Abstract
Daptomycin is an acidic, 13-amino acid, cyclic polypeptide that contains a number of non-proteinogenic residues and is modified at its N-terminus with a decanoyl chain. It has been in clinical use since 2003 against selected drug-resistant Staphylococcus aureus and Enterococcus spp infections. In vitro, daptomycin is active against Gram-positive pathogens at low concentrations but its antibiotic activity depends critically on the presence of calcium ions. This dependence has been thought to arise from binding of one or two Ca2+ ions to daptomycin as a required step in its interaction with the bacterial membrane. Here, we investigated the interaction of daptomycin with giant unilamellar vesicles (GUVs) composed 1-palmitoyl-2-oleoylphosphatidylcholine (POPC) and 1-palmitoyl-2-oleoylphosphatidylglycerol (POPG). We used fluorescence confocal microscopy to monitor binding of the peptide to GUVs and follow its effect on the membrane of the vesicle. We found that, in the absence of POPG or Ca2+, daptomycin does not bind measurably to the lipid membrane. In the presence of 20–30% PG in the membrane and 2 mM Ca2+, daptomycin induces the formation of membrane domains rich in acidic lipids. This effect is not induced by Ca2+ alone. In addition, daptomycin causes GUV collapse, but it does not translocate across the membrane to the inside of intact POPC:POPG vesicles. We conclude that pore formation is probably not the mechanism by which the peptide functions. On the other hand, we found that daptomycin coclusters with the anionic phospholipid POPG and the fluorescent probes used, leading to extensive formation of daptomycin–POPG domains in the membrane.
Graphical abstract
INTRODUCTION
Actinobacteria of the genus Streptomyces are a rich source of bioactive secondary metabolites. Roughly 50% of all natural antibiotics discovered in the last half-century have been isolated from streptomycetes (1). Only a few of these antibiotics have found their way into the clinic, but daptomycin, a lipopeptide produced by the soil bacterium Streptomyces roseosporus, has been in clinical use since 2003 against selected drug-resistant Staphylococcus aureus and Enterococcus spp infections. Daptomycin is an acidic, 13-amino acid (1.6 kDa), cyclic polypeptide (Figure 1A). It contains several non-proteinogenic residues, among which is kynurenine (Kyn), a highly fluorescent amino acid that accepts intramolecular Förster resonance energy transfer (FRET) from daptomycin’s tryptophan. The ten C-terminal residues are cyclized by the formation of an ester bond, between the alcohol group of threonine and kynurenine, which leads to the classification of daptomycin as a depsipeptide (2). The N-terminal tryptophan is modified with a decanoyl chain through an amide bond to its free nitrogen. Bacterial resistance against daptomycin exists but is not widespread, presumably because its principal target is the bacterial lipid membrane, rather than a protein receptor, and because of the recent introduction and limited use of daptomycin.
Figure 1.
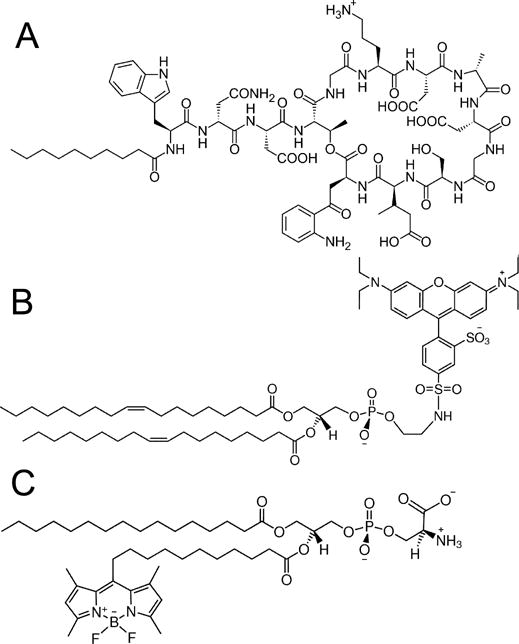
(A) Daptomycin (Source:Public Domain, Wikimedia Commons, https://en.wikipedia.org/wiki/Daptomycin), (B) Lissamine Rhodamine-DOPE (LRh-DOPE), (C) TopFluor-PS.
In vitro, daptomycin is active against a series of Gram–positive pathogens at low concentrations (≤ 1μg/mL−1, or nM–μM range) (2) but its antibiotic activity depends critically on the presence of calcium ions. This dependence is thought to arise from the binding of one or two Ca2+ ions to daptomycin as a required step in its interaction with the bacterial membrane (3–5). Binding of daptomycin occurs preferentially to anionic lipids, such as phosphatidylglycerol (PG) and other PG-derived lipids found in bacterial membranes. By contrast, binding to neutral phosphatidylcholine (PC) bilayers is still a matter of debate. Some researchers, including us, find very weak or undetectable binding to PC even in the presence of Ca2+ (6–8), while others detect binding, most often but not always in the presence of very high (unphysiological) Ca2+ concentrations (9–14).
In model membrane systems, daptomycin allows the flux of small cations (K+, Na+, and other alkali ions) across the bilayer, but not larger molecules (calcein, choline, hexamethonium) (6, 14–16). Daptomycin (fluorescently labeled on the peptide ring or the lipid moiety) has been reported to form small oligomers after associating with the lipid bilayer (5, 11, 12, 17, 18), and these oligomers have been implicated in the activity of daptomycin. In giant unilamellar vesicles (GUVs) doped with a rhodamine-labeled lipid, much larger clusters of bodipy-labeled daptomycin are detected that appear to colocalize with the labeled lipids (14). In S. aureus, addition of daptomycin leads to membrane depolarization (15). As well, fluorescently-labeled daptomycin has been shown to form clusters in bacterial cells (19–22).
Several questions remain regarding the interaction of daptomycin with the lipid bilayer. Here, we sought to address these questions in GUV preparations using the natural form of daptomycin, which eliminates possible artifacts arising from derivatization. We found that daptomycin binds well to PC:PG mixed membranes in the presence of Ca2+. But binding to pure PC membranes (in the presence of Ca2+) and binding to PC:PG membranes in the absence of Ca2+ could not be detected. Most striking, natural daptomycin forms extensive clusters on the membrane in the presence of PG, and it quantitatively colocalizes with anionic lipids. Finally, we found that daptomycin does not translocate across the lipid bilayer to any significant extent.
MATERIALS AND METHODS
Materials
Daptomycin was obtained from Cubist Pharmaceuticals, Inc. (Lexington, Ma). The chemical structure of daptomycin is shown in Figure 1A. The phospholipids and lipid fluorophores were purchased from Avanti Polar Lipids (Albaster, Al): 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-[1-rac-glycerol] (POPG), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[lissamine rhodamine B sulfonyl] (LRh-DOPE, see Figure 1B), and 1-palmitoyl-2-(dipyrrometheneboron difluoride)undecanoyl-sn-glycero-3-phospho-L-serine (TopFluor-PS, see Figure 1C). Carboxyfluorescein was purchased from Molecular Probes/Invitrogen (Carlsbad, Ca). Sucrose and calcium chloride (CaCl2) were purchased from Fisher Scientific (Pittsburg, Pa), and D(+)-glucose was purchased from ACROS Organics (NJ).
Preparation of Giant Unilamellar Vesicles
Giant unilamellar vesicles (GUVs) composed of POPC:POPG with molar ratios of 80:20 or 70:30, containing 0.1 mol % LRh-DOPE or 1 mol % TopFluor-PS were prepared by electroformation (23, 24), as previously described (25, 26), with some modifications. Aliquots of 7.5 μL of 1 mg/mL lipid were added to each electrode. The electrodes were then placed under vacuum for a minimum of 2 hours. Electroformation of the GUVs was performed in 0.1 M sucrose, for 2 hours at 2.4 V and 10 Hz, followed 30 minutes at 2.4–2.8 V and 2 Hz.
