Abstract
Ion beams have been used as an effective tool in mutation breeding for the creation of crops with novel characteristics. Recent analyses have revealed that ion beams induce large chromosomal alterations, in addition to small mutations comprising base changes or frameshifts. In an effort to understand the potential capability of ion beams, we analyzed an Arabidopsis mutant possessing an abnormal genetic trait. The Arabidopsis mutant uvh3-2 is hypersensitive to UVB radiation when photoreactivation is unavailable. uvh3-2 plants grow normally and produce seeds by self-pollination. SSLP and CAPS analyses of F2 plants showed abnormal recombination frequency on chromosomes 2 and 3. PCR-based analysis and sequencing revealed that one-third of chromosome 3 was translocated to chromosome 2 in uvh3-2. FISH analysis using a 180 bp centromeric repeat and 45S ribosomal DNA (rDNA) as probes showed that the 45S rDNA signal was positioned away from that of the 180 bp centromeric repeat in uvh3-2, suggesting the insertion of a large chromosome fragment into the chromosome with 45S rDNA clusters. F1 plants derived from a cross between uvh3-2 and wild-type showed reduced fertility. PCR-based analysis of F2 plants suggested that reproductive cells carrying normal chromosome 2 and uvh3-2–derived chromosome 3 are unable to survive and therefore produce zygote. These results showed that ion beams could induce marked genomic alterations, and could possibly lead to the generation of novel plant species and crop strains.
Keywords: ion beams, chromosomal rearrangement, Arabidopsis, FISH, NHEJ
INTRODUCTION
Ion beams have been applied to various farm and horticultural crops for the creation of novel plant characteristics [1]. The practical effectiveness of ion beams is based on their higher biological effect compared with other mutagens, such as γ-rays or chemical agents. Ion beams deposit energy in a high linear energy transfer (LET) manner, providing the bulk of energy around the ion tracks. Therefore, ion beams are more effective at inducing lethal damage such as double-strand breaks (DSBs) compared with other reagents [2–4]. Moreover, recent studies revealed that ion beams induce clustered damage on DNA [5–7], the repair of which is difficult to achieve with cellular repair systems [8]. Consequently, ion beam radiation kills cells more efficiently than the same dose of low-LET radiation [9–11].
Decades of analysis using the model plant Arabidopsis thaliana revealed that ion beams induce large DNA alterations, in addition to small mutations comprising base changes or frameshifts [12–14]. Furthermore, employing ion beams with higher LET for the irradiation results in larger deletions within the Arabidopsis genome [15, 16]. Analyses of mutations suggested that ion beam-induced damage was mainly repaired by non-homologous end joining (NHEJ), which sometimes rejoins the incorrect DSB ends [12, 15]. Thus, ion beams have the ability to induce marked mutations comprising large chromosome alterations, such as deletions, inversions, translocations, etc., thereby resulting in the production of new mutants or new varieties of crops.
Here we describe the creation of a new ion beam–induced mutant which possesses a large chromosomal alteration with novel character. The mutant grows normally, but has reduced fertility when crossed with wild-type plant, which is probably due to abnormal chromosome pairing at meiosis and loss of a chromosomal fragment. The results obtained here support the further use of ion beam breeding for the creation of novel plant species by means of chromosome engineering techniques.
MATERIALS AND METHODS
Plant material and mutagenesis
A. thaliana ecotype Columbia was the wild-type plant used in this study. For mutagenesis, wild-type seeds were irradiated with 220-MeV carbon ions from an azimuthally varying field (AVF) cyclotron (TARRI, QST, Takasaki, Japan) at a dose of 150 Gy, as previously described [17]. Approximately 750 M1 seeds were grown and self-pollinated to obtain M2. The offspring seeds derived from each M2 were pooled to establish ~3000 M2 lines.
Isolation of UVB-sensitive lines
For the isolation of UVB-sensitive mutants, M2 lines were screened using the root-bending assay under non-photoreactivating conditions, as previously described [17]. Seven to ten seeds per each M2 line were sown on a nutritive agar plate (2% sucrose and 0.1% [v/v] commercial nutrient; Hyponex, Osaka, Japan). The plate was placed vertically in a growth chamber (LH-200-RD; NK System, Osaka, Japan) at 23°C under continuous white light from fluorescent lamps for 3 days, during which time the roots were allowed to grow along the surface of the agar. The seedlings on the plate were placed under a UVB lamp (CSL-30B; COSMO BIO, Tokyo, Japan) and irradiated with a dose of 0.5 kJ m−2. The plate was then placed vertically, rotated 90° to change the direction of gravity, and kept under dark conditions in a growth chamber (LPH-200-RDS; NK System) at 23°C for another 3 days, after which time lines with reduced root growth were selected.
Analysis of root growth rate
About twenty Columbia and suv4/uvh3-2 (hereafter, we merely call uvh3-2 in this section) seedlings were grown on nutritive agar plates under continuous white light for 3 days. The seedlings were exposed to 0.125 to 0.625 kJ m−2 (for the dark condition) or 0.5 to 2 kJ m−2 (for the light condition) of UVB, and incubated in the dark or under continuous white light (~40 μE m−2 s−1), respectively, for another 3 days. The length of root growth after UVB irradiation was measured (using NIH Image J software version 1.47 m [18]) and expressed as a percentage of the mean length of non-irradiated roots in each line.
Segregation test
Segregation of the UVB-sensitive trait was examined by χ2 test using the following equation.
where Oi is the observed frequency count and Ei is the expected frequency count.
Analysis of meiotic recombination
F2 plants from a cross between uvh3-2 and Landsberg erecta (Ler) were grown, and a leaf disc from each plant was obtained and stored for DNA preparation. Plants were allowed to self-pollinate to prepare F2 line seeds, which were screened for UVB sensitivity by the root-bending assay. DNA was extracted from the leaf disc corresponding to homozygous UVB–sensitive lines. Polymorphism of chromosomes was detected using simple sequence length polymorphism (SSLP) [19] or cleaved amplified polymorphic sequence (CAPS) [20]. The sequence and location of markers are listed in the TAIR database (http://www.arabidopsis.org/).
