Abstract
PITX2 is a homeobox transcription factor involved in embryonic left/right signaling and more recently has been associated to cardiac arrhythmias. Genome wide association studies have pinpointed PITX2 as a major player underlying atrial fibrillation (AF). We have previously described that PITX2 expression is impaired in AF patients. Furthermore, distinct studies demonstrate that Pitx2 insufficiency leads to complex gene regulatory network remodeling, i.e. Wnt>microRNAs, leading to ion channel impairment and thus to arrhythmogenic events in mice. Whereas large body of evidences has been provided in recent years on PITX2 downstream signaling pathways, scarce information is available on upstream pathways influencing PITX2 in the context of AF. Multiple risk factors are associated to the onset of AF, such as e.g. hypertension (HTN), hyperthyroidism (HTD) and redox homeostasis impairment. In this study we have analyzed whether HTN, HTD and/or redox homeostasis impact on PITX2 and its downstream signaling pathways. Using rat models for spontaneous HTN (SHR) and experimentally-induced HTD we have observed that both cardiovascular risk factors lead to severe Pitx2 downregulation. Interesting HTD, but not SHR, leads to up-regulation of Wnt signaling as well as deregulation of multiple microRNAs and ion channels as previously described in Pitx2 insufficiency models. In addition, redox signaling is impaired in HTD but not SHR, in line with similar findings in atrial-specific Pitx2 deficient mice. In vitro cell culture analyses using gain- and loss-of-function strategies demonstrate that Pitx2, Zfhx3 and Wnt signaling influence redox homeostasis in cardiomyocytes. Thus, redox homeostasis seems to play a pivotal role in this setting, providing a regulatory feedback loop. Overall these data demonstrate that HTD, but not HTN, can impair Pitx2>>Wnt pathway providing thus a molecular link to AF.
Introduction
Atrial fibrillation (AF) is the most frequent arrhythmogenic defect in the human population, with an estimate incidence of 2–4% in the general population but rising up to 10% in the elderly [1]. Genetic mutations in a large array of ion channel encoding genes have been described, although only representing <10% of AF cases [2–5]. Recently, genome wide association studies (GWAS) have identified a discrete number of risk variants linked to AF. In particular, SNPs located in chromosome region 4q25, thus in the vicinity of PITX2/ENPEP, display highest association significance [6], while other SNPs linked to ZFHX3 (16q22)[7–8], KCNN3 (1q21) [9] and IL6R (16q13) [10] display more modest significance. Functional evidences demonstrated that 4q25 genomic region containing these risk variants can interact with PITX2 and ENPEP promoter sequences [11]. However it remains elusive how variation within other SNPs (ZFHX3 (16q22), KCNN3 (1q21) and IL6R (16q13)) is mechanistically linked to AF.
Experimental analyses demonstrated that Pitx2 insufficiency leads to atrial arrhythmias [12–14] by modulating distinct ion channels that contribute to the configuration of the cardiac action potential [13–15], as well as cell-cell gap junctional and calcium handling proteins [13,16]. In addition, Pitx2 modulates expression of several GWAS associated genes, such as IL6R, KCNN3 and ZFHX3. Importantly it also regulates WNT8 expression which, in turn, modulates a complex gene regulatory network, including multiple microRNAs, with a large impact on calcium homeostasis control and pro-arrhythmogenic events [16].
It is well-established that the onset of an AF episode triggers subsequent and more severe AF episodes, leading to electrical and structural remodeling of the diseased heart, a condition quoted as “AF begets AF” [17]. Electrical remodeling involves progressive changes in the cardiac electrical properties, leading to early afterdepolarization, delayed afterdepolizations and/or changes in the action potential duration configuration [17], culminating thus in rotor formation [18]. In this context, a pivotal role of reactive oxidative stress has been recently reported [19–20]. Structural remodeling involves atrial dilation, fibrosis and/or inflammation [21] indirectly promoting rotor formation and thus electrical re-entry circuitries [18].
Cardiovascular risk factors such hypertension (HTN), hyperthyroidism (HTD), diabetes and obesity have been repetitively demonstrated to promote onset of atrial fibrillation, respectively [22–24]. Furthermore, the occurrence of AF can be also triggered by preceding cardiovascular diseases such as hypertrophic cardiomyopathy and valvular heart diseases [23,25–26]. Whereas large body of evidences has been provided in recent years on PITX2 downstream signaling pathways, scarce information is available on upstream pathways influencing PITX2 in the context of AF. A seminal study demonstrated that prolonged hypertension is capable of decreasing PITX2 expression [27] but the functional consequences of such changes remain unexplored. Furthermore, PITX2 has been recently reported to control redox homeostasis in skeletal muscle [28] as well as in the regenerating heart [29] but not linked to redox homeostasis and cardiac arrhythmogenic defects has been reported.
In this study we demonstrate that Pitx2 expression is impaired in HTN and HTD experimental models. Interestingly, HTD but not HTN elicits a complex impairment of PITX2>Wnt>microRNA signaling which leads to abnormal ion channel expression. Importantly, ROS signaling is also altered in Pitx2 deficient mice and impaired ROS signaling regulates Pitx2>Wnt>microRNA cascade. Overall, our data demonstrate a complex regulatory network of AF risk factors and PITX2 downstream signaling providing additional molecular insights linking pro-arrhythmogenic substrates and AF.
Materials & methods
Experimental models
The Pitx2floxed and NppaCre transgenic mouse lines have been previously described [30–31]. Generation of conditional atrial (NppaCre) mutant mice has been previously described [14–15]. Three different conditions were used for the NppaCrePitx2 mice: wild-type Cre controls (NppaCre2Pitx2fl/fl), atrial-specific heterozygous (NppaCre+Pitx2fl/-), and atrial-specific homozygous (NppaCre+Pitx2-/-). This investigation conforms with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. The study was approved by the University of Jaén Bioethics Committee.
Male Wistar rats weighing 250–280 g were maintained on standard chow and tap water ad libitum. Two groups of animals were analyzed; a) control and b) hyperthyroid rats as previously described [32]. Briefly, hyperthyroidism was induced by injecting s.c. thyroxine (75 Mg/rat/day) for four weeks. Spontaneously hypertensive rats (SHR) and their corresponding Wistar Kyoto controls (WKY) were purchased to Harlan Laboratories with 8 weeks old, and maintained on standard chow and tap water ad libitum during 24 weeks. Tail systolic BP (SBP) and heart rate (HR) were recorded once a month by using tail-cuff plethysmography in unanaesthetized rats (LE 5001-Pressure Meter, Letica SA, Barcelona, Spain) as illustrated in S1 Fig.
Mouse genotyping
DNA for PCR screening was extracted from adult ear and/or tail samples and from embryonic yolk sacs. Screening of Cre and Pitx2 floxed alleles was routinely done using used specific primers as previously described [16]. Cycling conditions for Cre were as follows; 5 min at 95°C, 35 cycles of 30s at 95°C, 30s at 60°C and 90s at 72°C, and for Pitx2 as follows; 5 min at 95°C. 40 cycles of 30s at 95°C, 30s at 60°C and 90s at 72°C, followed by a final extension step of 10 min at 72°C, respectively.
Tissue and RNA isolation
When the experimental period was completed, rats were anaesthetized with thiobutabarbital (100 mg/kg IP, Inactin, Research Biochemicals International) and maintained at 37°C on a servo-controlled heated rodent operating table. Heart tissue samples corresponding to left (LA) and right (RA) atrial appendages and ventricular chambers were collected, processed accordingly to RNA isolation and stored at -80°C until used.
Genetically modified Pitx2 mice, and their corresponding controls, were sacrificed by cervical dislocation. Adult hearts were carefully dissected and briefly rinsed in Ringer’s solution. Tissue samples corresponding to the RA and LA were collected for each experimental condition, immediately snap-frozen in liquid nitrogen, and stored at -80°C until used. Pooled samples of at least three independent mice were processed for each condition, respectively. Three independent pooled samples were further processed for RNA isolation and qPCR analyses. Total RNA was isolated using Trizol (Roche) according to manufacture’s guidelines and DNase treated using RNase-Free DNase (Roche) for 1h at 30°C. In all cases, at least three distinct pooled samples were used to perform the corresponding qRT-PCR experiments.
First strand cDNA was synthesized at 50°C for 1h using 1 μg of RNA, oligo-dT primers and Superscript III Reverse Transcriptase (Invitrogen) according to manufacture’s guidelines. Negative controls to assess genomic contamination were performed for each sample, without reverse transcriptase, which resulted in all cases in no detectable amplification product.
qRT-PCR (mRNA)
RT-PCR was performed in Mx3005Tm QPCR System with an MxPro QPCR Software 3.00 (Stratagene) and SyBR Green detection system. Reactions were performed in 96-well plates with optical sealing tape (Cultek) in 20 μL total volume containing SYBR Green Mix (Finnzymes) and the corresponding cDNA. Three internal controls, mouse βactin, Gusb and GAPDH, were used in parallel for each run and represented as previously described [14,33]. Amplification conditions were as follows: denaturisation step of 95°C for 10 min, followed by 40 cycles of 95°C for 30s, 60°C for 30s, 72°C for 30s; with final elongation step of 72°C for 10 min. All primers were designed to span exon-exon boundaries using online Primer3 software Primer3input (primer3 www.Cgi v 0.2). Primer sequences are provided in S1 Table. No amplifications were observed in PCR control reactions containing only water as the template. Each PCR reaction was performed at least three times to obtain representative averages. The Livak method was used to analyze the relative quantification RT-PCR data [33] and normalized in all cases taking as 100% the wild-type (control) value, as previously described [14].
qRT-PCR (microRNA)
microRNA qRT-PCR was performed using Exiqon LNA microRNA qRT-PCR primers and detection kit according to manufacturer’s guidelines. All reactions were always run in triplicates using 5S as normalizing control, as recommended by the manufacturer. SyBR Green was used as quantification system on a Stratagene Q-Max 2005P qRT-PCR thermocycler. Relative measurements were calculated as described by Livak & Schmittgen [33] and control measurements were normalized to represent 100% as previously described [14].
