Abstract
Background and Aims
Enzymes belonging to the RNase T2 family are essential for normal rRNA turnover in eukaryotes. In Arabidopsis thaliana, this function is performed by RNS2. The null mutant rns2-2 has increased rRNA half-life and constitutive autophagy. The aim of this work was to determine the molecular changes that take place in the rns2-2 mutant that may lead to altered cellular homeostasis, manifested by the observed cellular phenotype.
Methods
To determine the effect of defective rRNA turnover on cellular homeostasis, comparative transcriptome and metabolome analyses of 10-day-old wild-type and rns2-2 seedlings were used to identify molecular processes affected in the mutant. Bioinformatics analyses suggested additional phenotypes that were confirmed through direct plant size measurements and microscopy.
Key Results
Few genes were differentially expressed in the rns2-2 mutant, indicating that control of autophagy in this genotype is mainly achieved at the post-transcriptional level. Among differentially expressed genes, transcripts related to carbon flux processes, particularly the pentose phosphate pathway (PPP), were identified. Metabolite analyses confirmed changes in the levels of PPP intermediates. Genes related to cell wall loosening were also differentially expressed in the mutant, and a decrease in monosaccharide components of cell wall hemicellulose were found. As a potential effect of weaker cell walls, rns2-2 plants are larger than wild-type controls, due to larger cells and increased water content. Elevated levels of reactive oxygen species (ROS) were also measured in rns2-2, and the constitutive autophagy phenotype was blocked by preventing ROS production via NADPH oxidase.
Conclusions
Lack of rRNA recycling in rns2-2 cells triggers a change in carbon flux, which is redirected through the PPP to produce ribose-5-phosphate for de novo nucleoside synthesis. rRNA or ribosome turnover is thus essential for cellular homeostasis, probably through maintenance of nucleoside levels as part of the salvage pathway.
Keywords: Arabidopsis thaliana, autophagy, carbon flux, cell wall, nucleoside homeostasis, RNS2, rRNA turnover
INTRODUCTION
As ribosomes account for a substantial proportion of cellular resources, ribosome turnover is a critical factor in maintaining homeostasis, particularly during nutrient deficiency (Klemperer and Pilley, 1985; Lardeux and Mortimore, 1987; Loffler et al., 1992; Warner, 1999). When yeast cells are subjected to starvation conditions, ribosomes are targeted for degradation through autophagy-related processes, in which cargo is taken up into the vacuole for degradation and recycling. A selective autophagy pathway termed ribophagy targets ribosomes in nitrogen-starved yeast cells (Kraft et al., 2008); ribosomes are preferentially targeted to the autophagic pathway compared with other cytoplasmic proteins under these conditions. The ubiquitin deconjugation enzyme Ubp3p and its cofactors Bre1p, Cdc48 and Ufd3 (Ossareh-Nazari et al., 2010) are essential factors for selective autophagy of 60S (but not 40S) ribosomal subunits (Kraft et al., 2008). A possible role in ribophagy for the ubiquitin ligase Rsp5 has also been proposed (Kraft and Peter, 2008), whereas the Ltn1 E3 ligase antagonizes ribophagy activation (Ossareh-Nazari et al., 2014). Non-selective autophagy has also been implicated in the degradation of ribosomes in yeast cells upon starvation (Huang et al., 2015).
The mechanism of autophagic RNA degradation was recently dissected in yeast (Huang et al., 2015). After transport of ribosomes to the vacuole through either ribophagy or non-selective autophagy, rRNA is hydrolysed by Rny1, a member of the RNase T2 family (MacIntosh, 2011), which is the main vacuolar RNase activity in yeast (MacIntosh et al., 2001; Huang et al., 2015). The turnover of rRNA yields 3ʹ-nucleoside monophosphates (NMPs) that are then converted to nucleosides by the vacuolar non-specific phosphatase Pho8. These nucleosides are transported back to the cytoplasm where they are further broken down by the nucleosidases Pnp1 and Urh1 to produce purine and pyrimidine bases that are either reused by the cell or secreted (Huang et al., 2015).
The turnover of functional ribosomes under non-stress conditions is less well characterized. In Arabidopsis thaliana, the vacuolar RNase RNS2, also a member of the RNase T2 family, is necessary for normal rRNA decay. Arabidopsis mutants lacking RNS2 activity have higher levels of total RNA (Floyd et al., 2015), and rRNA has a longer half-life in rns2 mutants than in wild-type (WT) plants (Hillwig et al., 2011). rRNA accumulates in the vacuole in the rns2-2 mutant, and this accumulation depends on the presence of the core autophagy protein ATG5 but not the core autophagy protein ATG9 (Floyd et al., 2015). Interestingly, lack of RNS2 activity results in an increased basal autophagy phenotype, with mutant cells displaying a level of autophagy under normal conditions similar to that observed in WT plants only under stress conditions (Hillwig et al., 2011; Floyd et al., 2015). These results suggested that normal ribosomes or rRNA are targeted for vacuolar degradation by a selective autophagic process, probably similar to ribophagy (MacIntosh and Bassham, 2011). Once in the vacuole, rRNA is hydrolysed by RNS2. We hypothesized that lack of rRNA degradation in the rns2 mutants leads to an imbalance in cellular homeostasis that triggers constitutive autophagy as a compensatory mechanism (Hillwig et al., 2011; Floyd et al., 2015).
This normal rRNA decay mechanism seems to be conserved in other eukaryotes. Zebrafish have two RNASET2 genes, but only one is constitutively expressed (Hillwig et al., 2009). This protein resides in lysosomes, and mutant zebrafish lacking RNASET2 activity also show accumulation of rRNA in the lytic organelle (Haud et al., 2011). Similar accumulation was observed in human cells in which RNASET2 expression was knocked down with artificial microRNAs (Haud et al., 2011). In both cases, electron microscopy showed that lysosomes are full of dense material, probably rRNA. This accumulation causes changes in cellular homeostasis that is particularly prominent in neuronal cells, and leads to white matter lesions in the brain, associated with familial cystic leukoencephalopathy (Henneke et al., 2009; Haud et al., 2011).
The constitutive autophagy phenotype observed in the arabidopsis rns2-2 mutant could be the result of different changes in cellular homeostasis. Since ribosomes are an important sink of cellular resources, lack of rRNA degradation could lead to a change in the energy balance or the availability of nitrogen in the cell. Alternatively, the phenotype could result from a decrease in available purine and pyrimidine bases. To gain insight into the type of imbalance that triggers constitutive autophagy in the rns2-2 mutant, we analysed changes in the transcriptome caused by the rns2-2 mutation and complemented this analysis with metabolome studies. We found a small number of differentially expressed genes (DEGs) in rns2-2, which indicated that the pentose phosphate pathway (PPP) and cell wall processes are affected in the mutant. We also observed changes in the level of some of the sugars that participate in the PPP and in the sugar composition of the cell wall. Transcriptome and metabolome data suggested high levels of NADPH production, which could lead to production of reactive oxygen species (ROS). After confirmation of elevated ROS levels in the mutant, we determined that inhibition of NADPH oxidase activity suppresses autophagy in rns2-2. Finally, we determined that the rns2-2 mutant is larger than WT plants due to an increase in cell growth, which could be caused by an increase in carbon availability and could explain the changes in cell wall observed in our analysis. Our results support the hypothesis that the mutant may use the PPP to divert carbon flux toward production of ribose-5-phosphate, an essential substrate for the de novo synthesis of nucleosides, and that constitutive autophagy in the mutant is likely to be triggered as a consequence of elevated ROS production in response to a deficiency in the nucleoside pool.
MATERIALS AND METHODS
Plant material for microarray and metabolite analysis, and RNA preparation
Seeds of A. thaliana ecotype Columbia-0 and the rns2-2 T-DNA insertion mutant were sterilized and stratified overnight as previously described (Hillwig et al., 2011). Seeds were plated on agar plates with Murashige and Skoog (MS) modified basal medium with Gamborg vitamins (Phytotech, M404). Plants were grown under 16 h light–8 h dark with a light intensity of approx. 120 μmol m–2 s–1 at 60 % humidity and 21 °C. Ten-day-old seedlings were then collected and frozen in liquid N2. Seedlings were ground using a mortar and pestle under liquid N2, and RNA was extracted from 50–100 μg of ground tissue using the Qiagen RNeasy plant mini kit (Qiagen 74903). RNA was then treated with TURBO DNase (Life Technologies, AM2238) to remove contaminating genomic DNA. RNA samples were tested for quality using the Agilent 2100 Bioanalyzer, and only samples with an RNA integrity number (RIN) >7 were used for microarray analysis.
Microarray analysis
RNA (250 ng) from rns2-2 mutant and WT seedlings was labelled using the GeneChip®3ʹ IVT Expression kit (Affymetrix, 901229) and hybridized to the Affymetrix Arabidopsis ATH1 Genome Array GeneChip (Affymetrix, 900385) using an Affymetrix hybridization kit (Affymetrix, 900720). Arrays were scanned on a GeneChip® scanner 30007G. Raw intensity data were generated by the Affymetrix Expression console software. Three independent biological replicates were analysed for each genotype. RNA labelling, hybridization and scanning were performed by the Microarray facility at Iowa State University.
