Key Points
Question
Is mitochondrial DNA copy number, measured from buffy coat/circulating leukocytes, associated with clinical cardiovascular disease (CVD) end points?
Findings
In this cohort analysis, among 21 870 participants from 3 cohort studies, the hazard ratio for incident CVD associated with a 1-SD decrease in mitochondrial DNA copy number was 1.23 (95% CI, 1.19-1.26). Addition of mitochondrial DNA copy number to the 2013 American College of Cardiology/American Heart Association Pooled Cohorts Equations for estimating 10-year atherosclerotic CVD risk was associated with significant improvement in risk classification and sensitivity and specificity for the 2013 American College of Cardiology/American Heart Association recommendations on initiating statin therapy.
Meaning
Mitochondrial DNA copy number is independently associated with incident CVD and may have clinical utility in improving CVD risk classification.
Abstract
Importance
Mitochondrial dysfunction is a core component of the aging process and may play a key role in atherosclerotic cardiovascular disease. Mitochondrial DNA copy number (mtDNA-CN), which represents the number of mitochondria per cell and number of mitochondrial genomes per mitochondrion, is an indirect biomarker of mitochondrial function.
Objective
To determine whether mtDNA-CN, measured in an easily accessible tissue (buffy coat/circulating leukocytes), can improve risk classification for cardiovascular disease (CVD) and help guide initiation of statin therapy for primary prevention of CVD.
Design, Setting, and Participants
Prospective, population-based cohort analysis including 21 870 participants (20 163 free from CVD at baseline) from 3 studies: Cardiovascular Health Study (CHS), Atherosclerosis Risk in Communities Study (ARIC), and Multiethnic Study of Atherosclerosis (MESA). The mean follow-up was 13.5 years. The study included 11 153 participants from ARIC, 4830 from CHS, and 5887 from MESA. Analysis of the data was conducted from March 10, 2014, to January 29, 2017.
Exposures
Mitochondrial DNA-CN measured from buffy coat/circulating leukocytes.
Main Outcomes and Measures
Incident CVD, which combines coronary heart disease, defined as the first incident myocardial infarction or death owing to coronary heart disease, and stroke, defined as the first nonfatal stroke or death owing to stroke.
Results
Of the 21 870 participants, the mean age was 62.4 years (ARIC, 57.9 years; MESA, 62.4 years; and CHS, 72.5 years), and 54.7% of participants were women. The hazard ratios for incident coronary heart disease, stroke, and CVD associated with a 1-SD decrease in mtDNA-CN were 1.29 (95% CI, 1.24-1.33), 1.11 (95% CI, 1.06-1.16), and 1.23 (95% CI, 1.19-1.26). The associations persisted after adjustment for traditional CVD risk factors. Addition of mtDNA-CN to the 2013 American College of Cardiology/American Heart Association Pooled Cohorts Equations for estimating 10-year hard atherosclerosis CVD risk was associated with improved risk classification (continuous net reclassification index, 0.194; 95% CI, 0.130-0.258; P < .001). Mitochondrial DNA-CN further improved sensitivity and specificity for the 2013 American College of Cardiology/American Heart Association recommendations on initiating statin therapy for primary prevention of ASCVD (net 221 individuals appropriately downclassified and net 15 individuals appropriately upclassified).
Conclusions and Relevance
Mitochondrial DNA-CN was independently associated with incident CVD in 3 large prospective studies and may have potential clinical utility in improving CVD risk classification.
This cohort study determines whether mitochondrial DNA copy number, measured in an easy accessible tissue (buffy coat/circulating leukocytes), is associated with improved risk classification for cardiovascular disease and help guide initiation of statin therapy for primary prevention of cardiovascular disease.
Introduction
Mitochondria play a critical role in energy homeostasis as the primary site of adenosine triphosphate production. Energy regulation is also dependent on proteins translated from genes encoded in mitochondrial DNA (mtDNA), a 16.7-kb circular DNA molecule. Mitochondrial dysfunction, a hallmark of the aging process, disrupts energy homeostasis and is believed to be a core component of several chronic conditions including cardiovascular disease (CVD). Mitochondrial dysfunction may also play a critical role in the initiation and progression of atherosclerosis, the primary pathological lesion underlying CVD. Evidence for this hypothesis is derived from both model organisms and human studies. First, in ApoE knockout mouse models of hyperlipidemia, mtDNA damage occurs prior to plaque formation, and there is significant correlation between mtDNA damage and extent of atherosclerosis in ApoE and human aorta specimens. Second, a study using polG knockout mice (which results in elevated rates of mtDNA mutations) and bone marrow transplants into ApoE knockout mice demonstrated that circulating cells increased the necrotic core and decreased the fibrous cap, which are key features of plaque instability. Finally, in humans, white blood cell (WBC) mtDNA damage has been associated with high-risk atherosclerotic plaques.
Mitochondrial DNA copy number (mtDNA-CN), while not a direct measure of mtDNA damage, is associated with mitochondrial enzyme activity and adenosine triphosphate production and can therefore serve as a biomarker of mitochondrial function. Mitochondrial DNA-CN, a measure of mtDNA levels per cell, is quantified using a low-cost scalable assay that allows for rapid determination of mitochondrial function in a large number of samples. Mitochondrial DNA-CN declines with age and is associated with all-cause mortality and chronic kidney disease in longitudinal studies. A 2017 small case-control study has also shown an association between peripheral blood mtDNA-CN and severity of coronary heart disease. We measured mtDNA-CN in DNA derived from blood samples in 21 870 individuals from 3 prospective cohort studies, the Atherosclerosis Risk in Communities (ARIC) study, the Cardiovascular Health Study (CHS), and the Multiethnic Study of Atherosclerosis (MESA), to evaluate the association of mtDNA-CN with prevalent and incident CVD events and the potential utility of mtDNA as a novel clinical biomarker of CVD risk.
Methods
Study Populations
The ARIC study recruited 15 792 individuals aged 45 to 65 years between 1987 and 1989 from 4 US communities. Twenty-seven percent of ARIC participants were African American. Our analysis was restricted to 11 455 unrelated participants who met quality control metrics implemented for genotyping arrays (used to generate the mtDNA-CN metric). We further excluded 302 individuals missing CVD risk factor or status information. The final sample size was 11 153 individuals.