Plasmid, microRNA and siRNA cell transfections
HL-1 cells (6 × 105 cells per well) were transfected with plasmids containing expression constructs for Pitx2, Wnt8a (Addgene), Wnt11a (Addgene, Cambridge, MA, USA), premiR-29a, pre-miR-200 (Exiqon) or siRNA-Pitx2c, siRNA-Zfhx3, siRNA-Enpep, siRNA-Sod2 (Sigma, Aldrich, Munich, Germany) as previously described [14,34]. Primary cultures of mouse fetal (E17.5) cardiomyocytes were isolated using standard procedures [35], cultures accordingly and treated with T4 hormone as previously reported [36]. siRNA sequences are provided in S1 Table.
Statistical analyses
For statistical analyses of datasets, unpaired Student’s t-tests were used. Significance levels or P values are stated in each corresponding figure legend. P < 0.05 was considered statistically significant.
Results
Pitx2c>Wnt>microRNA signaling is severely impaired in experimental hyperthyroidism (HTD) rat model
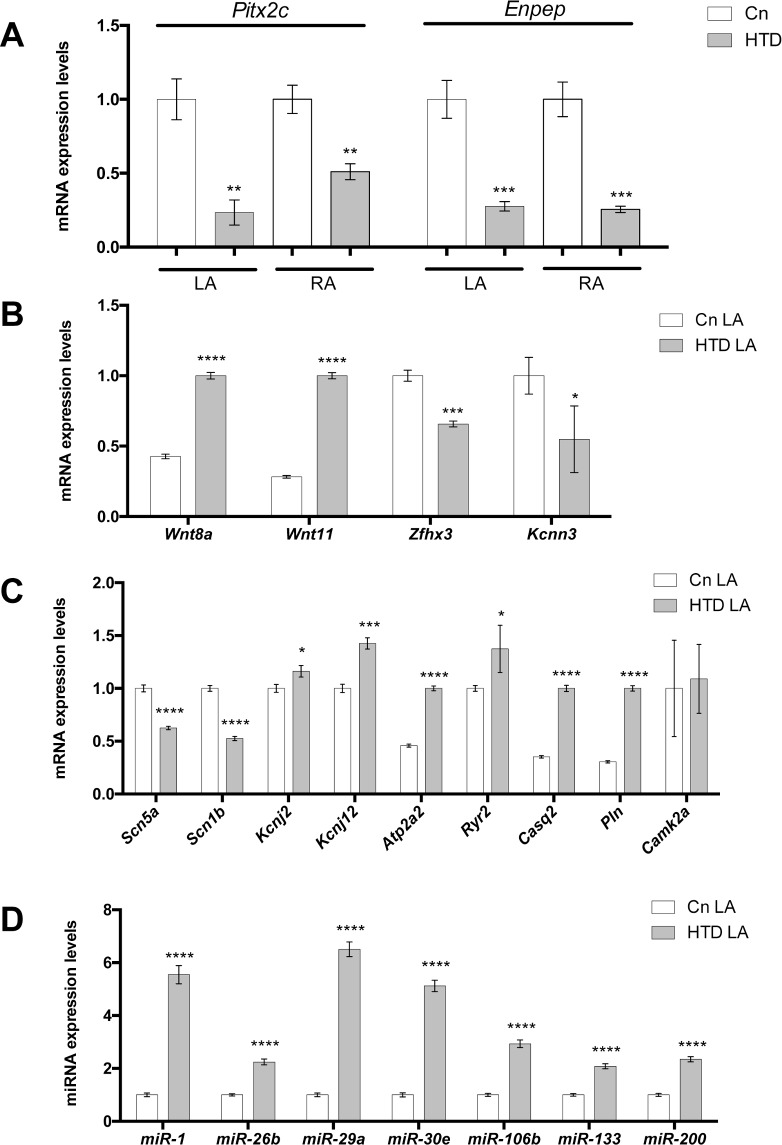
We have analyzed the expression levels of the homeobox transcription factor Pitx2c in an experimental rat model of induced hyperthyroidism (HTD) [32]. As a consequence of HTD, this experimental model also displays moderate hypertension as documented in S1 Fig. qPCR analyzed demonstrated that Pitx2c is severely down-regulated in right and left atrial chambers (Fig 1). Similar findings are also observed in an in vitro model of HTD (T4 administration) using fetal primary cultures of cardiomyocytes (S2 Fig). Curiously, Enpep is also severely impaired in both atrial chambers (Fig 1). Since multiple evidences have demonstrated that Pitx2 differentially modulates expression of AF GWAS associated genes as well as multiple components of the cardiac action potential in the left atrium [14,16], we analyzed their expression in the HTD model. Our data demonstrate that Wnt8 and Wnt11 were significantly increased whereas Zfhx3 and Kcnn3 were decreased in HTD rats as compared to age-matched controls (Fig 1). Importantly, INa encoding genes, i.e. Scn5a and Scn1b, are severely decreased whereas resting membrane potential IK1 encoding genes (Kcnj2 and Kcnj12) are up-regulated, as well as calcium handling proteins such as Atp2a2 (Serca), Ryr2, Casq2 and Pln while Camk2a display no significant differences (Fig 1). Overall these data suggest that HTD elicits down-regulation of Pitx2 and its downstream signaling pathway, mimicking the gene expression profile observed in atrial-specific Pitx2 deficient mice [14,16].
Fig 1.
(A) Pitx2 and Enpep gene expression in the left (LA) and right (RA) atrial chambers of experimental hyperthyroid rats (HTD) as compared to controls. Observe that Pitx2 and Enpep are significantly decreased in both atrial chambers in HTD hearts. (B) AF GWAS-associated gene expression in the LA chamber of HTD hearts. Wnt8 and Wnt11 are significantly increased whereas Zfhx3 and Kcnn3 are decreased in HTD hearts as compared to controls. (C) Ion channel encoding genes display significant differential expression in HTD as compared to controls, while Camk2a display no significant differences. (D) Pitx2-regulated microRNAs display significant increased expression in HTD as compared to controls. In all cases, n = 6.*p<0.01, **p<0.05, ***p<0.001,****p<0.0001.
A pivotal role for post-transcriptional regulation by non-coding RNAs in cardiac development and disease is emerging [37] and we have provided evidence that Pitx2 controls a large set of microRNAs with key functional roles in cardiac electrophysiology [15]. We therefore tested whether distinct Pitx2-regulated microRNAs were deregulated in our HTD rat model. qPCR of left atrial chambers demonstrated that miR-1, miR-26b, miR-29a, miR-30e, miR-106b, miR-133 and miR-200 are up-regulated in HTD rats as compared to controls (Fig 1), demonstrating a similar microRNA expression profile as in atrial-specific Pitx2 deficient mice [14,16].
Pitx2 alone is impaired in experimental spontaneous hypertension (HTN)
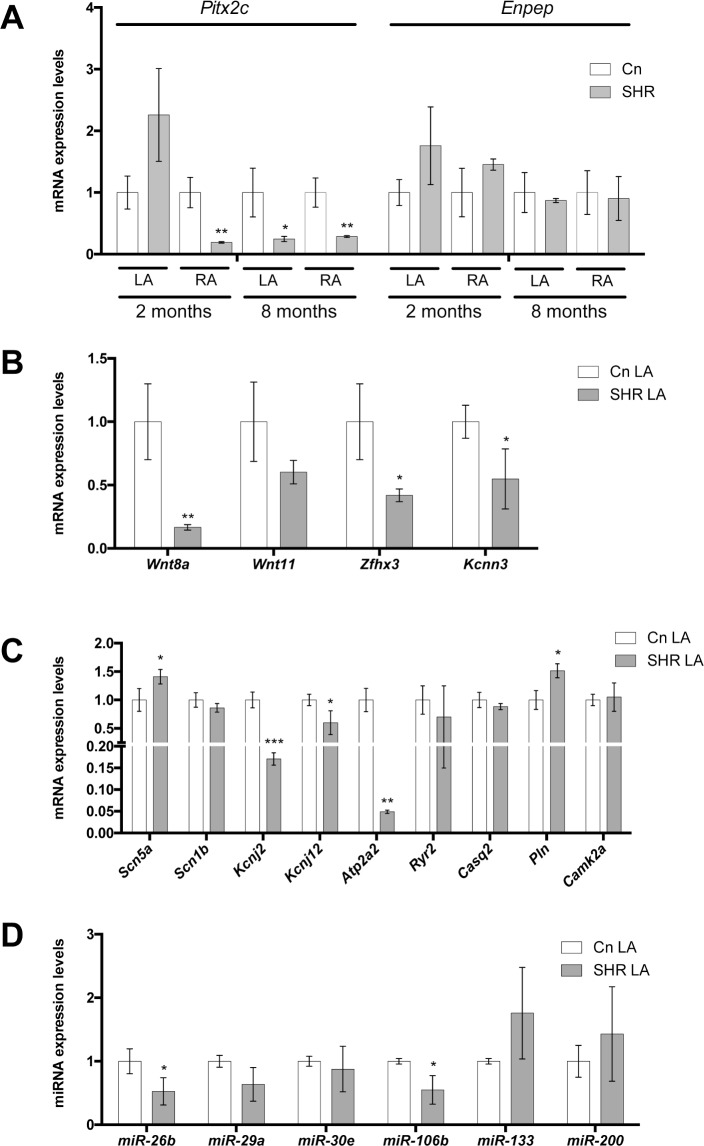
We further explored if Pitx2c is also altered in another AF risk factor experimental model, i.e. spontaneous hypertensive SHR rats [38]. We therefore analyzed Pitx2c expression levels at two distinct developmental stages, 2 and 8 month old rats, respectively. Interestingly, Pitx2c expression was significantly decreased in right atrium but not in the left atrium of 2 month old SHR rats, whereas down-regulation was equally observed at 8 months (Fig 2). Curiously, Enpep expression was not significantly different in left and right atrial chambers at any of these stages (Fig 2). Since most significant differences on Pitx2 expression were only observed at 8 months, subsequent analyses were only performed at this stage.
Fig 2.