TAIL-PCR and analysis of chromosomal rearrangements
TAIL-PCR was performed as previously described [21] with AD primers [AD1, 5′-(A/T)GTGNAG(A/T)ANCANAGA-3; AD2, 5′-NGTCGA(G/C)(A/T)GANA(A/T)GAA-3′; AD3, 5′-GTNCGA(G/C)(A/T)CANA(A/T)GTT-3′] and SP primers listed in Supplemental Table S1. The obtained sequence was compared with entries in the TAIR 10 database (http://www.arabidopsis.org/) to detect mutations in uvh3-2.
Fluorescence in situ hybridization
Young flower buds were used for the cytogenetic analysis. Fluorescence in situ hybridization (FISH) was performed essentially as previously described [22], with some modifications. Centromeric 180 bp repeats and 45S rDNA were amplified from the genomic DNA with sets of primers (180 bp-F: 5′-GATCAAGTCATATTCGACTC-3′, 180 bp-R: GTTGTCATGTGTATGATTGA and 45S rDNA-F: 5′-CAAGCAAGCCCATTCTCCTC-3′, 45S rDNA-R: 5′-CAACTAGACCATGAAAATCC-3′). Amplified 180 bp repeats and 45S rDNA were labeled by nick translation with biotin-16-dUTP (Roche, Basel, Switzerland) and digoxigenin-11-dUTP (Roche), respectively. Streptavidin-Alexa 488 (Invitrogen, Carlsbad, CA) was used for the detection of biotin-labeled probe, and anti-digoxigenin-Rhodamine Fab fragments (Roche) were used for detection of dig-labeled probe. Slides were counter-stained using 0.2 μg/ml DAPI and observed using fluorescent microscopy (BX53, Olympus, Tokyo, Japan) with a ×100 objective (UPLSAPO ×100, Olympus). Images were captured using a CCD camera (DOC CAM U3-50S5M-C, Molecular Devices, Sunnyvale, CA) controlled with MetaVue (Molecular Devices). The distances between 45S rDNA and 180 bp signals were measured using Fiji software [23].
Analysis of fertility
uvh3-2 plants were crossed with Columbia to generate uvh3-2/+. Columbia, uvh3-2 and uvh3-2/+ plants were grown on soil and allowed to self-pollinate. Fertility was scored as previously described [24], with slight modifications. In brief, the 10–12th siliques on the main stem were dissected at about 7–10 days after pollination, and the number of normal seeds (green colour), abnormal seeds (brown or white) and aborted ovules within one side of the septum was counted under a stereomicroscope.
Analysis of chromosome segregation
In an effort to analyze chromosome segregation in the offspring of heterozygous plants, uvh3-2 plants were crossed with gl1-5 plants in a Columbia background [12]. The F1 plant was self-pollinated to produce F2 seeds. Eighty-nine F2 plants were grown on soil, and DNA was extracted. Each plant was examined to determine whether it carried gl-derived chromosome 3 (3) and/or uvh3-2–derived chromosome 2 (2′) by PCR using the primer pairs 3-Top F/3-Top R and 3-Top R/2-Top R, respectively. Plants carrying both chromosomes (3 and 2′) were further examined to determine the presence of gl-derived chromosome 2 (2) and/or a uvh3-2–derived chromosome 3 (3′) using the primer pairs 2-Top F/2-Top R and 3-Top F/3-Bot R, respectively (Supplemental Figure S1A). The sequences of the primers are shown in Supplemental Table S1.
RESULTS
Isolation of a UVB-sensitive mutant with an abnormal recombination pattern
In order to isolate UVB-sensitive mutants, A. thaliana Columbia seeds (M1) were irradiated using carbon ion beams with a dose of 150 Gy. M2 plants derived from M1 by self-pollination were screened for UVB sensitivity, which identified six mutants named suv1–6 (sensitive to UV). Subsequent analyses revealed that SUV1 encodes the catalytic subunit of DNA polymeraseζ [17], while SUV2 encodes ATR-interacting protein (ATRIP) [25]. The suv4 mutant, the focus in this report, is hypersensitive to UVB when kept in the dark after irradiation (Fig. 1A and E), and has only a slightly shorter root when kept in the light after irradiation (Fig. 1A and D). Without UVB-treatment, however, the suv4 plant grows normally and is fertile (Fig. 1B and C).
Fig. 1.
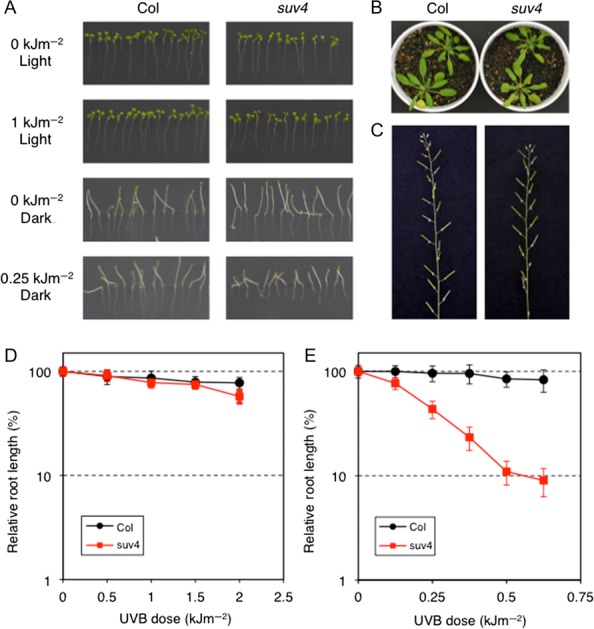
suv4 plants are hypersensitive to UVB radiation, but show no obvious phenotype under normal growth conditions. (A) Root growth inhibition of suv4 seedlings with UVB irradiation. Three-day-old seedlings were irradiated with 0 to 1 kJ m−2 of UVB and kept in the dark or under continuous white light for 3 days. The root growth of suv4 was significantly reduced by UVB when kept in the dark, but was not significantly reduced by UVB when kept under light conditions. (B, C) Plants were grown on soil at 23°C with a 16 h-light/8 h-dark photo-cycle in a growth chamber. (B) Columbia (left) and suv4 (right) plants grown at four weeks. (C) Inflorescences of Columbia (left) and suv4 (right) plants at 6 weeks. (D, E) Analysis of root growth rates after UVB irradiation. Three-day-old seedlings were irradiated with UVB and kept in the dark (D) or under continuous white light (E) for 3 days. The root growth after UVB treatment was measured using NIH Image, and expressed as a percentage of the mean length of non-irradiated root in each line. Each value is the mean ± SD of 14–20 measurements.