The CHS study recruited 5201 individuals older than 65 years between 1989 and 1990 from 4 US communities. In 1992 to 1993, CHS further recruited 687 African American participants to increase minority enrollment. Our analysis was restricted to 4892 individuals with nonmissing mtDNA-CN measurements. We further excluded 62 individuals missing CVD risk factor or status information. The final sample size was 4830 individuals.
The MESA study recruited 6814 individuals aged 45 to 85 years free of prevalent CVD between 2000 and 2001 from 6 US communities. The study included white individuals (38%), African American individuals (28%), Chinese American individuals (12%), and Hispanic individuals (22%). The present analysis was restricted to 6179 individuals with nonmissing mtDNA-CN measurements. We further excluded 263 individuals whose DNA was from the MESA family study because this DNA was derived from transformed cell lines. We further excluded 5 participants from racial/ethnic groups at centers in which there were fewer than 5 participants from the same racial/ethnic group as well as 24 individuals missing CVD risk factor or status information. The final sample size was 5887 individuals.
All centers obtained approval from their respective institutional review boards, and all participants provided written informed consent. This analysis was conducted with anonymized data and acknowledged as not human participants research by the Johns Hopkins Medicine institutional review board.
Measurement of mtDNA-CN
DNA for mtDNA-CN analysis was collected at different visits in each study. For this analysis, the visit of DNA collection was considered the baseline visit (eMethods in the Supplement). In ARIC, DNA was extracted using the Gentra Puregene Blood Kit (Qiagen) from buffy coat of whole blood collected in visits 1 (1987-1989), 2 (1990-1992), 3 (1993-1995), and 4 (1996-1998) in 4.2%, 79.6%, 15.6%, and 0.6% of participants, respectively. In CHS, DNA was extracted by salt precipitation following proteinase K digestion of buffy coat from whole blood collected at visit 1. In MESA, DNA was isolated from peripheral leukocytes from blood collected at visit 1 using the Gentra Puregene Blood Kit.
The methods for measuring mtDNA-CN have been previously described. Briefly, in ARIC and MESA, mtDNA-CN was calculated from probe intensities of mitochondrial single-nucleotide polymorphisms on the Affymetrix Genome-Wide Human SNP Array 6.0 (Genvisis), and batch effects, DNA quality, and starting DNA quantity were corrected using probe intensities from nuclear single-nucleotide polymorphisms (eMethods in the Supplement). In CHS, mtDNA-CN was measured using multiplexed TaqMan-based quantitative polymerase chain reaction as previously described, and batch effects were corrected using a linear mixed model (eMethods in the Supplement). Standardized residuals after adjusting for known influences on mtDNA-CN (age, sex, collection site, and race/ethnicity) were used as the mtDNA-CN metric for all analyses (ie, with a mean of 0 and SD of 1).
Cardiovascular Disease Risk Factors
Traditional CVD risk factors were measured across all 3 cohorts at each clinic visit. Race/ethnicity and smoking status were assessed by self-report. Details for measurements of total cholesterol, high-density lipoprotein cholesterol, blood pressure, and medication use for the 3 cohorts have been previously described. Diabetes was defined as fasting glucose level at least 126 mg/dL, nonfasting glucose level at least 200 mg/dL (to convert to millimoles per liter, multiply by 0.0555), self-reported physician diagnosis of diabetes, or diabetic medication use. Ten-year CVD risk was calculated using the Pooled Cohort Equation (PCE) from the 2013 American College of Cardiology/American Heart Association (ACC/AHA) Guideline on Assessment of Cardiovascular Risk.
Outcome Definition and Adjudication
The event adjudication process in CHS, ARIC, and MESA consisted of expert committee review of hospital records, telephone interviews, and death certificates. Prevalent coronary heart disease (CHD) was defined as self- or physician-reported history of myocardial infarction (MI) or cardiac procedures (coronary artery bypass grafting or coronary artery angioplasty) at baseline. In addition, adjudicated events between visit 1 and the baseline visit for this study were considered prevalent events. Prevalent stroke was defined as self- or physician-reported stroke at baseline.
Incident CHD was defined as the first incident MI or death owing to CHD. Incident stroke was defined as the first nonfatal stroke or death owing to stroke. Incident CVD included both incident CHD and stroke. Analyses for incident events were performed across all 3 studies after excluding participants with prevalent CVD at baseline in ARIC and CHS.
Statistical Analyses
For the primary analysis, we used mtDNA-CN as a continuous variable and evaluated the change in risk of study outcomes associated with a 1-SD decrease in mtDNA-CN. As secondary analysis, we categorized mtDNA-CN in quintiles based on the distribution within each cohort and compared the risk of study outcomes in quintiles 1 through 4 with quintile 5. We used logistic regression to model odds ratios (ORs) and 95% CIs for the association of mtDNA-CN with prevalent outcomes. For incident outcomes (adjudicated as described in the previous section), follow-up time was defined from baseline (DNA collection) through the development of a study end point, death, or loss to/end of follow-up (through 2011 in ARIC, 2014 in CHS, and 2012 in MESA). We used Cox proportional hazards regression to estimate the hazard ratios (HRs) and 95% CIs for the association between mtDNA and incident outcomes. For incident outcomes, we also calculated marginally adjusted incidence rates and adjusted incidence rate ratios using Poisson regression.
The model adjustments for both prevalent and incident disease included adding the following covariates: age, sex, collection center, and race/ethnicity in the base model (model 1) and additionally, total cholesterol, high-density lipoprotein cholesterol, systolic blood pressure, hypertension medication use, current smoking, and diabetes in the full model (model 2). Cox models were fit separately for each cohort and combined across cohorts using fixed-effects meta-analysis (“meta.summaries” function from the “rmeta” R package). Forest plots were generated using the “forestplot” R package (R Programming).
We conducted additional analyses to evaluate whether mtDNA-CN may improve discrimination and reclassification of study participants when added to the PCE (eMethods in the Supplement). Briefly, we compared the discrimination ability of the calculated 10-year risk score vs the combination of the 10-year risk score and mtDNA-CN using Harrell C statistics. We also examined whether adding mtDNA-CN could improve the net reclassification index (NRI) and/or integrated discrimination improvement for statin therapy based on the 2013 ACC/AHA recommendations for starting statin therapy, excluding individuals whose risk score would not affect therapy decisions based on the 2013 ACC/AHA guidelines (ie, prevalent CVD, older than 75 years, prevalent diabetes, high-density lipoprotein cholesterol levels <70 mg/dL, or low-density lipoprotein cholesterol levels ≥190 mg/dL [to convert to millimoles per liter, multiply by 0.0259]). The NRI reflects the net increment in prediction accuracy (defined as the proportion of patients classified to a correct category) comparing models with and without mtDNA-CN, while the integrated discrimination improvement reflects the change in calculated risk for each individual comparing models with and without mtDNA-CN.
