Abstract
Rationale: Nontuberculous mycobacteria are an important cause of morbidity in the United States, although patient outcomes vary greatly by species. Currently, nationally representative data on the distribution of mycobacterial species from clinical isolates are limited.
Objectives: Using a national hospitalization database capturing microbiologic data for nearly 6 million patient encounters, we describe the geographic distribution of, and patient demographic features associated with, clinical mycobacterial isolates in the United States.
Methods: Linked demographic and microbiologic data from the Premier Healthcare Database were extracted for all patient encounters from 2009 to 2013. Patients with at least one positive potentially pathogenic nontuberculous mycobacterial culture were identified as cases. The period prevalence was calculated, and patient-, encounter-, and hospital-level factors were analyzed. Regional differences in species distribution were analyzed; a subanalysis was conducted among patients with International Classification of Diseases, Ninth Revision, codes for pulmonary nontuberculous mycobacterial disease. Significant differences were assessed (P < 0.05).
Results: Of 5,928,830 unique patients included during the 5-year study period, 7,812 (0.13%) had at least one positive nontuberculous mycobacterial culture. The mean age of cases was 64 years (range, <1–89 yr), and most were female (52%) and white (70%). Hospitals with cases were more often labeled “urban” (96%), “teaching” (56%), and had at least 500 beds (78%). Species distribution differed significantly by geographic area. Mycobacterium avium complex ranged from 61 to 91% of isolates and were most frequent in the South and Northeast regions; M. abscessus/M. chelonae ranged from 2 to 18% of isolates and were most frequent in the West; and other species, including M. fortuitum and M. kansasii, ranged from 7 to 26% and were also most frequent in the West.
Conclusions: Significant geographic variation exists in the distribution of nontuberculous mycobacterial species in the United States. Whereas M. avium complex was the most common species isolated in the South, M. abscessus/M. chelonae was proportionately higher in the West. Greater clinical awareness in regions with increased levels of harder-to-treat mycobacteria are needed, given differences in treatment options and implications for patient outcomes.
Keywords: nontuberculous mycobacteria, United States, hospital database
Nontuberculous mycobacteria (NTM) represent a group of environmental bacteria commonly found in water and soil (1–4). Infections due to NTM are increasingly recognized as an important cause of morbidity and mortality in the United States; however, treatment options and subsequent patient outcomes vary greatly by species (4, 5). Although some studies have identified regional differences in NTM prevalence as well as associations with environmental factors (6), recent studies focused on variations in the distribution of NTM species in the general population are limited (7). Given the implications for therapy and infection control measures, knowledge of national species-specific patterns is important to further understanding the epidemiology of NTM in the United States.
In a recent study, researchers found higher NTM-related mortality in the southern United States (8), and studies of Medicare beneficiaries showed that pulmonary nontuberculous mycobacteria (PNTM) rates were highest in the West and Southeast and lowest in the Midwest (5, 6). However, the investigators lacked species-specific information to understand how mycobacterial species varied geographically. In another study, among patients with cystic fibrosis (CF), investigators observed geographic differences in the state-level distribution of Mycobacterium avium complex (MAC) versus Mycobacterium abscessus, with more M. abscessus identified in traditionally “high-prevalence” NTM areas, such as in the southern United States (9), although these findings in the high-risk CF population may not be generalizable. Two studies have provided estimates of national species distribution. The first, a study conducted in four health maintenance organizations, revealed that MAC predominated in all regions sampled (7). Authors of a national survey of physicians also found that the majority of species across all regions of the United States were MAC; however, the proportion of M. abscessus was slightly higher in the West than in other regions (10).
Using a large, nationally distributed, hospital-based patient dataset with patient-, isolate-, and facility-level information, we describe trends in the distribution of mycobacterial species obtained from clinical samples throughout the United States. Improving understanding of the distribution of mycobacterial species, as well as identifying geographic variations in NTM species, including regions with a higher burden of harder-to-treat mycobacteria such as M. abscessus, is critical for informing clinical practice and improving patient outcomes. Preliminary results of this work were previously presented in the form of a poster (11).