Fluorescence Confocal Experiments with LRh-DOPE and Carboxyfluorescein
The GUVs were viewed in a solution containing 0.1 M glucose, 2 μM daptomycin, 50 μM carboxyfluorescein, and various concentrations of CaCl2 (0.5, 2, and 20 mM). The samples we prepared by placing 200 μL of 0.115 M glucose solution containing the desired CaCl2 concentration in the center of a culture dish bounded by an O-ring. Then, 20 μL of a 22 μM solution of daptomycin in 1:1 (v/v) ethanol:water were added to the culture dish. The vesicles were added 5–10 minutes after the daptomycin. The images of the GUVs were taken as a function of time using a Fluoview FV 1000 confocal microscope. A laser diode was used to excite daptomycin at 405 nm. The kynurenine residue of daptomycin has an absorption maximum at about 365 nm (11), which is not suitable for confocal microscopy. However, daptomycin also has a lower but sufficient absorbance at 405 nm, which was used in these experiments. An Ar laser was used to excite TopFluor or carboxyfluorescein at 488 nm, and a He-Ne laser was used to excite rhodamine at 543 nm. Light from these sources was reflected by a dichroic mirror (DM 405/488/543). The photomultipliers were set to optimize the dynamic range of the response from the outer membrane without saturating it. For experiments in the absence of CaCl2, the glucose and sucrose solutions were run through a chelex column to eliminate trace amounts of Ca2+. All confocal microscopy experiments were performed for a minimum of 30 minutes.
Analysis of TopFluor-PS and Daptomycin Domains
The intensity of the daptomycin and TopFluor-PS fluorescence in the membranes of GUVs was measured using the Oval Profile plugin of the graphics software ImageJ (27). The analysis was performed on 16-bit grayscale images of the GUVs, after adjusting the maximum brightness to white on the brightest pixel of the image, and clearing the vesicle lumen for inner vesicles or other fluorescent spots. An illustration of this process is shown in Figure 2. The grayscale intensity was converted to green for TopFluor-PS and cyan for daptomycin, for the purpose of display. An oval enclosing the GUV was drawn (not shown in the figure). The radial sum function of Oval Profile was then used to add up the fluorescent pixels on the radial segment drawn from the center of the vesicle to the oval enclosing the vesicle as the end point of this segment moves along the oval. To use this function, the interior and the immediate exterior of the GUV must be cleared of any fluorescence associated with membrane debris or inner vesicles. The GUV images shown in the paper have been cleared in this manner. Images of the unaltered GUVs used in Figures 7 and 8 are shown in Figure S1. Because the fluorescence is only in the GUV membrane, the result is the intensity profile along the membrane, clockwise, from the right, as indicated by the arrow in Figure 2A. The correlation R between the fluorescence intensities of daptomycin (fD) and TopFluor (fT) was calculated according to Pearson’s r (28), as
| (1) |
where ⟨fD⟩ and ⟨fT⟩ are the averages of the daptomycin and TopFluor fluorescence intensities. In the example of Figure 2, the correlation is R = 0.609.
Figure 2.
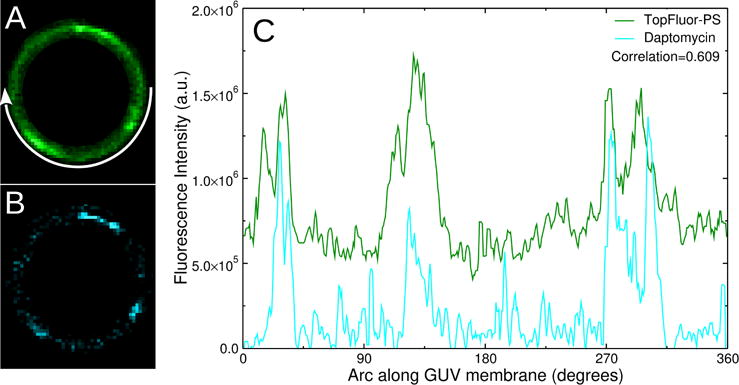
TopFluor-PS and daptomycin colocalize domains in a giant vesicle of POPC:POPG 80:20 at 2 mM Ca2+. (A) TopFluor-PS fluorescence; (B) Daptomycin fluorescence. (C) Fluorescence intensities along the circumference of the vesicle. As indicated by the arrow in A, the zero is at 3 o’clock and the angle moves clockwise.
RESULTS
Daptomycin binds to POPC:POPG GUVs in the presence of Ca2+ ions
We examined binding of daptomycin to GUVs of POPC and POPG (POPC:POPG 80:20 and 70:30) in solutions containing 2 μM daptomycin, 0.1 M glucose, and 50 μM carboxyfluorescein, in the presence and in the absence of Ca2+ ions (Figure 3). The water-soluble, membrane-impermeant dye carboxyfluorescein was added to the outside (green fluorescence in Figure 3A) and the lipid membrane was visualized with rhodamine-labeled DOPE (0.1 mol% LRh-DOPE), responsible for the red fluorescence in Figure 3A. The presence of daptomycin on the membranes of the GUVs is evident by the fluorescence of its kynurenine residue in Figure 3B (cyan). Similar daptomycin binding was observed to POPC:POPG 80:20 vesicles in the presence of 0.5 or 20 mM Ca2+. In these experiments, daptomycin was added to the solution 10 minutes before adding the vesicles. Daptomycin binding to the membranes was typically observed within 5 minutes after adding the vesicles. Note that no dye flux into the vesicles is observed in Figure 3A. There was no discernible difference between the daptomycin fluorescence observed on the membranes of POPC:POPG 70:30 and 80:20 vesicles. However, in the absence of Ca2+, no daptomycin binding was observed to vesicles of POPC:POPG 70:30 or 80:20. Finally, no daptomycin binding was observed to pure POPC vesicles (with or without lipid fluorophores) even in the presence of 2 or 20 mM Ca2+.
Figure 3.
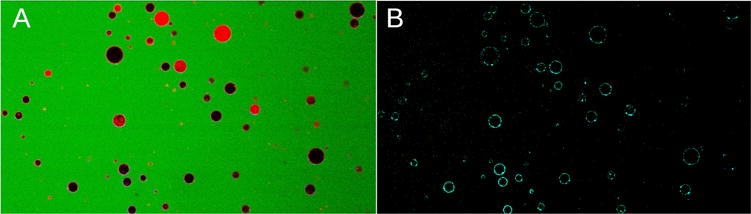
Binding of daptomycin (2 μM) to POPC:POPG 80:20, containing 0.1 mol% LRh-DOPE vesicles (GUVs) in the presence of 2 mM Ca2+. A, All channels: daptomycin (cyan), carboxyfluorescein (green), and LRh-DOPE (red). B, Daptomycin channel only (cyan). Eleven trials were performed on vesicles composed of POPC:POPG 80:20 or 70:30 POPC:POPG containing 0.1 mol% LRh-DOPE in the presence of daptomycin and either in the absence or presence of Ca2+. Most solid red vesicles are multilamellar vesicles (or are out of the focal plane), which were excluded from the analysis. But some are GUVs with large inner vesicles, which were included.
Daptomycin causes dye influx but does not cross the GUV membrane
For the sake of clarity, we begin by defining a few terms. In each lipid preparation, some of the GUVs contain smaller, inner vesicles. We refer to the larger GUVs, which may contain inner vesicles, as “parent GUVs.” The parent GUV is delimited by the outer membrane. Table 1 summarizes the results for influx and translocation in seven independent GUV preparations. In each experiment, we recorded the total number of parent GUVs (with or without inner vesicles); the number of parent GUVs with dye influx across their outer membrane (vesicles with partial or complete influx are counted into the same column); the number of parent GUVs that contained inner vesicles; and the number of parent GUVs that contained inner vesicles and showed dye influx across their outer membranes. The total number of inner vesicles, and the number of inner vesicles contained in parent GUVs that exhibited carboxyfluorescein influx were also recorded (Table 1).
Table 1.
