Abstract
Globally, there are approximately 240 million people chronically infected with hepatitis B virus (HBV) – a major cause of hepatocellular carcinoma. Ten different HBV genotypes (A-J) have been identified with distinct geographic distributions. Novel variants generated by recombination between different HBV genotypes have been documented worldwide and represent an important element of genetic variability with possible clinical implications. Here, the complete genome sequence of an HBV genotype D/E recombinant from Ghana is reported. The full-length sequence was obtained using rolling circle amplification followed by PCR and then sequenced using next generation sequencing (NGS). A consensus sequence was extracted from the NGS data and underwent phylogenetic analysis to determine genotype, as well as the recombination pattern. Subsequently, the sequence was compared to recombinants described previously in Africa. Based on MCMC phylogenetic analysis, SimPlot recombination analyses, and intragroup genetic distance, the isolate 007N full-length genome is unique compared to other reported D/E recombinants in Africa.
Keywords: Hepatitis B virus, Genotypes, Sub-genotypes, Recombinant/Recombination, Africa, Ghana
Introduction
Hepatitis B virus (HBV) remains a global health problem with approximately 240 million chronically infected people and 780,000 deaths per year due to acute infections, cirrhosis of the liver, and hepatocellular carcinoma [1, 2]. The prevalence of HBV is highest in sub-Saharan Africa and East Asia, where 5–10% of the adult population is chronically infected [1]. Hepatitis B is an enveloped, 3.2 kb partially double-stranded DNA virus, which belongs to the Hepadnaviridae family [3]. The virus is arranged in four overlapping open reading frames (ORFs): polymerase (P), surface (S), precore/core (C), and the X gene (X) [4]. Like retroviruses, HBV utilizes a reverse transcriptase enzyme during replication, resulting in high viral diversity [5].
There are ten HBV genotypes (A-J) classified by sequence divergence of at least 8% in the entire HBV genome [6–8]. Genotypes A-D and F have been further classified into subgenotypes with a genetic variation of 4–8% [9–11]. Genotypes A and D are prevalent in the European Union and in Central/South Asia; B and C in South/South East Asia and the Pacific region; E in West/Central Africa; F in South/Central America and Alaska; G and H in Europe and Japan; I in Vietnam and Laos; and J in Japan [11, 12].
Genotypes represent an invaluable tool for tracing the molecular evolution and transmission patterns of HBV [11]. Several studies suggest that clinical outcomes may be due to differences in pathogenic potential between genotypes [13–15]. HBV genotypes have been shown to differ according to the course of disease, development of mutations, and response to antiviral therapy [16]. Genotype determination is important in identifying patients who are at increased risk of disease progression and in optimizing treatment [15].
Recombination between genotypes occurs in regions where multiple genotypes co-circulate and facilitates diversification within individuals and in the general population. Novel variants generated by recombination between different HBV genotypes have been documented worldwide [17–21]. In West Africa, one or more A/B, A/C, A/E, C/E, D/E, and D/E/A recombinants have been identified [5, 22, 23]. Recombination is an important element of HBV genetic variability with possible clinical implications [24, 25]. The research reported here was aimed at further investigating an HBV variant from Ghana whose initial partial sequence was identified as a possible recombinant.
Methods
Patient Information
The newly identified recombinant is from a HIV-HBV coinfected, 52 year-old male from Northern Ghana who was previously enrolled in two cross-sectional studies. At the time of sample collection in 2013, he was antiretroviral treatment-naïve with an unknown CD4+ count, hepatitis B e-antigen negative, e-antibody positive, and had HBV DNA of 84,060,683 IU/mL. The original study investigated the proportion and factors associated with hepatitis B viremia in treatment-experienced and treatment-naïve HIV co-infected Ghanaians [26]. A subsequent sub-study examined HBV resistance mutations [27]. Phylogenetic analysis from this resistance study grouped the genotype D patient (sample 007N) with three D/E recombinants from Niger (FN594768-71), although it include only a 1,004 bp fragment corresponding to the HBV reverse transcriptase region. Because genotype D is relatively uncommon in West Africa and to determine if this variant was, in fact, a D/E recombinant, the entire full-length genome was analyzed.