For mapping the location of mutations and checking the segregation of the mutant phenotype, suv4 was crossed with Landsberg erecta (Ler) by the usual method, and the resultant F2 plants were grown. A part of the leaf of each F2 plant was sampled for subsequent DNA extraction (see below), and each plant was self-pollinated to prepare a seed pool (an F2 line). When the F2 lines were examined for UVB sensitivity, they were segregated as UVB-resistant (UVR) lines and UVB-sensitive (UVS) lines (Table 1). We first hypothesized that suv4 is a single recessive mutation and expected the segregation with ratio of 3 UVR: 1 UVS. However, the ratio of UVS lines to total lines was less than 25%, and the χ2 test revealed that the observed count was significantly different from the expected count (χ2 (1, n = 283) = 8.11, p < 0.01).
Table 1.
Segregation of UVB sensitivity in F2 lines
| Phenotype | Number of lines |
|---|---|
| UVB-sensitive | 50 |
| UVB-resistant | 133 |
| Total | 283 |
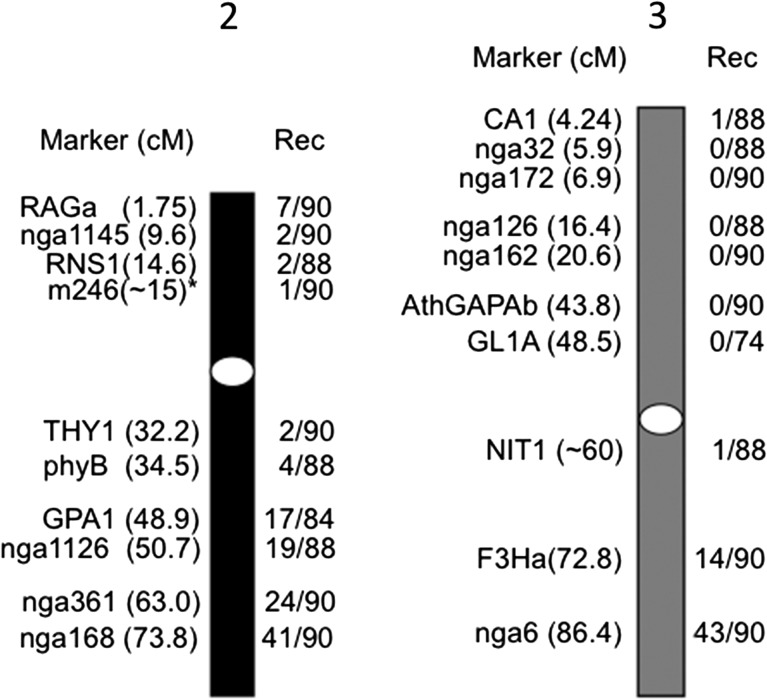
The DNA from UVS F2 plants was used to detect meiotic recombinations using SSLP and CAPS markers. In general, the recombination frequencies at sites near a given locus (for example, a recessive mutation locus) would be the lowest and as low as 0, although the frequencies must be ~0.5 at sites unlinked to the given locus. The results confirmed that the recombination frequencies in suv4 were totally abnormal (Fig. 2). In addition to a few recombinations detected by markers on chromosome 2, many markers located at the upper arm of chromosome 3 detected no recombination. From these results, we assumed a possibility that the suv4 genome comprises quite a large chromosomal rearrangement.
Fig. 2.
Abnormal recombination pattern in a cross between suv4 and Ler. The suv4 mutant was crossed with the Landsberg erecta (Ler) plant. F2 plants from this cross were screened for UVB sensitivity. The recombination events (presented by numbers of Ler genotype/numbers of analyzed chromosomes) in UVB-sensitive F2 were detected by the SSLP/CAPs method. Markers and the distance from the top (centimorgan; cM) are shown on the left side of chromosomes 2 (black bar) and 3 (gray bar), and recombination events are shown on the right side of the columns. White ovals indicate approximate positions of centromeres. *m246 is reported at 11.0 cM at TAIR, but is actually closer to the centromere than to RNS1.
Detection of chromosomal rearrangements by PCR
It is a quite common and well-known phenomenon that inversions prevent recombination [26–28]. Supporting this, we have previously observed that meiotic recombinations were prevented when a large inversion occurred, which had disrupted a gene at the endpoint(s) of the inversion to cause the mutation [12, 17, 29]. Based on the abnormal recombination frequency, we wondered whether a similar event had happened, and thus examined whether any disrupted genes were present that might be responsible for the suv4 mutation at around the top or centre of chromosome 3.
suv4 showed a UVB-hypersensitive phenotype under dark conditions much more clearly than under light conditions (Fig. 1A), which is reminiscent of mutants of ‘dark repair’ pathways such as nucleotide excision repair (NER) or base excision repair (BER). Since the A. thaliana XPG (AtXPG) gene encodes a component of NER, located at the centre of chromosome 3 [30], we hypothesized that a disruption of this gene could be responsible for the suv4 phenotype. To examine this hypothesis, we prepared five sets of primers covering the open reading frame of AtXPG. Our results showed that two of the primer pairs failed to amplify the DNA from suv4 (Fig. 3A), suggesting that the chromosome is discontinuous at a region between primers X-2F and X-3R. Therefore, we concluded that AtXPG gene is responsible for the majority of (if not all) the phenotype of suv4, and hereafter we renamed the suv4 as uvh3-2 after the reported AtXPG mutant uvh3 [30].