Daptomycin translocation and dye influx in POPC:POPG GUVs.
| Parent GUVs (outer membranes) | Inner Vesicles | |||||||
|---|---|---|---|---|---|---|---|---|
| Lipid PC:PG |
[Ca2+] (mM) |
Obs. time (min) | Total | With influx | With inner vesicles | With inner and influx vesicles | Total | Present in parent GUVs with influx |
| In the presence of 2 μM daptomycin | ||||||||
| 70:30 | 2 | 40 | 26 | 1 | 5 | 0 | 12 | 0 |
| 70:30 | 20 | 40 | 37 | 23 | 0 | 0 | 0 | 0 |
| 70:30 | 2 | 30 | 120 | 28 | 8 | 1 | 12 | 3 |
| 80:20 | 2 | 30 | 134 | 10 | 24 | 4 | 51 | 15 |
| 80:20 | 20 | 120 | 259 | 114 | 3 | 3 | 7 | 7 |
| 80:20 | 20 | 60 | 141 | 35 | 11 | 10 | 19 | 18 |
| 80:20 | 20 | 60 | 146 | 15 | 5 | 2 | 11 | 6 |
| total (all trials) | 863 | 226 | 56 | 20 | 112 | 49 | ||
| In the absence of daptomycin | ||||||||
| 70:30 | 20 | 90 | 14 | 0 | 3 | 0 | – | – |
| 80:20 | 20 | 40 | 210 | 21 | 18 | 2 | – | – |
| 80:20 | 20 | 40 | 108 | 4 | 5 | 0 | – | – |
| 80:20 | 2 | 50 | 141 | 0 | 15 | 0 | – | – |
| total (all trials) | 473 | 25 | 41 | 2 | – | – | ||
Dye influx, induced by daptomycin, was measured by the appearance of the carboxyfluorescein green fluorescence inside the GUVs. In the presence of daptomycin, only 26% of the parent GUVs exhibited significant influx across their outer membranes. (We typically consider influx to be significant if it occurs to at least 10% of the outside fluorescence intensity level.) It is also worth noting that experiments with 2 mM CaCl2 resulted in a lower dye influx (14% of GUVs) than those with 20 mM CaCl2 (32%). As a control, these experiments were performed with CaCl2 (2 mM and 20 mM) but without daptomycin (in its place 20 μL of 1:1 ethanol:water were added). In the absence of daptomycin, influx was only observed in 5% of the parent GUVs.
Daptomycin translocation across the membrane was measured as we previously described (26). The method takes advantage of parent GUVs that contain inner vesicles. If a peptide binds to the parent GUV and causes dye influx into its lumen, but the peptide does not cross the outer membrane, the inner vesicles remain dark. However, if the peptide translocates across the outer membrane, binds to the inner vesicles, and causes carboxyfluorescein influx into their lumens, the inner vesicles will show green fluorescence as well. In addition, the intrinsic kynurenine blue fluorescence of daptomycin will be detected on the membranes of the inner vesicles. With daptomycin, no dye flux into the inner vesicles was ever observed and no daptomycin fluorescence was observed in the membrane of the inner vesicles. Figure 4 shows examples of four parent GUVs with inner vesicles, from four different preparations. The membranes are visualized by LRh-DOPE (red) and daptomycin is visualized by its kynurenine fluorescence (cyan). No daptomycin is observed on the membranes of the inner vesicles. Thus, according to our interpretation of this experiment, daptomycin does not cross the GUV membrane.
Figure 4.
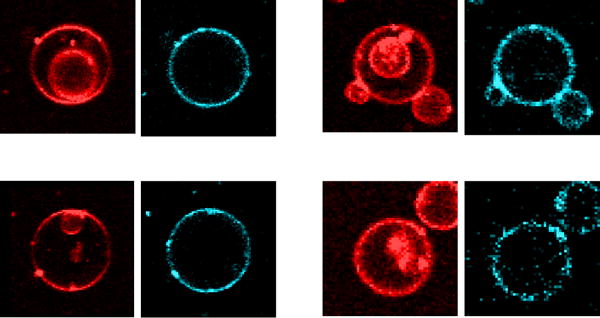
Four examples of parent GUVs containing inner vesicles, in the presence of daptomycin (2 μM, added outside). No daptomycin is observed on the inner vesicle membranes. In each example, the left panel shows the fluorescence of the lipid probe LRh-DOPE (red), which was incorporated in the membrane during the GUV preparation. The right panels show the daptomycin fluorescence, from its intrinsic kynurenine chromophore (cyan). Daptomycin is present in the outer membrane of the parent GUV, but not in the inner vesicles.
It is important to realize that, with our method, we conclude translocation occurred if a peptide is able to cross the outer membrane of a GUV, dissociate into its lumen, and bind to inner vesicles of that GUV. It is not possible to determine if a peptide simply crossed the bilayer but remained absorbed to it, now facing the GUV lumen. In this study, the GUVs were prepared without CaCl2 (initially, there is no Ca2+ inside the vesicles). Therefore, in order for daptomycin to bind significantly to the inner vesicles, Ca2+ must have entered the parent GUV. We expect Ca2+ flux to occur into the parent GUVs at least to the same extent as carboxyfluorescein influx, because Ca2+ is much smaller than carboxyfluorescein. Of the 112 inner vesicles recorded, 49 resided in parent GUVs with influx (Table 1). They should, therefore, have been exposed to Ca2+. However, none of those 49 inner vesicles had daptomycin bound to their membranes. This indicates that daptomycin translocation is not simply limited by the availability of Ca2+ in the lumen of the parent GUV. Rather, it appears that daptomycin is not able to translocate across the membrane.
Daptomycin induces GUV collapse
A high percentage (36%) of GUVs collapsed when exposed to daptomycin in the presence of Ca2+. Figure 5 shows the time scan of the collapse of a parent GUV that contains a single inner vesicle.
Figure 5.
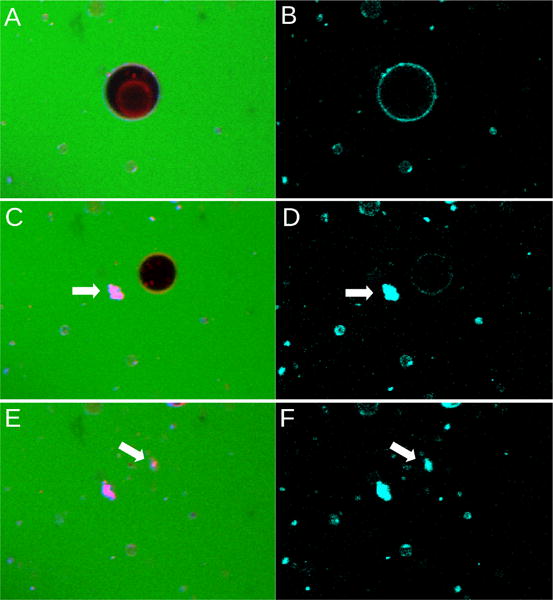
Daptomycin (2 μM) induces the collapse of a GUV composed of POPC:POPG 70:30, containing 0.1 mol% LRh-DOPE in the presence of 2 mM Ca2+. (A,B), Initial state of the same GUV. (A) All fluorescence channels (green, carboxyfluorescein; red, LRh-DOPE; cyan, daptomycin). (B) Daptomycin fluorescence channel only (cyan). (C,D) Very next image in the time scan (∼ 40 seconds between each image). The outer membrane of the GUV has now collapsed but its inner vesicle still remains (black in C). (E,F) Images taken ∼ 40 seconds later. These images were taken over a period of about 2 minutes.
The left panels (A,C,E) show the red fluorescence of the lipid probe LRh-DOPE incorporated in the membrane and the green background fluorescence of the aqueous dye carboxyfluorescein outside. The right panels (B,D,F) show the daptomycin fluorescence (cyan). The sequence of events is as follows. Panels (A,B) show the initial state of the GUV. In (A) the inner vesicle is visible (LRh-DOPE red fluorescence). In (B) the daptomycin fluorescence (cyan) is evident on the outer membrane of the parent GUV, but not on the membrane of the inner vesicle. The parent GUV collapses between images (A,B) and (C,D). Panels (C,D) show the next image in this sequence. The dark vesicle in (C) was the inner vesicle seen in (A). A very faint cyan fluorescence associated with the former inner vesicle is visible in D. The remnants of the outer membrane of the collapsed GUV appear as a mass indicated by the arrows (red in C, cyan in D). Thus, daptomycin remains associated with the lipid after the GUV collapse. The inner vesicle collapses shortly thereafter (E,F), about 2 minutes after the first image was taken. Its membrane appears as a mass, indicated by the arrows in (E,F), to the right of the mass resulting from the parent GUV.
Table 2 summarizes the vesicle collapse data for seven independent experiments in POPC:POPG 70:30 and 80:20, in the presence of 2 or 20 mM Ca2+, caused by 2 μM daptomycin (trials 1–7). A high percentage of vesicles collapsed in both 2 and 20 mM Ca2+. To determine if the vesicle collapse was due to an osmotic or charge density imbalance because of Ca2+ ions outside of the vesicles, controls were performed in the absence of daptomycin (Table 2, trials 8–11). The observation times for the control trials were similar to those used in the presence of daptomycin. However, the percent of collapsed vesicles is much lower for the experiments performed without daptomycin. Taking all trials into account, 36% of the vesicles collapsed in the presence of daptomycin whereas only 3% collapsed in the absence of daptomycin. The significantly higher vesicle collapse in the presence of daptomycin indicates that the peptide decreases the stability of the membranes. We observed GUVs for 30–120 minutes after their addition to a daptomycin solution. Little vesicle collapse occurred in the first minute. Typically, collapse happened after 5 minutes or longer. The time of collapse of the first vesicle in the samples examined varied from 1 to 9 minutes, with an average of 5 minutes.
