Key Points
Question
Do maternal breast milk and areolar skin bacterial communities transfer to the infant gut?
Findings
In this 12-month longitudinal study of 107 healthy mother-infant pairs, breastfed infants received 27.7% of their gut bacteria from breast milk and 10.4% from areolar skin during the first month of life. Bacterial diversity and composition changes were associated with the proportion of daily breast milk intake in a dose-dependent manner even after introduction of solid foods.
Meaning
Microbes in mother’s breast milk seed the infant gut, including those associated with beneficial effects, underscoring the importance of breastfeeding in maturation of the infant gut microbiome.
Abstract
Importance
Establishment of the infant microbiome has lifelong implications on health and immunity. Gut microbiota of breastfed compared with nonbreastfed individuals differ during infancy as well as into adulthood. Breast milk contains a diverse population of bacteria, but little is known about the vertical transfer of bacteria from mother to infant by breastfeeding.
Objective
To determine the association between the maternal breast milk and areolar skin and infant gut bacterial communities.
Design, Setting, and Participants
In a prospective, longitudinal study, bacterial composition was identified with sequencing of the 16S ribosomal RNA gene in breast milk, areolar skin, and infant stool samples of 107 healthy mother-infant pairs. The study was conducted in Los Angeles, California, and St Petersburg, Florida, between January 1, 2010, and February 28, 2015.
Exposures
Amount and duration of daily breastfeeding and timing of solid food introduction.
Main Outcomes and Measures
Bacterial composition in maternal breast milk, areolar skin, and infant stool by sequencing of the 16S ribosomal RNA gene.
Results
In the 107 healthy mother and infant pairs (median age at the time of specimen collection, 40 days; range, 1-331 days), 52 (43.0%) of the infants were male. Bacterial communities were distinct in milk, areolar skin, and stool, differing in both composition and diversity. The infant gut microbial communities were more closely related to an infant’s mother’s milk and skin compared with a random mother (mean difference in Bray-Curtis distances, 0.012 and 0.014, respectively; P < .001 for both). Source tracking analysis was used to estimate the contribution of the breast milk and areolar skin microbiomes to the infant gut microbiome. During the first 30 days of life, infants who breastfed to obtain 75% or more of their daily milk intake received a mean (SD) of 27.7% (15.2%) of the bacteria from breast milk and 10.3% (6.0%) from areolar skin. Bacterial diversity (Faith phylogenetic diversity, P = .003) and composition changes were associated with the proportion of daily breast milk intake in a dose-dependent manner, even after the introduction of solid foods.
Conclusions and Relevance
The results of this study indicate that bacteria in mother’s breast milk seed the infant gut, underscoring the importance of breastfeeding in the development of the infant gut microbiome.
This longitudinal cohort study evaluates the microbial colonization of the infant gut in breastfed compared with nonbreastfed infants.
Introduction
Microbial colonization of the infant gut plays an important role in lifelong health. In healthy newborns, the gut microbiome composition shows large-scale longitudinal changes until age 3 years, when it settles into an adult-like anaerobic pattern. Perturbations in the microbiome are associated with susceptibility to autoimmune diseases, such as diabetes, inflammatory bowel disease, atopy, and other conditions. The mechanisms of acquisition and progression toward a “normal” infant microbiome are poorly understood. Colonization of the infant gut is a complex process dependent on multiple overlapping factors, including age, mode of delivery, type of feeding, and environmental exposures.
Multiple studies have documented differences in the stool microbiota of breastfed compared with nonbreastfed individuals during infancy and adulthood. Studies show that breastfeeding confers protection against respiratory and gastrointestinal tract infections and allergic diseases in addition to reducing the risk of chronic diseases, such as diabetes, obesity, and inflammatory bowel disease. Most microbiota research on the breastfeeding effects to date has focused on infant stool. Little is known about the vertical transfer of breast milk microbes from mother to infant.
Breast milk contains a diverse population of bacteria that are hypothesized to seed the infant’s gut via breastfeeding. We set out to evaluate the association between the maternal bacterial community and the infant’s stool in healthy mother-infant pairs. In this report, we show the extent that maternal milk and areolar skin contribute to the developing infant microbiome. We also describe the predicted functional profile of milk and infant stool bacterial communities.
Methods
Study Participants
We recruited healthy mothers and their full-term infants in the community around Children’s Hospital Los Angeles, Los Angeles, California, and All Children’s Hospital, St Petersburg, Florida, between January 1, 2010, and February 28, 2015. We enrolled mother-infant pairs into the following infant age groups: 0 to 7 days (initial colonization period), 8 to 30 days (first period of full maternal milk supply), 31 to 90 days (before introduction of solids), 91 to 180 days (period of solid food introduction), and 181 to 365 days (after solid food introduction). Mother-infant pairs were excluded if either had a health condition or if the infant had received antibiotics. Institutional review boards at Children's Hospital Los Angeles and All Children's Hospital approved the study; written informed consent was obtained from all mothers, and financial compensation was provided.
At the initial study visit, study personnel interviewed mothers to collect clinical data, including basic demographics, pregnancy and delivery history, current or recent medications (including antibiotics), and infant feeding characteristics (age at formula and solid food introduction and frequency of breastfeeding, formula, and solid foods). On all follow-up visits, we asked about new medical conditions, antibiotic use, and feeding characteristics, using the Research Electronic Data Capture (REDCap Consortium; https://www.project-redcap.org/) database.