Data were normalized using robust multi-array average (RMA) (Irizarry et al., 2003). Data were further analysed to find DEGS using the affyLM GUI in R and a P-value cut-off of 0.005 (Wettenhall et al., 2006). The list of DEGs was analysed for over-represented Gene Ontology (GO) terms using Virtual Plant’s BioMaps analysis with a P-value cut-off of 0.01 (Katari et al., 2010). Network analysis was completed using Virtual Plant’s Gene Networks analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database, both the primary and secondary sub-types, allowing for zero hops (Kanehisa, 2000). The returned network analysis was further modified in Adobe Photoshop for visual clarity. Raw and normalized data were deposited in the GEO database (http://www.ncbi.nlm.nih.gov/geo/) to allow for public access (accession no. GSE81218).
qPCR
cDNA was synthesized from DNase-treated RNA using the qScript Flex cDNA kit (Quanta, 95049). Quantitative RT-PCR (qPCR) was performed using the Perfecta SYBR Green Supermix (Quanta, 95070) on a Stratagene MX4000, using 30 ng of sample cDNA template, and relative quantification was completed using a standard curve (Pfaffl, 2004). As a loading control to normalize the qPCR data, we used a transcript previously identified to be stably expressed, the TIP41-like (AT4G34270) transcript (forward primer sequence 5ʹ CCGGCGATTCAGATGGAGACGG 3ʹ and reverse 5ʹ TGCTGAGACGGCTTGCTCCTG 3ʹ) (Czechowski et al., 2005). Microarray targets verified by qPCR included AT3G45970, A. thaliana expansin-like A1 (forward primer 5ʹ GAGTTTCTTCGCCGGACA 3ʹ, reverse primer 5ʹ ATCGCAAGGAACTCTTTGGT 3ʹ); AT2G41640, glycosyl transferase (forward primer 5ʹ TGTGCTTCAAACGTCACCCA 3ʹ, reverse primer 5ʹ TGCGAAACGAATCTAGGAGGG 3ʹ); AT1G79530, glyceraldehyde 3-phosphate dehydrogenase of plastid 1 (forward primer 5ʹ ATGGGGTTACAGCAACCGAG 3ʹ, reverse primer 5ʹ CACGGGCAAAGCTAACGATG 3ʹ); and AT1G16300, glyceraldehyde 3-phosphate dehydrogenase of plastid 2 (forward primer 5ʹ ATGGGGTTACAGCAACCGAG 3ʹ, reverse primer 5ʹ ACGTTGGCGGGATATGGTTT 3ʹ).
Metabolite analysis
Ten-day-old seedlings were grown on MS vitamin and salt mixture phytoagar plates with 16 h light/8 h dark at 22 °C. Non-targeted metabolite profiling was carried out using gas chromatography–mass spectrometry (GC-MS). A 20 mg aliquot of sample tissue was extracted using 350 μL of hot methanol. Samples were incubated at 60 °C for 10 min followed by sonication for 10 min. A 350 μL aliquot of chloroform and 300 μL of water were added, samples mixed by vortexing, and polar and non-polar fractions separated by centrifugation. Both fractions were retained separately for metabolite analysis and dried in a nitrogen evaporator followed by a speed-vac concentrator. Samples were methoximated using 50 μL of 20 mg mL–1 methoxamine hydrochloride in dry pyridine at 30 °C for 1.5 h with shaking. Samples were treated with 70 μL of bis-trimethyl silyl trifluoroacetamide with 1 % trimethylchlorosilane for 30 min at 65 °C, dried under nitrogen, dissolved in 100 μL of pyridine, and analysed on an Agilent 7890A-GC using an HP5ms column equipped with a 5975C MSD detector (Agilent Technologies). Temperature was raised from 70 °C to 320 °C at a rate of 5 °C min–1 with a 2 mL min–1 helium flow rate. The gradient terminated at a hold at 320 °C. Ionization voltage was 70 eV and interface temperature was 280 °C. Data were deconvoluted and analysed using the Automatic Mass Spectral Deconvolution and Identification System (AMDIS) (D’Arcy and Mallard, 2004) and metabolites were identified using mass spectra referenced to authentic standards in the Iowa State University W. M. Keck Metabolomics Research Laboratory and the National Institute of Standards and Technology 05 mass spectra library (Schmidt et al., 2011).
Metabolites of the PPP were quantified from plants grown as for non-targeted analysis. Approximately 30 mg of fresh tissue from whole seedlings were collected and ground in liquid nitrogen. Samples were extracted with methanol/chloroform/water (2.5:1:1 v/v/v) followed by methanol/chloroform (1:1 v/v) (Weckwerth et al., 2004). Samples were combined and water was added, followed by centrifugation. Samples were then dried to a tenth volume. Extracts were labelled with aniline (Sigma) and EDC [N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide] (Sigma) as previously described (Yang et al., 2008; Jannasch et al., 2011). Standards (Sigma) were labelled with aniline-13C6 (Cambridge Isotope Laboratories) and EDC. Aniline-labelled standards and samples were analysed by liquid chromatography (LC)–mass spectrometry (LC-MS) in an Agilent QTOF 6540 using a Zorbax Eclipse XDB-C18 column (Agilent). Elution began in water and was raised to 50 % acetonitrile in 15 min, then raised to 90 % acentonitrile in 0.1 min with a flow rate of 0.8 mL min–1 while acquiring chromatograms in negative ion mode between m/z 100 and 1200. Data acquisition and processing were completed using the Agilent MassHunter software. Both GC-MS and LC-MS analyses were carried out at the Iowa State University W. M. Keck Metabolomics Research Laboratory.
Growth and cellular phenotype analysis
For rosette measurements, 66 arabidopsis plants of each genotype were grown on soil for 4 weeks. Basal rosettes were measured using Rosette Tracker software (De Vylder et al., 2012) for both diameter and area of the rosette. Water content was calculated by comparing the mass of adult basal rosette fresh tissue and after lyophilization for 4 d. Root lengths were measured on seedlings grown on vertical agar plates with MS modified basal medium with Gamborg vitamins. Roots were measured after removal from agarose and straightening on a solid surface. For the WT and rns2-2, 38 and 39 roots were measured representing three independent plates. The length of root cells was measured by confocal microscopy. Five-day-old roots grown on vertical agar plates as above were gently removed from the agar and stained for 10 min using 10 μg mL–1 Oregon Green 488 carboxylic acid diacetate (carboxy-DFFDA, stock 5 mg mL–1 in acetone) (Life Technologies, O6151) and 5 μg mL–1 propidium iodide (Molecular Probes, P3566) in half-strength MS. After three 5 min washes in half-strength MS medium, root cells just above the zone of elongation were imaged on a confocal microscope (Leica SP5×MP). For WT and rns2-2, 593 and 530 cell lengths were measured using ImageJ (Abràmoff et al., 2004), from 36 and 25 roots, respectively, taken from five independent plates. Supplementary Data Fig. S1 shows a representative image used for cell length analysis.
Cell wall analysis
For each genotype, six or seven samples were analysed. Four-week-old basal rosette tissue was ground in liquid nitrogen. Alcohol-insoluble cell wall was extracted using ethanol and acetone as previously described (Harholt et al., 2006) and treated with α-amylase. A 1 mg aliquot of alcohol-insoluble cell wall was hydrolysed using 2 n trifluoroacetic acid, and monosaccharide components were analysed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) using a CarboPac PA-20 (Dionex) column and gradient conditions as previously described (Zabotina et al., 2008; Pogorelko et al., 2011). Relative proportions and response factors were determined relative to a standard mixture of equimolar l-fucose, l-rhamnose, l-arabanose, l-galactose, d-glucose, d-xylose, d-mannose, d-galacturonic acid and d-glucuronic acid (Sigma).
Reactive oxygen species analysis and NADPH oxidase inhibition
Reactive oxygen species were visualized using 2ʹ, 7ʹ-dichlorofluorescin diacetate (DCFDA). Seven-day-old seedlings were stained and washed as previously described and visualized using fluorescence microscopy (Contento and Bassham, 2010). Root tips were imaged at the widest point to maintain consistency; images were quantified using ImageJ and normalized to the WT intensity. For inhibition of NADPH oxidase, 7-day-old seedlings were treated with either dimethylsulphoxide (DMSO) or 20 μM diphenyleneiodonium (DPI) (Liu et al., 2009). Samples were then stained with monodansylcadaverine (MDC), and autophagosomes (MDC-positive puncta) were visualized by microscopy as previously described (Floyd et al., 2015). The number of autophagosomes per frame was counted.
RESULTS
The rns2-2 mutation causes only minor transcriptome changes in arabidopsis seedlings
We hypothesized that comparison of gene expression in the rns2-2 null mutant with gene expression in WT plants could indicate how loss of RNase activity resulted in disruption of homeostasis, manifest as constitutive autophagy. Thus, as a first approach to understand the molecular processes that trigger this phenotype, we performed a transcriptome analysis of WT and rns2-2 mutant seedlings grown on plates, using the Affymetrix Arabidopsis ATH1 Genome Array GeneChip that provides almost full genome coverage.