Methods
Data Source
We conducted a retrospective cohort study using the Premier Healthcare Database, which is managed by Premier, Inc. (Charlotte, NC). The Premier Healthcare Database is a U.S. national database with demographic and microbiologic data, which are linked by unique patient identifiers, derived from 176 United States hospitals distributed throughout the country (38% in the South, 30% in the Midwest, and 16% each in the Northeast and West), which is similar to the overall distribution of U.S. hospitals (12). Information provided in the Premier Healthcare Database is collected as part of voluntary reporting done by participating hospitals, and patients are tracked over multiple encounters, including as both inpatients and outpatients, using unique patient identifiers, and it has been used in multiple epidemiologic studies to date (13). This dataset uniquely links microbiologic, demographic, and clinical data for millions of inpatient and outpatient encounters, facilitating the detection of epidemiologic trends of rare diseases using microbiologic results rather than billing codes. However, limitations of this dataset include the lack of information regarding the body site of specimen collection or the specific laboratory methods used.
Study Population and Case Definition
Our study population comprised all unique patients with an inpatient or outpatient encounter in the Premier Healthcare Database from 2009 to 2013 at hospitals with microbiologic data reported; detailed laboratory methods used at each facility for species identification were not available. For all unique patients present, we extracted demographic, comorbidity, facility-level, and microbiologic data; comorbidities were identified using International Classification of Diseases, Ninth Revision (ICD-9), codes. Patients with mycobacterial cultures performed were identified, and results were recorded, including any species identified. NTM-positive cases were defined as those with at least one mycobacterial culture growing a pathogenic NTM species, regardless of body site sampled for culture. Only the first positive NTM test per patient in the dataset was analyzed, and all species isolated from the first positive test were included. For noncases (including those patients who were tested and had a negative result for NTM as well as those not tested), we used data from their first encounter in the database.
Data Analysis
All mycobacterial species identified were classified into one of six groups: (1) MAC, (2) M. abscessus/M. chelonae, (3) M. fortuitum, (4) M. kansasii, (5) other rapidly growing mycobacteria, or (6) other potentially pathogenic NTM species (see Table E1 in the online supplement). Tests with mycobacterial species unidentified or considered nonpathogenic (M. gastri, M. gilvum, M. gordonae, M. kubicae, M. murale, M. paraffinicum, M. sphagni, and M. vaccae/vanbaalenii) were excluded. Patient, encounter, and hospital factors were analyzed by NTM case status and by mycobacterial species group. Geographic regions were defined according to United States Census boundaries (14). NTM-specific ICD-9 codes for pulmonary (031.0), cutaneous (031.1), and disseminated (031.2) NTM were analyzed, and a subgroup analysis was conducted including only patients with an ICD-9 code for PNTM. Differences were assessed using the chi-square test or Student’s t test, with significance evaluated at P < 0.05. All analyses were performed using SAS version 9.3 software (SAS Institute, Cary, NC). In this study, we used only de-identified patient data, and thus the study was considered exempt from National Institutes of Health Institutional Review Board review.
