Abstract
PIK3CA mutations are frequent in gastric cancer. However, their pathological and clinical implications are still unclear. We analyzed the clinicopathological characteristics according to the PIK3CA mutation status of patients with stage IB–IV disease who underwent gastrectomy between May 2003 and Dec. 2005 (cohort 1; n = 302) and of those with stage IV disease who received gastrectomy between Jul. 2006 and Dec. 2012 (cohort 2; n = 120). PIK3CA mutations were detected in 40 patients (13.2%) in cohort 1. In these patients, PIK3CA-mutant tumors were more frequently located in the upper third of the stomach (p = 0.021) and significantly showed poorly differentiated histology (p = 0.018) and increased lymphatic (p = 0.015), vascular (p = 0.005), and perineural invasion (p = 0.026). In addition, these tumors showed significantly increased lymphocyte and neutrophil infiltration in cancer stroma (p < 0.001), Epstein–Barr virus positivity (p < 0.001), and microsatellite instability (p = 0.015). Cytoplasmic Akt expression was significantly increased in these tumors (p = 0.001). In cohort 2, PIK3CA mutations were identified in 15 patients (12.5%). PIK3CA-mutant tumors showed significantly increased vascular invasion (p = 0.019) and microsatellite instability (p = 0.041). In addition, cytoplasmic Akt expression was also significantly increased (p = 0.018). However, in both cohorts, PIK3CA mutations were not associated with the prognosis of patients. In conclusion, PIK3CA mutations were associated with increased tumor aggressiveness, especially in locoregional disease, and Akt activation in gastric cancer. Our data suggest that PIK3CA-mutated gastric cancer is a distinct disease entity, which might need a different therapeutic approach.
Keywords: PIK3CA, mutation, AKT, expression, gastric cancer
INTRODUCTION
Gastric cancer (GC) is the fifth most common cancer and the third leading cause of death worldwide [1]. PIK3CA is the third most frequently mutated gene in GC [2, 3]. PIK3CA mutations are present in approximately 9%–12% patients with non-hypermutated tumors and 32% patients with hypermutated tumors.
Phosphatidylinositol 3-kinase (PI3K)/Akt signaling pathway is important in cancer cell proliferation and survival [4]. PI3K contains regulatory p85 and catalytic p110 subunits. PIK3CA encodes p110α, an isoform of the PI3K catalytic subunit. PIK3CA somatic mutations are frequently present in various human cancers such as liver, breast, colon, ovary, and GC [5, 6]. Mutant PIK3CA promotes the proliferation and invasion of human cancer cells [7] and confers resistance against HER2-targeted therapy for breast cancer [8, 9], thus warranting further investigation as a potential therapeutic target. However, only limited data on GC are available at present. Available evidence suggests that PIK3CA mutations in GC are more frequent in Epstein–Barr virus (EBV)-positive and MSI (microsatellite instability) subtypes than in the other subtypes [2, 3, 10, 11]. The most frequently detected PIK3CA mutation is H1047R in exon 20 especially in MSI subtype of GC [2, 12]. However, pathological or clinical implications of PIK3CA mutations are still unclear [13]. These unclear data might be attributed to the small sample size and the heterogeneous patient populations in previous studies. Moreover, as the detection of genetic alterations using next generation sequencing is more commonly conducted, targeted agents focusing on PIK3CA mutations are expected to be tested with high priority in metastatic GC. However, there has been no study focusing on clinicopathological characteristics of PIK3CA mutations in patients with stage IV GC. In addition, we still do not know whether PIK3CA mutations have the same clinicopathological implications on patients with locoregional disease and those with distant metastasis.
In this study, we aimed to elucidate the implications of PIK3CA mutations on detailed pathological characteristics and clinical outcomes in patients with GC using two independent cohorts consisting of patients with resected stage IB–IV GC and those with stage IV disease who underwent palliative resection.
RESULTS
Cohort 1
In all, 302 patients were consecutively enrolled in cohort 1. The male to female ratio was 202:100, and the median age was 61 years (range, 29–89 years). Of these, 273 patients (90.4%) had stage IB-III GC and 29 patients (9.6%) had stage IV GC. All the patients underwent curative or palliative gastrectomy.