Sample Collection, DNA Extraction, and Sequencing
We collected breast milk and areolar skin swabs from mothers and stool from infants’ soiled diapers during clinic or home study visits. Mothers expressed milk with a manual or electric breast pump using sterilized connectors into sterile bottles. Longitudinal samples were obtained when possible. The DNA extraction, library preparation, and sequencing of the V4 region of the 16S ribosomal RNA (rRNA) gene on a MiSeq sequencer (Illumina Inc) were performed using standard procedures with quality control (eMethods and eFigure 1 in the Supplement).
Statistical Analysis
We compared differences in α diversity using Faith phylogenetic diversity; β diversity, using unweighted UniFrac and Bray-Curtis; and community composition between groups based on feeding characteristics. To evaluate sources of infant stool microbiome variation, we modeled infant age, demographics, delivery type, geographic collection site, and feeding characteristics with the permutational multivariate analysis of variance using distance matrices (adonis, R vegan package; R Foundation). Interaction terms were tested and not statistically significant. To predict the source of bacterial communities in infant stool, we used SourceTracker, version 0.9.5, software in Quantitative Insights Into Microbial Ecology to compare sequences in breast milk and areolar skin as input samples (source samples) with the sequences in infant stool (sink sample). To compare paired breast milk and infant stool samples, we performed a Wilcoxon rank sum test on UniFrac, Jaccard index, and Bray-Curtis distances between true mother-infant pairs and randomly paired mothers and infants matched by infant age within each run. Operational taxonomic unit (OTU) sharing was defined as the percentage of mother-infant dyads in which a given OTU was found in both members; permutation testing with randomly shuffled mother-infant pairings was used to assess significance. Oligotyping analysis was performed using sequences mapped to OTUs shared within mother-infant dyads as previously described. Statistical analyses were performed using R statistical software, version 3.0.3 (R Foundation).
Random forest models were used to predict taxonomy that classified the infant stool bacterial community into classes based on feeding method. We used a random forest machine-learning algorithm to determine a ranked list of all bacterial taxa in the order of age-discriminatory importance. Stool from a randomly chosen subset of vaginally born, exclusively breastfed infants (n = 42) at multiple time points throughout the first year of life was used to train the model. A sparse model with 44 predictors (57.5% variance explained) was selected on the basis of 10-fold cross validation. Relative microbiota maturity was calculated by microbiota age of a child minus microbiota age of healthy children of a similar chronologic age from the model.
We used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) to predict metagenome function using 16S rRNA marker gene sequences and referenced to published complete genome sequences. Both R, version 3.0.3 and STAMP, version 2.0.9 were used for statistical analyses of the functional profiles. A false discovery rate P value ≤.10 was considered significant for any analyses with multiple comparisons. A standard P value ≤.05 was considered significant for all other analyses.
Results
Study Population