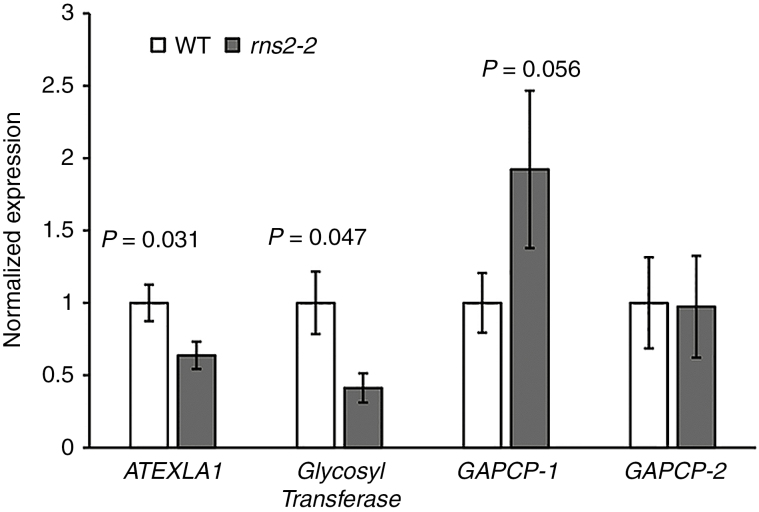
Initial data analysis using strict cut-offs for false discovery rate (FDR = 0.01) identified only RNS2 as a DEG. A more relaxed analysis identified 38 transcripts as differentially expressed (P < 0.005) between rns2-2 and WT seedlings (Table 1). Fifteen genes had increased expression (between 1.5- and 3.1-fold) in the mutant, and 22 genes, in addition to RNS2, had reduced expression (between 1.9- and 5.5-fold) in the rns2-2 plants compared with the WT. No autophagy-related genes were present in the DEG set, indicating that the upregulation of autophagy in the mutant is not due to increased autophagy gene expression. The small number of DEGs and the small magnitude of expression changes was unexpected, considering the strong cellular phenotype observed in the mutant (Hillwig et al., 2011; Floyd et al., 2015). We selected several representative genes to confirm the microarray results by qPCR analysis (Fig. 1). The expression of the expansin-like gene ATEXLA1 (AT3G45970) was determined to be 36 % lower in the mutant by qPCR and 52 % lower by microarray, and similar results were obtained for the glycosyltransferase gene AT2G41640. The microarray probe 262939_s_at matches two homologous arabidopsis genes, AT1G79530 and AT1G16300, corresponding to the plastidial glyceraldehyde-3-phosphate dehydrogenases GAPCP-1 and GAPCP-2. qPCR analysis showed that GAPCP-1 expression is almost 2-fold higher in rns2-2, close to the 2.29-fold increase observed for the probe 262939_s_at in the microarray. On the other hand, no change was observed for GAPCP-2 by qPCR.
Table 1.
List of genes differentially expressed in the rns2-2 mutant seedlings with respect to the wild type
| Locus | Fold change | P-value | Gene name and/or annotation |
|---|---|---|---|
| AT1G64390 | 3.11681 | 2.48 × 105 | ATGH9C2, glycosyl hydrolase |
| AT2G39700 | 3.09792 | 0.00044 | ATEXPA4, expansin |
| AT3G29030 | 2.648013 | 0.000256 | ATEXPA5, expansin |
| AT2G28790 | 2.591433 | 0.000145 | Osmotin-like protein |
| AT3G15650 | 2.438696 | 0.000443 | Phospholipase |
| AT2G03090 | 2.381119 | 0.000275 | ATEXPA15, expansin |
| AT1G79530; AT1G16300 | 2.289448 | 4.94 × 105 | GAPCP-1 and GAPCP-2, plastid glyceraldehyde-3-phosphate dehydrogenase |
| AT1G61580 | 2.278667 | 0.000421 | ARP2, ribosomal protein |
| AT3G14210 | 2.205033 | 5.22 × 105 | ESM1, carboxylesterase |
| AT1G74460 | 2.15002 | 0.000374 | GDSL-motif lipase |
| AT4G38660 | 2.100103 | 0.000249 | Thaumatin |
| AT2G45290 | 1.867839 | 0.000126 | Transketolase, putative |
| AT5G64070; AT5G09350 | 1.620917 | 0.000258 | Phosphatidylinositol 4-kinase |
| AT3G19540 | 1.583075 | 0.00039 | Unknown protein |
| AT1G01860 | 1.505633 | 0.000472 | PFC1, ribosomal RNA adenine methylase transferase |
| AT1G27980 | 0.530013 | 0.000331 | DPL1, dihydrosphingosine phosphate lyase |
| AT5G24800 | 0.524407 | 0.000204 | ATBZIP9, transcription factor |
| AT1G18270 | 0.522557 | 0.000196 | Ketose-bisphosphate aldolase/6- phosphogluconate dehydrogenase |
| AT3G45970 | 0.485587 | 0.000103 | ATEXLA1, expansin-like |
| AT1G13700 | 0.48172 | 0.000121 | Glucosamine/galactosamine-6- phosphate isomerase |
| AT3G52400 | 0.467186 | 7.67 × 105 | SYP122, syntaxin |
| AT3G62860 | 0.437221 | 0.00031 | Lipase |
| AT4G29950 | 0.394519 | 0.000183 | Microtubule-associated protein |
| AT3G10020 | 0.372676 | 8.84 × 105 | Unnamed protein |
| AT2G02710 | 0.365188 | 0.000418 | PAC motif-containing protein |
| AT2G28400 | 0.364799 | 0.00024 | Unknown protein |
| AT5G66650 | 0.323571 | 0.00012 | Unknown protein |
| AT5G51390 | 0.318702 | 0.000108 | Unknown protein |
| AT3G45730 | 0.31067 | 0.000349 | Unknown protein |
| AT2G41640 | 0.302148 | 0.000234 | Glycosyltransferase |
| AT5G49450; AT5G49448 | 0.285898 | 0.000191 | ATBZIP1, transcription activator |
| AT3G04640 | 0.26628 | 0.000295 | Glycine-rich protein |
| AT1G05575 | 0.246055 | 0.000199 | Unknown protein |
| AT1G02660 | 0.234144 | 0.00012 | Lipase class 3 |
| AT1G19530 | 0.221147 | 0.000481 | Unnamed protein |
| AT3G10930 | 0.204299 | 0.000385 | Unknown protein |
| AT5G59820 | 0.182259 | 0.000351 | RHL41, transcription factor |
| AT2G39780 | 0.025392 | 7.49 × 109 | RNS2 |
Fig. 1.
Verification of microarray results. Expression of genes identified as differentially expressed in the microarray analysis was analysed by quantitative RT-PCR (qPCR). RNA was extracted from WT and rns2-2 seedlings grown identically to the material used for transcriptome analysis. Genes selected for testing were ATEXLA1 (AT3G45970), glycosyl transferase (AT2G41640), GAPCP-1 (AT1G79530) and GAPCP-2 (AT1G16300). Results were normalized using the expression of TIP41-like (AT4G34270) as loading control and then to the average of the WT expression. The analysis was performed using four different RNA samples for each genotype. t-test. P-values are indicated above rns2-2–WT comparisons with a significant difference in expression level.
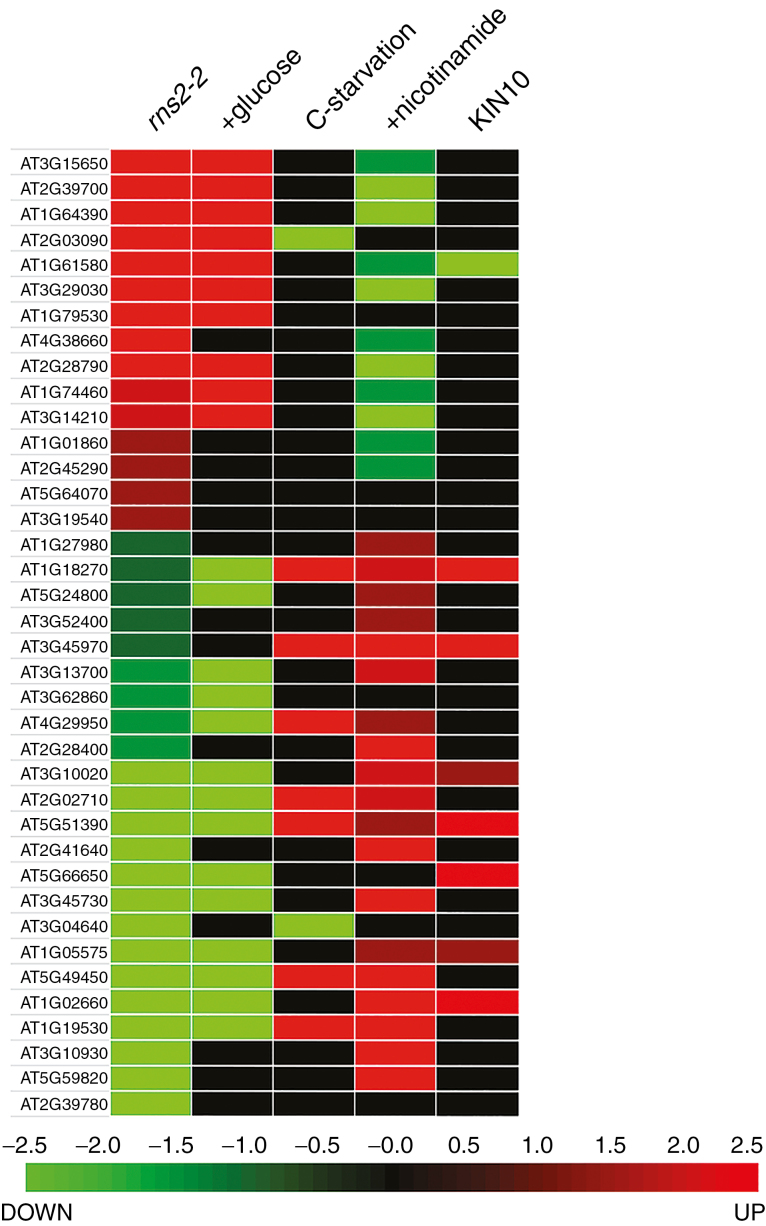
We used Genevestigator (Hruz et al., 2008) to identify stress conditions or treatments that co-regulate subsets of the rns2-2 DEGs. This analysis revealed that many DEGs in rns2-2 are also differentially regulated in plants grown under conditions that alter carbon availability (Fig. 2). Most DEGs were also regulated, in the same direction, in arabidopsis plants after transition from a sugar-restricted condition to a sugar-replete state by addition of glucose to the growth medium (Li et al., 2006). A similar correlation between differential expression in rns2-2 and response to carbon availability was observed in the data set corresponding to genes differentially expressed in response to moderate carbon depletion followed by addition of sucrose to replenish carbon availability (Osuna et al., 2007), with 18 genes regulated in the same direction by this treatment and in the rns2-2 mutant (all genes are included in the set regulated by glucose addition in Fig. 2). A number of rns2-2 DEGs were also regulated by carbon starvation (Contento et al., 2004; Nicolaï et al., 2006), particularly rns2-2-repressed genes, although in this case the regulation observed in rns2-2 is the reverse of that observed under starvation. A large proportion of DEGs are also regulated by nicotinamide treatment, with a pattern of regulation opposite to the pattern observed in rns2-2. Finally, a sub-set of rns2-2 DEGs were identified as KIN10 targets in an experiment that analysed changes in the transcriptome of protoplasts transiently expressing a KIN10 gene controlled by a constitutive promoter (Baena-Gonzalez et al., 2007). KIN10 is a protein kinase that activates gene expression and metabolic pathways in response to low cellular energy levels (Sheen, 2014).