In all, 45 PIK3CA mutations were detected in 40 patients (13.2%), with E545X in 22 patients (7.3%), E542K in 10 patients (3.3%), H1047X in 7 patients (2.3%), Q546X in 2 patients (0.7%), N345K in 2 patients (0.7%), R88Q in 1 patient (0.3%), and C420R in 1 patient (0.3%) (Supplementary Table 1). The clinical and pathological characteristics of cohort 1 according to their PIK3CA mutation status are compared in Table 1. PIK3CA-mutant tumors were more frequently located in the upper third of the stomach (36.8% vs. 17.7%; p = 0.021) and showed advanced T stage (pT4: 52.5% vs. 32.8%; p = 0.018). Thus, total gastrectomy was more frequently conducted in patients with PIK3CA-mutant tumors than in those with PIK3CA wild-type tumors. PIK3CA-mutant tumors significantly showed poorly differentiated histology (72.5% vs. 46.2%; p = 0.018). Further, use of Lauren classification system showed that most PIK3CA-mutant tumors in cohort 1 were not intestinal-type tumors (15.0% vs. 35.5%; p < 0.001). PIK3CA-mutant tumors also showed increased lymphatic (92.5% vs. 75.2%; p = 0.015), vascular (35.0% vs. 16.4%; p = 0.005), and perineural invasion (72.5% vs. 53.8%; p = 0.026). Moreover, these tumors showed increased lymphocytic or neutrophilic infiltration in cancer stroma (55.0% vs. 18.3%; p < 0.001), yielded significant positive results for EBV-encoded small RNA in situ hybridization (EBER ISH; 42.5% vs. 6.1%; p < 0.001), and were frequently classified as MSI-high (MSI-H; 20.0% vs. 7.3%; p = 0.015). Cytoplasmic Akt expression was significantly higher in PIK3CA-mutant tumors than in PIK3CA wild-type tumors (immunohistochemistry [IHC] 2+ or 3+: 42.8% vs. 23.3%; p = 0.001). However, overall survival (OS) was similar between patients with PIK3CA-mutant tumors and those with PIK3CA wild-type tumors (5-year OS rate: 60.0% [mutant] vs. 61.3% [wild-type]; p = 0.944; Figure 1A).
Table 1. Clinicopathological characteristics of patients in cohort 1 according to their PIK3CA mutation status.
| Characteristics | PIK3CA wild-type | PIK3CA mutant | p-value |
|---|---|---|---|
| (n = 262) (%) | (n = 40) (%) | ||
| Sex | |||
| Male | 174 (66.4) | 28 (70.0) | 0.653 |
| Female | 88 (33.6) | 12 (30.0) | |
| Age | |||
| < 70 years | 212 (80.9) | 29 (72.5) | 0.217 |
| ≥ 70 years | 50 (19.1) | 11 (27.5) | |
| Primary tumor location* | |||
| Lower third | 134 (53.8) | 17 (44.7) | 0.021 |
| Middle third | 71 (28.5) | 7 (18.4) | |
| Upper third | 44 (17.7) | 14 (36.8) | |
| Pathology | |||
| Well or moderately differentiated adenocarcinoma | 98 (37.4) | 8 (20.0) | 0.018 |
| Poorly differentiated adenocarcinoma | 121 (46.2) | 29 (72.5) | |
| Signet ring cell carcinoma | 32 (12.2) | 2 (5.0) | |
| Mucinous carcinoma | 11 (4.2) | 1 (2.5) | |
| Lauren classification | |||
| Intestinal type | 93 (35.5) | 6 (15.0) | < 0.001 |
| Diffuse type | 143 (54.6) | 23 (57.5) | |
| Mixed type | 26 (9.9) | 11 (27.5) | |
| T stage | |||
| pT1/T2 | 91 (34.7) | 6 (15.0) | 0.018 |
| pT3 | 85 (32.4) | 13 (32.5) | |
| pT4 | 86 (32.8) | 21 (52.5) | |
| N stage | |||
| pN0 | 53 (20.2) | 9 (22.5) | 0.774 |
| pN1/N2 | 114 (43.5) | 15 (37.5) | |
| pN3 | 95 (36.3) | 16 (40.0) | |
| Stage (by AJCC 7th edition) | |||
| I/II | 123 (46.9) | 13 (32.5) | 0.087 |
| III/IV | 139 (53.1) | 27 (67.5) | |
| Lymphatic invasion | |||
| Absent | 65 (24.8) | 3 (7.5) | 0.015 |
| Present | 197 (75.2) | 37 (92.5) | |
| Vascular invasion | |||
| Absent | 219 (83.6) | 26 (65.0) | 0.005 |
| Present | 43 (16.4) | 14 (35.0) | |
| Perineural invasion | |||
| Absent | 121 (46.2) | 11 (27.5) | 0.026 |
| Present | 141 (53.8) | 29 (72.5) | |
| Stroma reaction | |||
| Absent | 127 (48.5) | 11 (27.5) | < 0.001 |
| Desmoplasia | 87 (33.2) | 7 (17.5) | |
| Lymphoid or neutrophil | 48 (18.3) | 22 (55.0) | |
| EBER in situ hybridization | |||
| Negative | 246 (93.9) | 23 (57.5) | < 0.001 |
| Positive | 16 (6.1) | 17 (42.5) | |
| MSI | |||
| MSS | 243 (92.7) | 32 (80.0) | 0.015 |
| MSI-H | 19 (7.3) | 8 (20.0) | |
| Gastrectomy | |||
| Subtotal gastrectomy | 194 (74.0) | 21 (52.5) | 0.005 |
| Total gastrectomy | 68 (26.0) | 19 (47.5) | |
| Akt expression (cytoplasmic intensity) | |||
| Negative | 48 (20.7) | 3 (8.6) | 0.001 |
| 1+ | 130 (56.0) | 17 (48.6) | |
| 2+ | 49 (21.1) | 9 (25.7) | |
| 3+ | 5 (2.2) | 6 (17.1) |
Abbreviations: AJCC = American Joint Committee on Cancer; EBER = Epstein-Barr virus-encoded small RNA.
*Patients whose disease involved the entire stomach (n = 15) were excluded.
Figure 1. Survival analyses according to PIK3CA mutation status in cohort 1.