Results
Among the 176 hospitals in the Premier Healthcare Database with microbiologic data, 151 (86%) reported at least one NTM isolation. Overall, 5,928,830 unique patients were evaluated during the 5-year study period, of whom 7,812 (0.13%) had at least one NTM-positive culture (Table 1). Of these, 1,360 (17%) had at least two positive NTM cultures within the same encounter, and 7,118 (91%) had at least one positive acid-fast bacilli test result. When evaluating ICD-9 codes for other comorbidities occurring in five or more patients, 76% (n = 5,069) of NTM cases had a principal diagnosis associated with a respiratory condition. A minority of the NTM isolates identified had an ICD-9 code for PNTM (n = 487 [6%]) or nonpulmonary NTM (n = 397 [5%]). Of 7,812 patients with NTM infection, 7,941 potentially pathogenic NTM isolates representing 55 different NTM species were identified; 124 patients (2%) had more than one NTM species isolated from the same culture, and 111 isolates were nonpathogenic NTM species. MAC was most commonly identified (76%), followed by M. abscessus/M. chelonae (9%) (which were frequently grouped together in laboratory results), M. fortuitum (6%), M. kansasii (3%), other rapidly growing mycobacteria (1%), and other potentially pathogenic NTM species (5%). Among patients with an ICD-9 code for PNTM (n = 487), MAC was the most common NTM species identified (77%), followed by M. abscessus/M. chelonae (9%) (Table 2). For disseminated NTM, MAC was also the most common species identified (93%), followed by M. abscessus/M. chelonae (3%). For patients with an ICD-9 code for cutaneous NTM, M. abscessus/M. chelonae (50%) was most common. The 78 patients with an ICD-9 code for CF also had MAC most often identified (50%); however, M. abscessus/M. chelonae was also five times more common (45%) than in non-CF NTM cases. A majority of patients with M. fortuitum (88%) had this as their only species identified.
Table 1.
Baseline patient-, encounter-, and hospital-level proportions comparing nontuberculous mycobacteria-positive with non–nontuberculous mycobacteria–positive patients in Premier Healthcare Database, 2009–2013
| NTM-Positive Patients (n = 7,812) | Non–NTM-Positive Patients (n = 5,921,012) | |
|---|---|---|
| Patient-level characteristics | ||
| Year isolated | ||
| 2009 | 21 | 21 |
| 2010 | 23 | 23 |
| 2011 | 21 | 21 |
| 2012 | 19 | 18 |
| 2013 | 16 | 17 |
| Age categories* | ||
| <18 yr | 1 | 18 |
| 18–44 yr | 13 | 34 |
| 45–64 yr | 31 | 23 |
| ≥65 yr | 55 | 25 |
| Sex*† | ||
| Male | 48 | 38 |
| Female | 52 | 62† |
| Race* | ||
| White | 70 | 64 |
| Black | 14 | 16 |
| Hispanic/other | 16 | 20 |
| Encounter-level characteristics | ||
| Hospital encounter type* | ||
| Inpatient | 60 | 40 |
| Outpatient | 40 | 60 |
| Length of stay, d (among inpatients)* | ||
| Median (IQR) | 8 (5–13) | 4 (2–7) |
| Range | 1–439 | 1–1,463 |
| Discharge status (among inpatients)* | ||
| Death | 11 | 4 |
| Institution‡ | 35 | 30 |
| Other§ discharge status/not reported | 54 | 66 |
| Hospital-level characteristics | ||
| Geographic region* | ||
| New England‖ | 5 | 5 |
| Middle Atlantic¶ | 16 | 16 |
| East South Central** | 3 | 3 |
| South Atlantic†† | 29 | 27 |
| West South Central‡‡ | 12 | 10 |
| Mountain§§ | 3 | 3 |
| Pacific‖‖ | 13 | 10 |
| East North Central¶¶ | 14 | 16 |
| West North Central*** | 5 | 9 |
| Hospital location* | ||
| Rural | 4 | 9 |
| Urban | 96 | 91 |
| Hospital bed size* | ||
| 0–99 | 2 | 4 |
| 100–499 | 20 | 32 |
| ≥500 | 78 | 64 |
| Hospital teaching status* | ||
| Nonteaching | 44 | 52 |
| Teaching | 56 | 48 |
| Attending physician specialty (among inpatients)* | ||
| Pulmonary | 11 | 2 |
| Infectious disease | 2 | 0.4 |
| General/internal medicine | 44 | 23 |
| Other specialty | 43 | 75 |
Definition of abbreviations: IQR = interquartile range; NTM = nontuberculous mycobacteria.
Data are percentages unless otherwise indicated.
P < 0.05.
Sex was not reported for 940 patients in this group.
Patient was discharged or transferred to another hospital, healthcare facility, or home health organization.