Table 2.
Vesicle collapse caused by daptomycin in POPC:POPG GUVs.
| Lipid PC:PG |
[Ca2+] (mM) |
Observation time (min) |
GUVs | % GUVs | |
|---|---|---|---|---|---|
| Total | Collapsed | Collapsed | |||
| In the presence of 2 μM daptomycin | |||||
| 70:30 | 2 | 40 | 16 | 10 | 63 |
| 70:30 | 20 | 40 | 37 | 4 | 11 |
| 70:30 | 2 | 30 | 120 | 43 | 33 |
| 80:20 | 2 | 30 | 134 | 46 | 34 |
| 80:20 | 20 | 120 | 259 | 137 | 53 |
| 80:20 | 20 | 60 | 141 | 30 | 21 |
| 80:20 | 20 | 60 | 146 | 38 | 26 |
| total (all trials) | 863 | 308 | 36 | ||
| In the absence of daptomycin | |||||
| 70:30 | 20 | 90 | 14 | 0 | 0 |
| 80:20 | 20 | 40 | 210 | 12 | 6 |
| 80:20 | 20 | 40 | 108 | 1 | 1 |
| 80:20 | 2 | 50 | 141 | 0 | 0 |
| total (all trials) | 473 | 13 | 3 | ||
Daptomycin distribution coincides with that of anionic phospholipids
Nonuniform distributions of daptomycin were observed on the membrane of POPC:POPG GUVs (Figure 6). These clusters are not static, but move in the membrane, more so in the initial stages of the experiments. As will become clear below, the observation of daptomycin clustering is a consequence of the formation of POPG-daptomycin domains in the membrane. We also noticed that the anionic probe rhodamine-phosphatidylethanolamine (LRh-DOPE, Figure 1B), coclusters with daptomycin (Figure 6, B and C).
Figure 6.
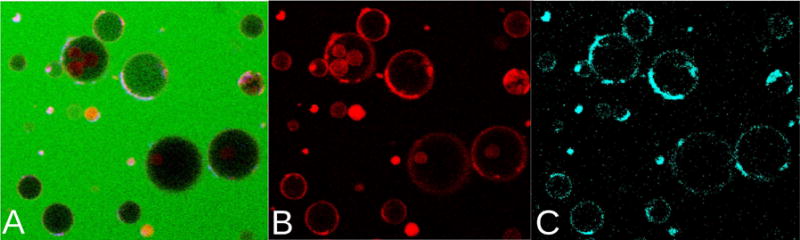
Rhodamine (LRh-DOPE) and daptomycin fluorescence distributions coincide on membranes of POPC:POPG (80:20), containing 0.1 mol% LRh-DOPE and 2 mM Ca2+. The daptomycin concentration in the outside solution was 2 μM. (A) All fluorescence channels (green, carboxyfluorescein; red, LRh-DOPE; cyan, daptomycin). (B) Rhodamine fluorescence only (red). (C) Daptomycin fluorescence channel only (cyan).
We further examined the formation of PG-daptomycin domains using GUVs composed of POPC:POPG 80:20 containing 1 mol% of the phosphatidylserine (PS) lipid fluorophore TopFluor-PS (Figure 1C). TopFluor-PS has a phosphatidylserine headgroup, which carries a negative charge in neutral solutions, and a bodipy-labeled acyl chain on the sn-2 position. TopFluor-PS, LRh-DOPE, and POPG are all anionic and contain multiple hydrogen-bonding groups. We therefore expect TopFluor-PS to behave similarly to POPG and work as a reporter for POPG clustering.
Indeed, daptomycin and TopFluor-PS coclustering was observed in the presence of 2 mM Ca2+ (Figure 7) or 20 mM Ca2+ (Figure 8). The TopFluor-PS and daptomycin domains are essentially coincident. This is evident in panels A and B of Figures 7 and 8, which show the location of TopFluor-PS (A) and daptomycin (B) fluorescence on the membrane of the vesicle. In the absence of daptomycin, TopFluor-PS domains were not observed (Figures 7C and 8C). A total of 13 vesicles were analyzed quantitatively, as shown in Figures 7 and 8, and hundreds were observed under the microscope to determine qualitatively if the daptomycin domains coincided with those of TopFluor-PS (or LRh-DOPE). In all GUV images examined TopFluor-PS coclustered with daptomycin in POPC:POPG membranes, in the presence of 2 or 20 mM Ca2+. To quantitatively determine the extent of coclustering, the TopFluor-PS and daptomycin fluorescence intensities on the membrane were measured. Plots of TopFluor-PS and daptomycin intensities as a function of the angle around the vesicle circumference are shown in panels D of Figures 7 and 8. In the example of Figure 7D, the correlation between the distributions of daptomycin and TopFluor-PS is R = 0.677, and in Figure 8D, the correlation is R = 0.831. Note that the correlation between the distributions of daptomycin and TopFluor-PS is strong but not perfect (Figures 2 and 7D). There are some TopFluor-PS clusters that appear to have little daptomycin.
Figure 7.
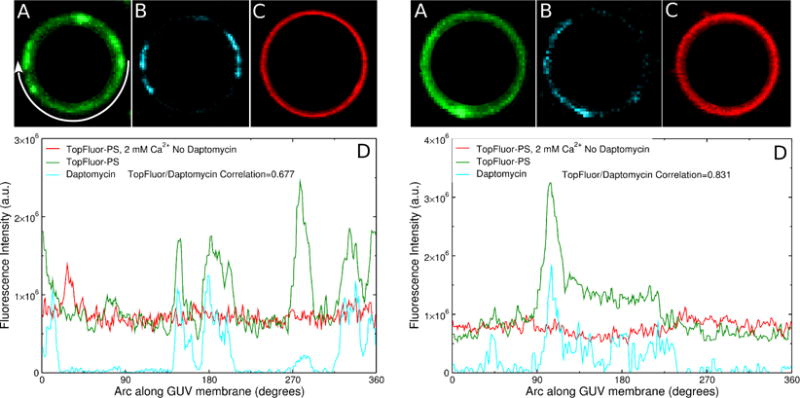
Examples of two GUVs of POPC:POPG 80:20 (left and right panels) showing TopFluor-PS and daptomycin domains in the presence of 2 mM Ca2+. The daptomycin concentration in the outside solution was 2 μM. Fluorescence of (A) TopFluor-PS and (B) daptomycin. (C) TopFluor-PS fluorescence in a GUV of POPC:POPG 80:20, without daptomycin, in the presence of 2 mM Ca2+. (D) Fluorescence intensities (same color code) along the circumference of the vesicles shown, as indicated by the arrow in (A) left panel.
Figure 8.
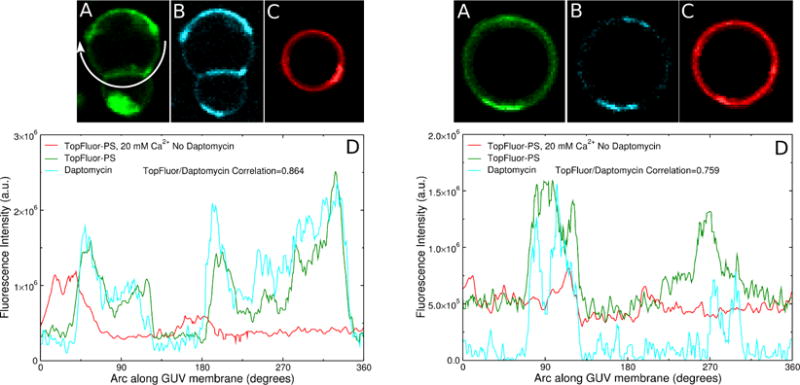
Examples of two GUVs of POPC:POPG 80:20 (left and right panels) showing TopFluor-PS and daptomycin domains in the presence of 20 mM Ca2+. The daptomycin concentration in the outside solution was 2 μM. Fluorescence of (A) TopFluor-PS and (B) daptomycin. (C) TopFluor-PS fluorescence in a GUV of POPC:POPG 80:20, without daptomycin, in the presence of 20 mM Ca2+. (D) Fluorescence intensities (same color code) along the circumference of the vesicles shown, as indicated by the arrow in (A) left panel.