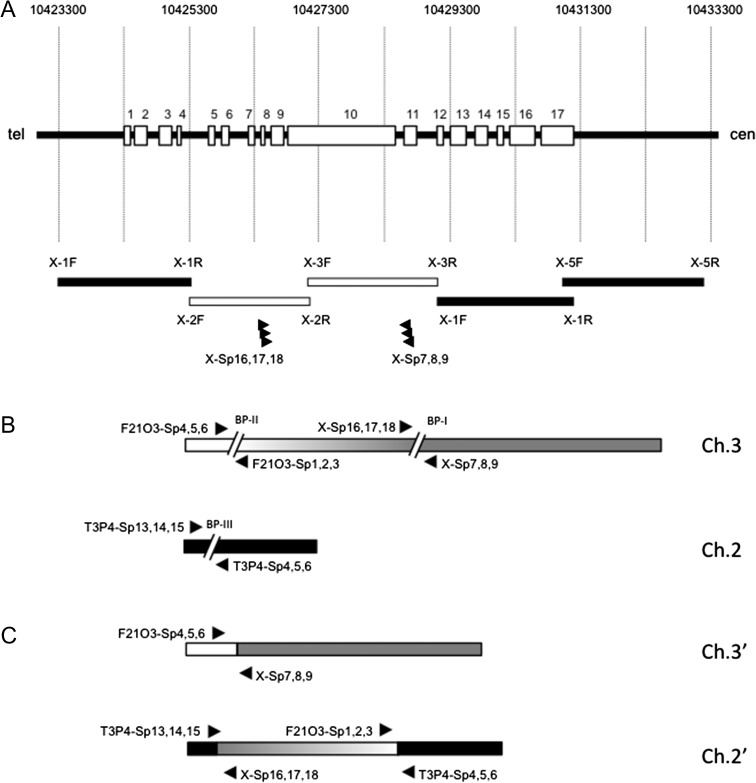
Fig. 3.
Detection of chromosomal rearrangement in uvh3-2 (suv4). (A) Schematic representation of the AtXPG gene and PCR analysis. Numbers at the top show the distances from the top of the chromosome. Open rectangles and numbers on the horizontal black bar indicate the exons of AtXPG. Black (amplified) and white (not amplified) bars at the bottom indicate the targets of PCR. Arrowheads indicate the positions and directions of TAIL-PCR primers. (B) Predicted chromosome breaks leading to rearrangements. The gray and black bars indicate wild-type chromosomes 3 and 2, respectively. Two regions shown by ‘//’ indicate predicted positions of chromosome breaks induced by ion beam irradiation. Arrowheads indicate the positions and directions of TAIL-PCR primers. (C) Rearranged chromosome 2 (2′) and chromosome 3 (3′) in uvh3-2. The ~8 Mbp fragment of chromosome 3 translocated into chromosome 2 in a reverse orientation, resulting in the two rearranged chromosomes 2′ and 3′. Arrowheads indicate the positions and directions of TAIL-PCR primers.
In an effort to identify the nucleotide changes in uvh3-2, we then tried to amplify the boundaries of the break points at the bottom of chromosome 3 by TAIL-PCR. The specific primers X-Sp7, 8 and 9 were designed based on the sequence from the 10th intron to the 11th exon of the AtXPG gene, reading from the right side of the possible break point (BP-I). The primers X-Sp16, 17 and 18 were designed based on the sequence from the 8th exon to the 8th intron, reading from the left side of BP-I. As a result, we found that the 3′-half of AtXPG is directly connected to the top of chromosome 3 (Fig. 3C, top), showing that there are two break points, at the centre (BP-I) and top (BP-II) of chromosome 3 (Fig. 3B, top). On the other hand, TAIL-PCR with X-SP16, 17 and 18 showed that the 5′-half of AtXPG is connected to the sequence present in chromosome 2, implying that there is a third break point (BP-III) on chromosome 2 (Fig. 3B, bottom). To confirm this idea, we repeated the TAIL-PCR assay using the specific primers F21O3-Sp1-6, based on both sides of predicted BP-II, and primers T3P4 –Sp4-15, based on both sides of BP-III. Taking all of the PCR results into account, it was revealed that a large fragment (~8 Mbp) of chromosome 3 was translocated to chromosome 2 with an inverted direction (Fig. 3C, bottom). Moreover, additional detailed PCR and sequencing analysis revealed that at least five break points are present within two chromosomes of uvh3-2, accompanied by a number of alterations such as deletion, insertion, duplication and inversion (Fig. 4).
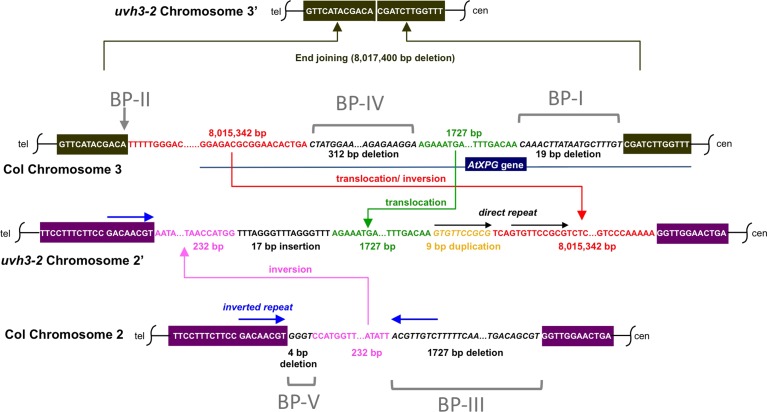
Fig. 4.