(A) Pitx2 and Enpep gene expression in the adult left (LA) and right (RA) atrial chambers of spontaneous hypertensive rats (SHR) at 2 and 8 months, respectively, as compared to WHK controls. Observe that Pitx2 is significantly decreased in both atrial chambers in SHR hearts at 8 months, whereas Enpep display no significant differences at any of the analyzed stages. (B) AF GWAS-associated gene expression in the LA chamber of HTD hearts. Wnt8, Wnt11, Zfhx3 and Kcnn3 are significantly decreased in HTD hearts as compared to controls. (C) Ion channel encoding genes such as Kcnj2, Kcnj12, Atp2a2 (decreased), Scn5a and Pln (increased) display significant differential expression in HTD as compared to controls, while Scn1b, Ryr2, Casq2 and Camk2a display no significant differences. (D) Pitx2-regulated microRNAs display no significant differences, except for miR-26b and miR-106b which are significantly decreased, in HTD as compared to controls. In all cases, n = 6. *p<0.01, ** p<0.05, *** p<0.001.
Analyses of AF GWAS associated genes demonstrate that Wnt8, Zfhx3 and Kcnn3 are significantly decreased in hypertensive rats whereas Wnt11 display no significant difference in the left atrial chambers of HTN rats as compared to controls (Fig 2). In addition, analyses of ion channel encoding genes demonstrate that Scn5a, but not Scn1b was significantly increased (Fig 2). Kcnj2, Kcnj12 and Atp2a2 (Serca2) are down-regulated whereas Ryr2, Casq2 and Camk2a display no significant differences (Fig 2) and only Pln is up-regulated in HTN left atrial chambers as compared to control normotensive rats (Fig 2). Thus, although Pitx2 is severely down-regulated in HTN rats, Pitx2-downstream signals, such as Wnt signaling or cardiac action potential determinants display either no changes or discordant changes as compared to atrial-specific Pitx2 deficient mice [14,16].
Following the same reasoning as for HTD experimental model, we assessed if Pitx2-downstream microRNA expression is impaired in HTN rats. Surprisingly none of the tested microRNAs, except for miR-26b and miR-106b, which in fact were decreased, display significant differences (Fig 2). Overall, these data demonstrate that while Pitx2 is impaired in HTN, its downstream pathways are mostly unaltered, demonstrating a discordant microRNA expression profile as compared to those revealed in atrial-specific Pitx2 deficient mice [14,16].
Enpep downregulation modulates PITX2 but not its downstream pathway
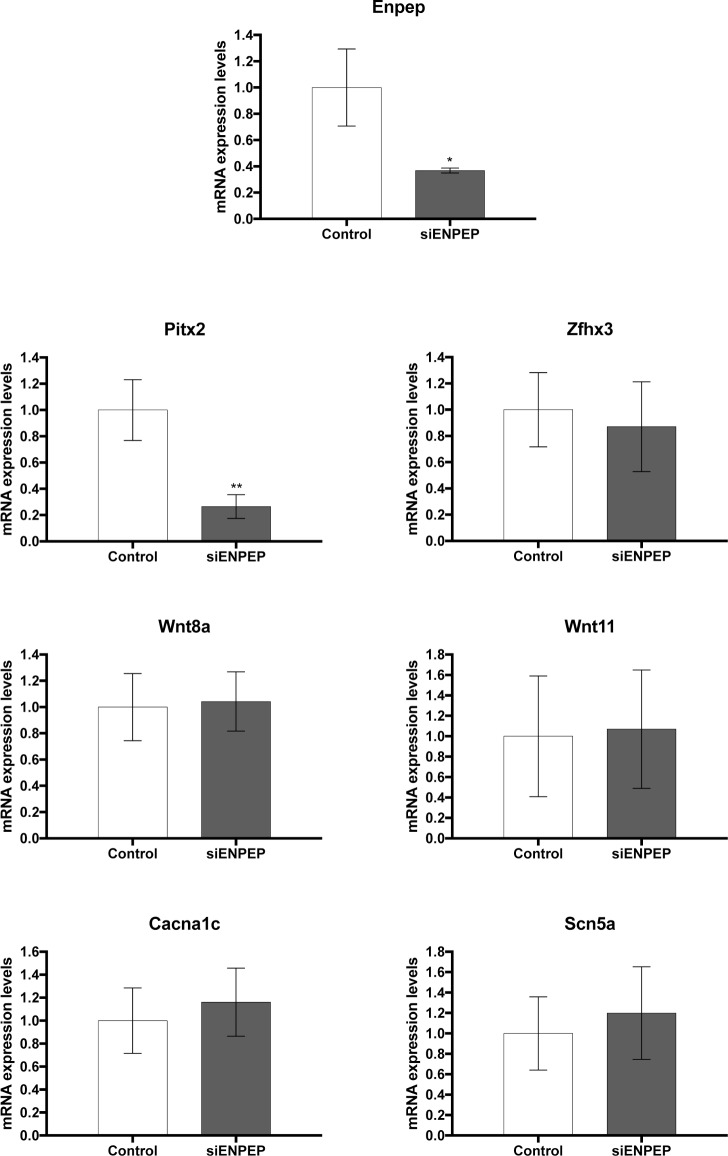
Given the fact that Enpep is deregulated in HTD experimental rats, it is widely expressed in cardiac regions that can contribute to AF and that genomic interaction between 4q25 AF risk variants containing sequences and the Enpep promoter have been reported in mice [11], it is plausible that ENPEP might have a role in AF predisposing factors. Furthermore, Pitx2 silencing in HL1 atrial cardiomyocytes also elicits Enpep downregulation (S2 Fig). We therefore silenced Enpep expression in HL1 atrial cardiomyocyte and evaluated expression of Pitx2c and Pitx2-Wnt downstream signaling. Our analyses demonstrated that Enpep silencing decreased Pitx2 expression whereas Zfhx3, Wnt8, Wnt11, Cacna1c and Scn5a were unaltered (Fig 3). Thus these data demonstrate that Enpep contribution on the Pitx2-Wnt signaling pathways is limited to Pitx2 regulation.
Fig 3. Gene expression analyses in Enpep silenced HL-1 atrial cardiomyocytes.
Note that Enpep is decreased (siEnpep), leading to decreased expression of Pitx2c but not of Wnt8a, Wnt11, Zfhx3, Cacna1c or Scn5a. Representative values of three pooled replicates on three independent biological transfections. *p<0.01, **p<0.05.
Impaired ROS signaling contributes to Pitx2 pathophysiology
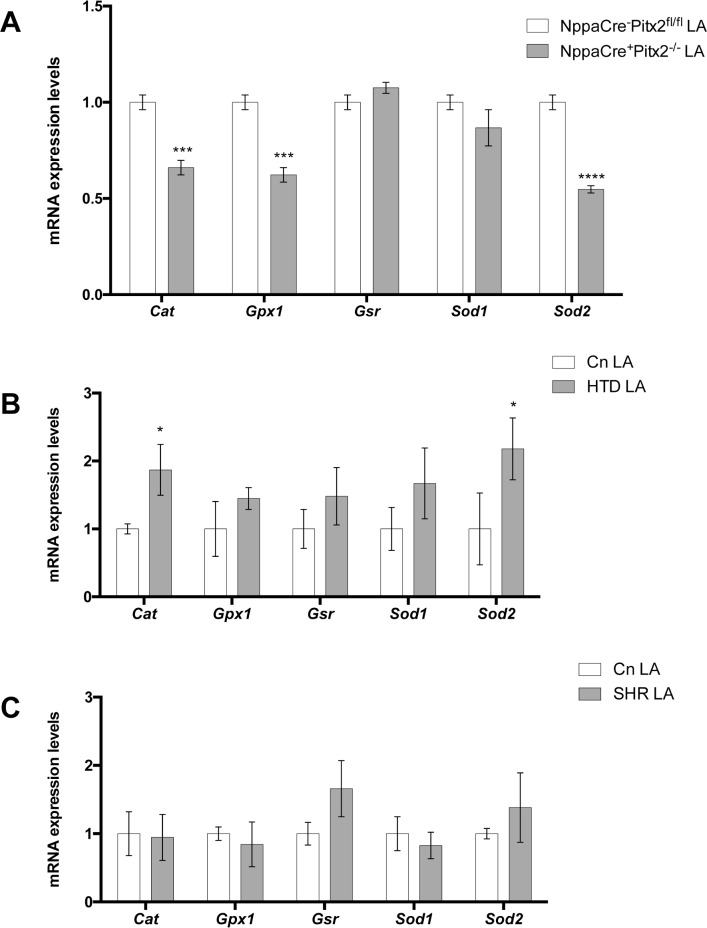
Reactive oxidative signaling (ROS) has been recently linked to AF [20,39] and experimental evidences demonstrated that impairing ROS can act as pro-arrhythmogenic factor [40]. We have therefore investigated if ROS is impaired in our atrial-specific Pitx2 mouse mutants as well as in AF risk factor HTD and SHR experimental models. qPCR demonstrate that several components of ROS signaling are impaired. In particular, we noticed that catalase (Cat), glutathione peroxidase (Gpx) and mitochondrial superoxide dysmutase (Sod2) were significantly down-regulated in the left atrial chamber of atrial-specific Pitx2 mouse mutants as compared to controls, whereas cytoplasmic superoxide dysmutase (Sod1) and glutathione reductase (Gsr) were unaltered (Fig 4). Subsequently we tested if ROS is impaired in SHR and/or HTD experimental models. Interestingly, HTD leads to up-regulation of Cat and Sod2 but not any of the other ROS analyzed components (Fig 4) whereas HTN (SHR) elicited no significant changes in any of the studied ROS components (Fig 4). These data illustrate that Pitx2 insufficiency leads to altered ROS signaling, a condition that is minimally (HTN) or partially (HTD) impaired, respectively.
Fig 4.
Redox signaling gene expression analyses of adult left atria (LA) corresponding to NppaCre+Pitx2-/- (A), HTD (B) and SHR (C), respectively, as compared to their corresponding controls. Note that Cat, Gpx1 and Sod2 are significantly decreased in NppaCre+Pitx2-/- as compared to NppaCre-Pitx2fl/fl controls, Cat and Sod2 are significantly increased in HTD mice whereas no changes are observed in SHR mice. In all cases, n = 6. *p<0.01, ***p<0.001, ****p<0.0001.