Fig. 2.
Genes differentially expressed in the rns2-2 mutant are regulated by the energy status of the cell. The heatmap shows the regulation of the DEGs identified in the microarray analysis and regulation by carbon starvation, glucose re-feeding or nicotinamide treatments, each of which changes the energy status of the cell, obtained from public databases and the literature. Expression in protoplasts overexpressing KIN10 is also included in the comparisons.
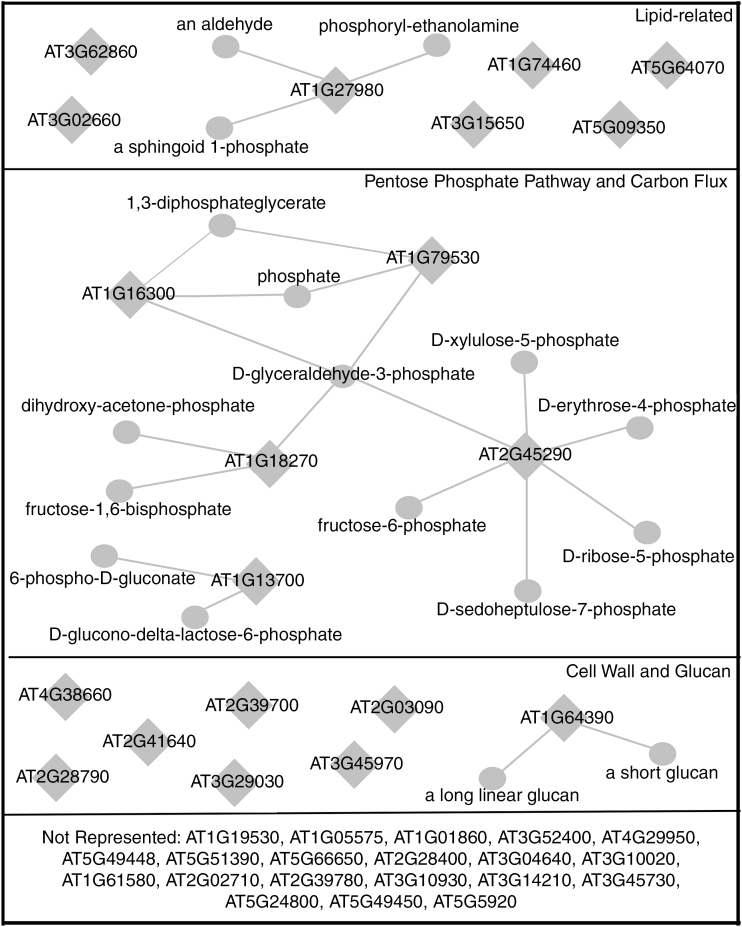
We carried out a GO enrichment analysis to identify functional categories over-represented in our data set of DEGs. We identified a significantly enriched (P > 10–8) number of genes involved in cell wall modification and in the PPP, among other terms (a full list of significant over- or under-represented GO terms is provided in Supplementary Data Table S1). Network analysis also revealed a clear node connected to the PPP and carbon flux (Fig. 3). This set of DEGs included three induced genes (the plastidial glyceraldehyde-3-phosphate dehydrogenases GAPCP-1 and GAPCP-2 and the transketolase AT2G45290) and two repressed genes (the glucosamine/galactosamine-6-phosphate isomerase family protein gene AT1G13700 and the ketose-bisphosphate aldolase class-II family protein gene AT1G18270). AT1G18270 is interesting in that the predicted protein product has both a ketose-bisphosphate aldolase domain and a 6-phosphogluconate dehydrogenase domain, potentially linking glycolysis/gluconeogenesis with the PPP. Less-organized groups of genes related to cell wall processes and lipid metabolism, signalling and transport were also identified. Cell wall-associated genes include several expansins (AT2G39700, AT3G29030 and AT2G03090) and expansin-like (AT3G45970) genes, a glycosyl hydrolase (AT1G64390) and a glycosyltransferase (AT2G41640), and two osmotin-like genes (AT2G28790 and AT4G38660). Lipid-related genes induced include a phospholipase/carboxylesterase (AT3G15650), two phosphatidylinositol 4-kinases not differentiated by the microarray probe (AT5G64070 and AT5G09350) and a GDSL-motif lipase (AT1G74460), while repressed genes are a dihydrosphingosine phosphate lyase (AT1G27980) and two lipases (AT3G62860 and AT1G02660).
Fig. 3.
Differentially expressed genes in the rns2-2 mutant suggest changes in carbon flux and cell wall modifications. Network analysis of DEGs was carried out using Virtual Plant and the Aracyc database. Diamonds represent genes, while circles represent metabolites. Not all DEGs were associated with an enriched category.
Metabolite analysis confirms changes in the pentose phosphate pathway and cell wall composition of the rns2-2 mutant
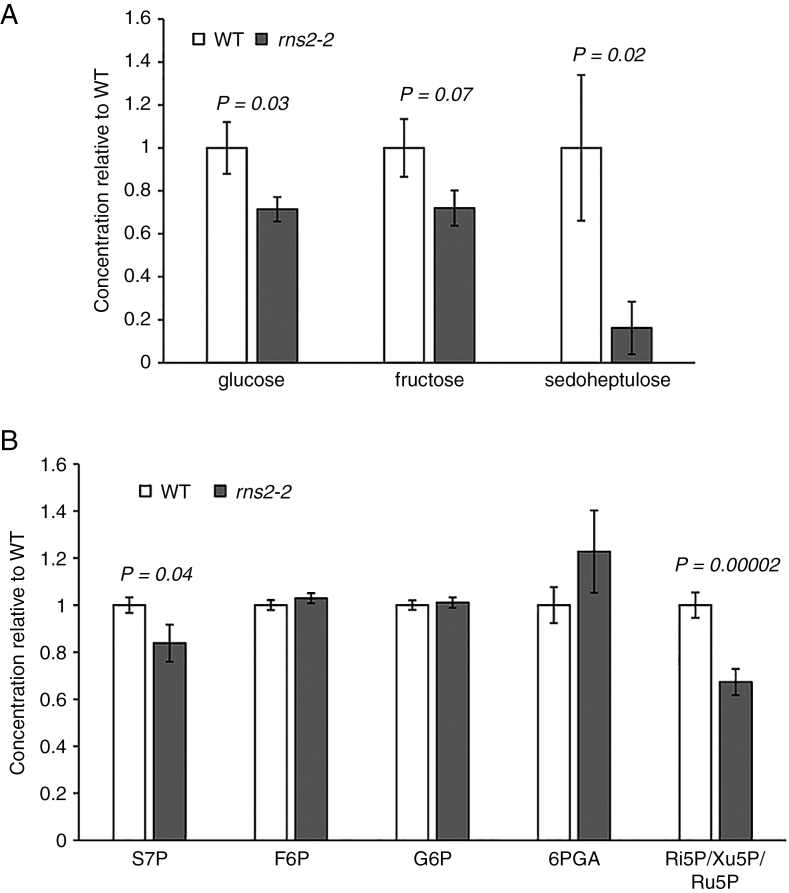
To confirm that the metabolic pathways identified by our transcriptome analysis were altered in the mutant, we performed non-targeted metabolome profiling. Seedlings grown exactly as the material used for transcriptome analysis were used to extract polar and non-polar metabolites. The resulting extracts were analysed by GC-MS. Few non-polar metabolites showed differential accumulation in the rns2-2 mutant, and none was unequivocally identified based on MS (not shown). Among the polar metabolites identified, glucose and sedoheptulose showed significantly reduced levels in the rns2-2 mutant compared with the WT (Fig. 4A). We also observed a decrease in fructose level, although it was not significant using a P < 0.05 cut-off (Fig. 4A). It is important to note that phosphate groups are lost with our analysis method; thus it is not possible to differentiate between phosphorylated and non-phosphorylated forms of these metabolites. While the changes in glucose and fructose levels could indicate general changes in carbon metabolism via flux through a number of pathways, the change in sedoheptulose levels combined with the gene expression analyses pointed to an effect of the rns2-2 mutation on the PPP. We therefore performed a direct analysis of PPP metabolites using LC-MS (Yang et al., 2008; Jannasch et al., 2011). This analysis demonstrated that the sedoheptulose-7-phosphate (S7P) level is significantly lower in the rns2-2 mutant. The LC method used did not efficiently separate ribose-5-phosphate (Ri5P), ribulose-5-phosphate (Ru5P) and xylulose-5-phosphate (Xu5P), but the combined peaks were also significantly lower in the rns2-2 mutant than in WT plants (Fig. 4B).