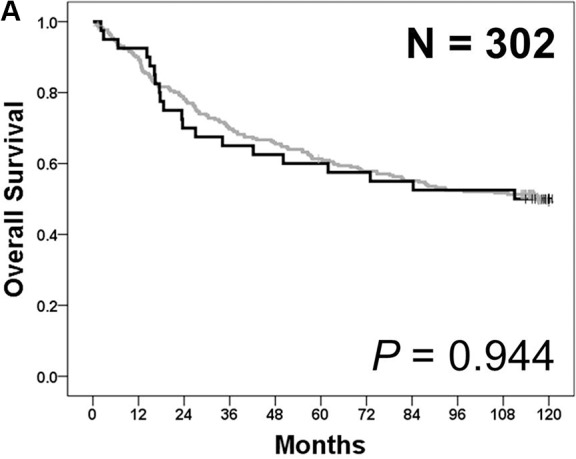
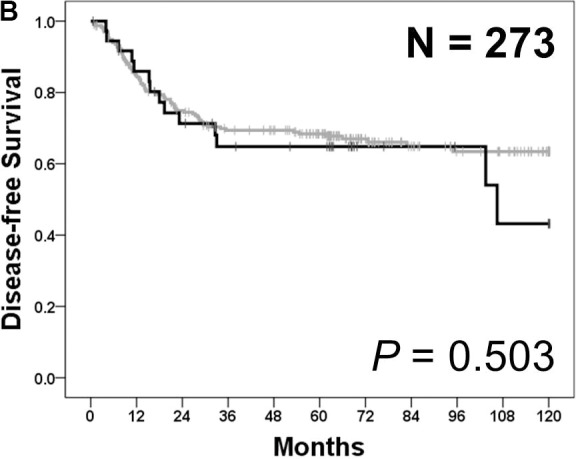
(A) Overall survival was similar between patients with PIK3CA-mutant tumors (black) and those with PIK3CA wild-type tumors (gray) (p = 0.944). (B) In patients with stage I–III gastric cancer (n = 273), disease-free survival was similar between patients with PIK3CA-mutant tumors (black) and those with PIK3CA wild-type tumors (gray) (p = 0.503).
The same analyses were performed for 273 patients with stage IB–III tumors who had received curative surgery. The clinical and pathological characteristics of these patients according to their PIK3CA mutation status are compared in Supplementary Table 2. The results obtained from these patients were almost the same as those obtained for all the patients in cohort 1. Disease-free survival (DFS) after the radical surgery was not significantly different between patients with PIK3CA-mutant tumors and those with PIK3CA wild-type tumors (5-year DFS rate: 64.8% [mutant] vs. 68.4% [wild-type]; p = 0.503; Figure 1B).
Cohort 2
In all, 120 patients were consecutively included. The male to female ratio was 77:43, and the median age was 58 years (range, 25–88 years). All the patients in cohort 2 had stage IV disease (distant metastasis) at diagnosis and underwent palliative gastrectomy. In addition, 104 patients (86.7%) in this cohort received palliative chemotherapy after the surgery.
In all, 16 PIK3CA mutations were detected in 15 patients (12.5%), with E545X in 6 (5.0%) patients, H1047X in 5 (4.2%) patients, Q546X in 2 (1.7%) patients, R88Q in 2 (1.7%) patients, and E542K in 1 (0.8%) patient (Supplementary Table 1). The clinicopathological characteristics of cohort 2 according to their PIK3CA mutation status are compared in Table 2. PIK3CA-mutant tumors frequently showed vascular invasion (80.0% vs. 47.6%; p = 0.019) and MSI-H (20.0% vs. 3.8%; p = 0.041). Cytoplasmic Akt expression was significantly increased in PIK3CA mutant tumors (IHC 2+ or 3+: 26.7% vs. 11.5%; p = 0.018). However, primary tumor location, histological subtypes, T stage, lymphatic or perineural invasion, stromal reaction, and EBER ISH positivity were not significantly different between PIK3CA-mutant and PIK3CA wild-type tumors, which was inconsistent with the results obtained for cohort 1. OS was not significantly different between patients with PIK3CA-mutant tumors and those with PIK3CA wild-type tumors (median OS: 16.8 months [range, 7.1–26.5 months] in PIK3CA-mutant patients vs. 20.8 months [range, 14.9–26.7 months] in PIK3CA wild-type patients; p = 0.416; Figure 2).
Table 2. Clinicopathological characteristics of patients in cohort 2 according to their PIK3CA mutation status.