Next-Generation Sequencing
HBV DNA was extracted from 500 uL of patient serum using the QIAamp UltraSens Virus Kit (QIAGEN, Valencia, CA, USA), and the full-length sequence was obtained using rolling circle amplification followed by PCR [18]. DNA-seq was performed by the Genomics, Epigenomics and Sequencing Core (GESC) at the University of Cincinnati. The PCR product was transferred to a microTUBE (Covaris, Woburn, MA) and sheared with the Covaris S2 focused-ultrasonicator. The sample was prepared for the sequencing library using the PrepX DNA Library kit, Apollo 324 NGS automatic library prep system (WaferGen, Fremont, CA), and a ChIP-seq script. The ligated library was indexed and enriched by 6 cycles of PCR using sample-specific index and universal PCR primers, followed by automated AMPure XP bead purification (Beckman Coulter, Brea, CA). Equal-amount (ng) libraries determined by Nanodrop were pooled, diluted, and qPCR quantified with NEBNext Library Quant Kit (New England Biolabs, Ipswich, MA) using ABI 9700HT real-time PCR system (Lifetech, Grand Island, NY).
The quantified pooled library was used for cluster generation in the cBot system (Illumina, San Diego, CA). A library at the final concentration of 16 pM was clustered onto a flow cell using Illumina TruSeq SR Cluster kit v3 and sequenced for 50 cycles using TruSeq SBS kit on Illumina HiSeq system. A total of 445,497 reads (51 bp each) were collected and imported into CLC Genomic Workbench 9 for quality control and consensus assembly. 366,512 (75.5%) reads passed the initial quality import filter. A consensus sequence was extracted from 348,145 (94.9%) of the remaining reads using the de novo assembler module v1.3 and complete HBV reference genomes from GenBank. The reads excluded from the consensus sequence did not meet the similarity cut off (similarity fraction set to 0.80) and only mapped to a small portion of the HBV genome.
Sequence Alignment & Bayesian Markov Chain Monte Carlo Analysis
The consensus sequence from this novel HBV variant was aligned with full-length references in ClustalX 2.1 [28]. Additional phylogenetic inference was performed using a Bayesian Markov chain Monte Carlo (MCMC) approach as implemented in the BEAST v1.8.0 program [29] under an uncorrelated log-normal relaxed molecular clock and the general time-reversible model with nucleotide site heterogeneity estimated using a gamma distribution. The initial MCMC analysis was run for a chain length of 500,000,000 with sampling every 50,000th generation. Results were visualized in Tracer v1.5 to confirm chain convergence, and the effective sample size (ESS) was calculated for each parameter. All ESS values were >1,000 indicating sufficient sampling. The maximum clade credibility tree was selected from the posterior tree distribution after a 10% burn-in using TreeAnnotator v1.8.0. An additional MCMC analysis was conducted using all recombinant sequences, genotype E references, and subgenotype D7-D10 references and a chain length of 200,000,000.
Recombination Analysis & Intragroup Genetic Distance
To identify possible recombination, bootscanning analysis of full-length sequences was performed in SimPlot version 3.5.1 using the Kimura 2-parameter with a 300 base pair (bp) window, a 30 bp step increment, and 1,000 bootstrap replicates [30]. The full-length HBV sequence was compared to consensus sequences generated using full-length GenBank references for HBV genotypes A-H. If >80 % of the permuted trees showed similarity to more than one genotype, the “parental” sequences were retained within a second bootscanning analysis along with the consensus Pan troglodytes sequence as an outlier. To eliminate the possibility of a dual infection, the full-length consensus sequence was aligned with a second sequence amplified by targeted PCR and Sanger sequencing [27]. Despite the use of different amplification and sequencing methods, the consensus sequences were identical, indicating that the patient was not infected with multiple HBV genotypes. Intragroup genetic distances were calculated by pairwise comparison of nucleotide sequences using the Kimura 2-parameter method of Molecular Evolutionary Genetics Analysis (MEGA) v7.0.18 [31, 32]. The complete genome of patient 007N was deposited in GenBank under the accession number KU711666.
Results
Bayesian Markov Chain Monte Carlo Analysis
The phylogenetic analyses included 13 full-length D/E recombinant references from different regions in Africa including Niger (FN594767, FN594768, FN594769, FN594770, and FN594771), Sudan (KU736916 and KU736917), Ethiopia (KU736914 and KU736915), Kenya (KP168420 and KP168421), Ghana (GQ161754), and Gabon (GU177079). As shown in Figure 1, all recombinants from East Africa grouped together, and most of the recombinants from West Africa – excluding GU177079, FN594767, and 007N – grouped together. D/E recombinant sequences – regardless of their country of origin – were most closely related to subgenotype D7 sequences but did not form a single monophyletic group.
Figure 1.