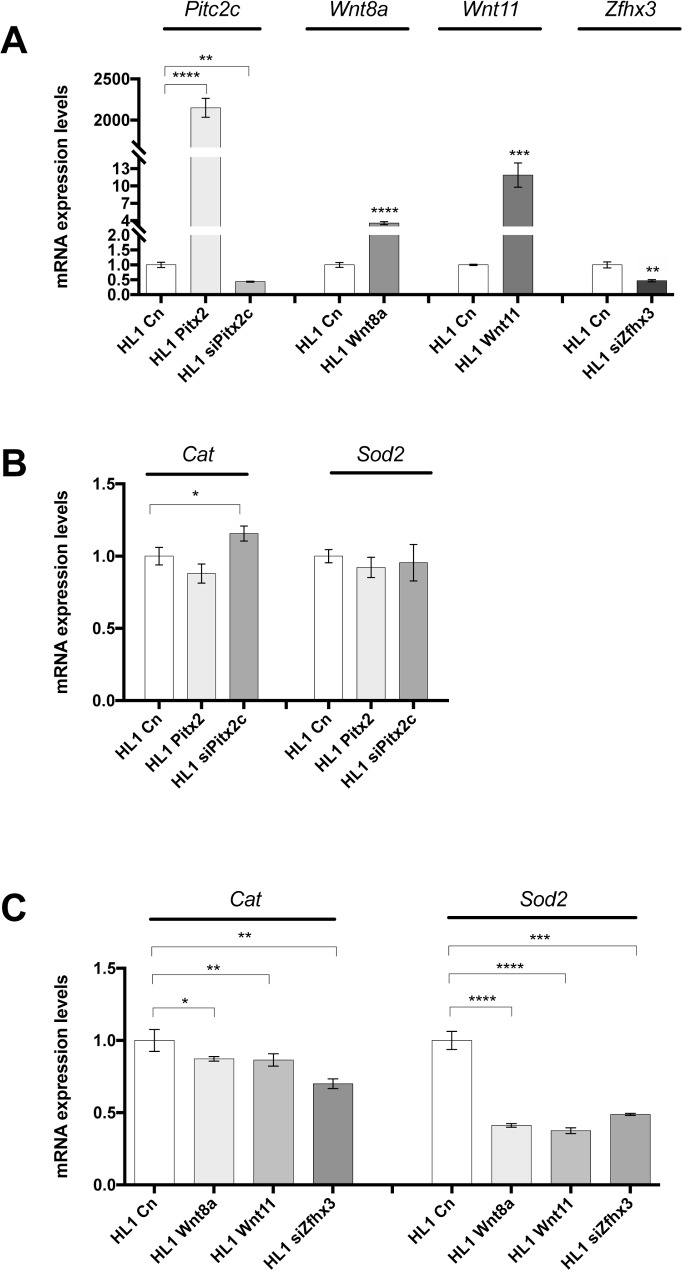
To test if Pitx2c can directly modulate ROS components, Pitx2 over-expression and silencing (siRNA) experiments were carried out in HL-1 atrial cardiomyocytes (Fig 5). Unexpectedly, Pitx2 over-expression did not influence Cat and Sod2 expression whereas Pitx2 silencing slightly up-regulated Cat but not Sod2, as depicted in Fig 5. These data are in contrast with our previous findings in atrial-specific Pitx2 mutant mice (Fig 4). Thus it could be possible that Pitx2 does not directly target Cat/Sod2 expression but else is modulated downstream of Pitx2. Mimicking Pitx2 insufficient model, we thus over-expressed Wnt8 and Wnt11, respectively and inhibited Zfhx3 by using siRNA in HL-1 atrial cardiomyocytes. Interestingly, both over-expression of Wnt signaling (Wnt8 and Wnt11) and inhibition of Zfhx3 (Fig 5), respectively, lead to significant downregulation of Cat and Sod2 expression (Fig 5). These data therefore demonstrate that ROS signaling is modulated by Wnt and Zfhx3 signaling downstream Pitx2.
Fig 5.
(A) Expression analyses of Pitx2c, Wnt8a, Wntt11 and Zfhx3 in gain- and loss-of-function experimental transfection of HL-1 atrial cardiomyocytes for Pitx2, Wnt8a, Wnt11 and Zfhx3, respectively. (B) Expression analyses of Cat and Sod2 in Pitx2 gain and loss-of-function experimental transfection of HL-1 atrial cardiomyocytes. Observe that only Pitx2c silencing (siPitx2c) significantly increases Cat expression. (C) Expression analyses of Cat and Sod2 in Wnt8a and Wnt11 gain of function transfections and Zfhx3 siRNA silencing (siZfhx3) of HL-1 atrial cardiomyocytes. Observe that Cat and Sod2 are significantly decreased in all cases, as compared to controls. Representative values of three pooled replicates on three independent biological transfections. *p<0.01, **p<0.05, ***p<0.001, ****p<0.0001.
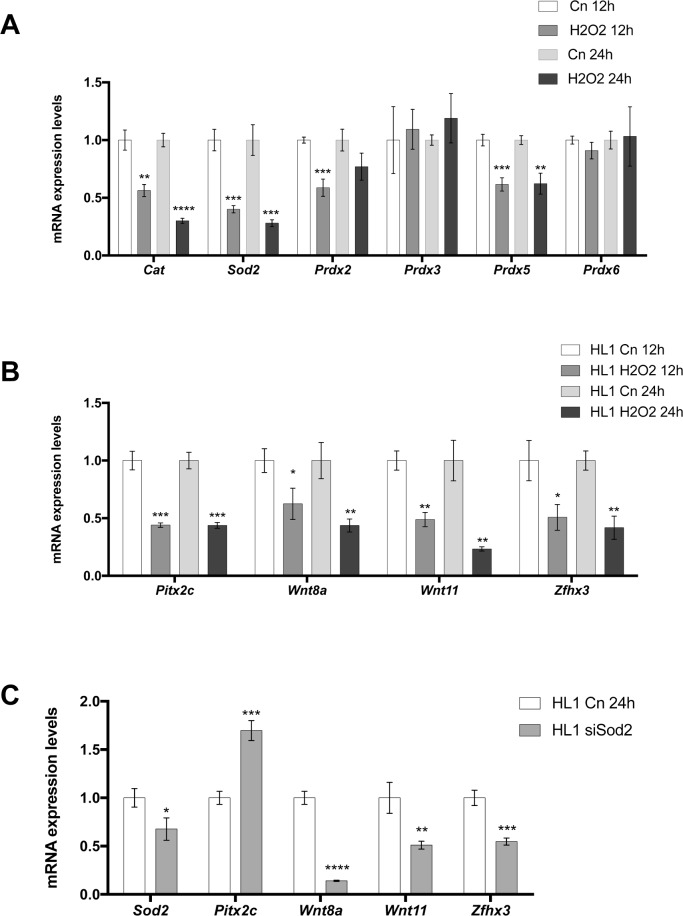
To test if the opposite pathways is operative, i.e. if ROS signaling alters Pitx2>Wnt pathway, we treated HL-1 atrial cardiomyocytes with hydrogen peroxidase (H2O2) at distinct incubation times ranging from 1h to 24h and we assessed if Pitx2, Wnt signaling and Zfhx3 is impaired. Cat, Sod2 and also peroxiredoxines (Prxd2, Prdx3, Prdx5 and Prdx6) were assayed in parallel as indicators of ROS signaling activity. Incubation times ranging from 1h to 6h display basically no significant changes in the expression level (data not shown), except for a transient up-regulation of Wnt11 (3h/6h; S3 Fig). Curiously, sustained down-regulation of Zfhx3 expression was observed at all time analyzed (S3 Fig). At 12h/24h of H202 administration Cat, Sod2, Pdrx2 (only 12h) and Prdx5 were significantly decreased in treated cells as compared to controls, whereas Prdx3 and Prdx6 display no significant differences (Fig 6). These data suggest therefore that impaired ROS signaling was successfully achieved at 12h/24h of H202 administration. Importantly, Pitx2, Wnt8, Wnt11 and Zfhx3 were severely down-regulated after H202 administration at both 12h/24h (Fig 6). Thus these data demonstrate that ROS impairment can influence Pitx2>Wnt signaling. To further support these findings we selectively inhibited Sod2 expression by siRNA in HL1 atrial cardiomyocyte and assayed Pitx2>Wnt signaling components by qPCR. Sod2 inhibition leads to down-regulation of Wnt8a, Wnt11 and Zfhx3 and surprisingly to up-regulation of Pitx2 as depicted in Fig 6. While up-regulation of Pitx2c after Sod2 siRNA remains to be fully understood, our data reveal a complex interplay between Pitx2>Wnt and ROS signaling in atrial cardiomyocytes.
Fig 6.
(A) Redox signaling gene expression analyses of H202 treated HL-1 atrial cardiomyocytes at 12 h and 24h. Observe that Cat, Sod2 and Prdx5 are significantly decreased at 12h and 24h after H202 administration, whereas Prdx2 is only decreased at 12h but not at 24h. (B) Gene expression analyses of Pitx2c, Wnt8a, Wnt11 and Zfhx3 after H202 administration. Observe that all of them are significantly decreased at 12h and 24h. (C) Gene expression analyses in Sod2 silenced HL-1 atrial cardiomyocytes. Note that Sod2 is decreased (siSod2), Pitx2c increases whereas Wnt8a, Wnt11 and Zfhx3 are significantly decreased. Representative values of three pooled replicates on three independent biological transfections. *p<0.01, **p<0.05, ***p<0.001, ****p<0.0001.