| Characteristics | PIK3CA wild-type | PIK3CA mutant | p-value |
|---|---|---|---|
| (n = 105) (%) | (n = 15) (%) | ||
| Sex | |||
| Male | 70 (66.7) | 7 (46.7) | 0.131 |
| Female | 35 (33.3) | 8 (53.3) | |
| Age | |||
| < 70 years | 83 (79.0) | 10 (66.7) | 0.324 |
| ≥ 70 years | 22 (21.0) | 5 (33.3) | |
| Primary tumor location* | |||
| Lower third | 47 (45.2) | 7 (46.7) | 0.395 |
| Middle third | 28 (26.9) | 6 (40.0) | |
| Upper third | 29 (27.9) | 2 (13.3) | |
| Pathology | |||
| Well or moderately differentiated adenocarcinoma | 28 (26.7) | 4 (26.7) | 0.801 |
| Poorly differentiated adenocarcinoma | 58 (55.2) | 9 (60.0) | |
| Signet ring cell carcinoma | 11 (10.5) | 1 (6.7) | |
| Mucinous carcinoma | 8 (7.6) | 1 (6.7) | |
| Lauren classification | |||
| Intestinal type | 29 (27.6) | 7 (46.7) | 0.107 |
| Diffuse type | 72 (68.6) | 8 (53.3) | |
| Mixed type | 4 (3.8) | 0 (0) | |
| T stage | |||
| pT1/T2/T3 | 17 (16.2) | 1 (6.7) | 0.464 |
| pT4 | 88 (83.8) | 14 (93.3) | |
| N stage | |||
| pN0/N1/N2 | 21 (20.0) | 2 (13.3) | 0.733 |
| pN3 | 84 (80.0) | 13 (86.7) | |
| Lymphatic invasion | |||
| Absent | 11 (10.5) | 2 (13.3) | 0.666 |
| Present | 94 (89.5) | 13 (86.7) | |
| Vascular invasion | |||
| Absent | 55 (52.4) | 3 (20.0) | 0.019 |
| Present | 50 (47.6) | 12 (80.0) | |
| Perineural invasion | |||
| Absent | 16 (15.2) | 2 (13.3) | 1.000 |
| Present | 89 (84.8) | 13 (86.7) | |
| Stroma reaction | |||
| Absent | 59 (56.2) | 8 (53.3) | 0.868 |
| Desmoplasia | 39 (37.2) | 6 (40.0) | |
| Lymphoid or neutrophil | 7 (6.7) | 1 (6.7) | |
| EBER in situ hybridization | |||
| Negative | 100 (95.2) | 14 (93.3) | 0.559 |
| Positive | 5 (4.8) | 1 (6.7) | |
| MSI | |||
| MSS | 101 (96.2) | 12 (80.0) | 0.041 |
| MSI-H | 4 (3.8) | 3 (20.0) | |
| Gastrectomy | |||
| Subtotal gastrectomy | 56 (53.3) | 9 (60.0) | 0.628 |
| Total gastrectomy | 49 (46.7) | 6 (40.0) | |
| Akt expression (cytoplasmic intensity) | |||
| Negative | 11 (11.5) | 1 (6.7) | 0.018 |
| 1+ | 74 (77.1) | 10 (66.7) | |
| 2+ | 9 (9.4) | 2 (13.3) | |
| 3+ | 2 (2.1) | 2 (13.3) |
Abbreviations: AJCC = American Joint Committee on Cancer; EBER = Epstein-Barr virus-encoded small RNA.
*Patients whose disease involved the entire stomach (n = 1) were excluded.
Figure 2. Overall survival according to PIK3CA mutation status in cohort 2.
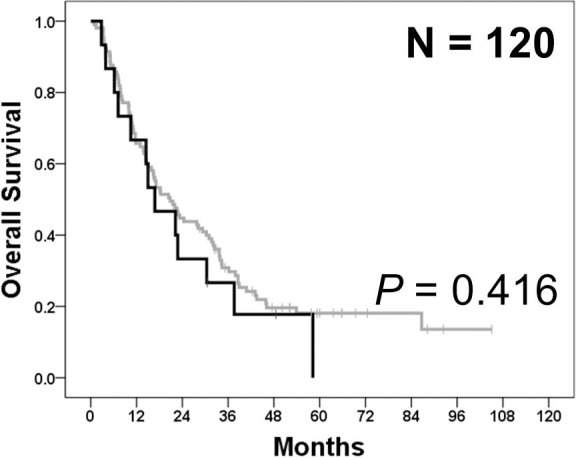
In cohort 2, overall survival was not significantly different between patients with PIK3CA-mutant tumors (black) and those with PIK3CA wild-type tumors (gray) (p = 0.416).
DISCUSSION
To the best of our knowledge, this is the largest study focused on the clinical implications of PIK3CA mutations in GC. In addition, this is the first and largest study that shows the clinical implication of PIK3CA mutations in GC patients with distant metastasis. In a previous study of Fang, et al., although more patients (n = 431) were included, they analyzed somatic mutations in the PI3K/Akt pathway genes altogether (including AKT1, AKT2, AKT3, PTEN, and PIK3CA), and did not conduct separate analyses focused on PIK3CA mutations [10]. Thus, their results did not provide any direct insights on the clinicopathological implications of PIK3CA mutations in GC patients. Moreover, they did not include GC patients with distant metastasis. In our study, the frequency of PIK3CA mutations was similar between 2 cohorts. In cohort 1, PIK3CA mutations were frequently detected in tumors located in the upper third of the stomach. In addition, these tumors significantly showed aggressive behavior such as advanced T stage; poorly differentiated histology; diffuse- or mixed-type tumors; and lymphatic, vascular, and perineural invasion. These findings were consistent when patients with stage IV GC were excluded (Supplementary Table 2). However, in cohort 2, primary tumor location and tumor behavior, except vascular invasion and MSI, were not significantly different according to PIK3CA mutation status.