Fig. 4.
Metabolite analysis indicates changes in levels of sugars in the rns2-2 mutant. (A) Non-targeted metabolite analysis. Polar metabolites were extracted from WT and rns2-2 mutant seedlings grown using the same conditions applied for microarray analysis. Metabolites were separated using gas chromatography and identified by mass spectrometry. Only metabolites with significantly (t-test) different levels are shown. At least four different samples were analysed for each genotype. (B) Targeted metabolite analysis to identify changes in the PPP. Ten-day-old WT and rns2-2 seedlings were grown as in (A). Metabolites were extracted, labelled using 13C, and analysed by RPLC-MS. Significant differences (t-test) are indicated. Results are the average of four independent experiments with at least three replicates per genotype. S7P, sedoheptulose-7-phosphate; F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; 6PGA, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; Xu5P, xylulose-5-phosphate, Ri5P, ribose-5-phosphate.
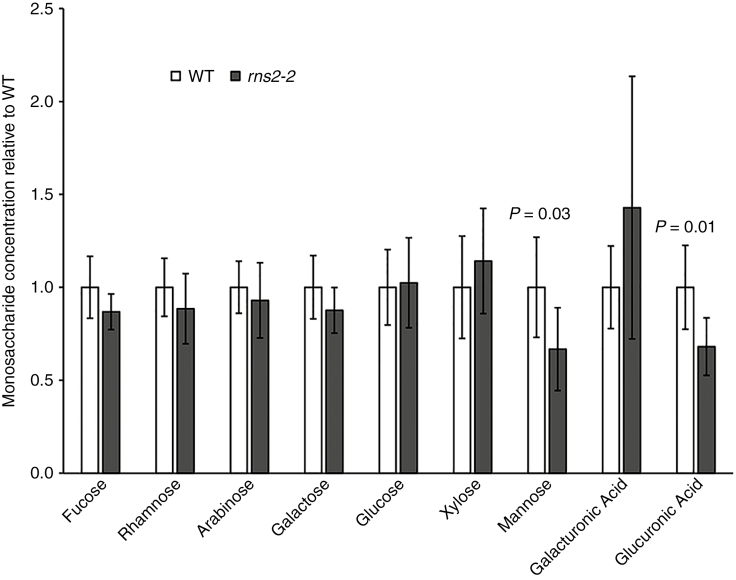
Our transcriptome data also indicated differential regulation of cell wall-modifying enzymes. Thus, we analysed the monosaccharide composition of cell walls from WT and mutant plants. Our results showed that mannose and glucuronic acid levels are significantly lower in the cell wall of rns2-2 plants (Fig. 5), suggesting that mutants have a potential defect in hemicellulose content (Scheller and Ulvskov, 2010).
Fig. 5.
Cell wall composition is affected in the rns2-2 mutant. Seedlings were grown as described in Fig. 4, and cell wall was extracted. The monosaccharide composition of WT and rns2-2 cell walls after α-amylase treatment was analysed by HPAEC-PAD. Results were obtained as mol % and then normalized to the average of the WT level for each metabolite. Significant differences (t-test) are indicated. At least six independent samples for each genotype were analysed.
The rns2-2 mutation causes a growth phenotype associated with cell length
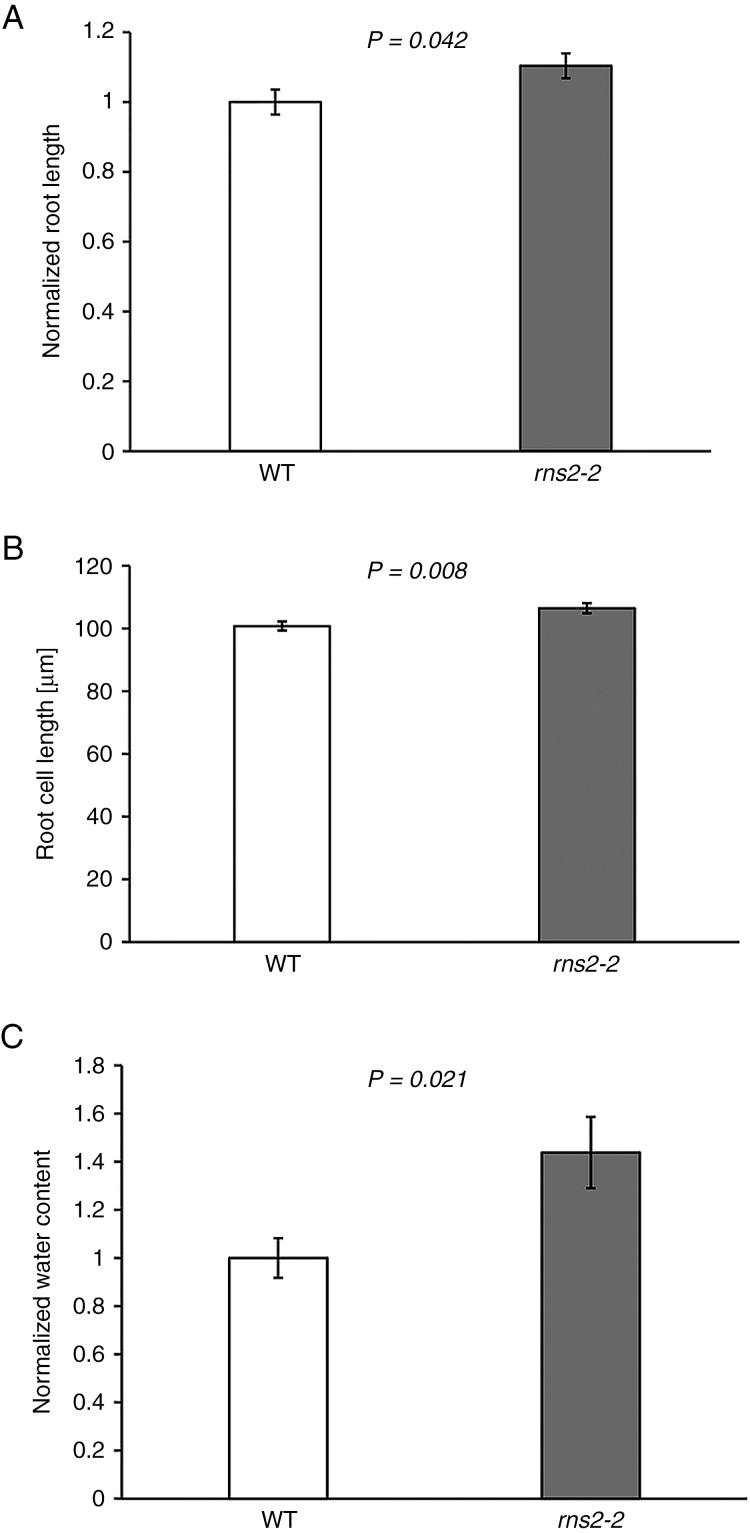
We previously observed that the rns2-2 mutant is larger than WT plants (Floyd et al., 2015). Differential expression of genes related to cell wall modification and changes in cell wall monosaccharide composition suggested weakened cell walls in the rns2-2 mutant. Thus, increased cell expansion may account for the mutant size difference. To test this hypothesis we first confirmed that rns2-2 basal rosettes are larger than WT rosettes (Supplementary Data Fig. S2). We also observed that root length is increased in mutant plants (Fig. 6A). The length of fully elongated root cells was analysed by confocal microscopy, and the root cells of the rns2-2 mutant were determined to have a small but significant increase in length relative to WT cells (Fig. 6B), with rns2-2 cells being on average 5.7 μm longer. Finally, the proportion of dry weight vs. fresh weight of mutant and WT plants was determined, and rns2-2 was found to have a higher water content than the WT (Fig. 6C).
Fig. 6.
Phenotypic characterization of rns2-2 plants. (A) Root length was measured in 10-day-old WT and rns2-2 seedlings grown on vertical agar plates. t-test P-value is indicated. Three independent plates containing the two genotypes (15 plants for each genotype per plate) were analysed. Both plant size and root length were normalized to the average of the WT values. (B) Roots were stained with propidium iodide and Oregon Green dyes, and imaged using a confocal microscope. After image capture, cell lengths were measured using ImageJ. t-test P-value is indicated. More than 500 cells were measured for each genotype. (C) Water content was determined as the weight difference of basal rosette tissue from WT and rns2-2 seedlings grown as in Fig. 4, before and after 4 d of lyophilization. The dry weight/wet weight ratio was normalized to the WT average. t-test P-value is indicated. Five independent samples for each genotype were analysed.
