E4033-E4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Picard M, Gu Z. Genetic evidence for elevated pathogenicity of mitochondrial DNA heteroplasmy in autism spectrum disorder. PLoS Genet. 2016;12(10):e1006391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace DC. A mitochondrial bioenergetic etiology of disease. J Clin Invest. 2013;123(4):1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace DC. Mitochondrial DNA variation in human radiation and disease. Cell. 2015;163(1):33-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geschwind DH, Sowinski J, Lord C, et al. ; AGRE Steering Committee . The Autism Genetic Resource Exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69(2):463-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Zhang H, Ma D, et al. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459(7246):528-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glessner JT, Wang K, Cai G, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459(7246):569-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge Reference Sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. [DOI] [PubMed] [Google Scholar]
- 29.Kloss-Brandstätter A, Pacher D, Schönherr S, et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 2011;32(1):25-32. [DOI] [PubMed] [Google Scholar]
- 30.van Oven M, Kayser M. Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Hum Mutat. 2009;30(2):E386-E394. [DOI] [PubMed] [Google Scholar]
- 31.Torroni A, Huoponen K, Francalacci P, et al. Classification of European mtDNAs from an analysis of three European populations. Genetics. 1996;144(4):1835-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torroni A, Schurr TG, Yang CC, et al. Native American mitochondrial DNA analysis indicates that the Amerind and the Nadene populations were founded by two independent migrations. Genetics. 1992;130(1):153-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42(1):121-130. [PubMed] [Google Scholar]
- 34.Courtenay MD, Gilbert JR, Jiang L, et al. Mitochondrial haplogroup X is associated with successful aging in the Amish. Hum Genet. 2012;131(2):201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanley JA, Negassa A, Edwardes MD, Forrester JE. Statistical analysis of correlated data using generalized estimating equations: an orientation. Am J Epidemiol. 2003;157(4):364-375. [DOI] [PubMed] [Google Scholar]
- 36.Hubbard AE, Ahern J, Fleischer NL, et al. . To GEE or not to GEE: comparing population average and mixed models for estimating the associations between neighborhood risk factors and health. Epidemiology. 2010;21(4):467-474. [DOI] [PubMed] [Google Scholar]
- 37.Stokes ME, Davis CS, Koch GG. Categorical Data Analysis Using the SAS System. 2nd ed Cary, NC: SAS Institute Inc; 2000. [Google Scholar]
- 38.Ziegler A, Vens M. Generalized estimating equations. Notes on the choice of the working correlation matrix. Methods Inf Med. 2010;49(5):421-425. [DOI] [PubMed] [Google Scholar]
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc (B). 1995;57(1):289-300. [Google Scholar]
- 40.Sadun AA, La Morgia C, Carelli V. Leber’s hereditary optic neuropathy. Curr Treat Options Neurol. 2011;13(1):109-117. [DOI] [PubMed] [Google Scholar]
- 41.Pisano A, Preziuso C, Iommarini L, et al. Targeting estrogen receptor β as preventive therapeutic strategy for Leber’s hereditary optic neuropathy. Hum Mol Genet. 2015;24(24):6921-6931. [DOI] [PubMed] [Google Scholar]
- 42.Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell. 2006;17(5):2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Argese E, Bettiol C, Marchetto D, et al. Study on the toxicity of phenolic and phenoxy herbicides using the submitochondrial particle assay. Toxicol In Vitro. 2005;19(8):1035-1043. [DOI] [PubMed] [Google Scholar]
- 44.Lin-Hendel EG, McManus MJ, Wallace DC, Anderson SA, Golden JA. Differential mitochondrial requirements for radially and non-radially migrating cortical neurons: implications for mitochondrial disorders. Cell Rep. 2016;15(2):229-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. List of 163 Illumina 550 Chip SNPs
eTable 2. Correspondence Between Haplogroup Calls Using 30,893 Whole mtDNA Sequences