Includes discharged to home, left against medical advice, discharged to law enforcement, still a patient and expected to return, and information not available.
Connecticut, Massachusetts, Maine, New Hampshire, Rhode Island, and Vermont.
New Jersey, New York, and Pennsylvania.
Alabama, Kentucky, Mississippi, and Tennessee.
District of Columbia, Delaware, Florida, Georgia, Maryland, North Carolina, South Carolina, Virginia, and West Virginia.
Arkansas, Louisiana, Oklahoma, and Texas.
Arizona, Colorado, Idaho, Montana, New Mexico, Nevada, Utah, and Wyoming.
Alaska, California, Hawaii, Oregon, and Washington.
Illinois, Indiana, Michigan, Ohio, and Wisconsin.
Iowa, Kansas, Minnesota, Missouri, North Dakota, Nebraska, and South Dakota.
Table 2.
Nontuberculous mycobacteria species distribution by age group, ICD-9 code, and hospital encounter type in Premier Healthcare Database, 2009–2013
| Mycobacterium avium Complex (%) | Mycobacterium abscessus/Mycobacterium chelonae (%) | Mycobacterium fortuitum (%) | Other Rapidly Growing Pathogenic Mycobacteria (%) | Mycobacterium kansasii (%) | Other Potentially Pathogenic NTM Species Not Otherwise Specified (%) | |
|---|---|---|---|---|---|---|
| All NTM isolates identified | 76 | 9 | 6 | 1 | 3 | 5 |
| Age category | ||||||
| <18 yr | 67 | 20 | 4 | 3 | 2 | 4 |
| 18–44 yr | 69 | 12 | 8 | 2 | 3 | 6 |
| 45–64 yr | 74 | 8 | 7 | 1 | 4 | 6 |
| ≥65 yr | 79 | 9 | 5 | 1 | 2 | 4 |
| Pulmonary NTM ICD-9 code status | ||||||
| Present (n = 487) | 77 | 9 | 5 | <1 | 6 | 3 |
| Absent (n = 397) | 74 | 16 | 5 | <1 | 1 | 4 |
| Hospital-encounter type | ||||||
| Inpatient | 76 | 8 | 7 | 1 | 3 | 5 |
| Outpatient | 77 | 11 | 4 | 1 | 2 | 5 |
Definition of abbreviations: ICD-9 = International Classification of Diseases, Ninth Revision; NTM = nontuberculous mycobacteria.
Characteristics of Patients with NTM Infection and Hospitals
The median age of patients with NTM infection was 67 years (range, <1–89 yr), and 55% were at least 65 years of age (Table 1). Most patients with NTM infection were white (70%), and half were female (52%). Patient encounters were most often inpatient (60%), and the median length of hospital stay for NTM-associated inpatient encounters was 8 days (interquartile range, 5–13), compared with 4 days (interquartile range, 2–7) for other inpatient encounters (P < 0.0001). Hospitals with patients with NTM infection were more often labeled as “urban” (96%) or “teaching” (56%) and had a hospital bed size greater than or equal to 500 (78%). Patients with NTM infection were significantly more likely to have a general medicine physician (44%) than a pulmonary specialist (11%) as their attending physician. Among patients with NTM infection, chronic obstructive pulmonary disease was the most commonly reported comorbidity (28%), followed by bronchiectasis (11%); malignant neoplasms of the trachea, bronchus, and lung (6%); and HIV (6%).
M. abscessus/M. chelonae was significantly more common among those younger than 18 years of age than among persons aged 45–64 years (20% vs. 8%), whereas MAC was significantly more common among patients older than 64 years of age than among those younger than 18 years of age (79% vs. 67%) (Table 2). M. fortuitum was slightly more common among persons aged 18–44 years than among those older than 64 years of age (8% vs. 5%), as were other potentially pathogenic NTM species (7% vs. 4%) (P = 0.003). Of the 105 patients aged less than 18 years, 23 (22%) had an ICD-9 code for CF. Among patients with an ICD-9 code for PNTM, M. kansasii was more common among 18- to 44-year-olds than among persons older than 64 years of age (15% vs. 2%).