Detail of chromosomal rearrangement in uvh3-2. In the uvh3-2 mutation, at least five chromosome breaks were induced. An 8 017 400 bp fragment containing a portion of the AtXPG gene was deleted from chromosome 3′ and subdivided into 8 015 342 bp and 1727 bp fragments. The former was inversely, while the latter was directly, inserted into chromosome 2′. Chromosome 2′ also possesses another inversion, an insertion, a duplication and deletions. The 17 bp insertion on chromosome 2′ is highly repeated in the A. thaliana genome, and thus its origin could not be determined. An inverted repeat (blue arrows) on chromosome 2 could account for inversion of a 232 bp fragment (shown in magenta) in between.
Visual detection of chromosomal rearrangements
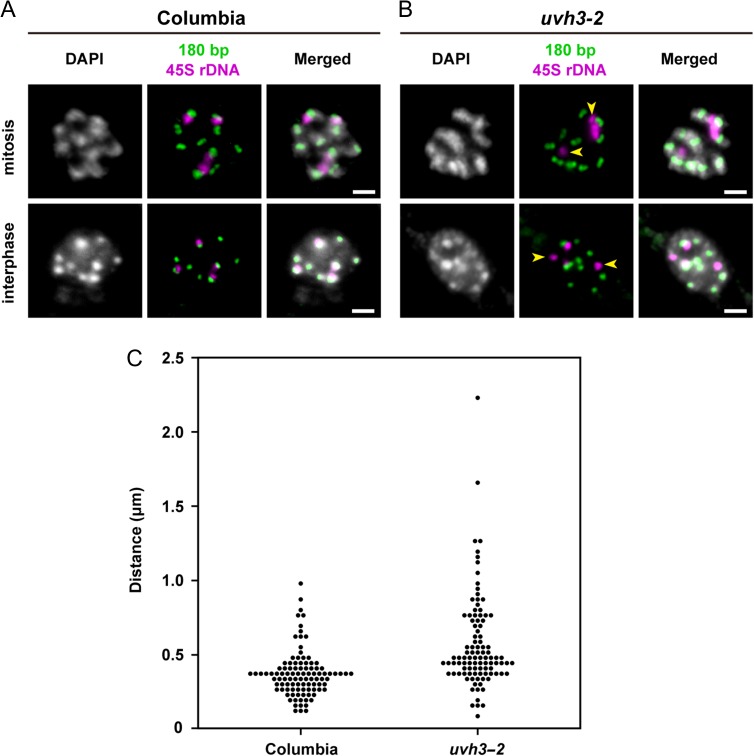
To confirm the chromosomal rearrangement in uvh3-2, we performed FISH analysis using centromeric 180 bp repeats and 45S rDNA as probes. The centromeric 180 bp repeats are present at the centromere of all chromosomes of A. thaliana and form a cluster of 2.7–3 Mb [22]. The 45S rDNA, which also forms a cluster, is present at the subtelomeric regions of the short arms of chromosomes 2 and 4 [31]. When mitotic cells in wild-type plants were hybridized with both probes, 45S rDNA signals were always detected at close proximity to those of the 180 bp repeats (Fig. 5A). By contrast, mitotic cells from uvh3-2 showed a different pattern, in that some 45S rDNA signals were detached from those of the 180 bp repeats (Fig. 5B, arrowheads). Similar patterns were observed with interphase cells of wild-type and uvh3-2 plants. To quantify the separation, the distances between both signals were measured and plotted (Fig. 5C). The results suggested that the 45S rDNA and centromeric 180 bp repeats were separated by a large chromosomal fragment in uvh3-2, which is consistent with the results of the sequencing analysis.
Fig. 5.
Detection of centromeric 180 bp repeats and 45S rDNA. (A) Columbia (B) uvh3-2. The upper and lower images show mitotic chromosomes and interphase nuclei, respectively. Arrowheads indicate 45S rDNA signals not associated with 180 bp signals. Scale bars, 2 μm. (C) Dot-density plot of distance between 45S rDNA and the nearest 180 bp repeat signals from interphase nuclei of Columbia (n = 100) and uvh3-2 (n = 100). A significant difference was detected between Columbia and uvh3-2 using Welch's t-test (P < 0.01).
Rearranged chromosomes reduced the fertility of uvh3-2/+ plants
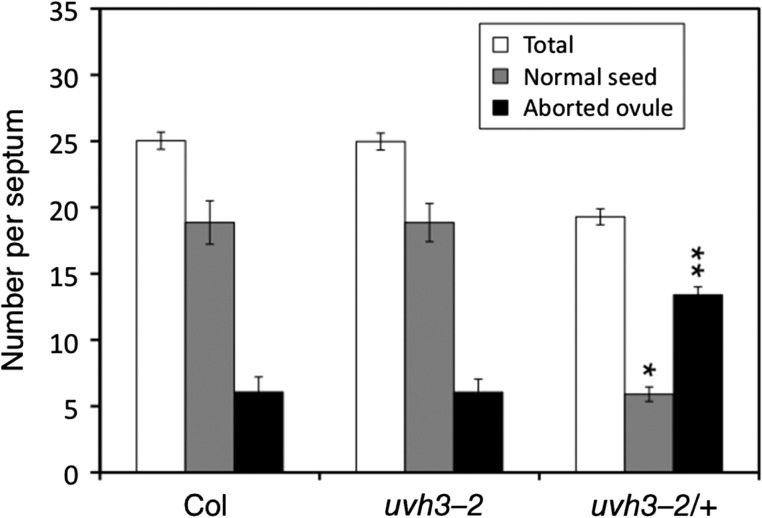
The marked changes in chromosome structures suggested that abnormal chromosomes affect chromosome segregation in meiosis. To examine this possibility, we crossed uvh3-2 plants with Columbia and obtained uvh3-2/+ seeds. Columbia, uvh3-2 and uvh3-2/+ plants were then grown on soil and self-pollinated, after which time normal seeds, abnormal seeds and aborted ovules in siliques were analyzed. In contrast to uvh3-2 siliques which look normal and are indistinguishable from those of wild-type, the siliques of uvh3-2/+ plants were slightly shorter in length (data not shown). The average number of total ovules (seeds and aborted ovules) per septum in Columbia, uvh3-2 and uvh3-2/+ siliques was 25, 25 and 19, respectively (Fig. 6). Only a few abnormal seeds, which involved chlorophyll mutation or embryonic lethality, were detected from all plants (data not shown). The number of normal seeds in Columbia and uvh3-2 was almost identical, while that in uvh3-2/+ plants was significantly lower (Fig. 6). The number of aborted ovules in uvh3-2/+ plants was significantly higher than that in Columbia or uvh3-2. These data suggested that uvh3-2/+ plants possess reduced fertility, probably due to a failure in ovule development.