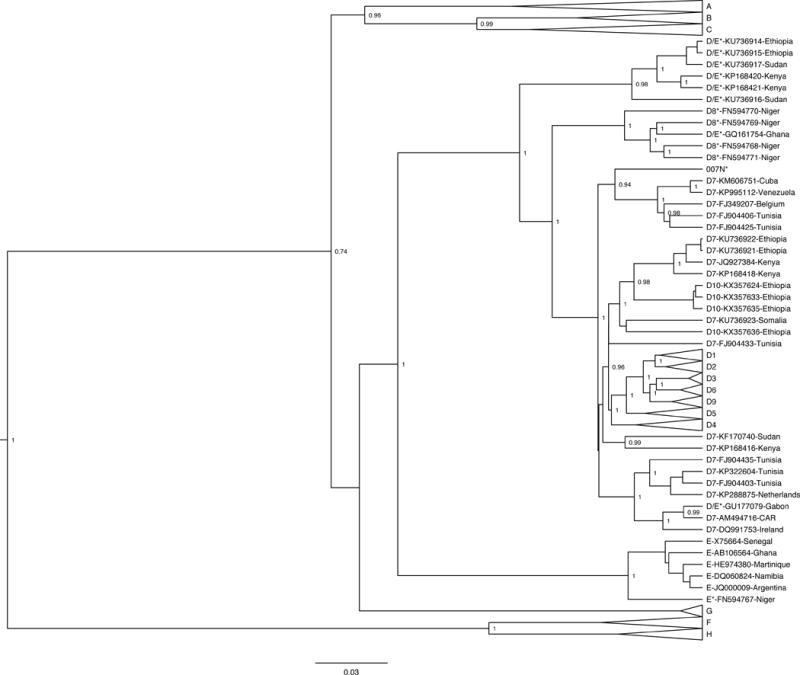
Bayesian phylogenetic tree of 87 full-genome HBV sequences generated with BEAST v1.8.0. The sample of interest is denoted as “007N”, while reference sequences are labeled “genotype – accession number – country of origin”. All reported recombinants are marked with an asterisk after their genotype designation, and relevant posterior values are shown.
A subsequent MCMC analysis was conducted focusing specifically on the D/E recombinant references, isolate 007N, genotype E references, and subgenotype D7-D10 references (Figure 2). All but three of the D/E recombinants – GU177079, FN594767, and 007N – clustered together. The D/E recombinant from Gabon (GU177079) clustered with the subgenotype D7 strains from Ireland, Central African Republic, the Netherlands, and Tunisia. The E/D recombinant from Niger (FN594767) clustered with the genotype E references. The isolate of interest – 007N – clustered with a different group of D7 references from Tunisia, as well as D7 strains from Belgium, Cuba, and Venezuela. There are multiple clusters of D7 reference strains – some grouping with known D/E recombinants and others grouping with D10 reference strains from Ethiopia. Thus, subgenotype D7 is not a well-defined monophyletic cluster based on the limited sequence data available currently.
Figure 2.
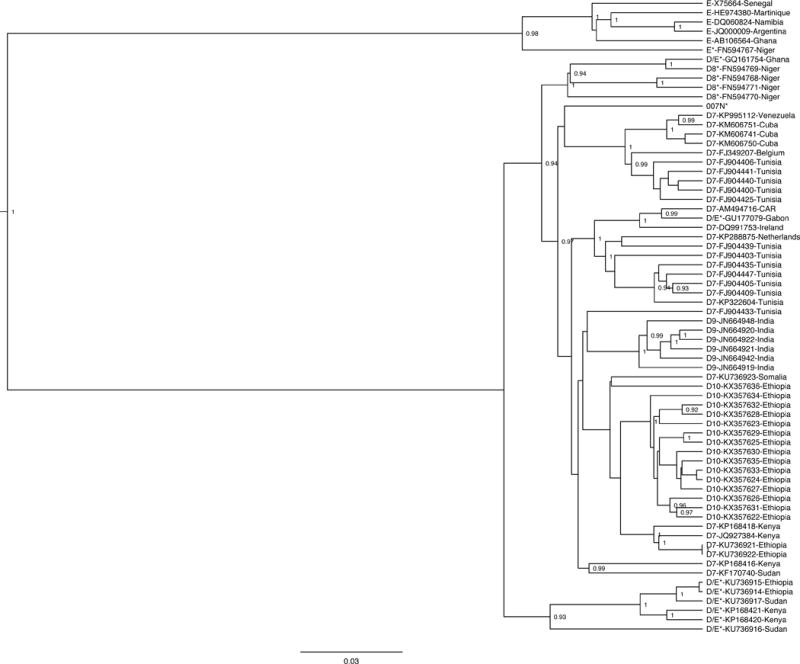
Bayesian phylogenetic tree of 68 full-genome HBV sequences generated with BEAST v1.8.0. Genotype E, subgenotype D7, subgenotype D8, subgenotype D9, subgenotype D10, and genotype D/E recombinant reference sequences are included. The sample of interest is denoted as “007N”, while reference sequences are labeled “genotype – accession number – country of origin”. All reported recombinants are marked with an asterisk after their genotype designation, and relevant posterior values are shown.