We enrolled 228 healthy individuals from 121 families, including 107 mother-infant pairs, 2 mothers only, and 12 infants only. Median infant age at the time of specimen collection was 40 days (range, 1-331 days). Family and feeding characteristics are reported in the Table and eTable 1 in the Supplement, respectively. Eighty (66.1%) of our families were Hispanic/Latino. Seventy-eight (64.5%) infants were born by vaginal delivery. We analyzed at least 2 longitudinal samples from 73 (60.3%) families. At the time of sample collection (n = 569), 484 (85.1%) samples showed that the infants were primarily breastfed (defined as receiving breast milk for >75% of their daily milk intake), including 298 (52.4%) who were exclusively breastfed (defined as never having received formula).
Table. Characteristics of 121 Families.
| Characteristic | No. (%) |
|---|---|
| Race/ethnicity | |
| African American | 5 (4.1) |
| Asian/Pacific Islander | 11 (9.1) |
| Hispanic/Latino | 80 (66.1) |
| White | 16 (13.2) |
| Mixed | 3 (2.5) |
| Unknown | 6 (5.0) |
| Infant sex | |
| Male | 52 (43.0) |
| Female | 50 (41.3) |
| Unknown | 19 (15.7) |
| Delivery method | |
| Vaginal | 78 (64.5) |
| Cesarean section | 31 (25.6) |
| Planned | 16 (51.6) |
| Unplanned | 15 (48.4) |
| Unknown | 12 (9.9) |
| Maternal antibiotics at delivery | |
| Yes | 37 (30.6) |
| No | 66 (54.5) |
| Unknown | 18 (14.9) |
| Infant age at specimen collection, d | |
| 0-7 | 90 (15.8) |
| 8-30 | 163 (28.6) |
| 31-90 | 127 (22.3) |
| 91-180 | 147 (25.8) |
| 181-365 | 42 (7.4) |
Breast Milk, Areolar, and Infant Stool Microbiomes
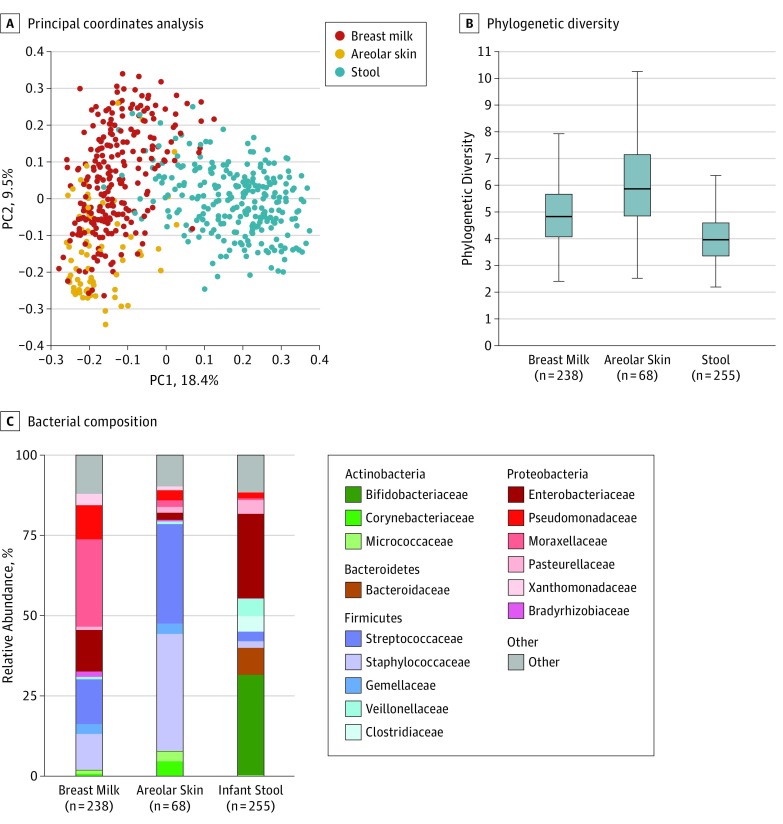
Distinct bacterial communities were present in milk, areolar skin, and stool, which differed in both composition and diversity (Figure 1). Proteobacteria (Moraxellaceae, Enterobacteriaceae, and Pseudomonadaceae) constituted the dominant phylum in milk, Firmicutes (Staphylococcaceae and Streptococcaceae) were dominant in areolar skin, and Proteobacteria (Enterobacteriaceae) and Actinobacteria (Bifidobacteriaceae) accounted for more than 50% of the community in infant stool. Milk and areolar samples collected from the right and left breast were similar (eFigure 2 in the Supplement); therefore, samples from the right breast were used for analyses if samples from both sides were collected.
Figure 1. Microbial Communities in Breast Milk, Areolar Skin, and Infant Stool .
A, Principal coordinate (PC) analysis of unweighted UniFrac distances revealed that breast milk, areolar skin swabs, and stool separate into 3 distinct groups. Percentages are the percent variation explained by each PC axis. B, Faith phylogenetic diversity was used to measure α diversity. The 3 groups all significantly differed from each other in 2-way comparisons of α diversity (P < .001). Infant stool was the least diverse in terms of community membership, likely reflecting the young age of study infants at the time of stool collection and their initial states of colonization following complete or near-sterility at birth. C, Breast milk had significantly higher amounts of Moraxellaceae and Pseudomonadaceae (false discovery rate [FDR] P < .001), whereas areolar skin swabs were enriched in Streptococcaceae and Staphylococcaceae (FDR P < .001) and infant stool comprised greater amounts of Bifidobacteriaceae (FDR P < .001). Boxes indicate the 25th to 75th percentile; line within the box, median; error bars, 1.5 × interquartile range.
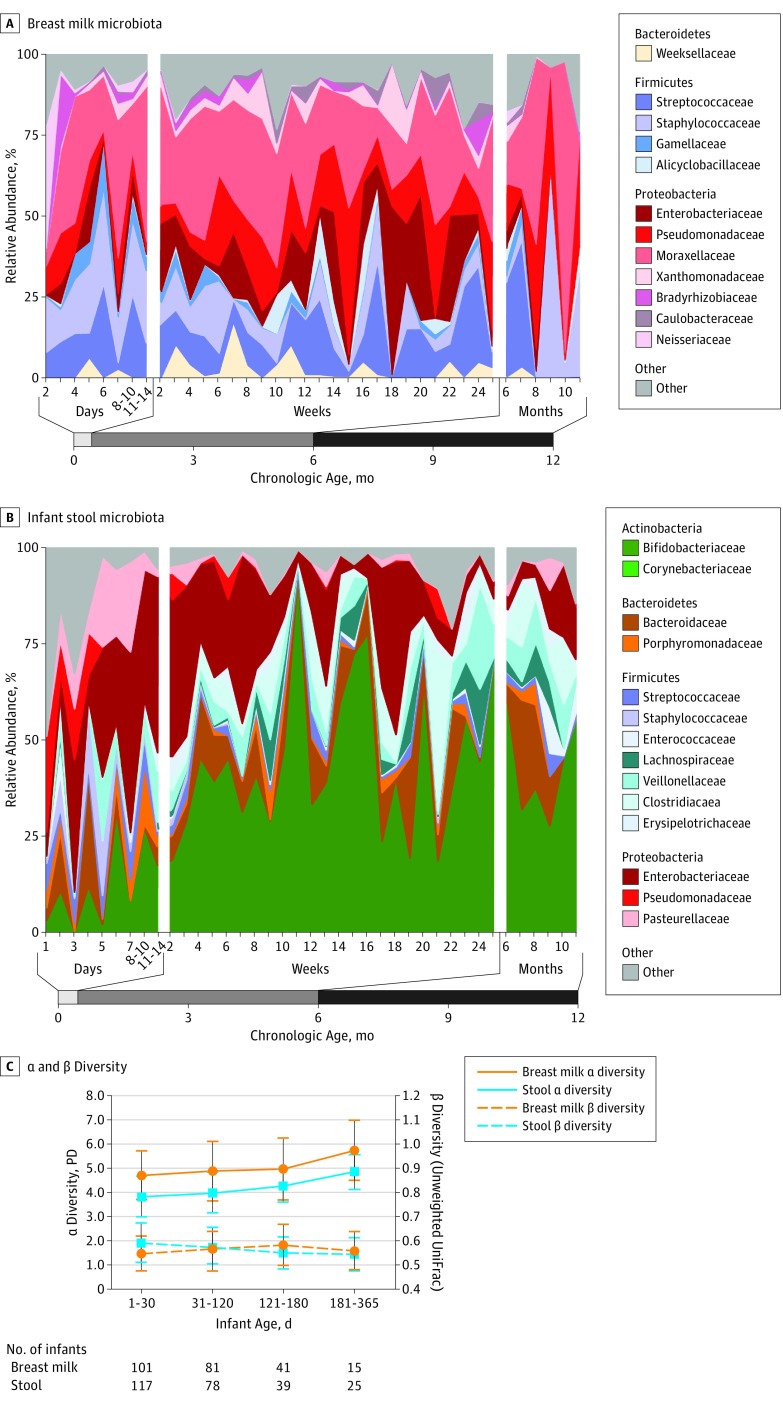