Previous studies could not find a significant association between PIK3CA mutations and primary tumor location in GC [11, 12]. Recently, Fang, et al. analyzed mutations in the PI3K/Akt pathway by using tumor tissues from 431 patients with stage I-III GC [10]. This study showed that approximately two-third mutations in the PI3K/Akt pathway were located in PIK3CA. In addition, they reported that the mutations in the PI3K/Akt pathway were not significantly associated with primary tumor location. However, in intestinal-type patients, tumors with mutated PI3K/Akt pathway were less frequently located in the upper third of the stomach. In contrast, in diffuse-type patients, tumors with mutated PI3K/Akt pathway were more frequently located in the upper third of the stomach. Because mutations in the PI3K/Akt pathway located in AKT1, AKT2, AKT3, PTEN, and PIK3CA were analyzed altogether [10], their results did not provide any direct insights on the association between PIK3CA mutation status and primary tumor location. In our study, we could not find a significant association between PIK3CA mutations and primary tumor location according to Lauren classification (Supplementary Table 3). However, in diffuse-type patients in cohort 1, PIK3CA-mutant tumors tended to be more frequently located in the upper third of the stomach than PIK3CA wild-type tumors.
In cohort 1, PIK3CA-mutated tumors had significantly more aggressive features than PIK3CA wild-type tumors. However, in cohort 2, except vascular invasion, other aggressive characteristics in PIK3CA-mutated tumors in cohort 1 were not observed. Because all the patients in cohort 2 had stage IV disease, the proportion of tumors showing lymphatic, vascular, and perineural invasion was higher compared with cohort 1. Therefore, the effects of PIK3CA mutations on tumor aggressiveness in cohort 2 appear less distinct than those in cohort 1. The results suggest that PIK3CA mutations might be involved in the aggressiveness of GC in the locoregional stage, rather than in the metastatic stage. Previous studies have failed to identify the association due in part to the relatively small sample size [14–17]. Fang and colleagues showed that mutations in the PI3K/Akt pathway were more frequently present in intestinal-type tumors [10]. However, other characteristics associated with tumor aggressiveness, including lymphovascular invasion, were not significantly different according to the PI3K/Akt pathway mutation status. Barbi, et al. reported that although PIK3CA-mutant tumors were mostly in advanced T-stage, other characteristics of tumor aggressiveness were not significantly different according to PIK3CA mutation status [12].
Previous studies have uniformly reported that PIK3CA mutations are not associated with the prognosis of patients with GC [10, 12–17]. Our study demonstrated again that the PIK3CA mutations were not associated with poor survival outcomes in patients with GC, although PIK3CA-mutated tumors had more aggressive pathologic features. It is known that lymphatic, vascular, and perineural invasions, and poorly differentiated histology are related to poor prognosis [18–21], while MSI-H is a good prognostic factor in patients with GC [22–24]. Therefore, the coexistence of both good and poor prognostic factors in patients with PIK3CA mutations might explain the discrepancy between pathologic features and survival outcomes. In addition, it is also possible that, if PIK3CA mutation increases the aggressiveness of GC mainly in the locoregional stage, the mutation may not influence the prognosis of patients after appropriate radical surgery and adjuvant chemotherapy.
Mutant PIK3CA activates Akt pathways, which in turn promote the growth and invasion of cancer cells [7]. This mechanism has also been explored in GC [25]. In line with these results, we confirmed that PIK3CA mutations are associated with Akt activation in GC, thus promoting tumor aggressiveness. An aberrantly activated PI3K/Akt pathway in PIK3CA-mutated GC could be a potential therapeutic target in these patients. The PI3K/Akt pathway activates mammalian target of rapamycin (mTOR). An mTOR inhibitor everolimus has been investigated in patients with GC regardless of PIK3CA mutation status. In phase II studies, everolimus has shown promising clinical activity in patients with previously treated GC [26, 27]. However, a phase III trial GRANITE-1 failed to show the superiority of everolimus over placebo with respect to OS [28]. Nevertheless, a recent case report showed good treatment response to everolimus in a patient with GC harboring PIK3CA mutation and showing pS6 overexpression [29]. In addition, in vitro studies involving various cancer cell lines suggest that PIK3CA mutations are predictive markers of everolimus sensitivity [30, 31]. BKM120, a direct PIK3CA inhibitor, and BEZ234, a dual PIK3CA and mTOR inhibitor, exert pro-apoptotic effects on gastric and colorectal cancer cell lines [32]. Moreover, the anti-proliferative effects of everolimus were more potent in PIK3CA-mutated cell lines than in PIK3CA wild-type cell lines. Therefore, further clinical trials targeting PI3K/Akt pathway in GC should be conducted in selected patients such as PIK3CA-mutated ones, not all comers.
Our study has some limitations. First, although this is the largest study focused on the clinicopathological and prognostic implications of PIK3CA mutations in GC, in some analyses, the statistical power is still insufficient to draw strong conclusions due to the limited sample size. For example, a non-inferiority log rank test of OS with 262 patients with PIK3CA wild-type and 40 patients with PIK3CA mutant achieved 52.1% power at a 0.05 type I error to detect an equivalence hazard ratio of 1.50 in cohort 1, when we conducted a post hoc power calculation. Therefore, in further studies, a larger sample size is required to robustly validate all findings we observed. Second, since we analyzed relatively frequent and well renowned hotspot mutations in exons 1, 4, 7, 9, and 20 of the PIK3CA gene, the implications of relatively rare or non-hotspot PIK3CA mutations that were not covered in this study are still unknown. This limitation could be overcome by using the next generation sequencing methods in further studies.