We also performed prespecified subgroup analyses by race/ethnicity, sex, and age and tested for potential interactions by adding an interaction term to the regression model. Finally, because mtDNA-CN from peripheral blood is associated with WBC count, we further adjusted the model by including log-transformed WBC count as a covariate among 8726 participants in ARIC in whom WBC measurements were available. All statistical analyses were performed using R, version 3.2.2 (R Programming). A 2-sided P value of less than .05 was considered significant.
Results
The study included 11 153 participants from ARIC, 4830 participants from CHS, and 5887 participants from MESA (total sample size, 21 870). The mean age of study participants was 62.4 years (ARIC, 57.9 years; MESA, 62.4 years; and CHS, 72.5 years), and 54.7% of participants were women (n = 11 967) (eTable 1 in the Supplement). Comparing age- and sex-adjusted quintiles of mtDNA-CN, individuals in the highest quintile of mtDNA had significantly more favorable CVD risk factor profiles, with the exception of total cholesterol levels (while not significant, higher total cholesterol levels were observed in quintile 5).
Prevalent Disease
One thousand seven hundred seven participants had prevalent CVD at baseline (1003 in ARIC and 704 in CHS; MESA recruitment excluded prevalent CVD). The meta-analysis ORs for prevalent CHD, stroke, and CVD associated with a 1-SD decrease in mtDNA-CN were 1.26 (95% CI, 1.20-1.33), 1.25 (95% CI, 1.14-1.36), and 1.27 (95% CI, 1.21-1.34), respectively, after adjusting for age, sex, race/ethnicity, and collection center (eFigure 1 in the Supplement and Table 1, model 1). Incorporating additional traditional CVD risk factors resulted in modest attenuation of the ORs (Table 1, model 2). The associations were consistent in the 4-study race/ethnicity and sex groups evaluated except in African American participants in CHS, although the sample size in this group was small (n = 753; 112 CVD events) (eFigures 2 and 3 in the Supplement).
Table 1. Odds Ratio for Prevalent Cardiovascular Disease by Quintiles of mtDNA Copy Number.
| Event | ARIC | CHS | ||||
|---|---|---|---|---|---|---|
| No./Total No. of Events | Model 1, OR (95% CI)a | Model 2, OR (95% CI) | No./Total No. of Events | Model 1, OR (95% CI) | Model 2, OR (95% CI) | |
| Coronary heart disease | ||||||
| Q1 | 236/2231 | 1.97 (1.57-2.48) | 1.66 (1.31-2.11) | 141/966 | 1.92 (1.43-2.59) | 1.80 (1.33-2.45) |
| Q2 | 181/2230 | 1.44 (1.14-1.83) | 1.31 (1.02-1.67) | 122/966 | 1.56 (1.16-2.11) | 1.57 (1.16-2.14) |
| Q3 | 146/2231 | 1.15 (0.89-1.47) | 1.15 (0.89-1.50) | 107/966 | 1.28 (0.95-1.74) | 1.30 (0.95-1.79) |
| Q4 | 149/2230 | 1.16 (0.91-1.49) | 1.23 (0.95-1.59) | 87/966 | 1.06 (0.77-1.45) | 1.03 (0.74-1.43) |
| Q5 | 130/2231 | 1 [Reference] | 1 [Reference] | 85/966 | 1 [Reference] | 1 [Reference] |
| Continuousb | 842/11 153 | 1.26 (1.18-1.35) | 1.18 (1.10-1.26) | 542/4830 | 1.26 (1.15-1.39) | 1.24 (1.12-1.37) |
| P trend | NA | <.001 | <.001 | NA | <.001 | <.001 |
| Stroke | ||||||
| Q1 | 73/2231 | 2.22 (1.48-3.40) | 1.84 (1.22-2.84) | 53/966 | 2.61 (1.59-4.41) | 2.47 (1.50-4.18) |
| Q2 | 44/2230 | 1.31 (0.83-2.07) | 1.17 (0.74-1.86) | 41/966 | 1.91 (1.14-3.27) | 1.89 (1.13-3.25) |
| Q3 | 44/2231 | 1.31 (0.83-2.08) | 1.29 (0.82-2.05) | 39/966 | 1.78 (1.06-3.06) | 1.76 (1.05-3.04) |
| Q4 | 42/2230 | 1.24 (0.79-1.97) | 1.27 (0.80-2.02) | 48/966 | 2.19 (1.33-3.69) | 2.15 (1.30-3.64) |
| Q5 | 34/2231 | 1 [Reference] | 1 [Reference] | 23/966 | 1 [Reference] | 1 [Reference] |
| Continuousb | 237/11 153 | 1.23 (1.09-1.37) | 1.14 (1.01-1.28) | 204/4830 | 1.28 (1.11-1.49) | 1.26 (1.09-1.46) |
| P trend | NA | <.001 | .03 | NA | <.001 | .002 |
| Cardiovascular disease | ||||||
| Q1 | 283/2231 | 2.01 (1.63-2.49) | 1.71 (1.37-2.13) | 188/966 | 2.24 (1.71-2.93) | 2.14 (1.62-2.83) |
| Q2 | 208/2230 | 1.39 (1.12-1.74) | 1.26 (1.00-1.59) | 157/966 | 1.71 (1.30-2.25) | 1.73 (1.31-2.30) |
| Q3 | 178/2231 | 1.18 (0.94-1.48) | 1.19 (0.94-1.51) | 134/966 | 1.36 (1.03-1.80) | 1.37 (1.03-1.83) |
| Q4 | 179/2230 | 1.18 (0.94-1.48) | 1.24 (0.98-1.57) | 122/966 | 1.25 (0.94-1.66) | 1.22 (0.91-1.64) |
| Q5 | 155/2231 | 1 [Reference] | 1 [Reference] | 103/966 | 1 [Reference] | 1 [Reference] |
| Continuousa | 1003/11 153 | 1.26 (1.18-1.33) | 1.17 (1.10-1.25) | 704/4830 | 1.31 (1.21-1.43) | 1.30 (1.19-1.42) |
| P trend | NA | <.001 | <.001 | NA | <.001 | <.001 |
Abbreviations: ARIC, Atherosclerosis Risk in Communities; CHS, Cardiovascular Health Study; mtDNA, mitochondrial DNA; NA, not applicable; OR, odds ratio; Q, quintile.