The breast milk bacterial community as the infant aged is shown in Figure 2. The breast milk bacterial communities remained unchanged in α diversity (within-sample diversity) throughout the first year of life. β Diversity (between-sample diversity) increased between mothers in the first 6 months after delivery and then decreased. There were no significant differences in breast milk or areolar bacteria by race/ethnicity, infant sex, mode of delivery, or geographic location of enrollment site.
Figure 2. Evaluation of Breast Milk and Infant Stool During the First Year of Life.
A, Relative abundance of most bacterial taxa in 238 breast milk samples from 94 mothers remained stable with age. B, Infant stool composition changed with increasing infant age in a nonrandom pattern. A total of 259 stool samples from 112 infants are shown here. C, α and β diversity (α diversity, Faith phylogenetic diversity) increased in infant stool (P = .01) but not breast milk (P = .06) with infant age. Differences between individuals (β diversity, unweighted UniFrac) in breast milk increased during the first 6 months of life (P < .001) and then decreased. β diversity of infant stool steadily decreased (P < .001) during the first 6 months and then began to level off after 6 months of life. Nonparametric t tests were used for comparisons of α and β diversity between age groups.
Taxonomic Changes in Infant Stool as a Function of Age, Feeding Type, and Other Factors
Bacterial diversity in infant stool increased with age, with convergence between individual infants near 12 months (Figure 2). Actinobacteria, specifically Bifidobacteriaceae (r = 0.31, P < .001), typically increased and Proteobacteria, specifically Enterobacteriaceae (r = 0.27, P < .001), decreased as the infant aged. Bacterial abundance changes occurred in direct or inverse relationships with each other in a complex network (eFigure 3 in the Supplement). Members of the Bifidobacteriaceae and Enterobacteriaceae families were dominant in the 3 observed clusters.
To evaluate factors contributing to the infant stool microbiome, we applied a permutational multivariate analysis of variance using the unweighted UniFrac and Bray-Curtis distance matrices (eTable 2 in the Supplement). Age accounted for 23% of the variation between infant stool samples (unweighted UniFrac, P = .001). The timing of formula introduction (R2 = 0.023, P = .001), solid food introduction (R2 = 0.029, P = .001), percentage of daily breastfeeding (R2 = 0.008, P = .002), and delivery method (R2 = 0.009, P = .001) also represented smaller, but significant, independent drivers of the microbiome trajectory in the developing infant. Race was a significant factor using unweighted UniFrac, but not Bray-Curtis, distances.
Infant Gut Microbiota Composition Association With Amount of Daily Breastfeeding
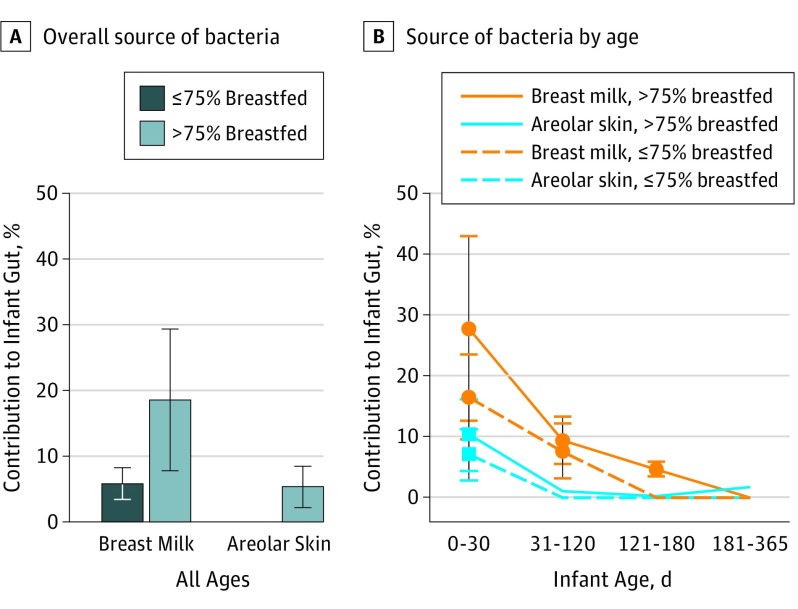
We used SourceTracker to estimate the proportion of bacteria in infant stool that originated from breast milk and areolar skin (Figure 3). Overall, primarily breastfed infants received more bacteria from breast milk and areolar skin compared with those not primarily breastfed (breast milk, 18.5% vs 5.7%, P < .001, Wilcoxon rank sum; areolar, 5.2% vs 0.001%, P = .01, Wilcoxon rank sum). During the first 30 days of life, infants who were primarily breastfed received mean (SD) 27.7% (15.2%) of the bacteria from breast milk and 10.4% (6.0%) from areolar skin. The remaining 61.9% (16.2%) came from other sources that we did not characterize. The contribution of bacteria from mother’s milk and areolar skin was highest during the first month of life and decreased as the infant aged.