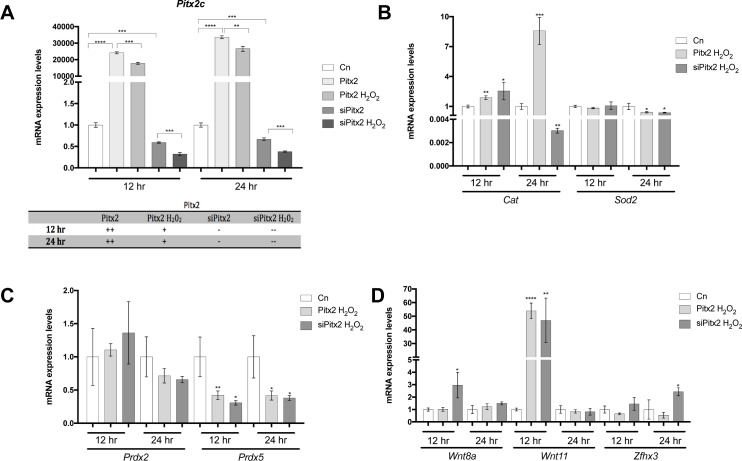
To further decipher the complex interplay between ROS and Pitx2 signaling, we experimentally test if Pitx2c gain- and loss-of-function was impaired by H202 administration in HL-1 atrial cardiomyocytes, aiming to decipher which is the overruling signaling pathway. Pitx2c transfection lead to over-expression of Pitx2c, which was significantly diminished by H202 administration at both experimental time points (12h/24h) (Fig 7). Similarly, Pitx2c expression was further inhibited by H202 administration after Pitx2c siRNA silencing experiments (Fig 7), further demonstrating that H202 administration significantly decreases Pitx2c expression.
Fig 7.
(A) Analyses of Pitx2c expression in Pitx2 gain and loss-of-function experiments with or without H202 administration for 12h and 24h, respectively. Observe that H202 administration significantly decreased Pitx2c expression in both Pitx2 overexpression and silencing conditions, at 12h and 24h. (B) Analyses of Cat and Sod2 expression (B), Prdx2 and Prdx5 (C) and Wnt8a, Wnt11 and Zfhx3 (D) in Pitx2 gain and loss-of-function setting with H202 administration for 12h and 24h, respectively. Observe that Cat is significantly increased by Pitx2 overexpression at 12h and 24h whereas is mildly increased and highly significantly decreased by Pitx2 siRNA treatment at 24h. On the other hand Sod2 is mildly decreased only at 24h in both conditions (B). Prdx5, but not Prdx2, is significantly decreased at 12h and 24h in both conditions (C). Wnt8a is only increased in Pitx2c siRNA conditions at 12h, Wnt11 is increased in both conditions only at 12h while Zfhx3 is significantly increased only by Pitx2c silencing at 24h. Representative values of three pooled replicates on three independent biological transfections. *p<0.01, **p<0.05, ***p<0.001, ****p<0.0001.
In this experimental context, H202 administration does not alter Sod2 expression in Pitx2c gain and loss-of-function settings at 12 hours but it significantly decreases it at 24 hours (Fig 7). On the contrary, Cat expression is enhanced by H202 administration in Pitx2 gain and loss-of-function settings at 12h but selectively decreased at 24hours only in Pitx2 loss-of-function conditions. These data suggest that H202 administration cannot overrule Pitx2 modulation of Cat. In line with these findings only Prdx5 is significantly decreased after H202 administration in Pitx2c gain and loss-of-function settings at both 12 hours and 24 hours (Fig 7; S4 Fig), further supporting that H202 administration overrules Pitx2c modulation of ROS signaling components.
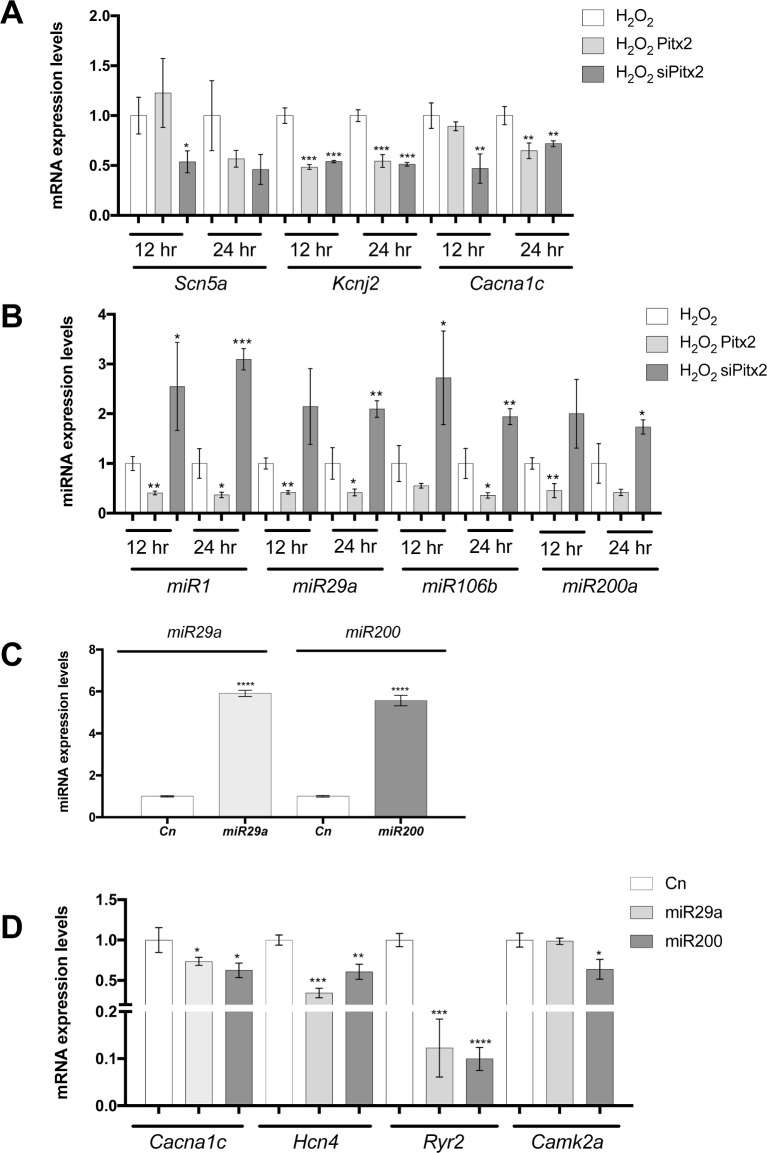
In order to investigate if H202 administration can also overrule Pitx2 modulation of its downstream targets, Wnt8a, Wnt11 and Zfhx3 expression was analyzed in this experimental setting. As it can be observed in Fig 7, H202 administration prevented Wnt8 and Wnt11 up-regulation in Pitx2c siRNA silenced conditions at 24h but not at 12hours, demonstrating that H202 administration can also overruled Wnt signaling up-regulation in absence of Pitx2c. On the other hand, Zfhx3 was up-regulated after H202 administration in Pitx2c silenced conditions at 24hours, demonstrating that H202 administration can directly affect Zfhx3 expression independently of Pitx2c expression levels. Overall these data further demonstrate that ROS signaling has a major impact on Pitx2>>Wnt signaling pathway. We further analyzed if ion channel expression would be affected in this experimental setting. qPCR analyses of Cacna1c, Scn5a and Kcnj2 expression demonstrated significant down-regulation at both 12 and 24 hours after H202 administration in Pitx2c silenced conditions (Fig 8). Importantly, such H202 mediated down-regulation was only observed for Kcnj2 in Pitx2c over-expressing cells, demonstrating a fine output balanced between ROS and Pitx2 function on the expression of distinct ion channels. Finally, we tested if microRNA expression would be also impaired in this context. A subset of Pitx2-modulated microRNA was assessed16. As depicted in Fig 8, miR-1, miR-29a, miR-106b and miR-200a was selectively inhibited by H202 treated Pitx2-overexpressing cells but up-regulated in H202 treated Pitx2 silenced cells at both time points (12h and 24h). These data illustrate that H202 administration does not overrule Pitx2 function as regulator of microRNA expression.
Fig 8.
Analyses of Scn5a, Kcnj2 and Cacna1c (A), miR-1, miR-29a, miR-106b and miR-200b (B) expression in Pitx2 gain and loss-of-function experiments after H202 administration for 12h and 24h, respectively. Observe that most ion channel are significantly decreased in both experimental conditions at both time points (A) whereas microRNAs are all significantly decreased after Pitx2 over-expression and increased following Pitx2c silencing at 12h and 24h after H202 administration. (C). miR-29a and miR-200 expression in HL-1 atrial cardiomyocytes transfected cells. Observe that each microRNAs is significantly increased after corresponding pre-miRNA transfection. (D) Ion channel gene expression in miR-29a and miR-200 transfected HL-1 atrial cardiomyocytes, respectively. Observe that miR-29 and miR-200 over-expression leads to significant decreased of Cacna1c, Hcn4, Ryr2 and Camk2a (except for miR-29a) expression. Representative values of three pooled replicates on three independent biological transfections. *p<0.01, **p<0.05, ***p<0.001, ****p<0.0001.
Modulation of miR-29 and miR-200 alters cardiac action potential determinants
We have previously demonstrated that Pitx2 modulates expression of miR-29 and miR-200, among other microRNAs [16] and furthermore we have demonstrated in this study that modulation of distinct ion channel is greatly influenced by H202 administration while microRNA signature is mostly dependent on Pitx2c but not H202 administration. We provide herein evidences that miR-29 and miR-200 over-expression also contributes to ion channel expression remodeling. HL-1 atrial cardiomyocytes transfected with miR-29 and miR-200 (Fig 8) significantly down-regulate Cacna1c, Hnc4 and Ryr2 expression, while Camk2a was significantly decreased with miR-200 but not miR-29 (Fig 8). Thus these data demonstrate that miR-29 and miR-200 impaired expression also contributes to develop pro-arrhythmogenic substrates.
Discussion
Multiple risk factors are associated with the onset of AF [23], however understanding of the molecular causative links remains poorly elucidated. Among these risk factors, hyperthyroidism is highly linked to increased prevalence of AF [41–43]. It is widely documented that electrical properties of the atrial cardiomyocytes are functional impaired in hyperthyroid patients [41] as well as in distinct experimental models of hyperthyroidism [44–47]. In particular, there are ample evidences that TH deregulation can influence calcium and potassium channels [44–47], whereas modulation of sodium channels is less documented [48]. Similarly, AF onset is also increased in HTN patients [23,49]. While extensive literature is available on the modulation of distinct ion channels in the vasculature of hypertensive patients [50–51], scarce evidence is available on cardiac ion channel deregulation and thus electrical impairment in the heart. Recently, generation of reactive oxygen species (ROS) has been associated to increase onset of AF [52–53] and dietary ROS reduction has been provided to be beneficial [54] on AF onset. Substantial evidence support ROS regulation of distinct cardiac ion channels [55–57], procuring thus a functional link between ROS signaling and impaired electrical activity leading to AF onset.