Intragroup Genetic Distance
Table 1 shows the mean genetic distance among all D/E recombinants including 007N. The mean genetic distance was 3.72%, with a range of 0.26% to 7.50%. All 14 full-length D/E recombinants were distinct from one another, with only one recombinant from Sudan and two from Ethiopia (KU736917, KU736914, and KU736915) having a genetic distance of less than 1% between them. Sequence 007N was most closely related to the other D/E recombinant from Ghana (GQ161754) with a genetic distance of 2.23%. The sequence from Niger (FN594767) had the genetic distance farthest from 007N at 7.50%. Among all the D/E recombinants, the most similar were the two recombinants from Ethiopia with a genetic distance of 0.26%, while the most divergent – aside from FN594767 and 007N – were FN594767 from Niger and FN594770 from Niger at 7.50%.
Table 1.
Genetic distances for all 14 D/E recombinants calculated by pairwise comparison of nucleotide sequences using the Kimura 2-parameter method.
| 007N | D/E*- GQ161754- Ghana |
D/E*- GU177079- Gabon |
D/E*- KU736914- Ethiopia |
D/E*- KU736915- Ethiopia |
D/E*- KU736916- Sudan |
D/E*- KU736917- Sudan |
D/E*- KP168420- Kenya |
D/E*- KP168421- Kenya |
E*- FN594767- Niger |
D8*- FN594768- Niger |
D8*- FN594769- Niger |
D8*- FN594770- Niger |
D8*- FN594771- Niger |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 007N | ||||||||||||||
| D/E*-GQ161754-Ghana | 2.23% | |||||||||||||
| D/E*-GU177079-Gabon | 2.74% | 2.77% | ||||||||||||
| D/E*-KU736914-Ethiopia | 3.33% | 2.98% | 3.40% | |||||||||||
| D/E*-KU736915-Ethiopia | 3.61% | 3.19% | 3.68% | 0.26% | ||||||||||
| D/E*-KU736916-Sudan | 3.40% | 3.05% | 3.51% | 2.23% | 2.34% | |||||||||
| D/E*-KU736917-Sudan | 3.16% | 2.81% | 3.37% | 0.58% | 0.84% | 2.20% | ||||||||
| D/E*-KP168420-Kenya | 3.65% | 3.61% | 4.04% | 2.41% | 2.48% | 2.61% | 2.27% | |||||||
| D/E*-KP168421-Kenya | 4.07% | 3.75% | 4.32% | 2.65% | 2.71% | 2.85% | 2.44% | 1.79% | ||||||
| E*-FN594767-Niger | 7.50% | 6.54% | 7.41% | 6.97% | 7.05% | 6.31% | 6.93% | 7.31% | 7.43% | |||||
| D8*-FN594768-Niger | 3.26% | 2.43% | 3.96% | 4.00% | 4.07% | 4.07% | 3.82% | 4.53% | 4.60% | 7.43% | ||||
| D8*-FN594769-Niger | 2.98% | 1.46% | 3.43% | 3.57% | 3.64% | 3.78% | 3.50% | 4.31% | 4.42% | 7.26% | 2.74% | |||
| D8*-FN594770-Niger | 3.23% | 2.13% | 3.61% | 3.79% | 4.00% | 3.68% | 3.61% | 4.25% | 4.32% | 7.50% | 3.12% | 2.81% | ||
| D8*-FN594771-Niger | 2.61% | 1.79% | 3.29% | 3.54% | 3.75% | 3.47% | 3.36% | 4.00% | 4.32% | 7.16% | 2.06% | 2.50% | 2.88% |
Recombination Analysis
All D/E recombinant sequences – including the 13 references and 007N – were analyzed further in SimPlot to determine recombination patterns. The bootscans of each recombinant were created using all full-length genotype D7 and E references used in the MCMC analysis with the full-length Pan troglodytes sequence as an outlier. Bootscans with a genome map to illustrate where the recombination breakpoints are located within the circular HBV genome are shown in Figure 3.
Figure 3.