Model 1 is adjusted for age, sex, collection center, and race/ethnicity. Model 2 is model 1 additionally adjusted for total cholesterol, high-density lipoprotein cholesterol, systolic blood pressure, current smoking status, hypertension medication status, and type 2 diabetes status.
Continuous OR is per 1-SD decrease in mtDNA copy number.
Incident Disease
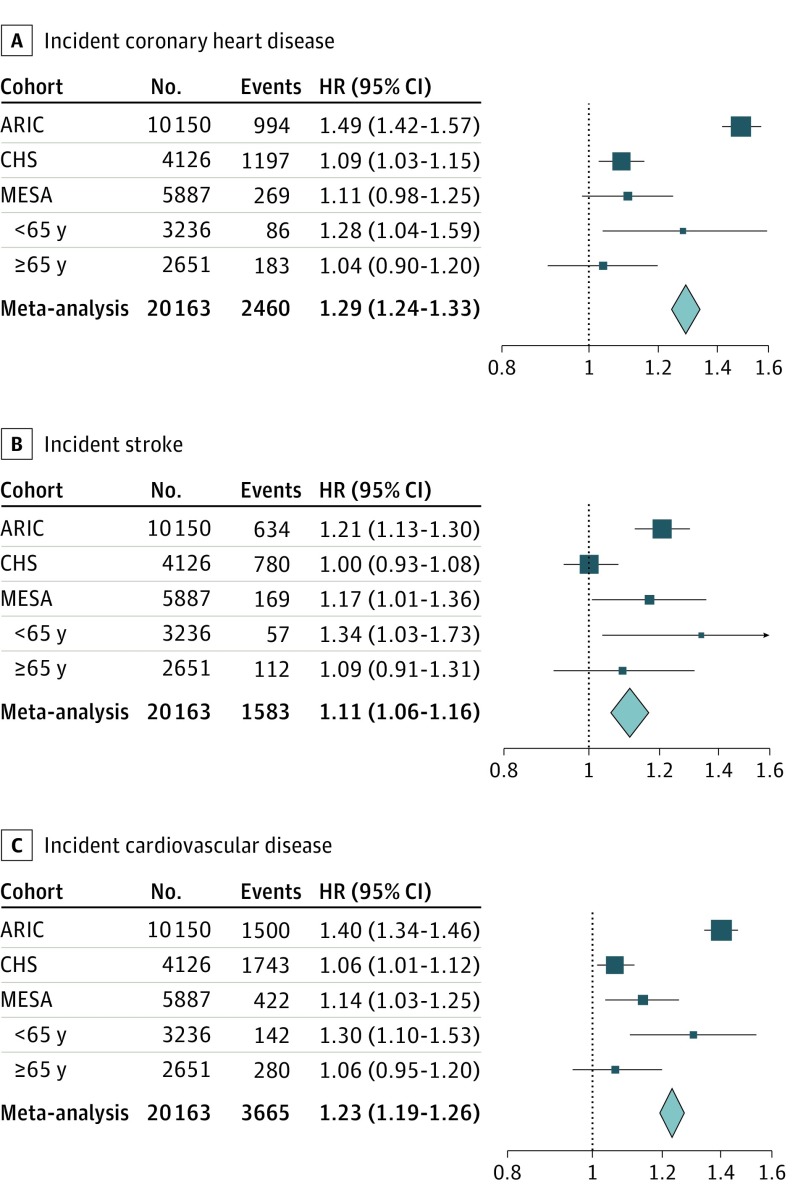
Participants free of CVD at baseline (n = 20 163) were followed up for a mean of 13.5 years, with 3665 incident CVD events (2460 incident CHD episodes and 1584 incident strokes). The pooled HRs for incident CHD, stroke, and CVD associated with a 1-SD decrease in mtDNA-CN were 1.29 (95% CI, 1.24-1.33), 1.11 (95% CI, 1.06-1.16), and 1.23 (95% CI, 1.19-1.26), respectively, after adjusting for age, sex, race/ethnicity, and study center (Figure). A modest attenuation of the HR was observed on the inclusion of traditional CVD risk factors (Table 2, model 2). The associations were consistent across study race/ethnicity and sex groups, although stronger associations were observed in ARIC compared with CHS (eFigures 4 and 5 and eTable 2 in the Supplement).
Figure. Association of Mitochondrial DNA Copy Number With Incident Cardiovascular Disease.
Forest plots are shown for meta-analyses of incident coronary heart disease (A), incident stroke (B), and incident cardiovascular disease (C). Age-stratified hazard ratios are shown for the Multiethnic Study of Atherosclerosis (MESA), but not included in the meta-analyses. ARIC indicates Atherosclerosis Risk in Communities; CHS, Cardiovascular Health Study; and HR, hazard ratio.
Table 2. Hazard Ratio for Incident CVD by Quintiles of mtDNA Copy Number.