Figure 3. Source of Bacterial Community in Infant Stool by Percent Daily Breastfeeding.
A, Among primarily breastfed infants of all ages, 18.5% of the microbial community found in the stool was derived from breast milk compared with only 5.7% in infants not primarily breastfed (P < .001, Wilcoxon rank sum). Similarly, 5.2% of the infant stool microbial community was derived from areolar bacteria in those who breastfed more than 75% of the time compared with 0.001% in infants who breastfed 75% or less of the time (P = .01, Wilcoxon rank sum). B, Amount of contribution from breast milk to the infant stool was highest during the first 30 days of life and decreased as the infant aged, even among those who continued to breastfeed more than 75% of the time. Error bars indicate SD.
Because bacterial communities in milk and areolar skin remained distinct for each mother, we hypothesized that an infant’s stool microbiota would be more similar to the infant’s mother’s microbiota compared with random mothers. Comparison using multiple distance metrics showed closer distances between infant stool and breast milk or areolar skin among true mother-infant pairs compared with random pairs (eTable 3 in the Supplement). In total, 26 of 478 OTUs were more significantly shared within mother-infant pairs vs random pairs (eTable 4 in the Supplement). Oligotyping analysis showed significant sharing of Streptococcus, Veillonella, and Rothia (eTable 5 in the Supplement).
Changes in the infant gut bacterial community were associated with the proportion of breastfeeding in a dose-dependent manner. The percentage of daily milk intake was associated with altered community composition, measured by unweighted UniFrac distance (R2 = 0.008, P = .002). Differences in gut phylogenetic diversity did not reach statistical significance (nonparametric t test, P = .06), but different compositions were observed between primarily and nonprimarily breastfed infants (eFigure 4 in the Supplement). We used random forest modeling to determine the bacterial taxa that best discriminate between the infant’s lifetime feeding pattern (ie, exclusively vs nonexclusively breastfed). Erysipelotrichaceae, Bacteroidaceae, and Ruminococcaceae were the most prominent families found in nonexclusively breastfed infants that discriminated between the 2 groups (eFigure 5A in the Supplement). Ruminococcaceae and Bacteroidaceae were more abundant among formula feeders at the time of the sample collection (eFigure 5B in the Supplement).
Age at Solid Food Introduction
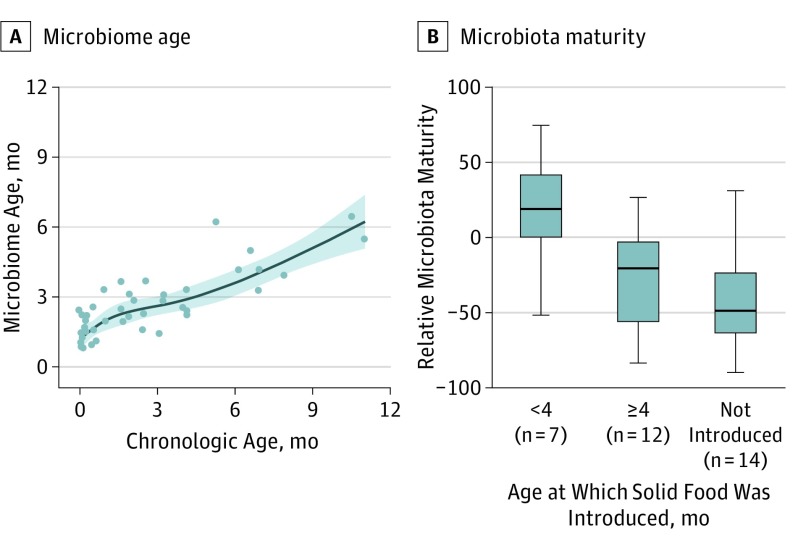
Following milk introduction, the next major change in the infant’s diet is the introduction of solid foods. Solid food introduction in infants aged 4 to 6 months was associated with a change in infant gut microbiome composition (eFigure 6 in the Supplement), but not phylogenetic diversity (nonparametric t test, P > .99). We interrogated our data to determine whether early solid food introduction changed the pattern of the microbiota trajectory, using a random forest regression model. Early solid food introduction (<4 months) in our infant cohort led to a rapid maturation of the infant stool microbiota (Kruskal-Wallis, P = .02) (Figure 4). The amount of daily breastfeeding as a proportion of total milk intake continued to influence the infant stool microbiome diversity and membership even after solid foods were introduced (eFigure 7 in the Supplement).