In conclusion, PIK3CA mutations were associated with increased tumor aggressiveness, especially in locoregional disease, and Akt activation in GC. However, PIK3CA mutation status was not related to the prognosis of patients. Our data suggest that PIK3CA-mutated GC is a distinct disease entity, which might need a different therapeutic approach.
MATERIALS AND METHODS
Patient cohorts and tissue microarrays
In cohort 1, patients with GC who underwent curative or palliative gastrectomy at Seoul National University Bundang Hospital (SNUBH) between May 2003 and Dec. 2005 were enrolled consecutively. Patients were excluded if they had stage IA (pT1N0M0) GC according to the 7th edition of the American Joint Committee on Cancer (AJCC) staging system. In cohort 2, patients with stage IV GC (distant metastasis) who underwent palliative gastrectomy at SNUBH between Jul. 2006 and Dec. 2012 were enrolled consecutively. Tissue microarrays were constructed by isolating representative 2-mm cores from surgical specimens by an experienced gastrointestinal pathologist. Baseline demographic, pathological, and clinical data, including DFS and OS, were collected by retrospectively reviewing electronic medical records of the patients. DFS was calculated from the date of the surgery to documented disease recurrence or death from any cause. OS was calculated from the date of the surgery to death from any cause.
PIK3CA mutation analyses by performing real-time polymerase chain reaction
All tumor samples were collected from surgical resection specimens. Hematoxylin-Eosin stained slides were reviewed by a pathologist (H.S.L.). Tumor areas were identified, and more than 1 × 1 cm area, which contained more than 60% tumor cells, was microscopically dissected. One or two 8-μm-thick formalin-fixed paraffin-embedded (FFPE) tumor tissue sections were deparaffinized. DNA was isolated using cobas® DNA Sample Preparation Kit (Roche, Branchburg, NJ, USA). Concentration of the isolated DNA was measured using NanoDrop UV spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA). The isolated DNA was diluted using DNA specimen diluent provided in cobas® 4800 Mutation Test Kit (Roche) to an optimal concentration of 2 ng/μL. Amplification and detection were performed using automated cobas® X480 analyzer (Roche). Real-time polymerase chain reaction (PCR) detected mutations in exons 1, 4, 7, 9, and 20 of PIK3CA. Results of real-time PCR are reported using the following 12 categories: (1) E542K, (2) N345K, (3) H1047X (L, R, or Y), (4) E545X (A, D, G, or K), (5) C420R, (6) G1049R, (7) Q546X (K, R, E, or L), (8) R88Q, (9) M1043I, (10) mutation not detected, (11) invalid (sample out of range/control failure), and (12) failed (hardware/software failure).
MSI analysis
DNA was extracted from tumor tissue and corresponding non-neoplastic gastric mucosa tissue by using InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA, USA). PCR was performed using fluorescent dye-labeled primers targeting five microsatellite markers, namely, BAT25, BAT26, D2S123, D5S346, and D17S250, as recommended in Bethesda guideline on MSI [33]. Fragment analysis was performed using ABI 3130 × l genetic analyzer and GeneMapper® software (Applied Biosystems, Foster City, CA, USA). Tumors were classified as MSI-H when at least two of the five markers yielded novel bands, MSI-low when additional alleles were observed with one of the five markers, and microsatellite stable when all the microsatellite markers examined showed identical patterns in both tumor and normal tissues.
EBER ISH
EBER ISH was performed using Ventana Benchmark XT (Ventana ISH iView kit, Ventana Corporation, Tucson, AZ, USA), according to manufacturer’s instructions. Briefly, 4-µm FFPE sections were deparaffinized and digested with protease II for 8 min. Next, probes were added to the samples, and the samples were denatured at 85°C for 12 min and were hybridized at 57°C for 1 h. After hybridization, the tissue samples were washed and incubated with anti-fluorescein monoclonal antibody for 20 min. Detection was performed using iView Blue Biotinylated Ig for 8 min (Ventana Corporation). Counterstaining was performed using Nuclear Fast Red II for 4 min (Ventana Corporation).
IHC for Akt
IHC for Akt was performed using an antibody against Akt1 (dilution, 1:100; catalog no. ab32505; Abcam, Burlingame, CA, USA). Immunostaining was performed using Leica Bond-Max Automation and Leica Bond Polymer Detection Kit (Leica Biosystems, Bannockburn, IL, USA), according to the manufacturer’s recommendations. Sections were deparaffinized, and antigens were retrieved using pH 9.0 Bond Epitope Retrieval Solution 2. Slides were incubated with the primary antibody for 60 min, followed by incubation with the polymer for 8 min. After counterstaining, Akt expression in the cytoplasm of tumor cells was examined. Staining intensity was graded as follows: 0, negative; 1+, weakly positive; 2+, moderately positive; and 3+, strongly positive. In addition to staining intensity, area of positive cancer cells was scored. For statistical analysis, cytoplasmic AKT staining was regarded as positive if 5% or more cancer cells expressed it.
Statistical analysis
Statistical analysis of categorical variables was performed using Pearson’s chi-square test, Fisher’s exact test, or linear-by-linear association, as appropriate. Median DFS and OS were calculated using Kaplan–Meier method. Survival data were compared using log rank test. All the statistical tests were two-sided, with level of significance defined at p < 0.05. All the analyses were performed using IBM SPSS version 22.0 (IBM, Armonk, NY, USA).