Fig. 6.
Reduction of fertility in F1 plants. Siliques on Columbia (+/+), uvh3-2 (uvh3-2/uvh3-2) and the F1 (uvh3-2/+) plants were dissected, and the number of normal seeds (gray columns) and aborted ovules (black columns) on one side of the septum were counted. The total number of normal seeds, abnormal seeds and aborted ovules is shown in the white columns. The number of normal seeds in uvh3-2/+ (*) was significantly lower compared with that in Columbia and uvh3-2 by Welch's t-test (P < 0.01). The number of aborted ovules in uvh3-2/+ (**) was significantly higher compared with that in Columbia or uvh3-2 (P < 0.01). Error bar shows SEM.
To examine the reason for this reduced fertility in uvh3-2/+ plants, we analyzed chromosome segregation in the offspring of heterozygous plants. To recognize the segregation visually, we crossed uvh3-2 with gl1-5 mutant, which develops no leaf hairs, in a Columbia background. About 90 of the F2 plants derived from the cross were analyzed by phenotype and/or PCR with primers specific to normal chromosomes 2 and 3, and to uvh3-2–derived chromosomes 2′ and 3′ (Figure S1). Based on the combination of chromosomes, F2 plants were classified into five groups (Table 2), comprising gl1 type (2, 2, 3, 3), uvh3-2 type (2′, 2′, 3′, 3′), heterozygote I (2, 2′, 3, 3′), heterozygote II (2, 2′, 3, 3) and heterozygote III (2′, 2′, 3, 3′). No other types of heterozygotes, such as (2, 2, 3′, 3) for example, were detected, suggesting that reproductive cells carrying normal chromosome 2 and uvh3-2–derived chromosome 3′ were unable to survive and thereby produce zygotes. Moreover, the smaller number of heterozygotes II and III compared with heterozygote I suggested that reproductive cells carrying uvh3-2–derived chromosome 2′ and normal chromosome 3 also affected development of the zygote, probably due to abnormal crossover on meiosis.
Table 2.
Analysis of F2 plants derived from a cross between gl1-5 and uvh3-2
| Genotype (chromosome combination) | Number (ratio) |
|---|---|
| gl1 (2, 2, 3, 3) | 17 (0.19) |
| Heterozygote I (2, 2′, 3, 3′) | 42 (0.70) |
| Heterozygote II (2, 2′, 3, 3) | 10 (0.11) |
| Heterozygote III (2′, 2′, 3, 3′) | 10 (0.11) |
| uvh3-2 (2′, 2′, 3′, 3′) | 10 (0.11) |
| Total | 89 (1.00) |
DISCUSSION
Ion beams have been utilized in mutation breeding since it was empirically determined that ion beams induce novel characters more effectively than other mutagens such as γ-rays or chemicals. Using model plants such as Arabidopsis and/or rice, the mechanisms responsible for the marked biological effects of ion beams have been investigated, and the results suggested that ion beams (i) deposit the bulk of their energy in small areas, and (ii) induce DSBs or clustered damage, the repair of which is negligible by cellular repair systems. To deal with DSBs, plant cells predominantly employ NHEJ pathways, which involve the risk of rejoining the incorrect break ends. Therefore, it is predicted that ion beams induce marked mutations, including deletions, insertions, inversions, duplications and/or translocations. Supporting this idea, several mutant plants carrying an inversion [12, 17, 29] or deletion [32, 33] within a given chromosome have been reported. Moreover, cytological analysis using tobacco cultured cells has shown that abnormal chromosomes such as minichromosomes or bridge structures are induced by ion beams [34, 35], suggesting that ion beams induced aberrations involving plural chromosomes. However, it is assumed that most of the cells or plants carrying a marked change could not survive or became sterile, thereby preventing further investigations in general. In this study, we have reported for the first time detailed analysis of a marked mutation involving two chromosomes. The uvh3-2 mutant involves a translocation of an ~8 Mbp fragment from chromosome 3 to chromosome 2. This suggested that ion beam irradiation induced plural DSBs on the two chromosomes, and that the break ends were rejoined incorrectly during the repair process. Sequence analysis showed that uvh3-2 also involved several deletions and insertions, suggesting that multiple fragments were rejoined randomly. The presence of inverted repeats and duplication near the rejoined sites suggested that some DSBs were rejoined using a short homologous sequence. Furthermore, the insertion of a short filler fragment is a typical characteristic of ion beam–induced mutations [12, 13]. These results suggested that the NHEJ pathway was predominantly employed in the process of uvh3-2 mutation. The marked and complicated changes in uvh3-2 indicated that plant cells are far more adaptable than predicted.
uvh3-2 plants possessed normal fertility when self-pollinated (Fig. 6), suggesting that disruption of AtXPG hardly affected the process of reproduction under normal growth conditions. In contrast, F1 plants derived from an outcross between uvh3-2 and wild-type plants possessed reduced fertility (Fig. 6). The PCR analysis of the F2 population showed uneven segregation of chromosomes (Table 2), probably due to some chromosome combinations being lethal and prohibiting the development of gametophytes. For example, a gametophyte cell, termed gametophyte ii (see Supplemental Figure 1B), carrying a normal chromosome 2 and a uvh3-2–derived chromosome 3′, has lost a large chromosome arm containing numerous genes. Therefore, gametophyte ii seems to have been almost lethal, which accounts for the fact that zygotes carrying (2, 2, 3′, 3) were not obtained. Moreover, the translocated fragment in chromosome 2 may have induced abnormal crossovers during meiosis. If a crossover between normal chromosome 3 and chromosome 2′ occurred, this would produce a dicentric or acentric chromosome and prohibit the development of gametophytes. Thus, the number of gametophyte iii (Supplemental Figure 1B) seems to have been less than that of other gametophytes. In summary, uvh3-2/+ showed low fertility due to (i) a partial loss of chromosome fragments, and (ii) abnormal meiotic recombinations caused by translocated chromosomes.