NTM species identified via an inpatient or outpatient encounter also differed. The proportion of patients with M. fortuitum was significantly higher for inpatient (7%) than for outpatient (4%) encounters (P < 0.05). In contrast, the proportion of M. abscessus/M. chelonae was significantly higher for outpatient (11%) than for inpatient (8%) isolates (P < 0.05). Among patients with an ICD-9 code for PNTM, MAC was more common among outpatients than among inpatients (83% vs. 74%), whereas M. kansasii was more common among inpatients than among outpatients (8% vs. 1%).
Geographic Distribution of NTM Species
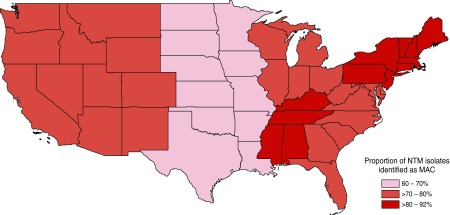
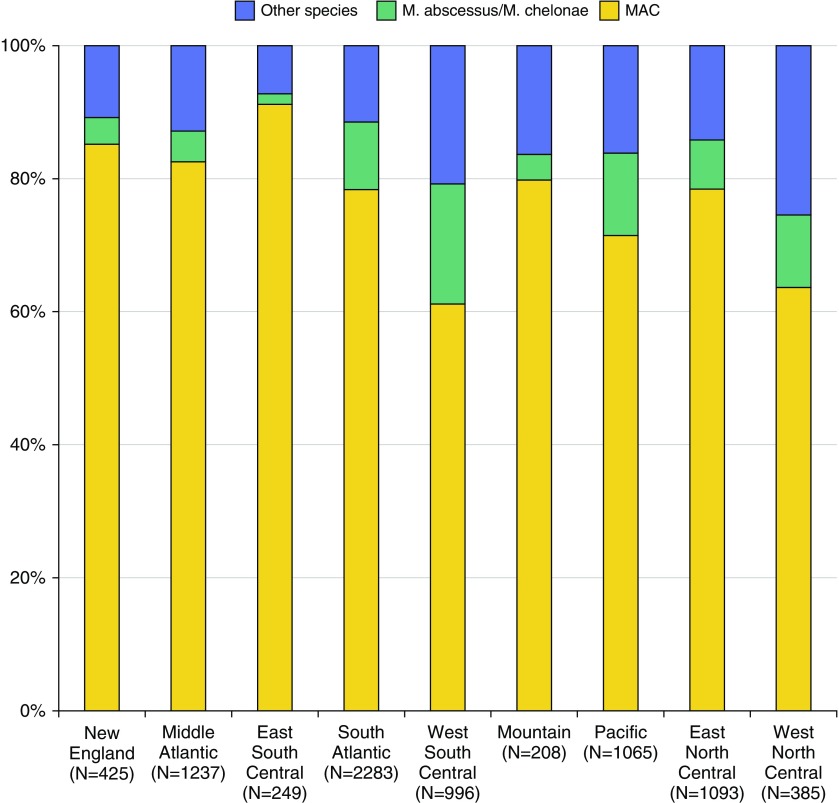
Species distribution differed significantly by geographic area (Figures 1 and 2). MAC ranged from 61% of NTM isolates in the West South Central (Arkansas, Louisiana, Oklahoma, and Texas), to 78% in the South Atlantic (Washington, DC; Delaware; Florida; Georgia; Maryland; North Carolina; South Carolina; Virginia; and West Virginia) and the East North Central (Illinois, Indiana, Michigan, Ohio, and Wisconsin), and to 91% in the East South Central (Alabama, Kentucky, Mississippi, and Tennessee). M. abscessus/M. chelonae ranged from 2% in the East South Central to 18% in the West South Central. Other NTM species, including M. fortuitum, M. kansasii, other rapidly growing mycobacteria, and other potentially pathogenic NTM species, comprised only 7% of NTM isolates in the East South Central, compared with 16% in the Pacific (Alaska, California, Hawaii, Oregon, and Washington), 21% in the West South Central, and 26% in the West North Central (Iowa, Kansas, Minnesota, Missouri, North Dakota, Nebraska, and South Dakota).