SUPPLEMENTARY MATERIALS TABLES
Footnotes
Ethical considerations
This study protocol was reviewed and approved by the institutional review board (IRB) of Seoul National University Hospital (IRB registration no. B-1407/260-305) and was conducted in accordance with the precepts established by the Declaration of Helsinki.
CONFLICTS OF INTEREST
The authors declare that they do not have any conflicts of interest.
GRANT SUPPORT
This study was supported in part by a research grant from the Seoul National University Bundang Hospital Research Fund (02-2015-009).
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, Ye XS, Do IG, Liu S, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21:449–456. doi: 10.1038/nm.3850. [DOI] [PubMed] [Google Scholar]
- 4.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- 5.Samuels Y, Wang Z, Bardelli A, Silliman N, Ptak J, Szabo S, Yan H, Gazdar A, Powell SM, Riggins GJ, Willson JK, Markowitz S, Kinzler KW, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 6.Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006;94:455–459. doi: 10.1038/sj.bjc.6602970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuels Y, Diaz LA, Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 2005;7:561–573. doi: 10.1016/j.ccr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 8.Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM, Stemke-Hale K, Hauptmann M, Beijersbergen RL, Mills GB, van de Vijver MJ, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12:395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Majewski IJ, Nuciforo P, Mittempergher L, Bosma AJ, Eidtmann H, Holmes E, Sotiriou C, Fumagalli D, Jimenez J, Aura C, Prudkin L, Diaz-Delgado MC, de la Pena L, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015;33:1334–1339. doi: 10.1200/JCO.2014.55.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fang WL, Huang KH, Lan YT, Lin CH, Chang SC, Chen MH, Chao Y, Lin WC, Lo SS, Li AF, Wu CW, Chiou SH, Shyr YM. Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget. 2016;7:6201–6220. doi: 10.18632/oncotarget.6641. https://doi.org/10.18632/oncotarget.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sukawa Y, Yamamoto H, Nosho K, Kunimoto H, Suzuki H, Adachi Y, Nakazawa M, Nobuoka T, Kawayama M, Mikami M, Matsuno T, Hasegawa T, Hirata K, et al. Alterations in the human epidermal growth factor receptor 2-phosphatidylinositol 3-kinase-v-Akt pathway in gastric cancer. World J Gastroenterol. 2012;18:6577–6586. doi: 10.3748/wjg.v18.i45.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barbi S, Cataldo I, De Manzoni G, Bersani S, Lamba S, Mattuzzi S, Bardelli A, Scarpa A. The analysis of PIK3CA mutations in gastric carcinoma and metanalysis of literature suggest that exon-selectivity is a signature of cancer type. J Exp Clin Cancer Res. 2010;29:32. doi: 10.1186/1756-9966-29-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bria E, De Manzoni G, Beghelli S, Tomezzoli A, Barbi S, Di Gregorio C, Scardoni M, Amato E, Frizziero M, Sperduti I, Corbo V, Brunelli M, Bersani S, et al. A clinical-biological risk stratification model for resected gastric cancer: prognostic impact of Her2, Fhit, and APC expression status. Ann Oncol. 2013;24:693–701. doi: 10.1093/annonc/mds506. [DOI] [PubMed] [Google Scholar]
- 14.Lee H, Hwang IS, Choi IJ, Kang YN, Park KU, Lee JH. Are PIK3CA Mutation and Amplification Associated with Clinicopathological Characteristics of Gastric Cancer? Asian Pac J Cancer Prev. 2015;16:4493–4496. doi: 10.7314/apjcp.2015.16.11.4493. [DOI] [PubMed] [Google Scholar]
- 15.Takahashi N, Yamada Y, Taniguchi H, Fukahori M, Sasaki Y, Shoji H, Honma Y, Iwasa S, Takashima A, Kato K, Hamaguchi T, Shimada Y. Clinicopathological features and prognostic roles of KRAS, BRAF, PIK3CA and NRAS mutations in advanced gastric cancer. BMC Res Notes. 2014;7:271. doi: 10.1186/1756-0500-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Yao D, Liu W, Wang N, Lv H, Zhang G, Ji M, Xu L, He N, Shi B, Hou P. Highly frequent PIK3CA amplification is associated with poor prognosis in gastric cancer. BMC Cancer. 2012;12:50. doi: 10.1186/1471-2407-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada K, Baba Y, Shigaki H, Ishimoto T, Miyake K, Kosumi K, Tokunaga R, Izumi D, Ohuchi M, Nakamura K, Kiyozumi Y, Kurashige J, Iwatsuki M, et al. Prognostic and clinical impact of PIK3CA mutation in gastric cancer: pyrosequencing technology and literature review. BMC Cancer. 2016;16:400. doi: 10.1186/s12885-016-2422-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dicken BJ, Graham K, Hamilton SM, Andrews S, Lai R, Listgarten J, Jhangri GS, Saunders LD, Damaraju S, Cass C. Lymphovascular invasion is associated with poor survival in gastric cancer: an application of gene-expression and tissue array techniques. Ann Surg. 2006;243:64–73. doi: 10.1097/01.sla.0000194087.96582.3e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Ling YH, Zhu CM, Hu WM, Zhang XK, Luo RZ, He JH, Yun JP, Li YF, Cai MY. Vascular invasion as an independent predictor of poor prognosis in nonmetastatic gastric cancer after curative resection. Int J Clin Exp Pathol. 2015;8:3910–3918. [PMC free article] [PubMed] [Google Scholar]