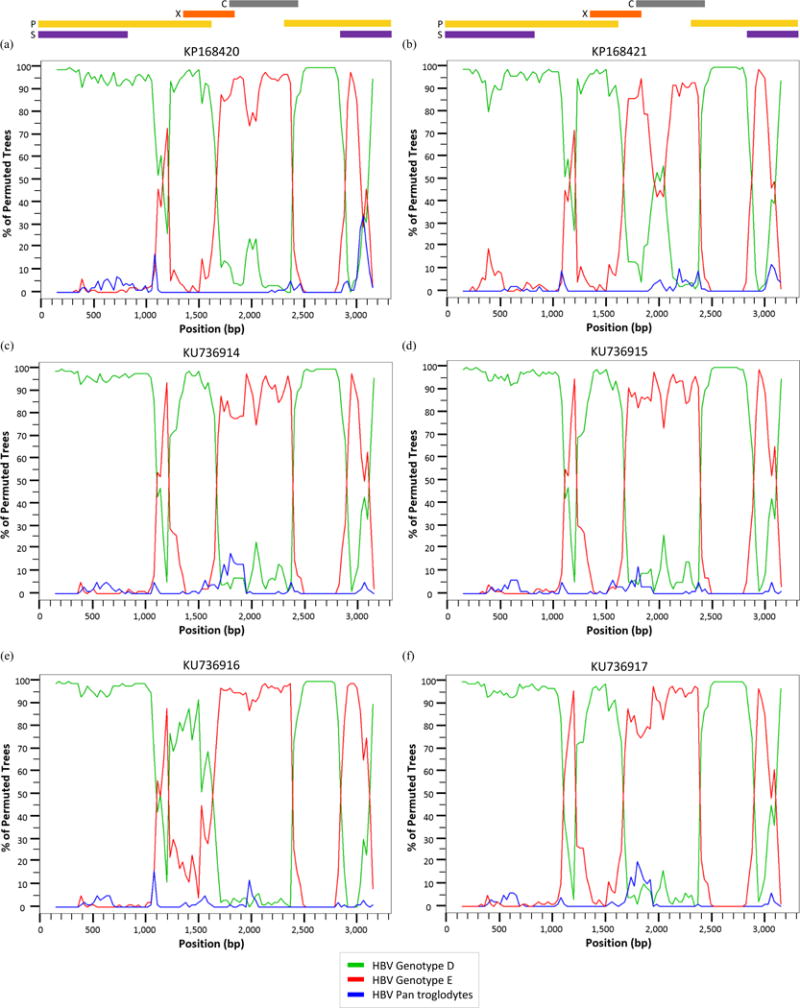
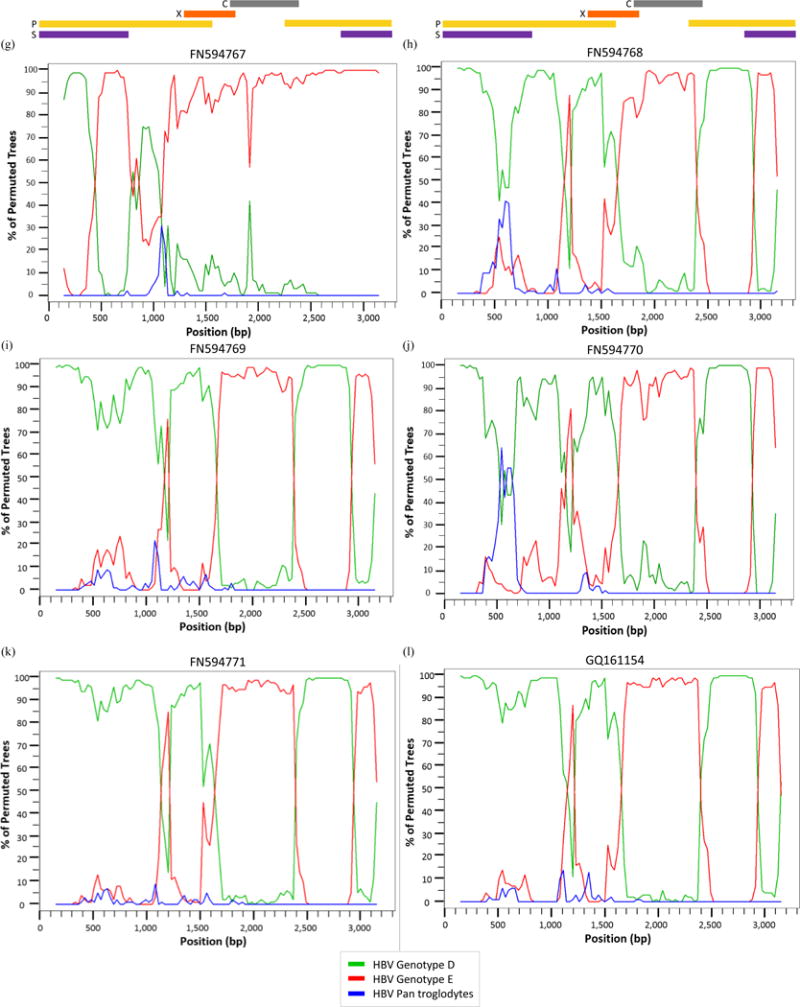
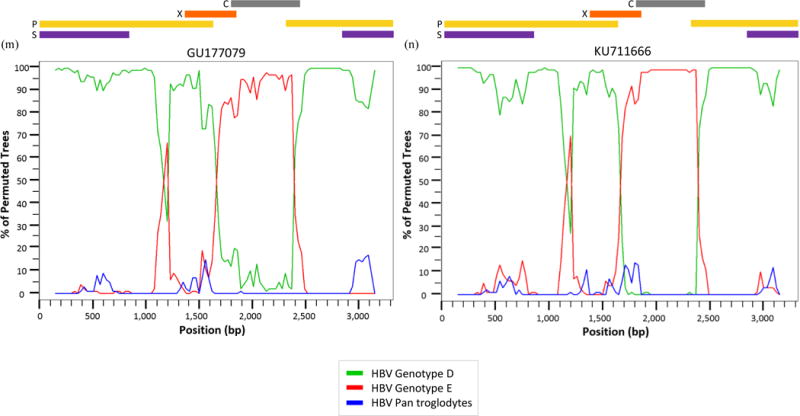
Bootscan analysis of reference (a) KP168420, (b) KP168421, (c) KU736914, (d) KU736915, (e) KU736916, (f) KU736917, (g) FN594767, (h) FN594768, (i) FN594769, (j) FN594770, (k) FN594771, (l) GQ161154, (m) GU177079, and (n) sample 007N (KU711666) using the Kimura 2-parameter with a 300 base pair (bp) window, a 30 bp step increment, and 1,000 bootstrap replicates.
Seven of the 14 sequences – KU736914-17, FN594768, FN594771, and GQ161754 – have a breakpoint between nt 1,100–1,200 (located in the P ORF) where >80% of permuted trees showed a shift from genotype D to genotype E. With the exception of sequence FN594767, all the recombinants showed a shift from genotype D to genotype E between nt 1,700–2,400 corresponding to the P, X, and C ORFs. Eleven recombinants from Kenya, Ethiopia, Sudan, Niger, and Ghana – KP168420-21, KU736914-17, FN594768-71, and GQ161154 – have an additional breakpoint between nt 2,900–3,100 which is located in the S and P overlapping ORF. One of the Niger samples – FN594767 – has a unique recombination pattern compared to other D/E recombinants with only one breakpoint between nt 500–900 in which >80% of permuted trees show a shift from genotype D to genotype E. There is a breakpoint between nt 900–1,100 – as seen in Fig. 3(g) – where the sequence shifts back from genotype E to genotype D, although the percentage of permuted trees is ~75%. This recombinant is referred to in its original publication as an E/D recombinant because a majority of the sequence data is similar to genotype E [33].