Chromosomes with multiple inversions have been used as ‘balancer chromosomes’ in Drosophila, which are useful in screening analyses and in maintaining heterozygous mutations [36]. Similar chromosomal techniques are utilized in mammals [37] and worms [38]. Chromosome engineering techniques are becoming increasingly popular in plant breeding. For example, a chromosome carrying pericentric inversion avoids crossover with the homologous normal chromosome, thereby preventing segregation in F2 and maintaining heterozygous offspring. Additionally, due to the presence of a chromosome with inversion, plural alleles on the opposite chromosome inherit together, which is useful in maintaining a multigenic trait.
At present, a large amount of F1 hybrid crops is being cultivated in the world, which utilizes ‘hybrid vigour’ to effect high yields or useful characters. Maize breeding programs exploiting heterosis began in the USA in the 1930s, and subsequent development of a double-cross hybrid scheme has successfully produced a large amount of hybrid seed corns [39]. In China, breeding systems were established in the 1970s, and hybrid rice has occupied >50% of the total rice area since the 1990s [40]. However, the process of creating hybrids often requires pollination control, such as the use of a male sterile strain or detasseling. Maintaining heterozygotes by means of engineered chromosomes would cut costs and therefore result in widespread use in the seed market. Interchromosome translocations, found in uvh3-2, affected the independence of chromosomes 2 and 3. This suggested that insertion of a homologous sequence into an ectopic site could regulate the distribution of plural chromosomes during meiosis. Such chromosome engineering techniques should continue to develop with the help of genome editing tools such as CRISPR/CAS.
The partial sterility of the F1 derived from uvh3-2 and Columbia is reminiscent of reproductive isolation observed in the wild. Noor and colleagues suggested that chromosome inversion caused reproductive isolation between two related Drosophila species [41]. It is believed that 17 translocations and 9 inversions have occurred over 10 million years since the divergence of Brassica oleracea and A. thaliana from a common ancestor [42]. The generation of uvh3-2 may have mimicked, at least in part, the creation of a new species by evolution.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Y. Hase for support with the ion beam irradiation, and C. Suzuki for technical assistance. Part of the results of this research were presented at the 56th annual meeting of the Japanese Society of Plant Physiologists, March 2015, as ‘An ion-beam induced balancer chromosome in Arabidopsis’, by Sakamoto et al.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Radiation Research online.
CONFLICT OF INTEREST
The authors declare that there are no conflicts of interest.
FUNDING
This work was partially supported by KAKENHI (Grant number 24241028) from Japan Society for the Promotion of Science (JSPS) to A.N.S. and by KAKENHI (Grant number 15H05955 and 15H05962) from The Ministry of Education, Culture, Sports, Science and Technology (MEXT)/JSPS to S.M.
REFERENCES
- 1. Tanaka A, Shikazono N, Hase Y. Studies on biological effects of ion beams on lethality, molecular nature of mutation, mutation rate, and spectrum of mutation phenotype for mutation breeding in higher plants. J Radiat Res 2010;51:223–33. [DOI] [PubMed] [Google Scholar]
- 2. Höglund E, Blomquist E, Carlsson J et al. DNA damage induced by radiation of different linear energy transfer: initial fragmentation. Int J Radiat Biol 2000;76:539–47. [DOI] [PubMed] [Google Scholar]
- 3. Kiefer J, Egenolf R, Ikpeme S. Heavy ion-induced DNA double-strand breaks in yeast. Radiat Res 2002;157:141–8. [DOI] [PubMed] [Google Scholar]
- 4. Yokota Y, Yamada S, Hase Y et al. Initial yields of DNA double-strand breaks and DNA fragmentation patterns depend on linear energy transfer in tobacco BY-2 protoplasts irradiated with helium, carbon and neon ions. Radiat Res 2007;167:94–101. [DOI] [PubMed] [Google Scholar]
- 5. Terato H, Tanaka R, Nakaarai Y et al. Quantitative analysis of isolated and clustered DNA damage induced by gamma-rays, carbon ion beams, and iron ion beams. J Radiat Res 2008;49:133–46. [DOI] [PubMed] [Google Scholar]
- 6. Tokuyama Y, Furusawa Y, Ide H et al. Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. J Radiat Res 2015;56:446–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiraishi I, Shikazono N, Suzuki M et al. Efficiency of radiation-induced base lesion excision and the order of enzymatic treatment. Int J Radiat Biol 2017;93:293–302. [DOI] [PubMed] [Google Scholar]
- 8. Eccles LJ, O'Neill P, Lomax ME. Delayed repair of radiation induced clustered DNA damage: friend or foe? Mutat Res 2011;711:134–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tanaka A, Shikazono N, Yokota Y et al. Effects of heavy ions on the germination and survival of Arabidopsis thaliana. Int J Radiat Biol 1997;72:121–7. [DOI] [PubMed] [Google Scholar]
- 10. Shikazono N, Tanaka A, Kitayama S et al. LET dependence of lethality in Arabidopsis thaliana irradiated by heavy ions. Radiat Environ Biophys 2002;41:159–62. [DOI] [PubMed] [Google Scholar]
- 11. Yokota Y, Hase Y, Shikazono N et al. LET dependence of lethality of carbon ion irradiation to single tobacco cells. Int J Radiat Biol 2003;79:681–5. [DOI] [PubMed] [Google Scholar]
- 12. Shikazono N, Suzuki C, Kitamura S et al. Analysis of mutations induced by carbon ions in Arabidopsis thaliana. J Exp Bot 2005;56:587–96. [DOI] [PubMed] [Google Scholar]