- 20.Deng J, You Q, Gao Y, Yu Q, Zhao P, Zheng Y, Fang W, Xu N, Teng L. Prognostic value of perineural invasion in gastric cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e88907. doi: 10.1371/journal.pone.0088907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bamboat ZM, Tang LH, Vinuela E, Kuk D, Gonen M, Shah MA, Brennan MF, Coit DG, Strong VE. Stage-stratified prognosis of signet ring cell histology in patients undergoing curative resection for gastric adenocarcinoma. Ann Surg Oncol. 2014;21:1678–1685. doi: 10.1245/s10434-013-3466-8. [DOI] [PubMed] [Google Scholar]
- 22.Falchetti M, Saieva C, Lupi R, Masala G, Rizzolo P, Zanna I, Ceccarelli K, Sera F, Mariani-Costantini R, Nesi G, Palli D, Ottini L. Gastric cancer with high-level microsatellite instability: target gene mutations, clinicopathologic features, and long-term survival. Hum Pathol. 2008;39:925–932. doi: 10.1016/j.humpath.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 23.Corso G, Pedrazzani C, Marrelli D, Pascale V, Pinto E, Roviello F. Correlation of microsatellite instability at multiple loci with long-term survival in advanced gastric carcinoma. Arch Surg. 2009;144:722–727. doi: 10.1001/archsurg.2009.42. [DOI] [PubMed] [Google Scholar]
- 24.Fang WL, Chang SC, Lan YT, Huang KH, Chen JH, Lo SS, Hsieh MC, Li AF, Wu CW, Chiou SH. Microsatellite instability is associated with a better prognosis for gastric cancer patients after curative surgery. World J Surg. 2012;36:2131–2138. doi: 10.1007/s00268-012-1652-7. [DOI] [PubMed] [Google Scholar]
- 25.Matsuoka T, Yashiro M. The Role of PI3K/Akt/mTOR Signaling in Gastric Carcinoma. Cancers (Basel) 2014;6:1441–1463. doi: 10.3390/cancers6031441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doi T, Muro K, Boku N, Yamada Y, Nishina T, Takiuchi H, Komatsu Y, Hamamoto Y, Ohno N, Fujita Y, Robson M, Ohtsu A. Multicenter phase II study of everolimus in patients with previously treated metastatic gastric cancer. J Clin Oncol. 2010;28:1904–1910. doi: 10.1200/JCO.2009.26.2923. [DOI] [PubMed] [Google Scholar]
- 27.Yoon DH, Ryu MH, Park YS, Lee HJ, Lee C, Ryoo BY, Lee JL, Chang HM, Kim TW, Kang YK. Phase II study of everolimus with biomarker exploration in patients with advanced gastric cancer refractory to chemotherapy including fluoropyrimidine and platinum. Br J Cancer. 2012;106:1039–1044. doi: 10.1038/bjc.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohtsu A, Ajani JA, Bai YX, Bang YJ, Chung HC, Pan HM, Sahmoud T, Shen L, Yeh KH, Chin K, Muro K, Kim YH, Ferry D, et al. Everolimus for previously treated advanced gastric cancer: results of the randomized, double-blind, phase III GRANITE-1 study. J Clin Oncol. 2013;31:3935–3943. doi: 10.1200/JCO.2012.48.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park JH, Ryu MH, Park YS, Park SR, Na YS, Rhoo BY, Kang YK. Successful control of heavily pretreated metastatic gastric cancer with the mTOR inhibitor everolimus (RAD001) in a patient with PIK3CA mutation and pS6 overexpression. BMC Cancer. 2015;15:119. doi: 10.1186/s12885-015-1139-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meric-Bernstam F, Akcakanat A, Chen H, Do KA, Sangai T, Adkins F, Gonzalez-Angulo AM, Rashid A, Crosby K, Dong M, Phan AT, Wolff RA, Gupta S, et al. PIK3CA/PTEN mutations and Akt activation as markers of sensitivity to allosteric mTOR inhibitors. Clin Cancer Res. 2012;18:1777–1789. doi: 10.1158/1078-0432.CCR-11-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weigelt B, Warne PH, Downward J. PIK3CA mutation, but not PTEN loss of function, determines the sensitivity of breast cancer cells to mTOR inhibitory drugs. Oncogene. 2011;30:3222–3233. doi: 10.1038/onc.2011.42. [DOI] [PubMed] [Google Scholar]
- 32.Mueller A, Bachmann E, Linnig M, Khillimberger K, Schimanski CC, Galle PR, Moehler M. Selective PI3K inhibition by BKM120 and BEZ235 alone or in combination with chemotherapy in wild-type and mutated human gastrointestinal cancer cell lines. Cancer Chemother Pharmacol. 2012;69:1601–1615. doi: 10.1007/s00280-012-1869-z. [DOI] [PubMed] [Google Scholar]
- 33.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, Hamilton SR, Hiatt RA, Jass J, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