| Event | ARIC | CHS | MESA | ||||||
|---|---|---|---|---|---|---|---|---|---|
| No./Total No. of Events | Model 1, HR (95% CI)a | Model 2, HR (95% CI) | No./Total No. of Events | Model 1, HR (95% CI) | Model 2, HR (95% CI) | No./Total No. of Events | Model 1, HR (95% CI) | Model 2, HR (95% CI) | |
| Coronary heart disease | |||||||||
| Q1 | 313/1948 | 2.55 (2.10-3.09) | 2.28 (1.88-2.77) | 220/778 | 1.31 (1.09-1.58) | 1.25 (1.04-1.51) | 59/1178 | 1.38 (0.93-2.04) | 1.30 (0.88-1.93) |
| Q2 | 214/2022 | 1.58 (1.28-1.95) | 1.46 (1.19-1.80) | 241/809 | 1.24 (1.03-1.49) | 1.22 (1.01-1.46) | 47/1177 | 1.13 (0.75-1.71) | 1.06 (0.70-1.61) |
| Q3 | 176/2053 | 1.25 (1.00-1.55) | 1.23 (0.99-1.53) | 250/832 | 1.21 (1.01-1.44) | 1.18 (0.98-1.41) | 64/1177 | 1.47 (1.00-2.16) | 1.40 (0.95-2.07) |
| Q4 | 139/2051 | 0.95 (0.76-1.20) | 0.97 (0.77-1.23) | 254/844 | 1.18 (0.99-1.41) | 1.15 (0.97-1.38) | 56/1177 | 1.28 (0.86-1.90) | 1.21 (0.81-1.80) |
| Q5 | 152/2076 | 1 [Reference] | 1 [Reference] | 232/863 | 1 [Reference] | 1 [Reference] | 43/1178 | 1 [Reference] | 1 [Reference] |
| Continuousb | 994/10 150 | 1.49 (1.42-1.57) | 1.40 (1.33-1.47) | 1197/4126 | 1.09 (1.03-1.15) | 1.07 (1.01-1.14) | 269/5887 | 1.11 (0.98-1.25) | 1.09 (0.97-1.23) |
| P trend | NA | <.001 | <.001 | NA | .005 | .02 | NA | .10 | .16 |
| Stroke | |||||||||
| Q1 | 158/1948 | 1.74 (1.36-2.22) | 1.52 (1.19-1.95) | 141/778 | 1.09 (0.87-1.38) | 1.05 (0.83-1.32) | 43/1178 | 1.67 (1.02-2.71) | 1.58 (0.97-2.57) |
| Q2 | 145/2022 | 1.48 (1.15-1.89) | 1.35 (1.05-1.73) | 150/809 | 1.05 (0.83-1.31) | 1.01 (0.81-1.27) | 37/1177 | 1.50 (0.91-2.47) | 1.38 (0.83-2.28) |
| Q3 | 122/2053 | 1.18 (0.91-1.53) | 1.15 (0.89-1.50) | 147/832 | 1.01 (0.80-1.26) | 0.99 (0.79-1.24) | 25/1177 | 0.97 (0.56-1.67) | 0.91 (0.53-1.58) |
| Q4 | 100/2051 | 0.96 (0.73-1.25) | 0.96 (0.73-1.26) | 187/844 | 1.25 (1.01-1.55) | 1.24 (1.00-1.53) | 38/1177 | 1.45 (0.88-2.39) | 1.39 (0.84-2.30) |
| Q5 | 109/2076 | 1 [Reference] | 1 [Reference] | 155/863 | 1 [Reference] | 1 [Reference] | 26/1178 | 1 [Reference] | 1 [Reference] |
| Continuousb | 634/10 150 | 1.21 (1.13-1.30) | 1.14 (1.06-1.22) | 780/4126 | 1.00 (0.93-1.08) | 0.98 (0.92-1.06) | 169/5887 | 1.17 (1.01-1.36) | 1.15 (0.99-1.34) |
| P trend | NA | <.001 | <.001 | NA | .95 | .67 | NA | .042 | .07 |
| CVD | |||||||||
| Q1 | 425/1948 | 2.21 (1.89-2.59) | 2.00 (1.70-2.34) | 329/778 | 1.27 (1.09-1.48) | 1.21 (1.04-1.41) | 98/1178 | 1.49 (1.09-2.03) | 1.41 (1.03-1.93) |
| Q2 | 331/2022 | 1.55 (1.31-1.83) | 1.44 (1.22-1.70) | 339/809 | 1.15 (0.99-1.34) | 1.12 (0.96-1.30) | 83/1177 | 1.30 (0.94-1.79) | 1.21 (0.87-1.67) |
| Q3 | 284/2053 | 1.26 (1.06-1.50) | 1.24 (1.05-1.48) | 353/832 | 1.12 (0.97-1.30) | 1.10 (0.94-1.27) | 84/1177 | 1.25 (0.90-1.72) | 1.18 (0.86-1.63) |
| Q4 | 219/2051 | 0.94 (0.79-1.13) | 0.96 (0.80-1.15) | 379/844 | 1.19 (1.03-1.38) | 1.17 (1.01-1.35) | 90/1177 | 1.34 (0.98-1.84) | 1.28 (0.93-1.75) |
| Q5 | 241/2076 | 1 [Reference] | 1 [Reference] | 343/863 | 1 [Reference] | 1 [Reference] | 67/1178 | 1 [Reference] | 1 [Reference] |
| Continuousb | 1500/10 150 | 1.40 (1.34-1.46) | 1.32 (1.27-1.38) | 1743/4126 | 1.06 (1.01-1.12) | 1.04 (0.99-1.10) | 422/5887 | 1.14 (1.03-1.25) | 1.12 (1.02-1.24) |
| P trend | NA | <.001 | <.001 | NA | .015 | .08 | NA | .009 | .02 |
Abbreviations: ARIC, Atherosclerosis Risk in Communities; CHS, Cardiovascular Health Study; CVD, cardiovascular disease; HR, hazard ratio; MESA, Multiethnic Study of Atherosclerosis; mtDNA, mitochondrial DNA; NA, not applicable; Q, quintile.
Model 1 is adjusted for age, sex, collection center, and race. Model 2 is model 1 additionally adjusted for total cholesterol, high-density lipoprotein cholesterol, systolic blood pressure, current smoking status, hypertension medication status, and type 2 diabetes status.
Continuous odds ratio is per 1-SD decrease in mtDNA copy number.
Given that mtDNA-CN is influenced by WBC count, we tested whether inclusion of log-transformed WBC as an additional covariate to model 1 attenuated the association with incident CVD in ARIC participants who had both mtDNA-CN and WBC count measured (8726, with 794 incident CVD events), with similar HRs observed (model 1, HR, 1.40; 95% CI, 1.34-1.46; model 1 + log(WBC), HR, 1.36; 95% CI, 1.30-1.43).