Thus, whereas there are evidence of functional impairment by these cardiovascular risk factors leading to AF, it largely remains unclear which are the molecular mechanisms driving AF in these contexts. Multiple point mutations in distinct ion channels have been reported to contribute to AF pathophysiology [58] yet covering less than 10% of AF cases. Genome-wide association studies have brought up novel candidate genes on AF pathophysiology among which most significantly associated risk variants are in the vicinity of the homeobox transcription factor PITX2 [6]. Genomic loci spanning within risk variants are capable of molecularly interacting with PITX2 as well as ENPEP regulatory elements [11], yet the specific role of these risk variants in AF remains elusive. Importantly, distinct Pitx2 loss-of-function experimental models demonstrated that this transcription factor controls a complex molecular signaling pathway that substantially modulates expression of multiple genes encoding ion channel and cell-cell proteins with pivotal role in cardiac electrophysiology [12–26]. We provide here first evidence that hyperthyroidism leads to decreased expression of PITX2 and ENPEP in the atrial chambers. Furthermore, we also demonstrated for the first time that impaired ROS signaling modulates PITX2 expression, while we corroborate previous findings that PITX2 expression, but not ENPEP, is significantly down-regulated in hypertensive rats [28]. Therefore these data demonstrate that PITX2 expression is impaired in HTD, a cardiovascular risk factor leading to AF, suggesting that PITX2 impairment could be pivotal provoking atrial arrhythmogenesis.
Pitx2 insufficiency in mice has been reported to distinctly modulate Sox2-Hcn4 expression [12] in the developing embryo leading to impaired conductive configuration and thus predisposition to AF. In addition, Pitx2 insufficiency in the adult heart has been proven to regulate INa and IK1 currents, leading to impaired ECG [14] and additionally to regulate calcium homeostasis by modulating Wnt signaling in a dose-dependent manner [16]. We provide herein evidences that HTD increases Wnt8/Wnt11 expression in the left atrial chamber while down-regulates Zfhx3 expression, mimicking thus similar results obtained in Pitx2 insufficient mice. Furthermore, genes encoding INa (Scn5a, Scn1b) were down-regulated, those encoding for IK1 (Kcnj2, Kcnj12) were up-regulated as well as those controlling calcium handling (Ryr2, Atp2a2, Casq2, Plb) were also up-regulated, in line with previous findings in atrial-specific conditional Pitx2 insufficient mice. Interestingly, such up-regulated Wnt signaling was not observed in the left atrial chamber of SHR rats (HTN model), neither such ion channel impaired expression. While Enpep is differentially expressed in HTN vs SHF left atrial chambers, Enpep provides no contribution to Wnt signaling or downstream ion channel expression. A plausible explanation might be that hypertension directly influences Wnt expression [59], counteracting Pitx2-mediated Wnt up-regulation. Importantly, HTD rats also course with mild hypertension, supporting the notion that the specific molecular changes observed in HTD but not in SHR (HTN) are modulated by thyroid hormones. Additional experiments will be needed to further dissect these pathways.
We and other have previously reported the pivotal role of microRNAs as post-transcriptional regulatory mechanism driven by Pitx2 in the context of atrial arrhythmogenesis [57–58], and we therefore analyzed if expression of distinct cardiac enriched microRNAs were impaired in HTD and SHR (HTN model) rats. Importantly, we have observed that microRNA profiling in the left atrium of HTD, but not in SHR, rats, mimicking previous findings in atrial-specific conditional Pitx2 insufficient mice [16]. Several lines of evidence have already reported the key regulatory role of miR-1 [60–62], miR-26 [63], miR-106b [64], miR-133 [65–66] and miR-200 [64] in arrhythmogenesis. We provide herein additional evidence that these Pitx2-modulated HTD-regulated microRNAs modulate distinct ion channel expression with relevance in atrial electrophysiology. miR-29 over-expression in HL1 atrial cardiomyocyte deregulate Cacna1c, Hnc4 and Ryr2, influencing therefore both the calcium handling and pacemaker activity, whereas miR-200 regulated Cacna1c, Ryr2 and Camk2a, in addition to Scn5a as previously reported [64], impacting therefore also in calcium handling. Importantly, miR-29 and miR-200 are not significantly impaired in SHR atrial chambers, suggesting that Wnt-microRNA might be a pivotal candidate establishing fundamental differences between HTD and HTN in atrial arrhythmogenesis susceptibility.
It has been demonstrated that Pitx2 play a pivotal role regulating redox homeostasis in the adult skeletal muscle as well as during skeletal muscle regeneration [28]. More recently, Pitx2 has been demonstrated to play a pivotal role promoting resistance to ischemia by activating an antioxidant response in the adult heart [29]. We provide herein evidences of a complex interplay between Pitx2 and redox signaling. On the one hand, Pitx2 insufficiency leads to significant expression impairment of key redox components, such as Cat (catalase) and Gpx (Glutathione reductase) and Sod2 (mitochondrial superoxide dismutase), in line with previous findings [28,29]. On the other hand, impaired redox homeostasis also significantly alters Pitx2 and its downstream AF signaling pathway, i.e. Wnt and Zfhx3 expression. Importantly, Wnt8/Wnt11 and Zfhx3 expression display significant decreased expression levels in both H2O2 and siRNA Sod2 atrial cardiomyocytes. Interestingly, redox signaling impairment is also observed in HTD but not in SHR (HTN model) rats, suggesting that redox impairment in the context of HTD can also contribute to atrial arrhythmogenesis in this setting.
Given the complex interplay between redox homeostasis and Pitx2 signaling, we have analyzed the molecular consequences of deregulating both pathways in atrial cardiomyocytes. We noticed that redox impairment significantly elicits down-regulation of Pitx2 independently of its expression levels, i.e. controls, Pitx2 over-expressing and Pitx2 silenced atrial cardiomyocytes. Furthermore, sustained H2O2 administration (24h) significantly blocked Pitx2-repressed Wnt expression and promotes Zfhx3 up-regulation. Whereas it is widely documented that redox signaling can compromise ion channel functioning and calcium homeostasis in cardiomyocytes [67], in our system we observed no influence of H2O2 administration on the regulatory impact of Pitx2 in distinct ion channels such as Scn5a, Kcnj2 and Cacna1c as well as multiple Pitx2-regulated microRNAs such as miR-1, miR-26, miR-29 and miR-200, in which redox impairment impact is less documented [68].
In summary, we provide herein evidences that Pitx2-Wnt signaling pathway is impaired in HTD rats (Fig 9). Comparative analyses with Pitx2 insufficient models highlights the molecular hallmark similarities between HTD and atrial-specific Pitx2 deficient hearts, in particular Wnt upregulation, Zfhx3 down-regulation and redox signaling impairment. These data therefore support the notion that AF onset in HTN patients is mediated by Pitx2 impairment. Furthermore, we provide evidences of a complex interplay between Pitx2 and redox signaling highlighting therefore the complex molecular interactions priming molecular substrates that contribute to atrial arrhythmogenesis.
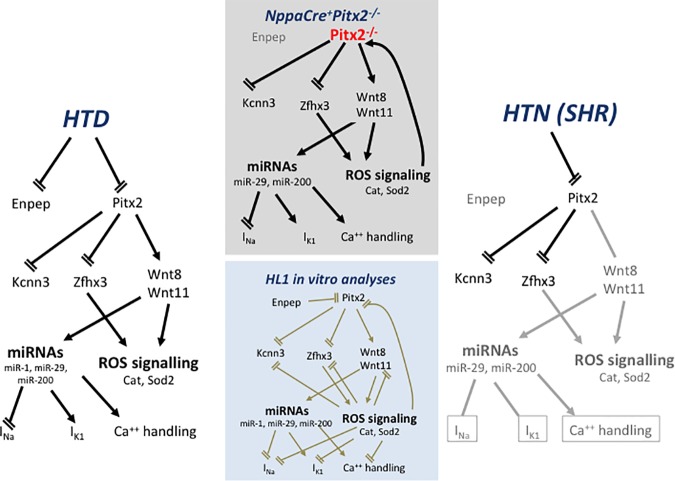
Fig 9. Schematic representation of the Pitx2>Wnt>ROS signaling pathway in NppaCrePitx2-/- insufficiency mice, as compared to experimental HTD and HTN (SHR) rat models.
Observe that Pitx2 insufficiency leads deregulation of Zfhx3 and Wnt signaling, which subsequently leads to microRNA and ROS signaling deregulation and thus ion channel impairment. In HTD but not in HTN, Pitx2>Wnt>ROS signaling is also impaired. Furthermore, in vitro gain- and loss-of-function analyses (brown arrows) further support a complex retroactive gene regulatory network.
Supporting information
(PDF)
A) Systolic blood pressure (SBP) in mmHg and heart rate (HR) in beats per minute (BPM) measured in control and hyperthyroid (HTD) rats at the end of the experiment (n = 10 each group). Mean ± SEM is displayed. *p<0.001 vs control group. B) Systolic blood pressure (SBP) in mmHg and heart rate (HR) in beats per minute (BPM) measured in Wistar-Kyoto (WKY) and spontaneously hypertensive rats (SHR) at the end of the experiment (n = 10 each group). Mean ± SEM is displayed. *p<0.01, **p<0.001 vs control group.
(TIFF)
Analyses of Pitx2 expression in primary culture of fetal cardiomyocytes treated with T4 as compared to controls (panel A). Observe that Pitx2 is significantly decreased after T4 administration. Analyses of Pitx2 (panel B) and Enpep (panel C) in HL1 atrial cardiomycytes after Pitx2 siRNA silencing. Observe that Pitx2 siRNA administration significantly decrease Pitx2 expression (panel B) and also Enpep expression (panel C). *p<0.01, ***p<0.001, ****p<0.0001.
(TIF)
Observe that H202 administration significantly increased Wnt11 at 3h and 6h while significantly decreased Zfhx3 expression at all experimental conditions analyzed. *p<0.01, **p<0.05, ****p<0.0001.