No daptomycin binding to pure POPC vesicles containing 1 mol % TopFluor-PS was detected (as no binding was detected to POPC vesicles containing 0.1 mol% LRh-DOPE). However, we wanted to be certain that the coclustering of daptomycin and TopFluor-PS observed in POPC:POPG vesicles was indeed a consequence of the formation of daptomycin-POPG domains. That is, TopFluor-PS should simply be a reporter of anionic lipid clustering with daptomycin, and the clustering should not be merely the result of interactions between daptomycin and TopFluor-PS. Therefore, we analyzed the effect of adding daptomycin to POPC vesicles containing 1 mol% TopFluor-PS (but no POPG) on the fluorescence distribution of this lipid probe in the membrane. As is evident in Figure 9, the TopFluor-PS distribution on the membrane is uniform (compare the signal/noise levels here with those in Figures 7 and 8). This shows that TopFluor-PS is a reporter of POPG clustering, but does not cluster by itself with daptomycin.
Figure 9.
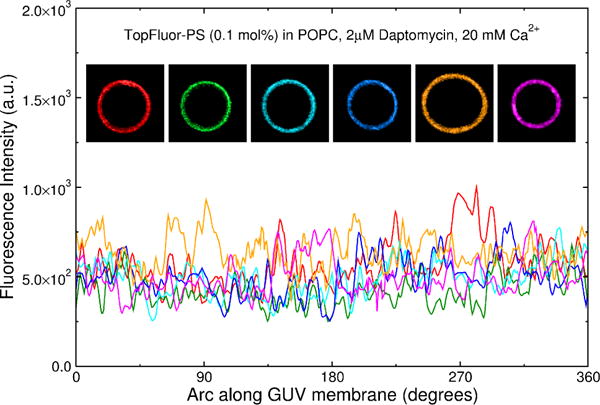
Examples of six GUVs of POPC containing 1 mol% TopFluor-PS in the presence of 2 μM daptomycin and 20 mM Ca2+. The fluorescence is uniform indicating that binding of daptomycin to TopFluor-PS does not induce the domains in these experiments. Rather, POPG is required to observe domain formation. The top panel shows 6 GUVs color coded in the same manner as the fluorescence intensity lines shown in the lower panel.
DISCUSSION
The purpose of this work was to better understand the physical chemical interactions of daptomycin with membranes that model the phospholipid bilayer of Gram-positive bacteria. The simplest model membrane must contain a significant fraction of the anionic lipid phosphatidylglycerol (PG). We chose to use GUVs of POPC:POPG with 70:30 and 80:20 molar ratios. We developed a method to quantitatively examine colocalization of membrane components and used it to demonstrate that daptomycin coclusters with anionic phospholipids. In addition, we investigated the unresolved controversy of whether daptomycin binds to zwitterionic lipids, namely, phosphatidylcholine (PC). We conclude that it does not bind to PC to any significant extent. A possible exception is discussed, which may occur if the PC membrane contains packing defects. Finally, we posit that PG clustering is at the root of the mechanism by which daptomycin kills bacterial cells, and discuss this hypothesis in light of the current knowledge of the interactions of daptomycin with bacteria in vivo.
We begin with a few words about the method to assess colocalization. The standard procedure to evaluate coclustering is to overlay two pictures of a GUV, each displaying one of two fluorescent reporter probes associated with two different membrane components, and judging the colocalization by eye, based on the overlap (or not) of the two fluorescent regions of the membrane. It is advantageous to use a more quantitative and objective approach. To this end, we used the freely available software ImageJ (27) in a way that, we hope, will be useful to other researchers.
Experimentally, to measure colocalization between daptomycin and POPG, we used a PS lipid fluorophore (TopFluor-PS, Figure 1C) to probe the distribution of PG. The evidence that TopFluor-PS reports on the PG distribution is strong. Namely, PG and TopFluor-PS do not cluster in the absence of daptomycin (panels C of Figures 7 and 8); and TopFluor-PS and daptomycin do not cluster in the absence of PG (Figure 9). Therefore, PG clearly appears to be involved in clustering with daptomycin. Most likely, this clustering is a consequence of a network of hydrogen bonding and electrostatic interactions of the PG mediated by Ca2+ and daptomycin. The choice of an anionic probe (PS) to report on an anionic bulk lipid (PG) is justified because there is a strong electrostatic component to the binding of daptomycin to lipid vesicles. Daptomycin binding to PC:PG membranes is strong, whereas binding to pure PC is very weak. For over 15 years, we have examined binding of various antimicrobial peptides to mixtures of PC:PG and PC:PS vesicles. A large component of the effect of PG or PS seems to be electrostatic (29–34). In addition, both PG and PS are capable of extensive hydrogen bonding, much more than PC.
Previously, a chain-labeled pyrene-PG was used as a mimic for PS to study domain formation in PC:PS membranes induced by the C2 domain of synaptotagmin I (35, 36). The results obtained provided strong evidence that the assumption that pyrene-PG was a reliable reporter for the PS behavior was essentially correct. Pyrene-PG, however, is not suitable for fluorescence microscopy experiments because of its emission in the UV and because it is unstable unless kept in an inert atmosphere, which is not possible in these experiments. We therefore used a PS-based probe (TopFluor-PS) as a mimic for PG (POPG), thus reversing the headgroups of probe and bulk phospholipid relative to the earlier experiments. If instead of TopFluor-PS, LRh-DOPE is present, which is also anionic (Figure 1B), the distribution of daptomycin is also correlated with that LRh-DOPE (Figure 6). This correlation is thus not restricted to a particular probe but must reflect a more general property of the membrane. We posit it reflects coclustering of daptomycin and PG.
In several vesicles, it appears that the clusters of daptomycin and the lipid fluorophore coincide with especially bright spots in the membrane. Similar observations have been made before for daptomycin (14) and mastoparans (37). It could be argued that these areas represent folds of the bilayer (37, 38), lipid–peptide aggregates on the membrane (14), or nonbilayer structures. Huang and coworkers (14) observed a change in the area of the GUV membrane upon interaction with daptomycin: the area of the GUV membrane first expands by ≈ 4% and then contracts to an area ≈ 1% smaller than the original membrane area (14). This corresponds to a maximal decrease in area of ≈ 5% relative to the largest area at the peak of the transient increase. Similar to us, Huang and coworkers (14) observe qualitative colocalization of bodipy-labeled daptomycin and LRh-DOPE on the membrane. However, we emphasize the clustering, whereas they emphasize what they call “lipid extraction.”
Caution should to be exercised when interpreting bright spots in fluorescence microscopy images (22). This is because the shape of a high-intensity fluorescent area does not faithfully reflect the shape of the structure from which that emission arises. That is, regions of high fluorescence intensity tend to appear larger (thicker) in the microscope, but that does not mean that those regions protrude out of the bilayer. There is no evidence that these bright spots correspond to nonbilayer structures (although that is possible). Most likely, the bright spots contain high local concentrations of daptomycin and TopFluor-PS (Figures 2, 7 and 8) or LRh-DOPE (Figure 6; see also (14)). However, we do not think that the decrease in area observed by Huang and coworkers (14) is in itself the explanation for the bright spots or the colocalization of daptomycin and lipid probes. The net area decrease is only ≈ 1% of the original GUV membrane (14). In contrast, the regions in which we observe colocalization of daptomycin and the anionic probe TopFluor-PS are extensive, about 20–50% of the membrane area. If about 1% of the area is “extracted,” then the remaining 20–50% of the membrane where colocalization is observed still needs to be accounted for.