Age Interaction of Association of mtDNA-CN on CVD
In our analysis, the effect of mtDNA-CN on incident CVD was significantly larger in ARIC compared with CHS (Table 2), despite observing moderately larger effect sizes for prevalent CVD in CHS (Table 1). A major difference in these cohorts is the age distribution; on average, ARIC participants were 14.6 years younger (mean for ARIC, 57.9 years; range, 44.9-74.1 years vs mean CHS, 72.5 years; range, 65.0-100.0 years). The MESA cohort (mean, 62.4 years; range, 44-84 years) included participants that spanned the age distribution of both cohorts. Stratifying MESA to those younger than 65 years and 65 years or older, there was a consistent trend of mtDNA-CN having a stronger association for all 3 incident outcomes in younger vs older MESA participants (eTable 3 in the Supplement, Figure). While a formal test for interaction between mtDNA-CN and age was not significant in any individual study, the interaction term was highly significant in the combined analysis (OR, 1.0106; 95% CI, 1.0072-1.0140; P < .001), and significant even when excluding the oldest cohort, CHS (OR, 1.0061; 95% CI, 1.0008-1.0115; P = .03).
Mitochondrial DNA-CN and Risk Discrimination and Reclassification
Adding mtDNA-CN to the ACC/AHA 2013 PCE risk equation improved discrimination for CVD events in the pooled ARIC, CHS, and MESA cohorts (ΔC statistic, 0.7%; 95% CI, 0.4 to 1.0), although this was largely driven by the improvement for CHD risk (ΔC statistic, 1.2%; 95% CI, 0.8 to 1.6) because no improvement was seen for estimating stroke risk (ΔC statistic, −0.2%; 95% CI, −0.5 to 0.2) (Table 3 and eTable 4 in the Supplement).
Table 3. Change in C Statistic for CVD With mtDNA-CN Over Base AHA Risk Score Model in the Pooled Cohorts.
| Risk Score | C Statistic (95% CI) | ||
|---|---|---|---|
| CHD | Stroke | CVD | |
| AHA Risk Score | 0.747 (0.733-0.759) | 0.763 (0.747-0.780) | 0.752 (0.742-0.761) |
| AHA Risk Score and mtDNA-CN | 0.760 (0.745-0.774) | 0.762 (0.745-0.777) | 0.759 (0.751-0.769) |
| Δ C statistic, % | 1.2 (0.8-1.6) | −0.2 (−0.5 to 0.2) | 0.7 (0.4-1.0) |
Abbreviations: AHA, American Heart Association; CHD, coronary heart disease; CVD, cardiovascular disease; mtDNA-CN, mitochondrial DNA copy number.
We also assessed the ability of mtDNA-CN to improve risk reclassification in the 14 084 individuals who met the ACC/AHA risk evaluation criteria for primary prevention (age 40 to 75 years, free of type 2 diabetes, no CVD history, and high-density lipoprotein cholesterol levels ≥70 mg/dL and low-density lipoprotein cholesterol levels <190 mg/dL). Adding mtDNA-CN to the PCE improved risk reclassification for CVD events, measured by continuous NRI (0.194; 95% CI, 0.130-0.258; P < .001) and by integrated discrimination improvement (0.009; 95% CI, 0.005-0.012; P < .001) (Table 4). Looking at the overall categorical NRI with cutoffs at 5% and 7.5%, we also observed a significant improvement for CVD risk reclassification (overall NRI, 0.032; 95% CI, 0.015-0.049; P < .001) (Table 4), although this is largely driven by CHD (eTable 5 in the Supplement). Overall, a net of 15 individuals with events were appropriately upclassified, and 221 individuals without events were appropriately downclassified. Using a hard cutoff of 7.5% 10-year ASCVD risk for initiating statin therapy, a net of 6 additional individuals would appropriately be statin eligible, and 139 would be appropriately not recommended to initiate statin therapy. However, using the PCE for this analysis could be biased by poor calibration. Indeed, the observed risk in those estimated to have 5% to 7.5% risk was 5.0%, suggesting that the PCE overestimated risk in our sample. In recalibrated models, the observed risk in the 5% to 7.5% risk category was 5.6%. Comparing the recalibrated model with and without mtDNA-CN, we still observed significant improvement for both overall categorical and continuous NRI as well as integrated discrimination improvement (Table 4).
Table 4. Net Reclassification Indexa and IDI Comparing AHA Risk Score With and Without mtDNA-CN in the Pooled Cohorts.
| Event | AHA Risk Score + mtDNA-CN | Recalibrated AHA Risk Score + mtDNA-CN | ||||||
|---|---|---|---|---|---|---|---|---|
| <5% Risk | 5% to <7.5% Risk | ≥7.5% Risk | Total | <5% Risk | 5% to <7.5% Risk | ≥7.5% Risk | Total | |
| Persons without event, No. | ||||||||
| <5% Risk | 4833 | 241 | 13 | 5087 | 6236 | 202 | 9 | 6447 |
| 5% to <7.5% Risk | 323 | 1286 | 287 | 1896 | 346 | 1347 | 256 | 1949 |
| ≥7.5% Risk | 26 | 413 | 5655 | 6094 | 11 | 343 | 4327 | 4681 |
| Total | 5182 | 1940 | 5955 | 13 077 | 6593 | 1892 | 4592 | 13 077 |
| Persons with event, No. | ||||||||
| <5% Risk | 72 | 19 | 1 | 92 | 128 | 11 | 6 | 145 |
| 5%-<7.5% Risk | 10 | 66 | 22 | 98 | 15 | 81 | 22 | 118 |
| ≥7.5% Risk | 0 | 17 | 800 | 817 | 0 | 20 | 724 | 744 |
| Total | 82 | 102 | 823 | 1007 | 143 | 112 | 752 | 1007 |
| Estimate (95% CI) | P Value | Estimate (95% CI) | P Value | |||||
| NRI Categorical | 0.032 (0.015-0.049) | <.001 | 0.022 (0.004-0.039) | .02 | ||||
| NRI Continuous | 0.194 (0.130-0.258) | <.001 | 0.156 (0.092-0.220) | <.001 | ||||
| IDI | 0.009 (0.005-0.012) | <.001 | 0.004 (0.002-0.007) | <.001 | ||||
Abbreviations: ACC, American College of Cardiology; AHA, American Heart Association; IDI, integrated discrimination improvement; mtDNA-CN, mitochondrial DNA copy number; NRI, net reclassification index.
Categorical NRI with <5.0% and 5% to <7.5% risk cutoffs. Ten-year hard atherosclerotic cardiovascular disease risk was estimated in the pooled cohorts using Cox proportional hazards regression, using similar criteria as the 2013 AHA/ACC guidelines.