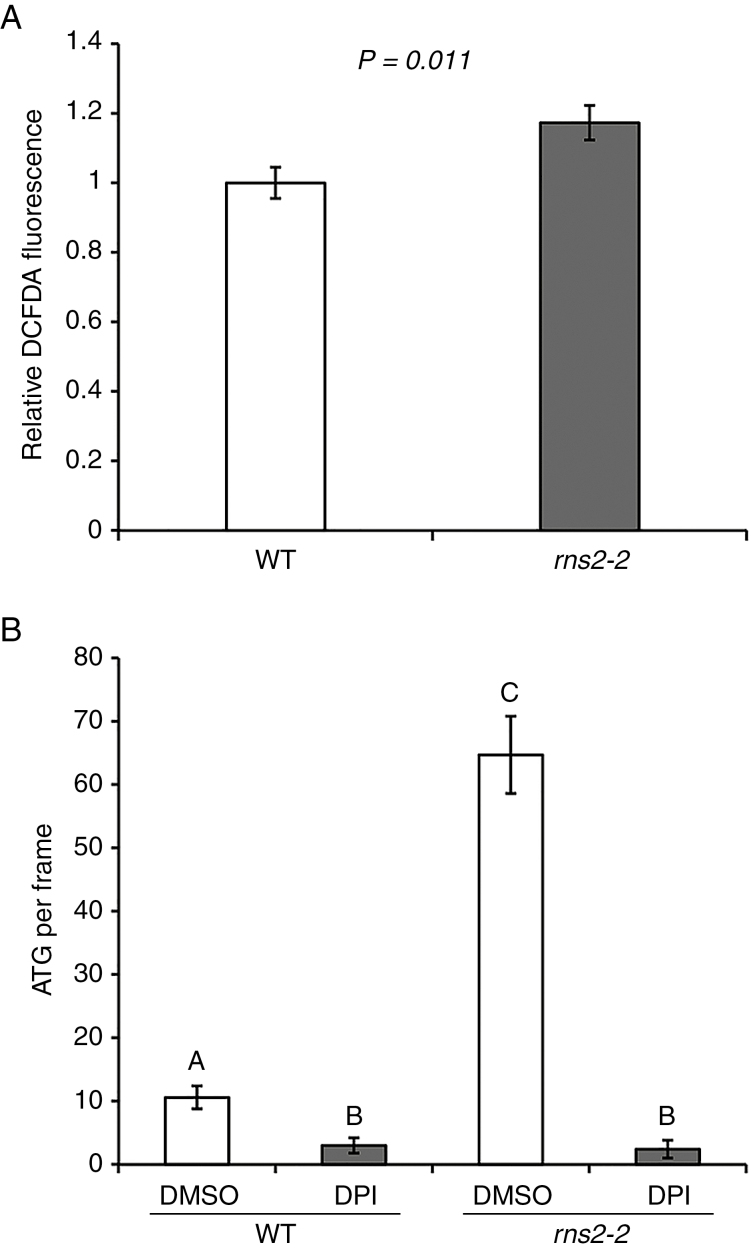
Inhibiting ROS production eliminates the rns2-2 autophagy phenotype
Our transcriptome and metabolome results led to the hypothesis that rns2-2 plants have increased production of NADPH (see the Discussion), which could lead to an increase in the production of ROS via NADPH oxidase. NADPH oxidases are conserved proteins whose enzymatic function is to provide ROS for physiological and developmental processes in plants and animals (Sagi and Fluhr, 2006). Using DCFDA staining, we determined that, indeed, levels of ROS are significantly higher in rns2-2 seedlings that in WT plants (Fig. 7A). Since ROS act as signalling molecules that trigger activation of autophagy (Liu and Bassham, 2012), we tested whether blocking ROS production using the NADPH oxidase inhibitor DPI would affect the constitutive autophagy phenotype observed in the rns2-2 mutant. We found that the constitutive autophagy phenotype of rns2-2 plants is lost upon DPI treatment, with the number of MDC-stained structures indistinguishable from that of DPI-treated WT plants (Fig. 7B; Supplementary Data Fig. S3).
Fig. 7.
ROS signalling controls autophagy in rns2-2 plants. (A) Measurement of ROS. Seven-day-old seedlings were stained with DCFDA to detect ROS. Fluorescence was measured using microscopy. and statistical comparison between WT and mutant was performed using Student’s t-test (P < 0.05). From 44 to 60 total measurements per genotype were taken from six independently grown sets. (B) Effect of DPI treatment on the rns2-2 autophagy phenotype. Roots of WT or rns2-2 seedlings were treated with DPI (NADPH oxidase inhibitor) or DMSO and then stained with MDC and imaged using fluorescence microscopy. The number of autophagosomes per frame was quantified. Statistical analysis was performed using Student’s t-test (P > 0.05) in comparisons against each treatment and genotype
DISCUSSION
Ribosome turnover is essential to maintain cellular homeostasis in arabidopsis. Mutations that inactivate the main vacuolar RNase, RNS2, lead to an increase in rRNA half-life (Hillwig et al., 2011). rns2 mutants accumulate higher levels of total RNA than WT plants, and 28S and 18S rRNAs accumulate in mutant vacuoles, indicating that rRNA recycling mainly occurs in the plant vacuole (Floyd et al., 2015). In addition to this increase in RNA accumulation, rns2 mutants have a constitutive autophagy phenotype, indicating that RNS2 function is needed to maintain normal cellular homeostasis (Hillwig et al., 2011; Floyd et al., 2015). Surprisingly, we found that this strong cellular phenotype was not accompanied by a large reprogramming of the transcriptome. A small number of genes showed differential expression in our microarray analysis, and none was part of the core autophagy machinery, even though we have shown that at least ATG5 and ATG9 are necessary for the autophagy phenotype displayed by rns2 plants (Floyd et al., 2015). Remarkably, about 20 % of the genes differentially expressed in the rns2-2 mutant have been identified as targets of KIN10 regulation, indicating that the KIN10 pathway is repressed in the mutant. KIN10 is a central integrator of energy signalling in plants (Baena-González et al., 2008), activating multiple signalling pathways as a response to low energy levels.
Consistent with the KIN10 comparison, our transcriptome analysis suggests that the rns2-2 plants are not in a nutritional deficit status, as DEGs show regulation consistent with normal levels of energy, based on the comparison with starvation or glucose/sucrose replenishing treatments. However, it is clear that carbon flux is affected in the mutants, as both expression of genes in the PPP pathway and PPP metabolites showed altered levels. The inverse correlation between regulation of DEGs in the rns2-2 mutant and regulation by nicotinamide treatments also deserves attention. Nicotinamide is a by-product of NAD degradation, and in plants it is normally salvaged to synthesize new NAD (Hunt et al., 2004; Gerdes et al., 2012). In other organisms, nicotinamide treatments cause an increase in NAD+ and NADP+ levels and decrease the redox ratio (NADH/total NAD) (Higashida et al., 1995; Benavente and Jacobson, 2008; Wilhelm and Hirrlinger, 2011), and the same effect could be expected in plants. Thus, inverse regulation by the rns2-2 mutation and nicotinamide treatments would suggest that the mutant has an elevated level of NAD(P)H, also consistent with our hypothesis that rns2-2 plants are not under energy starvation.
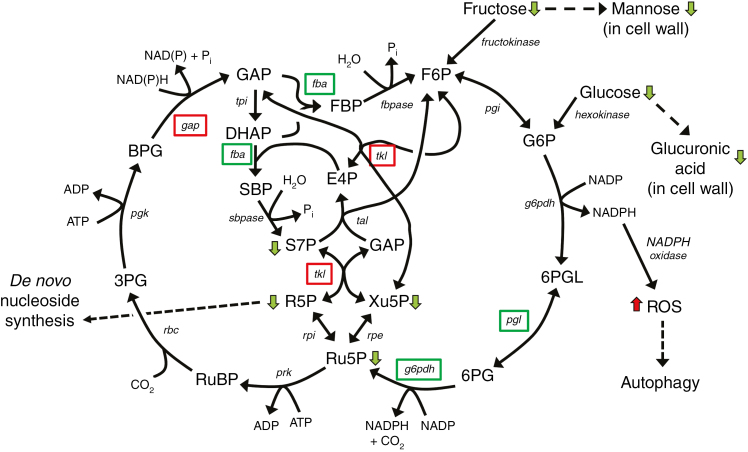
Most of the reactions catalysed by enzymes encoded by DE genes associated with carbon flux identified in our analysis are reversible, and changes in steady-state levels of metabolites are difficult to interpret because a decrease in an intermediary metabolite could equally be explained as a reduction in the activity of the pathway, or as an increase in its activity with increased consumption of intermediates. However, the indication that rns2-2 mutants are not starved, together with the PPP gene expression analysis, allows us to hypothesize a putative flux for the PPP (Fig. 8). Our model suggests that the oxidative phase of the PPP is downregulated (also suggesting that NADPH levels are high), while the non-oxidative phase is activated to channel glyceraldehyde-3-phosphate and fructose-6-phosphate towards the production of ribose-5-phosphate. The same conditions would favour production of glyceraldehyde-3-phosphate by glyceraldehyde-3-phosphate dehydrogenase, further pushing carbon flux to the generation of ribose-5-phosphate. By analogy with the mechanism of rRNA recycling described in yeast cells undergoing nitrogen starvation (Huang et al., 2015), we can expect that the plant 3ʹ-nucleoside monophosphates produced in the vacuole by RNS2 as a result of housekeeping rRNA hydrolysis are then further processed to nucleosides by an as yet uncharacterized vacuolar activity and transported to the cytoplasm for reutilization. We hypothesize that lack of rRNA recycling in the rns2-2 mutant directly impacts the nucleoside and/or nucleobase cellular pool, triggering compensatory mechanisms, including constitutive autophagy and redirection of the carbon flux for de novo nucleoside synthesis. Equilibrative Nucleoside Transporter 1 (ENT1) is the main exporter of nucleosides from the vacuole to the cytoplasm in arabidopsis (Girke et al., 2014). Transgenic plants with altered levels of this transporter show significant changes in nucleotide metabolism (Bernard et al., 2011; Girke et al., 2014), consistent with our model indicating that vacuolar RNA recycling is important to maintain nucleotide and cellular homeostasis.