Figure 1.
Distribution of Mycobacterium avium complex (MAC) by U.S. geographic region in the Premier Healthcare Database, 2009–2013 (11). NTM = nontuberculous mycobacteria.
Figure 2.
Distribution of nontuberculous mycobacteria species identified by United States geographic area in Premier Healthcare Database, 2009–2013. MAC = Mycobacterium avium complex.
Among patients aged 65 years or older, the distribution of MAC was the same as in the overall population, ranging from 66% of NTM isolates in the West South Central to up to 93% of isolates in the East South Central, and the distribution of M. abscessus/M. chelonae was also the same, ranging from 2% in the East South Central to 18% in the West South Central. Among patients with an ICD-9 code for PNTM, MAC ranged from 57% in the East South Central, to 94% in the Mountain (Arizona, Colorado, Idaho, Montana, New Mexico, Nevada, Utah, and Wyoming), and to 69% in the West North Central and Pacific. M. abscessus/M. chelonae ranged from no cases identified in the East South Central and West North Central regions to 15% in the Pacific and 11% in the South Atlantic.
Discussion
We provide current estimates of the distribution of NTM species in the United States using a large national sample of hospital-based encounters with microbiologic test results. Specifically, we identified different geographic patterns by NTM species, as well as variations among demographic and facility-level features of the affected patients. Geographic variations may be due to a number of factors, including climatic and environmental differences affecting the types of mycobacteria present in soil and water sources (6, 15).
In the present study, MAC was the most commonly isolated NTM species, followed by M. abscessus/M. chelonae and M. fortuitum; this pattern is consistent with previous findings (7, 16). However, NTM species frequencies varied by U.S. region. Although MAC was most commonly isolated overall, M. abscessus/M. chelonae was proportionately highest in the South. Other studies looking at NTM isolated from all body sites found similar species distributions in the United States (17, 18), although in state-specific studies, the proportion of NTM isolates identified as MAC ranged greatly, from only 40% in Virginia to over 84% in New York City (19–22). M. abscessus/M. chelonae was the second most common isolate in our study (9%), as well as in studies conducted nationwide (5%) (17) and in Oregon (5%) (20, 23), North Carolina (18%) (21), New York City (6%) (19), and Virginia (7%). It is important to note also that because M. abscessus is harder to treat (4), clinical outcomes may be disproportionally impacted in areas with higher frequencies of this species.
Most patients had only a single positive NTM culture, and few had corresponding ICD-9 codes for NTM, which is consistent with an estimated sensitivity of 27 to 50% for these codes (7, 24). Also consistent with prior studies were the reported frequencies of chronic obstructive pulmonary disease; bronchiectasis; malignant neoplasms of the trachea, bronchus, and lung; and HIV (7). A higher proportion of patients with NTM infection younger than 18 years of age in this study population also had an ICD-9 code for CF (22%) than in a previous study (25). Notably, only 11% of inpatients with NTM infection had a pulmonologist as their attending physician. Pulmonologists may have been involved in a consultant capacity, which was not discernible in the dataset. Hospitals with NTM-positive patients were more likely to be larger, to be located in urban areas, and to be teaching hospitals than were hospitals without NTM cases. This may be reflective of NTM being more common in urban areas or may potentially be due in part to differences in referral patterns, with more ill patients being referred to larger medical centers in urban areas, as well as those with a greater capacity for diagnostic mycobacterial testing and species identification.