Discussion
We explored the role of mtDNA-CN in cardiovascular disease in 21 870 participants from the ARIC, CHS, and MESA studies, including self-identified white, black, Hispanic, and Chinese individuals. Mitochondrial DNA-CN was inversely associated with both prevalent and incident CVD events, and it also demonstrated potential as a clinically useful predictor of CVD by improving risk prediction and reclassification for primary hard ASCVD prevention compared with the 2013 ACC/AHA PCE risk prediction. While an association between prevalent CHD and mtDNA-CN has been reported in a small case-control study, to our knowledge, this is the first time that mtDNA-CN has been shown to be associated with incident CVD.
In our analyses, the association of mtDNA-CN with incident CHD was stronger than with incident stroke. While CHD and stroke have overlapping risk factors, the risk factors play different roles in precipitating these outcomes. Indeed, while most CHD occurs in the setting of coronary atherosclerosis, the etiology of stroke is more diverse, including a confluence of atherosclerosis, cardiac embolism, and small vessel disease. Thus, one possible explanation for the stronger association with CHD is that mtDNA-CN is associated with the atherosclerotic processes that precipitate CHD, although additional work is required to validate this hypothesis.
We also observed significantly larger effect estimates of mtDNA-CN on CVD risk in younger individuals. One concern was that this result was driven by the different methods used to measure mtDNA-CN across the cohorts (which have different recruitment age ranges). We have some evidence suggesting that genotyping array-based methods to measure mtDNA-CN (ARIC and MESA) are more biologically relevant than quantitative polymerase chain reaction (CHS) (eTable 6 in the Supplement). However, the observation that the association with prevalent CVD was actually stronger in the oldest cohort (CHS) suggests that excluding prevalent disease may induce selection (survivor) bias by excluding from the longitudinal analyses individuals at highest risk for CVD owing to reduced mtDNA-CN by virtue of having prevalent disease. Indeed, this age-related attenuation of CVD risk factors has been observed for other risk factors.
While this study was not designed to address the mechanism by which levels of mtDNA could affect CVD risk, our results are in line with evidence supporting a role of mitochondria in cardiovascular disease. A key question remains as to whether mtDNA-CN has a causal effect, which would suggest new avenues for preventive intervention, or is a biomarker for another CVD risk factor. However, given that a significant association is observed after the inclusion of traditional CVD risk factors, such as sex, blood lipids, blood pressure, smoking status, and hypertension medication use, as covariates, our data support a role for mtDNA-CN as an independent risk factor associated with CVD events.
Strengths and Limitations
This study has several strengths and limitations. We used a large sample size combining 3 well-characterized prospective studies with long-term follow-up to assess the role of mtDNA-CN in CVD. However, the attenuation of the association with advancing age, the potential contribution of mtDNA-CN assessment method to the heterogeneity of the results, and the relatively small number of observed stroke events require additional replication of our findings in cohorts with sufficient sample size before mtDNA-CN can be formally incorporated to standard risk prediction scores. In addition, we only had a measurement of mtDNA-CN at a single point. This measurement is subject to analytical random error and to individual variability over time, and both types of errors may have contributed to underestimating the association between mtDNA-CN and CVD outcomes.
Conclusions
Mitochondrial DNA copy number was inversely associated with incident and prevalent CVD outcomes in 21 870 participants from 3 well-characterized cohorts. In addition, mtDNA-CN was significantly associated with improved risk discrimination and reclassification for primary prevention of hard ASCVD.
eMethods
eFigure 1: Association of mtDNA-CN With Prevalent CVD
eFigure 2: Association of mtDNA-CN With Prevalent CVD, Stratified by Race
eFigure 3: Association of mtDNA-CN With Prevalent CVD, Stratified by Sex
eFigure 4: Association of mtDNA-CN With Incident CVD, Stratified by Race
eFigure 5: Association of mtDNA-CN With Incident CVD, Stratified by Sex
eTable 1 Baseline Characteristics of Pooled ARIC, CHS, and MESA Study Participants by Quintiles of mtDNA-CN.
eTable 2. Adjusted Incidence Rates, Incidence Rate Ratios, and Incidence Rate Differences for Cardiovascular Disease (CVD) End Points by Quintiles of mtDNA Copy Number in the ARIC, CHS, and MESA studies.
eTable 3. Hazards Ratio for Incident Cardiovascular Disease (CVD) by Quintiles of mtDNA Copy Number in MESA.
eTable 4: Change in C Statistic With mtDNA-CN Over Base AHA Risk Score Model.
eTable 5: Net Reclassification Index (NRI) and Integrated Discrimination Improvement (IDI) Comparing AHA Risk Score with and without mtDNA-CN in the Pooled Cohorts Stratified by CVD Type.
eTable 6: Comparison of mtDNA-CN Derived From Affymetrix 6.0 Arrays vs. qPCR in 5460 MESA Participants for Variables Known to be Associated With mtDNA-CN.