Figure 4. Microbiome Age Estimation.
A, A subset of healthy, vaginally born, exclusively breastfed infants (n = 42) was used to train a random forest regression model that was then applied to estimate microbiota maturity. Relative microbiota maturity was calculated by microbiota age of a child relative to the microbiota age of healthy children of the same chronologic age from the spline fit line shown here. B, Differences in relative microbiota maturity based on the age at which solid food was introduced in our cohort. Early solid food introduction was associated with early microbiota maturity (<4 months vs ≥4 months, P = .02; ≥4 months vs not introduced, P = .30, Kruskal-Wallis). Boxes indicate the 25th to 75th percentile; line within the box, median; error bars, 1.5 × interquartile range.
Predicted Metagenome Function of Breast Milk and Infant Gut Bacterial Community
Given the effect of breast milk microbiota on the infant gut community, we sought to gain insight into the genes present in breast milk bacterial communities using PICRUSt. Breast milk contains bacteria with predicted high abundance in gene families associated with membrane transport and carbohydrate, amino acid, and energy metabolism (eFigure 8 in the Supplement). We compared metagenome predictions of infant stool microbiota based on the amount of breast milk intake. Infants who were primarily breastfed had a lower representation of genes involved in energy metabolism, sphingolipid metabolism, and glycan biosynthesis and metabolism (eFigure 9 in the Supplement). Infants with earlier introduction of solid food had microbiota with increased function related to xenobiotics biodegradation and metabolism.
Discussion
We characterized bacterial communities in mother-infant pairs and provide data suggesting that bacteria from mothers’ breast milk and areolar skin are transferred to their infants’ guts. Key support for this idea comes from comparisons of bacterial lineages in the infants’ guts with their mothers vs random mothers, which showed more shared lineages in the correct pairs. Our data do not exclude the possibility that bacteria from infants’ stool seed the mother’s microbiome. It is likely that bacteria cycle between the mother and infant in a constant exchange. However, we favor the idea that transfer of bacteria primarily occurs from mother to infant. Breast milk is an early source of bacteria and nutrition introduced to the infant gut within a few hours of birth. Bacteria from mother’s milk and skin are most prominent in their infants’ guts in the first month of life, accounting for nearly 40% of the gut bacteria in primarily breastfed infants. Mother to infant microbe transmission was compromised in infants who were not primarily breastfed. Other sources of the infant microbiome include the mother’s gut and vaginal bacteria as well as the environment. In mice, cross-fostering experiments initiated within 48 hours of birth have shown that the nursing mother, rather than the birth mother, dictates the infant microbiome composition, which persists after weaning and for life. In the Human Microbiome Project, Ding and Schloss found breastfeeding during infancy to be a major life-history characteristic that affects bacterial composition in adults. Early life may represent a critical window for bacterial imprinting of breast milk bacteria leading to nonrandom community assembly. Our model showed that the complex ecologic network that forms during the first 6 months of life is built on interdependent bacterial relationships in the infant gut. Breast milk bacteria that seed the gut first influence and select for bacteria that follow, leaving a footprint that can be detected even in adulthood. Our oligotype analysis indicated that breast milk provides Veillonella and Rothia, bacteria genera that have been associated with a lower incidence of asthma. These early bacterial seeding events may be a mechanism by which breastfeeding protects children.
The breast milk microbiota become more divergent between mothers during the first 6 months of her infant’s life. The divergence lessens after 6 months when breast milk is typically no longer an infant’s sole food source. The infant gut microbiota display increasing α diversity and reduced β diversity as the infant ages—a trajectory also observed by others. Mother-infant sharing decreases as the infants age. Milk bacteria are detectable in infant stool at least through 6 months of life, but differences in the gut microbiota and their function are evident throughout the entire first year. Some studies have suggested that even small amounts of formula supplementation would shift the microbiota from a breastfed pattern to a formula pattern. Our multivariate and diversity analyses indicate that the shift occurs in a dose-dependent manner. The proportion of breast milk intake also decreases with introduction of solid foods. A recent study suggests that cessation of breastfeeding, rather than introduction of solid foods, is the major driver in the development of an adult microbiota. In our cohort, introduction of solid foods prior to 6 months led to early maturation of the microbiota. Furthermore, continued breastfeeding after solid food introduction suppressed the diversification and enrichment of bacteria typically associated with solid foods. These findings further support the current World Health Organization and American Academy of Pediatrics recommendation for exclusive breastfeeding during the first 6 months with continued breastfeeding until at least 12 months.
Our predicted metagenome functional analysis shows that breast milk harbors bacteria with prominent carbohydrate, amino acid, and energy metabolism functions. Breastfeeding has been shown to strongly protect against obesity. Microbiota harvest energy from the diet, and energy storage in the host is divergent between obese and lean humans and mice. Infants in our cohort who were not primarily breastfed had a higher abundance of the Bacteroidaceae, which has been associated with subsequent obesity in most pediatric studies of early gut colonization, although not all studies agreed. Erysipelotrichaceae and Ruminococcaceae were also increased in infants not exclusively breastfed. Both families are enriched in older children and adults with higher body mass indexes. Furthermore, continued breastfeeding as the primary source of milk intake after introduction of solids appeared to suppress increases in multiple families within the phylum Firmicutes that are butyrate producers. Butyrate can serve as an energy substrate or signaling molecule to contribute to the de novo production of lipids. Firmicutes and butyrate have also been associated with an obese phenotype in mice and humans. These pathways need to be explored further.