(TIF)
Observe that no significant differences are observed in Prdx3 and Prdx6 expression, except for Prdx6 at 12h after treatment in Pitx2 silencing conditions. *p<0.01.
(TIF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was partially supported by grants BFU2012-38111, BFU2015-67131 (Ministerio de Economía y Competitividad, Gobierno de España), and CTS-1614 (Junta de Andalucia Regional Council) to DF and AA. This study was supported by the grant PI3/02384 from Carlos III Health Institute of Spain “Feder una manera de hacer Europa” to RW. No additional external funding was received for this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hakim FA, Shen WK. Atrial fibrillation in the elderly: a review. Future Cardiol. 2014; 10: 745–758. doi: 10.2217/fca.14.32 [DOI] [PubMed] [Google Scholar]
- 2.Brugada R, Tapscott T, Czernuszewicz GZ, Marian AJ, Iglesias A, Mont L, et al. Identification of a genetic locus for familial atrial fibrillation. N Engl J Med. 1997; 336: 905–911. doi: 10.1056/NEJM199703273361302 [DOI] [PubMed] [Google Scholar]
- 3.Chen WJ, Yeh YH, Lin KH, Chang GJ, Kuo CT. Molecular characterization of thyroid hormone-inhibited atrial L-type calcium channel expression: implication for atrial fibrillation in hyperthyroidism. Basic Res Cardiol. 2011;106:163–74. doi: 10.1007/s00395-010-0149-5 [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Xia M, Jin Q, Bendahhou S, Shi J, Chen Y, et al. Identification of a KCNE2 gain-of-function mutation in patients with familial atrial fibrillation. Am J Hum Genet. 2004;75:899–905. doi: 10.1086/425342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia M, Jin Q, Bendahhou S, He Y, Larroque MM, Chen Y, et al. A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem Biophys Res Commun. 2005;332:1012–1019. doi: 10.1016/j.bbrc.2005.05.054 [DOI] [PubMed] [Google Scholar]
- 6.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007; 448:353–357. doi: 10.1038/nature06007 [DOI] [PubMed] [Google Scholar]
- 7.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnabel RB, Kerr KF, Lubitz SA, Alkylbekova EL, Marcus GM, Sinner MF, et al. Atrial Fibrillation/Electrocardiography Working Group. Large-scale candidate gene analysis in whites and African Americans identifies IL6R polymorphism in relation to atrial fibrillation: the National Heart, Lung, and Blood Institute's Candidate Gene Association Resource (CARe) project. Circ Cardiovasc Genet. 2011;4:557–564. doi: 10.1161/CIRCGENETICS.110.959197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aguirre LA, Alonso ME, Badía-Careaga C, Rollán I, Arias C, Fernández-Miñán A, et al. Long-range regulatory interactions at the 4q25 atrial fibrillation risk locus involve PITX2c and ENPEP. BMC Biol. 2015;13:26–. doi: 10.1186/s12915-015-0138-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Bai Y, Li N, Ye W, Zhang M, Greene SB,et al. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. PNAS 2014;111:9181–9186. doi: 10.1073/pnas.1405411111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirchhof P, Kahr PC, Kaese S, Piccini I, Vokshi I, Scheld HH, et al. PITX2c is expressed in the adult left atrium, and reducing Pitx2c expression promotes atrial fibrillation inducibility and complex changes in gene expression. Circ Cardiovasc Genet. 2011;4:123–133. doi: 10.1161/CIRCGENETICS.110.958058 [DOI] [PubMed] [Google Scholar]
- 14.Chinchilla A, Daimi H, Lozano-Velasco E, Dominguez JN, Caballero R, Delpón E, et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ Cardiovasc Genet. 2011;4:269–279. doi: 10.1161/CIRCGENETICS.110.958116 [DOI] [PubMed] [Google Scholar]
- 15.Tao Y, Zhang M, Li L, Bai Y, Zhou Y, Moon AM, et al. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ Cardiovasc Genet. 2014;7:23–32. doi: 10.1161/CIRCGENETICS.113.000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lozano-Velasco E, Hernández-Torres F, Daimi H, Serra SA, Herraiz A, Hove-Madsen L, et al. Pitx2 impairs calcium handling in a dose-dependent manner by modulating Wnt signalling. Cardiovasc Res. 2016;109:55–66. doi: 10.1093/cvr/cvv207 [DOI] [PubMed] [Google Scholar]
- 17.Berenfeld O, Jalife J. Mechanisms of atrial fibrillation: rotors, ionic determinants, and excitation frequency. Cardiol Clin. 2014;32:495–506. doi: 10.1016/j.ccl.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jalife J, Kaur K. Atrial remodeling, fibrosis, and atrial fibrillation. Trends Cardiovasc Med. 2015; 25:475–484. doi: 10.1016/j.tcm.2014.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolke C, Bukowska A, Goette A, Lendeckel U. Redox control of cardiac remodeling in atrial fibrillation. Biochim Biophys Acta. 2015;1850:1555–1565 doi: 10.1016/j.bbagen.2014.12.012 [DOI] [PubMed] [Google Scholar]
- 20.Reilly SN, Liu X, Carnicer R, Recalde A, Muszkiewicz A, Jayaram R, et al. Up-regulation of miR-31 in human atrial fibrillation begets the arrhythmia by depleting dystrophin and neuronal nitric oxide synthase. Sci Transl Med. 2016;8:340ra74 doi: 10.1126/scitranslmed.aac4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dzeshka MS, Lip GY, Snezhitskiy V, Shantsila E. Cardiac Fibrosis in Patients With Atrial Fibrillation: Mechanisms and Clinical Implications. J Am Coll Cardiol. 2015;66:943–959. doi: 10.1016/j.jacc.2015.06.1313 [DOI] [PubMed] [Google Scholar]
- 22.Zhang Q, Liu T, Ng CY, Li G. Diabetes mellitus and atrial remodeling: mechanisms and potential upstream therapies. Cardiovasc Ther. 2014;32:233–41. doi: 10.1111/1755-5922.12089 [DOI] [PubMed] [Google Scholar]
- 23.Anumonwo JM, Kalifa J. Risk factors and genetics of atrial fibrillation. Cardiol Clin. 2014;32:485–494. doi: 10.1016/j.ccl.2014.07.007 [DOI] [PubMed] [Google Scholar]
- 24.Goudis CA, Korantzopoulos P, Ntalas IV, Kallergis EM, Ketikoglou DG. Obesity and atrial fibrillation: A comprehensive review of the pathophysiological mechanisms and links. J Cardiol. 2015;S0914–5087 [DOI] [PubMed] [Google Scholar]
- 25.Yadava M, Hughey AB, Crawford TC. Postoperative atrial fibrillation: incidence, mechanisms, and clinical correlates. Cardiol Clin. 2014;32:627–636. doi: 10.1016/j.ccl.2014.07.002 [DOI] [PubMed] [Google Scholar]
- 26.Kumar KR, Mandleywala SN, Link MS. Atrial and ventricular arrhythmias in hypertrophic cardiomyopathy. Card Electrophysiol Clin. 2015;7:173–186. doi: 10.1016/j.ccep.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 27.Scridon A, Fouilloux-Meugnier E, Loizon E, Rome S, Julien C, Barrès C, et al. Long-standing arterial hypertension is associated with Pitx2 down-regulation in a rat model of spontaneous atrial tachyarrhythmias. Europace 2015;17:160–165. doi: 10.1093/europace/euu139 [DOI] [PubMed] [Google Scholar]
- 28.L'honoré A, Commère PH, Ouimette JF, Montarras D, Drouin J, Buckingham M. Redox regulation by Pitx2 and Pitx3 is critical for fetal myogenesis. Dev Cell. 2014;29:392–405. doi: 10.1016/j.devcel.2014.04.006 [DOI] [PubMed] [Google Scholar]
- 29.Tao G, Kahr PC, Morikawa Y, Zhang M, Rahmani M, Heallen TR, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 2016;534:119–23. doi: 10.1038/nature17959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gage PJ, Suh H, Camper SA. Dosage requirement of Pitx2 for development of multiple organs. Development. 1999;126:4643–4651. [DOI] [PubMed] [Google Scholar]
- 31.de Lange FJ, Moorman AF, Christoffels VM. Atrial cardiomyocyte-specific expression of Cre recombinase driven by an Nppa gene fragment. Genesis. 2003;37:1–4. doi: 10.1002/gene.10220 [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Gómez I, Banegas I, Wangensteen R, Quesada A, Jiménez R, Gómez-Morales M, et al. Influence of thyroid state on cardiac and renal capillary density and glomerular morphology in rats. J Endocrinol. 2013;216:43–51. doi: 10.1530/JOE-12-0208 [DOI] [PubMed] [Google Scholar]
- 33.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 34.Lozano-Velasco E, Chinchilla A, Martínez-Fernández S, Hernández-Torres F, Navarro F, Lyons GE, et al. Pitx2c modulates cardiac-specific transcription factors networks in differentiating cardiomyocytes from murine embryonic stem cells. Cells Tissues Organs. 2011;194:349–62. doi: 10.1159/000323533 [DOI] [PubMed] [Google Scholar]
- 35.Daimi H, Lozano-Velasco E, Haj Khelil A, Chibani JB, Barana A, Amorós I, et al. Regulation of SCN5A by microRNAs: miR-219 modulates SCN5A transcript expression and the effects of flecainide intoxication in mice. Heart Rhythm. 2015;12:1333–42. doi: 10.1016/j.hrthm.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 36.Wang YG, Dedkova EN, Fiening JP, Ojamaa K, Blatter LA, Lipsius SL. Acute exposure to thyroid hormone increases Na+ current and intracellular Ca2+ in cat atrial myocytes. J Physiol. 2003. January 15;546(Pt 2):491–9. doi: 10.1113/jphysiol.2002.032847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philippen LE, Dirkx E, da Costa-Martins PA, De Windt LJ. Non-coding RNA in control of gene regulatory programs in cardiac development and disease. J Mol Cell Cardiol. 2015;89:51–58. doi: 10.1016/j.yjmcc.2015.03.014 [DOI] [PubMed] [Google Scholar]