The question of daptomycin binding to pure PC membranes is a still matter of controversy. Here we offer an explanation to resolve the issue. We do not observe binding of daptomycin to pure POPC vesicles (with or without Ca2+). There is evidence of daptomycin binding to dimyritoylphosphatidylcholine (DMPC) small unilamellar vesicles (SUVs) prepared by sonication (which have a diameter of about 25 nm) in the presence of Ca2+, and the fluorescence emission of the kynurenine residue is sensitive to the DMPC phase transition at about 24°C (10). This indicates that the peptide binds to the vesicles. However, SUVs, because of their high curvature and tight lipid packing, are strained structures with packing defects. Usually, they accommodate partitioning of small molecules much better than their more relaxed counterparts, large unilamellar vesicles (LUVs) and GUVs. In addition, the phase transition of DMPC occurs at about room temperature, where fluid and gel phase coexist in the same vesicle. This leads to a significant increase in the partitioning of most substances into the membrane, as the compressibility and area fluctuations of the bilayer reach a maximum at the temperature of the main phase transition (39–41). The permeability of the membrane also increases dramatically at the phase transition (42–49). It is unfortunate that DMPC was used in so many studies of daptomycin binding to PC membranes, often in conjunction with large concentrations of Ca2+ (up to 200 mM) (10–13). Huang and coworkers (14) infer daptomycin binding to DOPC through the increase in membrane area observed when the vesicles are exposed to the peptide. However, with 1 μM daptomycin and 1 mM Ca2+, that area increase is only of 3%. The DOPC membrane is generally well packed, but the regions of the GUV adjacent to the tip of the micropipette in the aspiration experiments (14), where the curvature changes sign twice, may contain packing defects or curvature strain that leads to daptomycin binding. In contrast, a study using surface plasmon resonance (8) found no binding of daptomycin to POPC small extruded vesicles (50 nm diameter) in the presence of 0.45 μM CaCl2, under conditions in which binding to POPG was evident. The question of daptomycin binding to PC may be put to rest by studying binding of daptomycin to LUVs of POPC or DOPC, which are entirely fluid at room temperature and whose membranes are very well packed. Preliminary measurements (Figure S2) using the same method that demonstrated daptomycin binding to DMPC (10), show that the fluorescence emission of the kynurenine residue does not increase significantly in the presence of POPC LUVs compared to its value in the presence of Ca2+ only. The slight difference observed is probably due mostly to scattering. In contrast, binding to POPC:POPG LUVs results in an enormous in increase in the kynurenine fluorescence emission. Thus, we do not observe significant binding to POPC by this method either, confirming the result of the GUV experiments.
There is also some disagreement regarding the Ca2+ requirement. We, and most other researchers, do not observe binding of daptomycin to POPC:POPG (80:20 or 70:30) in the absence of Ca2+ whereas Huang and coworkers (14) infer that binding occurs again from the increase in the membrane area of a GUV exposed to daptomycin. However, the increase in area observed in DOPC/DOPG 70:30 GUVs in the absence of Ca2+ is close to zero at 1 μM daptomycin and of only ∼ 1% at 5 μM daptomycin (14). It is necessary to increase the concentration of daptomycin to 50 μM to observe an area increase (followed by decrease) similar to that observed in DOPC:DOPG in the presence of 1 μM daptomycin and 1 mM Ca2+.
Let us now consider GUV collapse induced by daptomycin. We monitored the GUVs for 30–120 minutes after addition to a daptomycin solution (Table 2). Typically, GUV collapse only began after 5 minutes or longer. Huang and coworkers (14) monitored GUV area changes for about 1 minute. Not much collapse occurred in that time, but neither did we observe much vesicle collapse in the first minute of the experiments. However, in one case, GUV rupture was noted to occur in 10 seconds when exposed to 5 μM daptomycin (14). Very likely, more GUV collapse would have been observed in those experiments at longer times. An interesting question is why a GUV population in a given sample shows all-or-none behavior in terms of influx or collapse. That is, some vesicles show influx or collapse while others in the same sample do not. The amount of daptomycin bound to the outer membranes of GUVs is similar for all GUVs in a given sample. In other words, the daptomycin distribution is uniform. However, all-or-none behavior in vesicle populations is a frequent aspect of antimicrobial and cell-penetrating peptides. We have studied this problem extensively in GUVs (25, 26) and LUVs (50), using a variety of peptides, including classical antimicrobials, such as cecropin A and magainin 2, and many of their synthetic variants, as well as amphipathic cell-penetrating peptides based on transportan 10 and its variants, and synthetic variants of the cytolytic staphylococcal peptide δ-lysin. What we found is that the onset of influx in a particular GUV is stochastic, as is its collapse. Some peptides cause a graded influx, which corresponds to a unimodal distribution of dye influx levels in the GUVs, but others cause all-or-none influx, which results in a bimodal distribution of dye in the vesicles. Those bimodal distributions are not the result of a bimodal peptide distribution, but a consequence of the stochastic response of the vesicles when peptide is bound to their membranes. As far as we can tell, there is no trigger mechanism for influx or collapse. We should note that daptomycin is inefficient at causing influx compared to the set of α-helical, amphipathic, antimicrobial and cell-penetrating peptides we have previously examined. Only 26% of GUVs show influx across their outer membranes in the presence of 2 μM daptomycin (Table 1), whereas more typical antimicrobial peptides cause significant influx in most GUVs in the sample, after similar times, even at lower concentrations (0.5–1 μM) (25, 26, 51).
Some membrane-active peptides, such as mastoparans, also form denser regions, with higher peptide concentration on the membrane, which appear as bright spots under the microscope if the peptides are fluorescent (37), similar to the daptomycin and TopFluor-PS bright spots described here. Those regions have been interpreted as folds, or invaginations, of the membrane, where a bilayer patch slides over the main surface of the GUV membrane (37, 38). In addition, membrane deformations have been reported, for example in the cases of mastoparans and gomesin (37, 38, 52), in which a portion of the membrane protrudes, or the entire GUV significantly changes shape. We have noticed such deformations as well with several of the α-helical, amphipathic, membrane-active peptides we have studied in GUVs (26, 51), but we have not examined those deformations systematically. However, we have not observed those kinds of membrane deformations with daptomycin. The different behavior of daptomycin also extends to translocation. With most variants of α-helical, amphipathic, membrane-active peptides, we have observed translocation across the outer membrane of the GUV. But with daptomycin this appears not to be the case.
In summary, daptomycin is inefficient in the kinds of actions that have been proposed to constitute the mechanisms of antimicrobial peptides: causing flux across membranes, causing membrane collapse, translocating to the inside of vesicles and possibly binding to intracellular targets. We cannot exclude that, though inefficient, daptomycin uses one or more of those processes in its antimicrobial activity in vivo, but the most striking aspect of the interactions of daptomycin with mixed POPC:POPG GUVs is the formation of large domains rich both in daptomycin and anionic lipids (PG). It is thus tempting to hypothesize that clustering of PG is an essential part of the antimicrobial mechanism of daptomycin.
In bacteria, it appears that clustering of PG impairs the synthesis of the cell wall. Several lines of evidence from in vivo studies support this idea. First, the distribution of fluorescently labeled daptomycin in the Gram-positive bacterium Bacillus subtilis is not uniform, but the peptide is concentrated in division septa and along the cell wall (19). The importance of PG for daptomycin function became evident as depletion of PG from a B. subtilis strain led to the delocalization of daptomycin from those regions and greatly decreased the bacterial susceptibility to the peptide (19). Second, the preferential distribution of daptomycin to active division septa and foci along the cylindrical cell envelope was linked to the mechanism of cell death in B. subtilis (20). Daptomycin partitions to active membrane division sites, causing defects and deformations that culminate with membrane collapse. In addition, DivIVA, a protein essential for cell division, changes its distribution and colocalizes with daptomycin in cells treated with sub-lethal concentrations of the peptide. These observations led to the suggestion that daptomycin disrupts peptidoglycan synthesis by causing mislocalization of proteins involved in cell division (20). Interestingly, PG is found at higher concentration in the membrane in those locations (21).
More recently, Müller et al. (22) confirmed that daptomycin disrupts cell synthesis in E. feacalis. However, daptomycin does not dissipate the membrane potential at concentrations that inhibit bacterial growth; lytic concentrations are required for complete membrane depolarization. It thus seems unlikely that pore formation is the main mechanism of the antimicrobial activity of daptomycin. Rather, it was confirmed that daptomycin causes mislocalization of proteins involved in cell division, namely DivIVA, in addition to proteins important for cell shape. The clustering of membrane proteins appears to be related to the lipid distribution. Müller et al. (22) proposed that daptomycin partitions to regions of the membrane rich in fluid lipids and coclusters with these lipids, causing them to coalesce into large patches. When labeled with fluorophores, those clusters are visible as highly fluorescent patches, coincident with fluorescently labeled daptomycin. The simultaneous and immediate delocalization of different proteins suggests that the effect of daptomycin on protein is mediated by its effect on the lipid (22). Our hypothesis that daptomycin functions by clustering PG is entirely consistent with these results. Moreover, it specifies that those fluid lipids patches are PG clusters.