The reference recombinant from Gabon and 007N – Fig. 3(m) and 3(n) – both have only one breakpoint above 80%. Thus, the two sequences from Ghana – Fig. 3(l) and 3(n) – have distinctly different recombination patterns despite GQ161754 being the closest reference sequence to 007N based on genetic distance.
Discussion
Over 400 complete HBV intergenotypic recombinant genomes have been identified around the world [34]. HBV genotypes can help elucidate the transmission patterns of HBV because variants have a distinct geographic distribution genotype. Overall, intergenotypic recombinants exhibit similar circulation patterns as their original genotypes [34]. Hepatitis B virus genotype A is predominant in South/East Africa, D in North Africa, and E in West/Central Africa [12]. Genotype E hybrids are generally found in African countries with A/E recombinants in Cameroon, Ghana, and Guinea [35, 36] and D/E recombinants in Gabon, Ghana, Niger, Sudan, Kenya, and Ethiopia [33, 35, 37, 38]. There are two documented E hybrids reported from France (A/E) and Ireland (D/E); however, both involve patients originating from Africa [18, 39].
When all full-length intergenotypic recombinant genomes were compared, the majority (73%) consisted of HBV variants with identical recombination sites [34]. Similar breakpoint locations among several sequences occur due to recombination hot spots or a single ancestral recombination event circulating within a population. An analysis of breakpoint positions was conducted on 117 sequences with different breakpoints and/or genotype composition. Favored recombination sites were identified within nt 1,700–1,900, nt 1,800–2,000, and nt 2,100–2,300 [34]. Isolate 007N from this study has two breakpoints which span these favored recombination sites between nt 1,700–2,400. The other D/E recombinants analyzed in this study share this breakpoint and have an additional breakpoint between nt 1,100–1,200.