- 38.Okamoto AK.Development of a strain of spontaneously hypertensive rat. Jap Circ J 1963;27:282–293. [DOI] [PubMed] [Google Scholar]
- 39.Youn JY, Zhang J, Zhang Y, Chen H, Liu D, Ping P, et al. Oxidative stress in atrial fibrillation: an emerging role of NADPH oxidase. J Mol Cell Cardiol. 2013;62:72–9. doi: 10.1016/j.yjmcc.2013.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Violi F, Pastori D, Pignatelli P, Loffredo L. Antioxidants for prevention of atrial fibrillation: a potentially useful future therapeutic approach? A review of the literature and meta-analysis. Europace. 2014;16:1107–1116. doi: 10.1093/europace/euu040 [DOI] [PubMed] [Google Scholar]
- 41.Biondi B, Kahaly GJ. Cardiovascular involvement in patients with different causes of hyperthyroidism. Nat Rev Endocrinol. 2010;6:431–443. doi: 10.1038/nrendo.2010.105 [DOI] [PubMed] [Google Scholar]
- 42.Grais IM, Sowers JR. Thyroid and the heart. Am J Med. 2014;127:691–8. doi: 10.1016/j.amjmed.2014.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kreuzberg U, Theissen P, Schicha H, Schröder F, Mehlhorn U, de Vivie ER, et al. Single-channel activity and expression of atrial L-type Ca(2+) channels in patients with latent hyperthyroidism. Am J Physiol Heart Circ Physiol. 2000;278:H723–30. [DOI] [PubMed] [Google Scholar]
- 44.Arai M, Otsu K, MacLennan DH, Alpert NR, Periasamy M. Effect of thyroid hormone on the expression of mRNA encoding sarcoplasmic reticulum proteins. Circ Res. 1991;69:266–276. [DOI] [PubMed] [Google Scholar]
- 45.Shimoni Y, Fiset C, Clark RB, Dixon JE, McKinnon D, Giles WR. Thyroid hormone regulates postnatal expression of transient K+ channel isoforms in rat ventricle. J Physiol. 1997;500:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mager S, Palti Y, Binah O. Mechanism of hyperthyroidism-induced modulation of the L-type Ca2+ current in guinea pig ventricular myocytes. Pflugers Arch. 1992;421:425–430. [DOI] [PubMed] [Google Scholar]
- 47.Jiang M, Xu A, Tokmakejian S, Narayanan N. Thyroid hormone-induced overexpression of functional ryanodine receptors in the rabbit heart. Am J Physiol Heart Circ Physiol. 2000;278:H1429–1438. [DOI] [PubMed] [Google Scholar]
- 48.Dudley SC Jr, Baumgarten CM. Bursting of cardiac sodium channels after acute exposure to 3,5,3'-triiodo-L-thyronine. Circ Res. 1993;73:301–313. [DOI] [PubMed] [Google Scholar]
- 49.Emdin CA, Callender T, Cao J, Rahimi K. Effect of antihypertensive agents on risk of atrial fibrillation: a meta-analysis of large-scale randomized trials. Europace. 2015;17:701–710. doi: 10.1093/europace/euv021 [DOI] [PubMed] [Google Scholar]
- 50.Joseph BK, Thakali KM, Moore CL, Rhee SW. Ion channel remodeling in vascular smooth muscle during hypertension: Implications for novel therapeutic approaches. Pharmacol Res. 2013;70:126–138. doi: 10.1016/j.phrs.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pabbidi MR, Mazur O, Fan F, Farley JM, Gebremedhin D, Harder DR, et al. Enhanced large conductance K+ channel activity contributes to the impaired myogenic response in the cerebral vasculature of Fawn Hooded Hypertensive rats. Am J Physiol Heart Circ Physiol. 2014;306:H989–H1000. doi: 10.1152/ajpheart.00636.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montaigne D, Marechal X, Lefebvre P, Modine T, Fayad G, Dehondt H, et al. Mitochondrial dysfunction as an arrhythmogenic substrate: a translational proof-of-concept study in patients with metabolic syndrome in whom post-operative atrial fibrillation develops. JACC. 2013;62:1466–1473. doi: 10.1016/j.jacc.2013.03.061 [DOI] [PubMed] [Google Scholar]
- 53.Anderson EJ, Efird JT, Davies SW, O'Neal WT, Darden TM, Thayne KA, et al. Monoamine oxidase is a major determinant of redox balancein human atrial myocardium and is associated with postoperative atrial fibrillation. JAHA. 2014;3:e000713 doi: 10.1161/JAHA.113.000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martínez-González MÁ, Toledo E, Arós F, Fiol M, Corella D, Salas-Salvadó J, et al. Extravirgin olive oil consumption reduces risk of atrial fibrillation: the PREDIMED (Prevención con Dieta Mediterránea) trial. Circulation. 2014;130:18–26. doi: 10.1161/CIRCULATIONAHA.113.006921 [DOI] [PubMed] [Google Scholar]
- 55.Zima AV, Blatter LA. Redox regulation of cardiac calcium channels and transporters. Cardiovasc Res. 2006;71:310–21. doi: 10.1016/j.cardiores.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 56.Wagner S, Rokita AG, Anderson ME, Maier LS. Redox regulation of sodium and calcium handling. Antioxid Redox Signal. 2013;18:1063–77. doi: 10.1089/ars.2012.4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aggarwal NT, Makielski JC. Redox control of cardiac excitability. Antioxid Redox Signal. 2013;18:432–68. doi: 10.1089/ars.2011.4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Franco D, Chinchilla A, Daimi H, Dominguez JN, Aránega A. Modulation of conductive elements by Pitx2 and their impact on atrial arrhythmogenesis. Cardiovasc Res. 2011;91:223–31. doi: 10.1093/cvr/cvr078 [DOI] [PubMed] [Google Scholar]
- 59.Laumanns IP, Fink L, Wilhelm J, Wolff JC, Mitnacht-Kraus R, Graef-Hoechst S, et al. The noncanonical WNT pathway is operative in idiopathic pulmonary arterial hypertension. Am J Respir Cell Mol Biol. 2009. 40(6):683–91. doi: 10.1165/rcmb.2008-0153OC [DOI] [PubMed] [Google Scholar]
- 60.Terentyev D, Belevych AE, Terentyeva R, Martin MM, Malana GE, Kuhn DE, et al. miR-1 overexpression enhances Ca(2+) release and promotes cardiac arrhythmogenesis by targeting PP2A regulatory subunit B56alpha and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ Res. 2009;104:514–21. doi: 10.1161/CIRCRESAHA.108.181651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jia X, Zheng S, Xie X, Zhang Y, Wang W, Wang Z, et al. MicroRNA-1 accelerates the shortening of atrial effective refractory period by regulating KCNE1 and KCNB2 expression: an atrial tachypacing rabbit model. PLoS One. 2013;8:e85639 doi: 10.1371/journal.pone.0085639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao C, Gui Y, Guo Y, Xu D. The regulatory function of microRNA-1 in arrhythmias. Mol Biosyst. 2016;12:328–33. doi: 10.1039/c5mb00806a [DOI] [PubMed] [Google Scholar]
- 63.Luo X, Pan Z, Shan H, Xiao J, Sun X, Wang N, et al. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J Clin Invest. 2013;123:1939–1951. doi: 10.1172/JCI62185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Daimi H, Lozano-Velasco E, Haj Khelil A, Chibani JB, Barana A, Amorós I, et al. Regulation of SCN5A by microRNAs: miR-219 modulates SCN5A transcript expression and the effects of flecainide intoxication in mice. Heart Rhythm. 2015;12:1333–1342. doi: 10.1016/j.hrthm.2015.02.018 [DOI] [PubMed] [Google Scholar]
- 65.Shan H, Zhang Y, Cai B, Chen X, Fan Y, Yang L, et al. Upregulation of microRNA-1 and microRNA-133 contributes to arsenic-induced cardiac electrical remodeling. Int J Cardiol. 2013;167:2798–2805. doi: 10.1016/j.ijcard.2012.07.009 [DOI] [PubMed] [Google Scholar]
- 66.Li YD, Hong YF, Yusufuaji Y, Tang BP, Zhou XH, Xu GJ, et al. Altered expression of hyperpolarization-activated cyclic nucleotide-gated channels and microRNA-1 and -133 in patients with age-associated atrial fibrillation. Mol Med Rep. 2015;12:3243–8. doi: 10.3892/mmr.2015.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Köhler AC, Sag CM, Maier LS. Reactive oxygen species and excitation-contraction coupling in the context of cardiac pathology. J Mol Cell Cardiol. 2014;73:92–102. doi: 10.1016/j.yjmcc.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 68.Cheng Y, Liu X, Zhang S, Lin Y, Yang J, Zhang C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol. 2009;47:5–14. doi: 10.1016/j.yjmcc.2009.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
A) Systolic blood pressure (SBP) in mmHg and heart rate (HR) in beats per minute (BPM) measured in control and hyperthyroid (HTD) rats at the end of the experiment (n = 10 each group). Mean ± SEM is displayed. *p<0.001 vs control group. B) Systolic blood pressure (SBP) in mmHg and heart rate (HR) in beats per minute (BPM) measured in Wistar-Kyoto (WKY) and spontaneously hypertensive rats (SHR) at the end of the experiment (n = 10 each group). Mean ± SEM is displayed. *p<0.01, **p<0.001 vs control group.
(TIFF)
Analyses of Pitx2 expression in primary culture of fetal cardiomyocytes treated with T4 as compared to controls (panel A). Observe that Pitx2 is significantly decreased after T4 administration. Analyses of Pitx2 (panel B) and Enpep (panel C) in HL1 atrial cardiomycytes after Pitx2 siRNA silencing. Observe that Pitx2 siRNA administration significantly decrease Pitx2 expression (panel B) and also Enpep expression (panel C). *p<0.01, ***p<0.001, ****p<0.0001.
(TIF)
Observe that H202 administration significantly increased Wnt11 at 3h and 6h while significantly decreased Zfhx3 expression at all experimental conditions analyzed. *p<0.01, **p<0.05, ****p<0.0001.
(TIF)
Observe that no significant differences are observed in Prdx3 and Prdx6 expression, except for Prdx6 at 12h after treatment in Pitx2 silencing conditions. *p<0.01.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.