More generally, the targeting of cell division septa and consequent disruption of peptidoglycan synthesis is not unique to daptomycin. Similar observations have been reported for other antimicrobial peptides, such as human β-defensins (hBDs), LL-37, and cecropin A (21, 53). In Enterococcus feacalis, the translocase SecA and several sortases are concentrated in membrane foci in division septa and in the cell envelope along a helical pattern. The function of the Sec pathway depends on the presence PG, and hBDs were found to disrupt the localization of SecA and sortases leading to their dispersion. It was suggested that anionic lipids, namely PG, form microdomains in the membrane, which are essential for proper function of the secretion apparatus. Further, localization of cationic antimicrobial peptides to the foci involved in secretion, mediated by PG and other anionic lipids, would lead to the disruption of these domains and protein mislocalization (53). Weisshaar and coworkers (21) have suggested that it is the disruption of peptidoglycan synthesis by LL-37 and cecropin A, not membrane permeabilization and consequent dissipation of membrane potential, that is responsible for the antimicrobial activity of these peptides.
CONCLUSION
We investigated the interactions of the peptide antibiotic daptomycin with GUVs of POPC:POPG in the presence of Ca2+. In agreement with previous reports, we found that the presence of Ca2+, in solution, and the negatively charged phospholipid POPG, in the lipid vesicles, are necessary for daptomycin binding to the membrane. Upon binding, the peptide perturbs the membrane to a significant extent. It is inefficient at causing influx of the water-soluble dye carboxyfluorescein, but causes about one-third of the GUVs to collapse. Furthermore, daptomycin does not appear to translocate across the lipid membrane. Thus, pore formation is unlikely to be the mechanism by which the antibiotic functions. On the other hand, we found that daptomycin coclusters with POPG and anionic fluorescent lipid probes, leading to formation of extensive daptomycin/POPG domains in the membrane. We posit that this major membrane reorganization is essential to the function of daptomycin.
Supplementary Material
Acknowledgments
This work was supported in part by grant CHE-1464769 from the National Science Foundation and grant GM072507 from the National Institutes of Health. We thank Alison Taylor and Mark Gay for their help with the confocal fluorescence microscopy, a facility funded by National Science Foundation grant DBI 0420948.
Footnotes
Supporting Information: Figure S1 shows TopFluor fluorescence in the same GUVs shown in Figures 7 and 8, but without clearing the inside of the vesicles or their immediate outside vicinity. Figure S2 shows the fluorescence intensity of the intrinsic daptomycin chromophore kynurenine, upon excitation of the tryptophan residue, in the presence of Ca2+, POPC, and POPC:POPG LUVs.
ORCID
Paulo Almeida: 0000-0003-4591-938X
Notes
The authors declare no competing financial interest.
References
- 1.Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Baltz RH. Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. Curr Opin Chem Biol. 2009;13:144–151. doi: 10.1016/j.cbpa.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 3.Ball LJ, Goult CM, Donarski JA, Micklefield J, Ramesh V. NMR structure determination and calcium binding effects of lipopeptide antibiotic daptomycin. Org Biomol Chem. 2004;2:1872–1878. doi: 10.1039/b402722a. [DOI] [PubMed] [Google Scholar]
- 4.Ho SW, Jung D, Clahoun JR, Lear JD, Okong M, Scott WRP, Hancock RE, Straus SK. Effect of divalent cations on the structure of the antibiotic daptomycin. Eur Biophys J. 2008;37:421–433. doi: 10.1007/s00249-007-0227-2. [DOI] [PubMed] [Google Scholar]
- 5.Taylor R, Butt K, Scott B, Zhang T, Muraih JK, Mintzer E, Taylor S, Palmer M. Two successive calcium-dependent transitions mediate membrane binding and oligomerization of daptomycin the related antibiotic A54145. Biochim Biophys Acta. 2016;1858:1999–2005. doi: 10.1016/j.bbamem.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Jung D, Rozek A, Okon M, Hancock REW. Structural transitions as determinants of the action of the calcium-dependent antibiotic daptomycin. Chem Biol. 2004;11:949–957. doi: 10.1016/j.chembiol.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Jung D, Powers JP, Straus SK, Hancock REW. Lipid-specific binding of the calcium-dependent antibiotic daptomycin leads to changes in lipid polymorphism of model membranes. Chem Phys Lipids. 2008;154:120–128. doi: 10.1016/j.chemphyslip.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 8.Kinouchi H, Onishi M, Kamimori H. Lipid membrane-binding properties of daptomycin using surface plasmon resonance. Anal Sci. 2013;29:297–301. doi: 10.2116/analsci.29.297. [DOI] [PubMed] [Google Scholar]
- 9.Lakey JH, Lear EJA. The role of acyl chain character and other determinants on the bilayer activity of A21978C an acidic lipopeptide antibiotic. Biochim Biophys Acta. 1986;859:219–226. doi: 10.1016/0005-2736(86)90217-8. [DOI] [PubMed] [Google Scholar]
- 10.Lakey JH, Ptak M. Fluorescence indicates a calcium-dependent interaction between the lipopeptide antibiotic LY146032 and phospholipid membranes. Biochemistry. 1988;27:4639–4645. doi: 10.1021/bi00413a009. [DOI] [PubMed] [Google Scholar]
- 11.Muraih JK, Pearson A, Silverman J, Palmer M. Oligomerization of daptomycin on membranes. Biochim Biophys Acta. 2011;1808:1154–1160. doi: 10.1016/j.bbamem.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Muraih JK, Harris J, Taylor SD, Palmer M. Characterization of daptomycin oligomerization with perylene excimer fluorescence: Stoichiometric binding of phosphatidylglycerol triggers oligomer formation. Biochim Biophys Acta. 2012;1818:673–678. doi: 10.1016/j.bbamem.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 13.Zhang T, Muraih JK, MacCormick B, Silverman J, Palmer M. Daptomycin forms cation- and size-selective pores in model membranes. Biochim Biophys Acta. 2014;1838:2425–2430. doi: 10.1016/j.bbamem.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 14.Chen YF, Sun TL, Sun Y, Huang HW. Interaction of daptomycin with lipid bilayers: a lipid extracting effect. Biochemistry. 2014;53:5384–5392. doi: 10.1021/bi500779g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silverman JA, Perlmutter NG, Shapiro HM. Correlation of daptomycin bactericidal activity and membrane depolarization in Staphylococcus aureus. Antimicrob Agents Chemother. 2003;47:2538–2544. doi: 10.1128/AAC.47.8.2538-2544.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Scoten K, Straus SK. Daptomycin leakage is selective. ACS Infect Dis. 2016;2:682–687. doi: 10.1021/acsinfecdis.6b00152. [DOI] [PubMed] [Google Scholar]
- 17.Muraih JK, Palmer M. Estimation of the subunit stoichiometry of the membrane-associated daptomycin oligomer by FRET. Biochim Biophys Acta. 2012;1818:1642–1647. doi: 10.1016/j.bbamem.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 18.Zhang T, Taylor SD, Palmer M, Duhamel J. Membrane binding and oligomerization of the lipopeptide A54145 studied by pyrene fluorescence. Biophys J. 2016;111:1267–1277. doi: 10.1016/j.bpj.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hachmann AB, Angert ER, Helmann JD. Genetic factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53:1598–1609. doi: 10.1128/AAC.01329-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pogliano J, Pogliano N, Silverman JA. Daptomycin-mediated reorganization of membrane architecture causes mislocalization of essential cell division proteins. J Bacteriol. 2012;194:4494–4504. doi: 10.1128/JB.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi H, Rangarajan N, Weisshaar JC. Lights, camera, action! Antimicrobial peptide mechanisms imaged in space and time. Trends Microbiol. 2016;24:111–122. doi: 10.1016/j.tim.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Müller A, Wenzel M, Strahl H, Grein F, Saaki TN, Kohl B, Siersma T, Bandow JE, Sahl HG, Schneider T, Hamoen LW. Daptomycin inhibits cell envelope synthesis by interfering with fluid membrane microdomains. Proc Natl Acad Sci USA. 2016;110:E7077–E7086. doi: 10.1073/pnas.1611173113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angelova MI, Soléau S, Méléard P, Faucon JF, Bothorel P. Preparation of giant vesicles by external AC electric fields. Kinetics and applications. Prog Colloid Polym Sci. 1992;89:127–131. [Google Scholar]