Limitations
This study has limitations. We did not sequence communities from maternal mouth, skin other than areolar, vagina, or stool, which may have contributed additional bacteria to infants. The origin of breast milk bacteria is unclear. One hypothesis is that milk bacteria come from the infant’s oral cavity. We did not collect infant oral specimens. However, we did not see differences in the microbiota of breast milk by delivery type in our cohort, as has been similarly reported by other investigators. Our recruitment focused on breastfeeding participants, and we did not enroll large numbers of strictly formula-fed infants. Some participants were unable to adhere to our longitudinal collection schedule. Sequencing of the 16S rRNA gene is limited in analysis at the strain level. SourceTracker is an estimation/prediction tool; the results may differ if the model included all potential sources of microbiota or additional longitudinal samples. We used PICRUSt to estimate bacterial gene content; because the input is 16S rRNA gene data, we did not capture any eukaryotic or viral contributions to the metagenome. PICRUSt also cannot distinguish variation at the strain level and different strains of 1 bacteria can vary in gene counts. Metagenomic shotgun sequencing would permit strain and more accurate functional analysis, but is costly. Finally, we focused on bacterial communities, yet milk is a complex substance with many bioactive components, including human milk oligosaccharides, which may promote the persistence of specific bacterial lineages.
Conclusions
Our study confirms a bacterial community in breast milk and tracks that community from mothers into the infant gut. Breast milk bacteria influence the establishment and development of the infant microbiome with continued impact after solid food introduction. Furthermore, breast milk contributes bacteria associated with a decreased risk for developing allergic diseases. Our results emphasize the importance of breastfeeding in the assembly of the infant gut microbiome.
eMethods. Detailed Methodology
eFigure 1. Quality Control
eFigure 2. Right vs Left Breast
eFigure 3. Modeling of OTU Networks at Early Development (1st Month of Life) and Between 4 and 6 Months of Life
eFigure 4. Breast Milk Influences Composition in a Dose-Dependent Manner
eFigure 5. Bacterial Taxa That Discriminate Microbial Communities in Infant Stool Using a Random Forest Algorithm Based on Feeding Status
eFigure 6. Introduction of Solid Foods Into the Infant Diet Results in Changes in the Microbial Community
eFigure 7. Differences in the Infant Stool Microbiome Arising From the Amount of Daily Breastfeeding Persist Even After Solid Foods Are Introduced
eFigure 8. Functional Capabilities of Breast Milk Microbial Community Predicted Using PICRUSt
eFigure 9. Differences in Predicted Metagenomic Functional Analysis of Infant Stool Dependent on Feeding Method
eTable 1. Characteristics of the Infant’s Feeding Type at the Time of Each Unique Sample Collection
eTable 2. Factors Contributing To The Variation In The Microbial Community Of Infant Stool From Adonis Multivariate Analysis of Variance Using the Bray-Curtis and Unweighted UniFrac Distance Matrices
eTable 3. Distance Comparison of Microbial Communities Between True Compared With Random Mother-Infant Pairs (Wilcoxon Rank-Sum Test)
eTable 4. OTUs With Significant Difference in Sharing Rate Between True Mother-Infant Pairs Compared to Random Mother-Infant Pairs When Comparing Bacterial Communities in Breast Milk and Infant Stool
eTable 5. Oligotyping Analysis Performed Using Sequences Mapped to Otus Shared Within Mother-Infant Dyads
References
- 1.Ahern PP, Faith JJ, Gordon JI. Mining the human gut microbiota for effector strains that shape the immune system. Immunity. 2014;40(6):815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(suppl 1):4578-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subramanian S, Huq S, Yatsunenko T, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510(7505):417-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrieta MC, Stiemsma LT, Dimitriu PA, et al. ; CHILD Study Investigators . Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):307ra152. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22(10):1187-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Rosa PS, Warner BB, Zhou Y, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci U S A. 2014;111(34):12522-12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azad MB, Konya T, Maughan H, et al. ; CHILD Study Investigators . Gut microbiota of healthy Canadian infants: profiles by mode of delivery and infant diet at 4 months. CMAJ. 2013;185(5):385-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971-11975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madan JC, Hoen AG, Lundgren SN, et al. Association of cesarean delivery and formula supplementation with the intestinal microbiome of 6-week-old infants. JAMA Pediatr. 2016;170(3):212-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Llorente C, Plaza-Diaz J, Aguilera M, et al. Three main factors define changes in fecal microbiota associated with feeding modality in infants. J Pediatr Gastroenterol Nutr. 2013;57(4):461-466. [DOI] [PubMed] [Google Scholar]
- 14.Lax S, Smith DP, Hampton-Marcell J, et al. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345(6200):1048-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bäckhed F, Roswall J, Peng Y, et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe. 2015;17(5):690-703. [DOI] [PubMed] [Google Scholar]