Based on the MCMC phylogenetic analysis, SimPlot recombination analysis, and intragroup genetic distance calculations, the isolate 007N full-length genome is unique compared to other reported D/E recombinants in Africa. As shown in Figures 1 and 2, isolate 007N – as well as other published, full-length D/E recombinant strains – clustered with HBV subgenotype D7. This may indicate that the D/E recombinants are more specifically D7/E recombinant strains. However, this conclusion cannot be confidently drawn from the data available given that multiple clusters of subgenotype D7 sequences were observed in Figure 2. Despite many of the subgenotype D7 strains coming from the same country of origin, they did not form a monophyletic group, suggesting that the D7 subgenotype is not accurately defined to date and requires further analysis of additional samples from a wider geographic distribution of patients.
Available data on HBV genotypes and their effect on disease profile are currently limited to specific genotypes – mostly A, B, C, and D. However, evidence suggests that HBV genotypes and subgenotypes play a critical role in host-viral interactions. Knowledge of HBV genotype will enable clinicians to determine patients’ response to treatment and their risk of future complications [13–15]. More expansive epidemiological and in vitro studies are needed to better understand the pathogenic effects of all HBV genotypes. These data would then help to determine if HBV genotype D differs from genotype E in its recombinogenic potential and/or if a particular recombinant HBV genotype differs from a non-recombinant HBV genotype in terms of pathogenic mechanisms and patient outcomes.
Acknowledgments
The initial study was funded by a 2012 International Developmental Grant from the Lifespan/Tufts/Brown CFAR (P30AI042853) and the Brown/Tufts AIDS International Training and Research Program (D43TW000237) to Drs. Archampong and Kwara. Additional support was provided by Brown University – University of Ghana partnership through a USAID/HED grant (Award # AEG-A-00-05-00007). Dr. Kwara received additional support from Fogarty international Center (D43TW010055). HBV amplicon-seq was conducted by Genomics, Epigenomics and Sequencing Core at Department of Environmental Health, University of Cincinnati, which is supported in part through CEG grant (NIEHS P30-ES006096).
Footnotes
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. For this type of case study, formal consent is not required.
References
- 1.WHO. Hepatitis B Fact Sheet No 204. 2016 URL: http://www.who.int/mediacentre/factsheets/fs204/en/
- 2.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Vaccine. 2012;30:2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 3.Faure E. Virus Res. 2006;117:185–201. doi: 10.1016/j.virusres.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 4.Kramvis A. Intervirology. 2014;57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- 5.Olinger CM, Venard V, Njayou M, Oyefolu AO, Maiga I, Kemp AJ, Omilabu SA, le Faou A, Muller CP. J Gen Virol. 2006;87:1163–1173. doi: 10.1099/vir.0.81614-0. [DOI] [PubMed] [Google Scholar]
- 6.Schaefer S. World J Gastroenterol. 2007;13:14–21. doi: 10.3748/wjg.v13.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olinger CM, Jutavijittum P, Hübschen JM, Yousukh A, Samountry B, Thammavong T, Toriyama K, Muller CP. Emerg Infect Dis. 2008:1777–1780. doi: 10.3201/eid1411.080437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tatematsu K, Tanaka Y, Kurbanov F, Sugauchi F, Mano S, Maeshiro T, Nakayoshi T, Wakuta M, Miyakawa Y, Mizokami M. J Virol. 2009:10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramvis A, Kew M, François G. Vaccine. 2005;23:2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 10.Stanojević B, Osiowy C, Schaefer S, Bojović K, Blagojević J, Nešić M, Yamashita S, Stamenković G. Infection, Genetics and Evolution. 2011;11:1475–1480. doi: 10.1016/j.meegid.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Kurbanov F, Tanaka Y, Mizokami M. Hepatology Research. 2010;40:14–30. doi: 10.1111/j.1872-034X.2009.00601.x. [DOI] [PubMed] [Google Scholar]