- 24.Apellániz B, Nieva JL, Schwille P, García-Sáez AJ. All-or-none versus graded: Single-vesicle analysis reveals lipid composition effects on membrane permeabilization. Biophys J. 2010;99:3619–3628. doi: 10.1016/j.bpj.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wheaten SA, Lakshmanan A, Almeida PF. Statistical analysis of peptide-induced graded and all-or-none fluxes in giant vesicles. Biophys J. 2013;105:432–443. doi: 10.1016/j.bpj.2013.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheaten SA, Ablan FDO, Spaller BL, Trieu JM, Almeida PF. Translocation of cationic amphipathic peptides across the membranes of pure phospholipid giant vesicles. J Am Chem Soc. 2013;135:16517–16525. doi: 10.1021/ja407451c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasband WS. ImageJ. U.S. National Institutes of Health; Bethesda, Maryland, USA: 1997–2016. https://imagej.nih.gov/ij/ [Google Scholar]
- 28.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in FORTRAN: The Art of Scientific Computing. 2nd. Cambridge University Press; Cambridge: 1994. [Google Scholar]
- 29.Almeida PF, Pokorny A. Interactions of antimicrobial peptides with lipid bilayers. In: Egelman Edward H., editor. Comprehensive Biophysics. Vol. 5. Academic Press; 2012. pp. 189–222. Membranes. [Google Scholar]
- 30.Yandek LE, Pokorny A, Floren A, Knoelke K, Langel U, Almeida PFF. Mechanism of the cell-penetrating peptide Tp10 permeation of lipid bilayers. Biophys J. 2007;92:2434–2444. doi: 10.1529/biophysj.106.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregory SM, Cavenaugh AC, Journigan V, Pokorny A, Almeida PFF. A quantitative model for the all-or-none permeabilization of phospholipid vesicles by the antimicrobial peptide cecropin A. Biophys J. 2008;94:1667–1680. doi: 10.1529/biophysj.107.118760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory SM, Pokorny A, Almeida PFF. Magainin 2 revisited: A test of the quantitative model for the all-or-none permeabilization of phospholipid vesicles. Biophys J. 2009;96:116–131. doi: 10.1016/j.bpj.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yandek LE, Pokorny A, Almeida PFF. Small changes in the primary structure of transportan 10 alter the thermodynamics and kinetics of its interaction with phospholipid vesicles. Biochemistry. 2008;47:3051–3060. doi: 10.1021/bi702205r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yandek LE, Pokorny A, Almeida PFF. Wasp mastoparans follow the same mechanism as the cell-penetrating peptide transportan 10. Biochemistry. 2009;48:7342–7351. doi: 10.1021/bi9008243. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hinderliter AK, Almeida PFF, Creutz CE, Biltonen RL. Domain formation in a fluid mixed bilayer modulated through binding of the C2 protein motif. Biochemistry. 2001;40:4181–4191. doi: 10.1021/bi0024299. [DOI] [PubMed] [Google Scholar]
- 36.Hinderliter A, Biltonen RL, Almeida PFF. Lipid modulation of protein-induced membrane domains as a mechanism for controlling signal transduction. Biochemistry. 2004;43:7102–7110. doi: 10.1021/bi036334t. [DOI] [PubMed] [Google Scholar]
- 37.Cabrera MPS, Alvares DS, Leite NB, Souza BM, Palma MS, Riske KA, Neto JR. New insight into the mechanism of action of wasp mastoparan peptides: Lytic activity and clustering observed with giant vesicles. Langmuir. 2011;27:10805–10813. doi: 10.1021/la202608r. [DOI] [PubMed] [Google Scholar]
- 38.Riske KA. Optical microscopy of giant vesicles as a tool to reveal the mechanism of action of antimicrobial peptides and the specific case of gomesin. Adv Planar Lipid Bilayers and Liposomes. 2015;21:99–129. [Google Scholar]
- 39.Heimburg T. Mechanical aspects of membrane thermodynamics. Estimation of the mechanical properties of lipid membranes close to the chain melting transition from calorimetry. Biochim Biophys Acta. 1998;1415:147–162. doi: 10.1016/s0005-2736(98)00189-8. [DOI] [PubMed] [Google Scholar]
- 40.Thomas Heimburg T, Jackson AD. On soliton propagation in biomembranes and nerves. Proc Natl Acad Sci USA. 2005;102:9790–9795. doi: 10.1073/pnas.0503823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanova VP, Heimburg T. Histogram method to obtain heat capacities in lipid monolayers, curved bilayers, and membranes containing peptides. Phys Rev E. 1914;2001(63):04. doi: 10.1103/PhysRevE.63.041914. [DOI] [PubMed] [Google Scholar]
- 42.Papahadjopoulos D, Jacobson K, Nir S, Isac T. Phase transitions in phospholipid vesicles: Fluorescence polarization and permeability measurements concerning the effect of temperature and cholesterol. Biochim Biophys Acta. 1973;311:330–348. doi: 10.1016/0005-2736(73)90314-3. [DOI] [PubMed] [Google Scholar]
- 43.Tsong TY. Effect of phase transition on the kinetics of dye transport in phospholipid bilayer structures. Biochemistry. 1975;14:5409–5414. doi: 10.1021/bi00696a004. [DOI] [PubMed] [Google Scholar]
- 44.Tsong TY, Kanehisa MI. Relaxation phenomena in aqueous dispersions of synthetic lecithins. Biochemistry. 1977;16:2674–268. doi: 10.1021/bi00631a014. [DOI] [PubMed] [Google Scholar]
- 45.Tsong TY, Greenberg M, Kanehisa MI. Anesthetic action on membrane lipids. Biochemistry. 1977;16:3115–3121. doi: 10.1021/bi00633a012. [DOI] [PubMed] [Google Scholar]
- 46.Blok MC, van der Neut-Kok ECM, van Deenen LLM, De Gier J. The effect of chain length and lipid phase transitions on the selective permeability properties of liposomes. Biochim Biophys Acta. 1975;406:187–196. doi: 10.1016/0005-2736(75)90003-6. [DOI] [PubMed] [Google Scholar]
- 47.Marsh D, Watts A, Knowles PF. Evidence for phase boundary lipid: Permeability of tempo-choline into dimyristoylphosphatidylcholine vesicles at the phase transition. Biochemistry. 1976;15:3570–3578. doi: 10.1021/bi00661a027. [DOI] [PubMed] [Google Scholar]
- 48.Corvera E, Mouritsen OG, Singer MA, Zuckermann MJ. The permeability and the effect of acyl-chain length for phospholipid bilayers containing cholesterol: theory and experiment. Biochim Biophys Acta. 1992;1107:261–270. doi: 10.1016/0005-2736(92)90413-g. 1992. [DOI] [PubMed] [Google Scholar]
- 49.Cruzeiro-Hansson L, Mouritsen OG. Passive ion permeability of lipid membranes modeled via lipid-domain interfacial area. Biochim Biophys Acta. 1988;944:63–72. doi: 10.1016/0005-2736(88)90316-1. [DOI] [PubMed] [Google Scholar]
- 50.Almeida PF, Pokorny A. Mechanism of antimicrobial, cytolytic, and cell-penetrating peptides: From kinetics to thermodynamics. Biochemistry. 2009;48:8083–8093. doi: 10.1021/bi900914g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ablan FD, Spaller BL, Abdo KI, Almeida PF. Charge distribution fine-tunes the translocation of α-helical amphipathic peptides across membranes. Biophys J. 2016;111:1738–1749. doi: 10.1016/j.bpj.2016.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domingues TM, Riske KA, Miranda A. Revealing the lytic mechanism of the antimicrobial peptide gomesin by observing giant unilamellar vesicles. Langmuir. 2010;26:11077–11084. doi: 10.1021/la100662a. [DOI] [PubMed] [Google Scholar]
- 53.Kandaswamy K, Liew TH, Wang CY, Huston-Warren E, Meyer-Hoffert U, Hultenby K, Schröder JM, Caparon MG, Normark S, Henriques-Normark B, Hultgren SJ, Kline KA. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci USA. 2013;110:20230–20235. doi: 10.1073/pnas.1319066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