References
- 1.Dai D-F, Rabinovitch PS, Ungvari Z. Mitochondria and cardiovascular aging. Circ Res. 2012;110(8):1109-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballinger SW, Patterson C, Knight-Lozano CA, et al. Mitochondrial integrity and function in atherogenesis. Circulation. 2002;106(5):544-549. [DOI] [PubMed] [Google Scholar]
- 3.Yu E, Calvert PA, Mercer JR, et al. Mitochondrial DNA damage can promote atherosclerosis independently of reactive oxygen species through effects on smooth muscle cells and monocytes and correlates with higher-risk plaques in humans. Circulation. 2013;128(7):702-712. [DOI] [PubMed] [Google Scholar]
- 4.Jeng J-Y, Yeh TS, Lee JW, Lin SH, Fong TH, Hsieh RH. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem. 2008;103(2):347-357. [DOI] [PubMed] [Google Scholar]
- 5.Mengel-From J, Thinggaard M, Dalgård C, Kyvik KO, Christensen K, Christiansen L. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum Genet. 2014;133(9):1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashar FN, Moes A, Moore AZ, et al. Association of mitochondrial DNA levels with frailty and all-cause mortality. J Mol Med (Berl). 2015;93(2):177-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tin A, Grams ME, Ashar FN, et al. Association between mitochondrial DNA copy number in peripheral blood and incident CKD in the atherosclerosis risk in communities study. J Am Soc Nephrol. 2016;27(8):2467-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L-P, Cheng K, Ning MA, et al. Association between peripheral blood cells mitochondrial DNA content and severity of coronary heart disease. Atherosclerosis. 2017;261:105-110. [DOI] [PubMed] [Google Scholar]
- 9.The ARIC Investigators The Atherosclerosis Risk in Communities (ARIC) study: design and objectives. Am J Epidemiol. 1989;129(4):687-702. [PubMed] [Google Scholar]
- 10.Arking DE, Pfeufer A, Post W, et al. A common genetic variant in the NOS1 regulator NOS1AP modulates cardiac repolarization. Nat Genet. 2006;38(6):644-651. [DOI] [PubMed] [Google Scholar]
- 11.Fried LP, Borhani NO, Enright P, et al. The cardiovascular health study: design and rationale. Ann Epidemiol. 1991;1(3):263-276. [DOI] [PubMed] [Google Scholar]
- 12.Bild DE, Bluemke DA, Burke GL, et al. Multi-Ethnic Study of Atherosclerosis: objectives and design. Am J Epidemiol. 2002;156(9):871-881. [DOI] [PubMed] [Google Scholar]
- 13.Psaty BM, Kuller LH, Bild D, et al. Methods of assessing prevalent cardiovascular disease in the Cardiovascular Health Study. Ann Epidemiol. 1995;5(4):270-277. [DOI] [PubMed] [Google Scholar]
- 14.Genuth S, Alberti KG, Bennett P, et al. ; Expert Committee on the Diagnosis and Classification of Diabetes Mellitus . Follow-up report on the diagnosis of diabetes mellitus. Diabetes Care. 2003;26(11):3160-3167. [DOI] [PubMed] [Google Scholar]
- 15.Goff DC Jr, Lloyd-Jones DM, Bennett G, et al. ; American College of Cardiology/American Heart Association Task Force on Practice Guidelines . 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129(25)(suppl 2):S49-S73. [DOI] [PubMed] [Google Scholar]
- 16.Ives DG, Fitzpatrick AL, Bild DE, et al. Surveillance and ascertainment of cardiovascular events: the Cardiovascular Health Study. Ann Epidemiol. 1995;5(4):278-285. [DOI] [PubMed] [Google Scholar]
- 17.White AD, Folsom AR, Chambless LE, et al. Community surveillance of coronary heart disease in the Atherosclerosis Risk in Communities (ARIC) Study: methods and initial two years’ experience. J Clin Epidemiol. 1996;49(2):223-233. [DOI] [PubMed] [Google Scholar]
- 18.Lumley T. rmeta: Meta-analysis. https://cran.r-project.org/web/packages/rmeta/rmeta.pdf. Published 2012.
- 19.Gordon M, Lumley T forestplot: advanced forest plot using ‘grid’ graphics. https://cran.r-project.org/web/packages/forestplot/forestplot.pdf. Published 2016.
- 20.Uno H, Cai T survIDINRI: IDI and NRI for comparing competing risk prediction models with censored survival data. https://cran.r-project.org/web/packages/survIDINRI/survIDINRI.pdf. Published 2013.
- 21.Chen S, Xie X, Wang Y, et al. Association between leukocyte mitochondrial DNA content and risk of coronary heart disease: a case-control study. Atherosclerosis. 2014;237(1):220-226. [DOI] [PubMed] [Google Scholar]
- 22.Stoekenbroek RM, Boekholdt SM, Luben R, et al. Heterogeneous impact of classic atherosclerotic risk factors on different arterial territories: the EPIC-Norfolk prospective population study. Eur Heart J. 2016;37(11):880-889. [DOI] [PubMed] [Google Scholar]
- 23.Kannel WB, McGee D, Gordon T. A general cardiovascular risk profile: the Framingham Study. Am J Cardiol. 1976;38(1):46-51. [DOI] [PubMed] [Google Scholar]
- 24.Kannel WB, Neaton JD, Wentworth D, et al. Overall and coronary heart disease mortality rates in relation to major risk factors in 325,348 men screened for the MRFIT. Multiple Risk Factor Intervention Trial. Am Heart J. 1986;112(4):825-836. [DOI] [PubMed] [Google Scholar]
- 25.Howard G, Manolio TA, Burke GL, Wolfson SK, O’Leary DH. Does the association of risk factors and atherosclerosis change with age? an analysis of the combined ARIC and CHS cohorts: the Atherosclerosis Risk in Communities (ARIC) and Cardiovascular Health Study (CHS) investigators. Stroke. 1997;28(9):1693-1701. [DOI] [PubMed] [Google Scholar]
- 26.Abbott RD, Curb JD, Rodriguez BL, et al. Age-related changes in risk factor effects on the incidence of coronary heart disease. Ann Epidemiol. 2002;12(3):173-181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
eFigure 1: Association of mtDNA-CN With Prevalent CVD
eFigure 2: Association of mtDNA-CN With Prevalent CVD, Stratified by Race
eFigure 3: Association of mtDNA-CN With Prevalent CVD, Stratified by Sex
eFigure 4: Association of mtDNA-CN With Incident CVD, Stratified by Race
eFigure 5: Association of mtDNA-CN With Incident CVD, Stratified by Sex
eTable 1 Baseline Characteristics of Pooled ARIC, CHS, and MESA Study Participants by Quintiles of mtDNA-CN.
eTable 2. Adjusted Incidence Rates, Incidence Rate Ratios, and Incidence Rate Differences for Cardiovascular Disease (CVD) End Points by Quintiles of mtDNA Copy Number in the ARIC, CHS, and MESA studies.
eTable 3. Hazards Ratio for Incident Cardiovascular Disease (CVD) by Quintiles of mtDNA Copy Number in MESA.
eTable 4: Change in C Statistic With mtDNA-CN Over Base AHA Risk Score Model.
eTable 5: Net Reclassification Index (NRI) and Integrated Discrimination Improvement (IDI) Comparing AHA Risk Score with and without mtDNA-CN in the Pooled Cohorts Stratified by CVD Type.
eTable 6: Comparison of mtDNA-CN Derived From Affymetrix 6.0 Arrays vs. qPCR in 5460 MESA Participants for Variables Known to be Associated With mtDNA-CN.