- 16.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Section on Breastfeeding Breastfeeding and the use of human milk. Pediatrics. 2012;129(3):e827-e841. [DOI] [PubMed] [Google Scholar]
- 18.Jost T, Lacroix C, Braegger CP, Rochat F, Chassard C. Vertical mother-neonate transfer of maternal gut bacteria via breastfeeding. Environ Microbiol. 2014;16(9):2891-2904. [DOI] [PubMed] [Google Scholar]
- 19.Hunt KM, Foster JA, Forney LJ, et al. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One. 2011;6(6):e21313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martín V, Maldonado-Barragán A, Moles L, et al. Sharing of bacterial strains between breast milk and infant feces. J Hum Lact. 2012;28(1):36-44. [DOI] [PubMed] [Google Scholar]
- 21.Faith DP. Conservation evaluation and phylogenetic diversity. Biol Conservation. 1992;61(1):1-10. [Google Scholar]
- 22.Knights D, Kuczynski J, Charlson ES, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8(9):761-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eren AM, Maignien L, Sul WJ, et al. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4(12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langille MG, Zaneveld J, Caporaso JG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: Statistical Analysis of Taxonomic and Functional Profiles. Bioinformatics. 2014;30(21):3123-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daft JG, Ptacek T, Kumar R, Morrow C, Lorenz RG. Cross-fostering immediately after birth induces a permanent microbiota shift that is shaped by the nursing mother. Microbiome. 2015;3:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez PF, Doré J, Leclerc M, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics. 2007;119(3):e724-e732. [DOI] [PubMed] [Google Scholar]
- 28.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am J Clin Nutr. 1999;69(5):1035S-1045S. [DOI] [PubMed] [Google Scholar]
- 29.Harder T, Bergmann R, Kallischnigg G, Plagemann A. Duration of breastfeeding and risk of overweight: a meta-analysis. Am J Epidemiol. 2005;162(5):397-403. [DOI] [PubMed] [Google Scholar]
- 30.Bäckhed F, Ding H, Wang T, et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci U S A. 2004;101(44):15718-15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalliomäki M, Collado MC, Salminen S, Isolauri E. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr. 2008;87(3):534-538. [DOI] [PubMed] [Google Scholar]
- 32.Scheepers LE, Penders J, Mbakwa CA, Thijs C, Mommers M, Arts IC. The intestinal microbiota composition and weight development in children: the KOALA Birth Cohort Study. Int J Obes (Lond). 2015;39(1):16-25. [DOI] [PubMed] [Google Scholar]
- 33.Vael C, Verhulst SL, Nelen V, Goossens H, Desager KN. Intestinal microflora and body mass index during the first three years of life: an observational study. Gut Pathog. 2011;3(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White RA, Bjørnholt JV, Baird DD, et al. Novel developmental analyses identify longitudinal patterns of early gut microbiota that affect infant growth. PLoS Comput Biol. 2013;9(5):e1003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balamurugan R, George G, Kabeerdoss J, Hepsiba J, Chandragunasekaran AM, Ramakrishna BS. Quantitative differences in intestinal Faecalibacterium prausnitzii in obese Indian children. Br J Nutr. 2010;103(3):335-338. [DOI] [PubMed] [Google Scholar]
- 36.Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A. 2009;106(7):2365-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koleva PT, Bridgman SL, Kozyrskyj AL. The infant gut microbiome: evidence for obesity risk and dietary intervention. Nutrients. 2015;7(4):2237-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5-11. [DOI] [PubMed] [Google Scholar]
- 39.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027-1031. [DOI] [PubMed] [Google Scholar]
- 40.Urbaniak C, Angelini M, Gloor GB, Reid G. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome. 2016;4(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods. Detailed Methodology
eFigure 1. Quality Control
eFigure 2. Right vs Left Breast
eFigure 3. Modeling of OTU Networks at Early Development (1st Month of Life) and Between 4 and 6 Months of Life
eFigure 4. Breast Milk Influences Composition in a Dose-Dependent Manner
eFigure 5. Bacterial Taxa That Discriminate Microbial Communities in Infant Stool Using a Random Forest Algorithm Based on Feeding Status
eFigure 6. Introduction of Solid Foods Into the Infant Diet Results in Changes in the Microbial Community
eFigure 7. Differences in the Infant Stool Microbiome Arising From the Amount of Daily Breastfeeding Persist Even After Solid Foods Are Introduced
eFigure 8. Functional Capabilities of Breast Milk Microbial Community Predicted Using PICRUSt
eFigure 9. Differences in Predicted Metagenomic Functional Analysis of Infant Stool Dependent on Feeding Method
eTable 1. Characteristics of the Infant’s Feeding Type at the Time of Each Unique Sample Collection
eTable 2. Factors Contributing To The Variation In The Microbial Community Of Infant Stool From Adonis Multivariate Analysis of Variance Using the Bray-Curtis and Unweighted UniFrac Distance Matrices
eTable 3. Distance Comparison of Microbial Communities Between True Compared With Random Mother-Infant Pairs (Wilcoxon Rank-Sum Test)
eTable 4. OTUs With Significant Difference in Sharing Rate Between True Mother-Infant Pairs Compared to Random Mother-Infant Pairs When Comparing Bacterial Communities in Breast Milk and Infant Stool
eTable 5. Oligotyping Analysis Performed Using Sequences Mapped to Otus Shared Within Mother-Infant Dyads