Fig. 8.
Model of the effect of the rns2-2 mutation on arabidopsis metabolism. Integration of transcriptome and metabolome data suggests that carbon flux is shuttled through the PPP. Since rns2-2 plants are deficient in rRNA turnover and nucleotide salvage, the increased carbon flux through the PPP pathway may be needed to generate ribose-5-phosphate for de novo synthesis of nucleotides to maintain cellular homeostasis. Changes in metabolism also result in increased NADPH levels. This increase causes accumulation of reactive oxygen species (ROS) that in turn signal the activation of the autophagy machinery. Green boxes indicate enzymes encoded by a gene downregulated in rns2-2, and red boxes upregulated. Green and red arrows indicate metabolites with lower and higher levels in rns2-2, respectively. F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; Xu5P, xylulose-5-phosphate; GAP, glyceraldehyde-3-phosphate; E4P, erythrose-4-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; RuBP, ribulose-1,5-bisphosphate; 3PG, 3-phosphoglycerate; BPG, 1,3-bisphosphoglycerate; DHAP, dihydroxyacetone-phosphate; SBP, sedoheptulose-1,7-bisphosphate; FBP, fructose-1,6-bisphosphate; pgi, glucose phosphate isomerase; g6pdh, glucose-6-phosphate dehydrogenase; pgl, 6-phosphogluconolactonase; rpe, ribulose-5-phosphate epimerase; tal, transaldolase; tkl, transketolase; fbpase, fructose-2,6-bisphosphatase; fba, fructose-bisphosphate aldolase; rpi, ribose-5-phosphate isomerase; sbpase, sedoheptulose-1,7-bisphosphatase; tpi, triosephosphate isomerase; gap, glyceraldehyde-3-phosphate dehydrogenase; pgk, phosphoglycerate kinase; rbc, ribulose-1,5-bisphosphate carboxylase/oxygenase; prk, phosphoribulokinase.
We observed increased expression of several expansins in the rns2-2 mutant. Expansins cause loosening of the cell wall, probably by disrupting non-covalent interactions between cellulose fibrils or between cellulose and xyloglucans (hemicellulose) (Cosgrove, 2015). We also found changes in the monosaccharide composition of the cell wall in mutant plants, specifically a decrease in mannose and glucuronic acid, two common components of hemicellulose (Scheller and Ulvskov, 2010). Decreased levels of these sugars could reduce the degree of hemicellulose cross-linking and cause further loosening of the cell wall. Cell wall loosening results in a reduction in cell wall stress and turgor pressure, which is followed by increased water flow into the cell. This water influx elastically expands the wall until turgor and wall stress are restored, and as a result cells elongate (Cosgrove, 2016). Thus, reduced strength of the cell wall could explain the increased cell and plant size phenotypes observed in the rns2-2 mutant, and the increase in water content in mutant plants. The reduction in cell wall monosaccharide components could potentially also be a consequence of the alteration in carbon flux (Fig. 8). We observed reduced levels of fructose and glucose in the mutant, and these sugars are precursors of mannose and glucuronic acid, respectively (Seifert, 2004; Reboul et al., 2011). Our microarray results identified the glycosyl hydrolase ATGH9C2 gene (AT1G64390) as the top induced gene in the mutant; AT1G64390 was previously proposed to facilitate cell elongation (Markakis et al., 2012), supporting our hypothesis.
Why do rns2-2 mutants show a constitutive autophagy phenotype? It is clear that autophagy is connected to nutrient recycling and maintenance of cellular energy status (Singh and Cuervo, 2011; Liu and Bassham, 2012; Avin-Wittenberg et al., 2015). It has also been shown that autophagy contributes to starch breakdown in leaves (Wang et al., 2013). The relatively low number of genes with alterations in transcript level in the rns2-2 mutant indicates that the regulation of the autophagy phenotype is mainly achieved at the post-transcriptional level in the mutant. There is strong evidence that autophagy is, at least in part, regulated post-transcriptionally in plants, particularly through the TOR (target of rapamycin) pathway (Liu and Bassham, 2012; Xiong and Sheen, 2014). TOR is a protein kinase that phosphorylates components of the ATG1 kinase complex (Kamada et al., 2000), which in turn represses autophagy activation. TOR is therefore a negative regulator of autophagy, and decreased TOR expression leads to constitutive autophagy in arabidopsis (Liu and Bassham, 2010). In addition, the mammalian homologue of KIN10, AMPK, is a positive regulator of autophagy via post-transcriptional mechanisms, in pathways both upstream of and independent from TOR signalling (Dobrenel et al., 2016). Our microarray and metabolome results suggest, however, that the rns2-2 mutant is not in a low energy state, which should maintain TOR signalling and repress the KIN10 pathway, as also observed in our results. Thus, it is evident that the autophagy phenotype in rns2-2 is uncoupled from the signalling pathways sensing sugar or energy status.
It is possible that a mechanism sensing a decrease in the level of nucleosides or related metabolites may trigger autophagy to increase the turnover of macromolecules needed to provide carbon backbones which can be channelled through the PPP for de novo nucleoside synthesis. Alternatively, autophagy could be triggered as a side effect of changes in carbon flux. Our analysis suggests that rns2-2 plants might have high NADPH levels, which in turn could result in production of ROS. Transformation of tobacco cells with a mutant calmodulin that hyperactivates NAD kinase results in an increase in NADPH levels and a concomitant increase in ROS accumulation through the activity of NADPH oxidase (Harding et al., 1997). This indicates that NADPH oxidase is not the rate-limiting step in ROS production, and that an increase in NADPH level in the rns2-2 mutant could in fact trigger production of ROS. Consistent with this hypothesis, an increase in ROS levels was observed in rns2-2, and ROS accumulation is a known inducer of autophagy in plants (Liu and Bassham, 2012). DPI inhibition of NADPH oxidase activity in the rns2-2 mutant results in complete reversal of the constitutive autophagy phenotype, confirming the involvement of NADPH oxidase-dependent ROS in this process, and providing support for a potential link between NADPH levels and the rns2-2 phenotype. Interestingly, ROS produced by NADPH oxidase activity have also been shown to participate in cell growth, and DPI treatment inhibits root cell elongation (Foreman et al., 2003). Thus, ROS signalling could also contribute to the morphological phenotype of rns2-2 plants.
Clearly, more detailed metabolome studies combined with genetic approaches and, eventually, direct carbon flux analyses will be necessary to understand fully the molecular bases for the rns2-2 cellular and morphological phenotypes. However, our results strongly support an essential role for rRNA recycling in maintenance of cellular homeostasis and primary metabolism, probably by providing nucleosides or related metabolites for cellular function.
SUPPLEMENTARY DATA
Supplementary data are available online at https://academic.oup.com/aob and consist of the following. Table S1: GO term analysis of transcriptome data. Figure S1: representative image used for cell length analysis. Figure S2: rosette size phenotype of the rns2-2 mutant. Figure S3: effect of DPI treatment on the rns2-2 autophagy phenotype.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Zhihong Song and Dr Ann Perera from the W. M. Keck Metabolomics Research Laboratory at Iowa State University for technical assistance with GC-MS and LC-MS and identification of compounds. Andrew Severin from the Iowa State University Genome Informatics Facility provided valuable help with microarray analysis. We also thank Dr Olga Zabotina for assistance with cell wall analysis, helpful discussions and for critical reading of the manuscript. This work was supported by a grant from the United States National Science Foundation [MCB-1051818] to G.C.M. and D.C.B.