- 12.Kramvis A, Kew MC. Hepatol Res. 2007;37:S9–s19. doi: 10.1111/j.1872-034X.2007.00098.x. [DOI] [PubMed] [Google Scholar]
- 13.Chan HL, Wong GL, Tse CH, Chim AM, Yiu KK, Chan HY, Sung JJ, Wong VW. Clin Gastroenterol Hepatol. 2009;7:1361–1366. doi: 10.1016/j.cgh.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 14.Zhong YW, Li J, Song HB, Duan ZP, Dong Y, Xing XY, Li XD, Gu ML, Han YK, Zhu SS, Zhang HF. BMC Infect Dis. 2011;11:262. doi: 10.1186/1471-2334-11-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanwar S, Dusheiko G. Curr Gastroenterol Rep. 2012;14:37–46. doi: 10.1007/s11894-011-0233-5. [DOI] [PubMed] [Google Scholar]
- 16.Shi YH. Jpn J Infect Dis. 2012;65:476–482. doi: 10.7883/yoken.65.476. [DOI] [PubMed] [Google Scholar]
- 17.Araujo NM, Araujo OC, Silva EM, Villela-Nogueira CA, Nabuco LC, Parana R, Bessone F, Gomes SA, Trepo C, Kay A. J Gen Virol. 2013;94:150–158. doi: 10.1099/vir.0.047324-0. [DOI] [PubMed] [Google Scholar]
- 18.Martel N, Gomes S, Chemin I, Trépo C, Kay A. Journal of Virological Methods. 2013;193:653–659. doi: 10.1016/j.jviromet.2013.07.045. [DOI] [PubMed] [Google Scholar]
- 19.Osiowy C, Gordon D, Borlang J, Giles E, Villeneuve JP. J Gen Virol. 2008;89:3009–3015. doi: 10.1099/vir.0.2008/005124-0. [DOI] [PubMed] [Google Scholar]
- 20.Owiredu WK, Kramvis A, Kew MC. J Med Virol. 2001;64:441–454. doi: 10.1002/jmv.1070. [DOI] [PubMed] [Google Scholar]
- 21.Su H, Liu Y, Xu Z, Cheng S, Ye H, Xu Q, Liu Q, Tan S, Xu D. PLoS One. 2014;9:e84005. doi: 10.1371/journal.pone.0084005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tufon KA, Meriki HD, Anong DN, Mbunkah HA, Nkuo-Akenji T. BMC Res Notes. London: 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Candotti D, Diarra B, Bisseye C, Tao I, Quang KP, Sanou M, Laperche S, Sanogo R, Allain JP, Simpore J. J Med Virol. 2016 doi: 10.1002/jmv.24589. [DOI] [PubMed] [Google Scholar]
- 24.Kay A, Zoulim F. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds P, Midgley S. J Virol. 2005;79:15467–15476. doi: 10.1128/JVI.79.24.15467-15476.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Archampong TN, Lartey M, Sagoe KW, Obo-Akwa A, Kenu E, Gillani FS, Yang H, Boamah I, Flanigan T, Kwara A. BMC Infect Dis. 2016;16:14. doi: 10.1186/s12879-016-1342-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Archampong TN, Boyce CL, Lartey M, Sagoe KW, Obo-Akwa A, Kenu E, Blackard JT, Kwara A. Antivir Ther. 2016 doi: 10.3851/IMP3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 29.Drummond AJ, Suchard MA, Xie D, Rambaut A. Mol Biol Evol. 2012:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura M. Journal of Molecular Evolution. 1980 [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K. Molecular Biology and Evolution. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chekaraou MA, Brichler S, Mansour W, Gal FL, Garba A, Deny P, Gordien E. J Gen Virol. 2010;91:1609–1620. doi: 10.1099/vir.0.018127-0. [DOI] [PubMed] [Google Scholar]
- 34.Araujo NM. Infect Genet Evol. 2015;36:500–510. doi: 10.1016/j.meegid.2015.08.024. [DOI] [PubMed] [Google Scholar]
- 35.Garmiri P, Loua A, Haba N, Candotti D, Allain JP. J Gen Virol. 2009;90:2442–2451. doi: 10.1099/vir.0.012013-0. [DOI] [PubMed] [Google Scholar]
- 36.Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, Ndembi N, Ngansop C, Kaptue L, Miura T, Ido E, Hayami M, Ichimura H, Mizokami M. J Gen Virol. 2005;86:2047–2056. doi: 10.1099/vir.0.80922-0. [DOI] [PubMed] [Google Scholar]
- 37.Yousif M, Mudawi H, Bakhiet S, Glebe D, Kramvis A. BMC Infect Dis. 2013:328. doi: 10.1186/1471-2334-13-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ochwoto M, Kimotho JH, Oyugi J, Okoth F, Kioko H, Mining S, Budambula NL, Giles E, Andonov A, Songok E, Osiowy C. BMC Infect Dis. 2016;16:101. doi: 10.1186/s12879-016-1409-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laoi BN, Crowley B. J Med Virol. 2008;80:1554–1564. doi: 10.1002/jmv.21273. [DOI] [PubMed] [Google Scholar]