- 13. Yoshihara R, Hase Y, Sato R et al. Mutational effects of different LET radiations in rpsL transgenic Arabidopsis. Int J Radiat Biol 2010;86:125–31. [DOI] [PubMed] [Google Scholar]
- 14. Yoshihara R, Nozawa S, Hase Y et al. Mutational effects of γ-rays and carbon ion beams in Arabidopsis seedlings. J Radiat Res 2013;54: 1050–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hase Y, Yoshihara R, Nozawa S et al. Mutagenic effects of carbon ions near the range end in plants. Mutat Res 2012;731:41–7. [DOI] [PubMed] [Google Scholar]
- 16. Hase Y, Nozawa S, Narumi I et al. Effects of ion beam irradiation on size of mutant sector and genetic damage in Arabidopsis. Nucl Instrum Methods Phys Res B 2016;391:14–9. [Google Scholar]
- 17. Sakamoto A, Vo LTT, Hase Y et al. Disruption of the AtREV3 gene causes hypersensitivity to ultraviolet B light and γ-rays in Arabidopsis: implication of the presence of a translesion synthesis mechanism in plants. Plant Cell 2003;15:2042–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 2012;9:671–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bell CJ, Ecker JR. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 1994;19:137–44. [DOI] [PubMed] [Google Scholar]
- 20. Konieczny A, Ausubel FM. A procedure for mapping Arabidopsis mutations using co-dominant ecotype specific PCR-based markers. Plant J 1993;4:403–10. [DOI] [PubMed] [Google Scholar]
- 21. Liu YG, Mitsukawa N, Oosumi T et al. Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J 1995;8:457–63. [DOI] [PubMed] [Google Scholar]
- 22. Murata M, Yokota E, Shibata F et al. Functional analysis of the Arabidopsis centromere by T-DNA insertion-induced centromere breakage. Proc Natl Acad Sci U S A 2008;105:7511–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schindelin J, Arganda-Carreras I, Frise E et al. Fiji: an open-source platform for biological-image analysis, Nat Methods 2012;9:676–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shikazono N, Yokota Y, Kitamura S et al. Mutation rate and novel tt mutants of Arabidopsis thaliana induced by carbon ions. Genetics 2003;163:1449–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sakamoto AN, Vo TTL, Puripunyavanich V et al. A UVB-hypersensitive mutant in Arabidopsis thaliana is defective in the DNA damage response. Plant J 2009;60:509–17. [DOI] [PubMed] [Google Scholar]
- 26. Dobzhansky T, Epling C. The suppression of crossing over in inversion heterozygotes of Drosophila pseudoobscura. Proc Natl Acad Sci U S A 1948;34:137–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schulz-Schaeffer J. Cytogenetics: Plants, Animals, Humans. New York: Springer-Verlag, 1980. [Google Scholar]
- 28. Griffiths AJF, Miller JH, Suzuki DT et al. An Introduction to Genetic Analysis. 8th edn New York: W. H. Freeman & Co, 2004. [Google Scholar]
- 29. Kitamura S, Shikazono N, Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J 2004;37:104–14. [DOI] [PubMed] [Google Scholar]
- 30. Liu Z, Hall JD, Mount DW. Arabidopsis UVH3 gene is a homolog of the Saccharomyces cerevisiae RAD2 and human XPG DNA repair genes. Plant J 2001;26:329–38. [DOI] [PubMed] [Google Scholar]
- 31. Murata M, Heslop-Harrison JS, Motoyoshi F. Physical mapping of the 5S ribosomal RNA genes in Arabidopsis thaliana by multi-color fluorescence in situ hybridization with cosmid clones. Plant J 1997;12:31–7. [DOI] [PubMed] [Google Scholar]
- 32. Rahman A, Nakasone A, Chhun T et al. A small acidic protein 1 (SMAP1) mediates responses of the Arabidopsis root to the synthetic auxin 2,4-dichlorophenoxyacetic acid. Plant J 2006;47:788–801. [DOI] [PubMed] [Google Scholar]
- 33. Takano N, Takahashi Y, Yamamoto M et al. Isolation of a novel UVB-tolerant rice mutant obtained by exposure to carbon-ion beams. J Radiat Res 2013;54:637–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hase Y, Shimono K, Inoue M et al. Biological effects of ion beams in Nicotiana tabacum L. Radiat Environ Biophys 1999;38:111–5. [DOI] [PubMed] [Google Scholar]
- 35. Hase Y, Yamaguchi M, Inoue M et al. Reduction of survival and induction of chromosome aberrations in tobacco irradiated by carbon ions with different linear energy transfers. Int J Radiat Biol 2002;78:799–806. [DOI] [PubMed] [Google Scholar]
- 36. Muller HJ. Genetic variability, twin hybrids and constant hybrids, in a case of balanced lethal factors. Genetics 1918;3:422–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hentges KE, Justice MJ. Checks and balancers: balancer chromosomes to facilitate genome annotation. Trends Genet 2004;20:252–9. [DOI] [PubMed] [Google Scholar]
- 38. Edgley ML, Baillie DL, Riddle DL et al. Genetic balancers, In: The C. elegans Research Community (ed.). WormBook (doi/10.1895/wormbook.1.89.1), 2006. http://www.wormbook.org (30 March 2017, date last accessed). [DOI] [PMC free article] [PubMed]
- 39. Schnable PS, Springer NM. Progress toward understanding heterosis in crop plants. Annu Rev Plant Biol 2013;64:71–88. [DOI] [PubMed] [Google Scholar]
- 40. Cheng SH, Zhuang JY, Fan YY et al. Progress in research and development on hybrid rice: a super-domesticate in China. Ann Bot 2007;100:959–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Noor MA, Grams KL, Bertucci LA et al. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A 2001;98:12084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kowalski SP, Lan TH, Feldmann KA et al. Comparative mapping of Arabidopsis thaliana and Brassica oleracea chromosomes reveals islands of conserved organization. Genetics 1994;138:499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.