LITERATURE CITED
- Abràmoff MD, Magalhães PJ, Ram SJ. 2004. Image processing with ImageJ. Biophotonics International 11: 36–42. [Google Scholar]
- Avin-Wittenberg T, Bajdzienko K, Wittenberg G et al. . 2015. Global analysis of the role of autophagy in cellular metabolism and energy homeostasis in Arabidopsis seedlings under carbon starvation. The Plant Cell 27: 306–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. 2007. A central integrator of transcription networks in plant stress and energy signalling. Nature 448: 938–942. [DOI] [PubMed] [Google Scholar]
- Baena-González E, Rolland F, Sheen J. 2008. KIN10/11 are master regulators of the convergent stress transcriptome. In: Allen JF, Gantt E, Golbeck JH, Osmond B, eds. Photosynthesis. Energy from the sun: 14th International Congress on Photosynthesis. Dordrecht: Springer Netherlands, 1331–1337. [Google Scholar]
- Benavente CA, Jacobson EL. 2008. Niacin restriction upregulates NADPH oxidase and reactive oxygen species (ROS) in human keratinocytes. Free Radical Biology and Medicine 44: 527–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard C, Traub M, Kunz HH, Hach S, Trentmann O, Möhlmann T. 2011. Equilibrative nucleoside transporter 1 (ENT1) is critical for pollen germination and vegetative growth in Arabidopsis. Journal of Experimental Botany 62: 4627–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contento AL, Bassham DC. 2010. Increase in catalase-3 activity as a response to use of alternative catabolic substrates during sucrose starvation. Plant Physiology and Biochemistry 48: 232–238. [DOI] [PubMed] [Google Scholar]
- Contento AL, Kim S-J, Bassham DC. 2004. Transcriptome profiling of the response of Arabidopsis suspension culture cells to Suc starvation. Plant Physiology 135: 2330–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2015. Plant expansins: diversity and interactions with plant cell walls. Current Opinion in Plant Biology 25: 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ. 2016. Plant cell wall extensibility: connecting plant cell growth with cell wall structure, mechanics, and the action of wall-modifying enzymes. Journal of Experimental Botany 67: 463–476. [DOI] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Arcy P, Mallard WG. 2004. AMDIS – User Guide.US Department of Commerce, Technology Administration, National Institute of Standards and Technology (NIST), Gaithersburg, MD, USA. [Google Scholar]
- De Vylder J, Vandenbussche F, Hu Y, Philips W, Van Der Straeten D. 2012. Rosette Tracker: an open source image analysis tool for automatic quantification of genotype effects. Plant Physiology 160: 1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J et al. . 2016. TOR signaling and nutrient sensing. Annual Review of Plant Biology 67: 261–285. [DOI] [PubMed] [Google Scholar]
- Floyd BE, Morriss S, MacIntosh GC, Bassham DC. 2015. Evidence for autophagy-dependent pathways of rRNA turnover in Arabidopsis. Autophagy 11: 2199–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JHF et al. . 2003. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422: 442–446. [DOI] [PubMed] [Google Scholar]
- Gerdes S, Lerma-Ortiz C, Frelin O et al. . 2012. Plant B vitamin pathways and their compartmentation: a guide for the perplexed. Journal of Experimental Botany 63: 5379–5395. [DOI] [PubMed] [Google Scholar]
- Girke C, Daumann M, Niopek-Witz S, Möhlmann T. 2014. Nucleobase and nucleoside transport and integration into plant metabolism. Frontiers in Plant Science 5: 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding SA, Oh SH, Roberts DM. 1997. Transgenic tobacco expressing a foreign calmodulin gene shows an enhanced production of active oxygen species. EMBO Journal 16: 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV. 2006. ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiology 140: 49–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haud N, Kara F, Diekmann S et al. . 2011. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proceedings of the National Academy of Sciences, USA 108: 1099–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke M, Diekmann S, Ohlenbusch A et al. . 2009. RNASET2-deficient cystic leukoencephalopathy resembles congenital cytomegalovirus brain infection. Nature Genetics 41: 773–775. [DOI] [PubMed] [Google Scholar]
- Higashida H, Robbins J, Egorova A et al. . 1995. Nicotinamide-adenine dinucleotide regulates muscarinic receptor-coupled K+ (M) channels in rodent NG108-15 cells. Journal of Physiology 482: 317–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, Rizhsky L, Wang Y, Umanskaya A, Essner JJ, Macintosh GC. 2009. Zebrafish RNase T2 genes and the evolution of secretory ribonucleases in animals. BMC Evolutionary Biology 9: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig MS, Contento AL, Meyer A, Ebany D, Bassham DC, MacIntosh GC. 2011. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proceedings of the National Academy of Sciences, USA 108, 1093–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G et al. . 2008. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Kawamata T, Horie T et al. . 2015. Bulk RNA degradation by nitrogen starvation-induced autophagy in yeast. EMBO Journal 34: 154–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt L, Lerner F, Ziegler M. 2004. NAD – new roles in signalling and gene regulation in plants. New Phytologist 163: 31–44. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F et al. . 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Jannasch A, Sedlak M, Adamec J. 2011. Quantification of pentose phosphate pathway (PPP) metabolites by liquid chromatography–mass spectrometry (LC-MS). In: Metz OT, ed. Metabolic profiling: methods and protocols. Totowa, NJ: Humana Press, 159–171. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. Journal of Cell Biology 150: 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M. 2000. Post-genome informatics. Oxford: Oxford University Press. [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF et al. . 2010. VirtualPlant: a software platform to support systems biology research. Plant Physiology 152: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemperer HG, Pilley DJ. 1985. The breakdown of Tetrahymena ribosomes by lysosomal enzymes: inhibition by cytosol. International Journal of Biochemistry 17: 399–404. [Google Scholar]
- Kraft C, Peter M. 2008. Is the Rsp5 ubiquitin ligase involved in the regulation of ribophagy?Autophagy 4: 838–840. [DOI] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nature Cell Biology 10: 602–610. [DOI] [PubMed] [Google Scholar]
- Lardeux BR, Mortimore GE. 1987. Amino acid and hormonal control of macromolecular turnover in perfused rat liver. Evidence for selective autophagy. Journal of Biological Chemistry 262: 14514–14519. [PubMed] [Google Scholar]
- Li Y, Lee KK, Walsh S et al. . 2006. Establishing glucose- and ABA-regulated transcription networks in Arabidopsis by microarray analysis and promoter classification using a Relevance Vector Machine. Genome Research 16: 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. 2010. TOR is a negative regulator of autophagy in Arabidopsis thaliana. PLoS One 5: e11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Bassham DC. 2012. Autophagy: pathways for self-eating in plant cells. Annual Review of Plant Biology 63: 215–237. [DOI] [PubMed] [Google Scholar]
- Liu Y, Xiong Y, Bassham DC. 2009. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 5: 954–963. [DOI] [PubMed] [Google Scholar]
- Loffler A, Abel S, Jost W, Beintema JJ, Glund K. 1992. Phosphate-regulated induction of intracellular ribonucleases in cultured tomato (Lycopersicon esculentum) cells. Plant Physiology 98: 1472–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC. 2011. RNase T2 family: enzymatic properties, functional diversity, and evolution of ancient ribonucleases. In: Nicholson AWW, ed. Ribonucleases. Berlin: Springer, 89–114. [Google Scholar]
- MacIntosh GC, Bassham DC. 2011. The connection between ribophagy, autophagy and ribosomal RNA decay. Autophagy 7: 662–663. [DOI] [PubMed] [Google Scholar]
- MacIntosh GC, Bariola PA, Newbigin E, Green PJ. 2001. Characterization of Rny1, the Saccharomyces cerevisiae member of the T2 RNase family of RNases: unexpected functions for ancient enzymes?Proceedings of the National Academy of Sciences, USA 98: 1018–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markakis MN, De Cnodder T, Lewandowski M et al. . 2012. Identification of genes involved in the ACC-mediated control of root cell elongation in Arabidopsis thaliana. BMC Plant Biology 12: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaï M, Roncato MA, Canoy AS et al. . 2006. Large-scale analysis of mRNA translation states during sucrose starvation in Arabidopsis cells identifies cell proliferation and chromatin structure as targets of translational control. Plant Physiology 141: 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Bonizec M, Cohen M et al. . 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Reports 11: 548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B, Niño CA, Bengtson MH, Lee J-W, Joazeiro CAP, Dargemont C. 2014. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. Journal of Cell Biology 204: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osuna D, Usadel B, Morcuende R et al. . 2007. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. The Plant Journal 49: 463–491. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2004. Quantification strategies in real-time PCR. In: Bustin SJ, ed. AZ of quantitative PCR. La Jolla, CA, International University Line Press, 89–113. [Google Scholar]
- Pogorelko G, Fursova O, Lin M, Pyle E, Jass J, Zabotina OA. 2011. Post-synthetic modification of plant cell walls by expression of microbial hydrolases in the apoplast. Plant Molecular Biology 77: 433–445. [DOI] [PubMed] [Google Scholar]
- Reboul R, Geserick C, Pabst M et al. . 2011. Down-regulation of UDP-glucuronic acid biosynthesis leads to swollen plant cell walls and severe developmental defects associated with changes in pectic polysaccharides. Journal of Biological Chemistry 286, 39982–39992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi M, Fluhr R. 2006. Production of reactive oxygen species by plant NADPH oxidases. Plant Physiology 141, 336–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annual Review of Plant Biology 61: 263–289. [DOI] [PubMed] [Google Scholar]
- Schmidt MA, Barbazuk WB, Sandford M et al. . 2011. Silencing of soybean seed storage proteins results in a rebalanced protein composition preserving seed protein content without major collateral changes in the metabolome and transcriptome. Plant Physiology 156: 330–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ. 2004. Nucleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Current Opinion in Plant Biology 7: 277–284. [DOI] [PubMed] [Google Scholar]
- Sheen J. 2014. Master regulators in plant glucose signaling networks. Journal of Plant Biology 57: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Cuervo Ana M. 2011. Autophagy in the cellular energetic balance. Cell Metabolism 13: 495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yu B, Zhao J et al. . 2013. Autophagy contributes to leaf starch degradation. The Plant Cell 25: 1383–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends in Biochemical Sciences 24: 437–440. [DOI] [PubMed] [Google Scholar]
- Weckwerth W, Wenzel K, Fiehn O. 2004. Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics 4: 78–83. [DOI] [PubMed] [Google Scholar]
- Wettenhall JM, Simpson KM, Satterley K, Smyth GK. 2006. affylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics 22: 897–899. [DOI] [PubMed] [Google Scholar]
- Wilhelm F, Hirrlinger J. 2011. The NAD+/NADH redox state in astrocytes: independent control of the NAD+ and NADH content. Journal of Neuroscience Research 89: 1956–1964. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Sheen J. 2014. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiology 164: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WC, Sedlak M, Regnier FE, Mosier N, Ho N, Adamec J. 2008. Simultaneous quantification of metabolites involved in central carbon and energy metabolism using reversed-phase liquid chromatography–mass spectrometry and in vitro13C labeling. Analytical Chemistry 80: 9508–9516. [DOI] [PubMed] [Google Scholar]
- Zabotina O, Malm E, Drakakaki G, Bulone V, Raikhel N. 2008. Identification and preliminary characterization of a new chemical affecting glucosyltransferase activities involved in plant cell wall biosynthesis. Molecular Plant 1: 977–989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.