This study was subject to several limitations. We were unable to identify the source of the NTM infection as respiratory, skin and soft tissue, or disseminated in nature, owing to the lack of data available on specimen collection site. However, the distribution of species that we report is similar to what is often detected in other studies on PNTM cases (18–23). Additionally, we conducted a subanalysis limited to patients with NTM infection with the ICD-9 code for PNTM and found that the species distribution and other patient and hospital factors were similar between patients with and those without a corresponding NTM ICD-9 code, suggesting that most isolates were likely respiratory isolates. We also were limited to species results as reported in the database, which frequently grouped M. abscessus and M. chelonae together despite their clinical differences. We also lacked data on laboratory procedures used for species identification, limiting our ability to assess the impact of changes in methodology over time on species-level patterns and trends. It is also possible that some patients classified as noncases did in fact have NTM infection and remained untested. Conversely, patients with a single positive NTM culture may not have clinical disease, though these results still reflect potential geographic differences in isolates circulating in the environment. Another key limitation is that smaller facilities or those located in more rural areas may differ from larger and more urban institutions in ways that impact the data available for analysis and epidemiologic trends observed, including differences in referral patterns, testing procedures, patient access to care, and clinical awareness of NTM. However, this would likely have a greater impact on prevalence assessments by demographic features and less so on geographic differences in mycobacterial species isolated.
In summary, significant geographic variation exists in the distribution of NTM species in the United States. Whereas MAC is the most frequently isolated species in the South and Northeast, M. abscessus/M. chelonae is proportionately higher in the West and South relative to other geographic regions of the country. Greater clinical awareness in geographic areas with increased levels of harder-to-treat mycobacteria are needed, given differences in treatment options and implications for patient outcomes.
Supplementary Material
Footnotes
Supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Heart, Lung, and Blood Institute, and the National Institutes of Health Clinical Center. The opinions expressed in this article are the authors’ own and do not reflect the views of the National Institutes of Health, the U.S. Department of Health and Human Services, or the U.S. government.
Author Contributions: A.B.S.: contributed to the concept of the work as well as the analysis and interpretation of the data for the work, drafted the work, and approved the final version to be published; Y.L.L. and J.A.: contributed to the analysis and interpretation of the data for the work, critically revised the draft of the work, and approved the final version to be published; and A.M.Z., K.N.O., S.S.K., and D.R.P.: contributed to the design of the work and interpretation of the data for the work, critically revised the draft of the work, and approved the final version to be published.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Maekawa K, Ito Y, Hirai T, Kubo T, Imai S, Tatsumi S, Fujita K, Takakura S, Niimi A, Iinuma Y, et al. Environmental risk factors for pulmonary Mycobacterium avium-intracellulare complex disease. Chest. 2011;140:723–729. doi: 10.1378/chest.10-2315. [DOI] [PubMed] [Google Scholar]
- 2.Primm TP, Lucero CA, Falkinham JO., III Health impacts of environmental mycobacteria. Clin Microbiol Rev. 2004;17:98–106. doi: 10.1128/CMR.17.1.98-106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falkinham JO., III Current epidemiologic trends of the nontuberculous mycobacteria (NTM) Curr Environ Health Rep. 2016;3:161–167. doi: 10.1007/s40572-016-0086-z. [DOI] [PubMed] [Google Scholar]
- 4.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, et al. ATS Mycobacterial Diseases Subcommittee American Thoracic Society Infectious Disease Society of America An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases Am J Respir Crit Care Med 2007175367–416.[Published erratum appears in Am J Respir Crit Care Med 2007;175:744–745.] [DOI] [PubMed] [Google Scholar]
- 5.Adjemian J, Olivier KN, Seitz AE, Holland SM, Prevots DR. Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am J Respir Crit Care Med. 2012;185:881–886. doi: 10.1164/rccm.201111-2016OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adjemian J, Olivier KN, Seitz AE, Falkinham JO, III, Holland SM, Prevots DR. Spatial clusters of nontuberculous mycobacterial lung disease in the United States. Am J Respir Crit Care Med. 2012;186:553–558. doi: 10.1164/rccm.201205-0913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, Blosky MA, Montes de Oca R, Shea YR, Seitz AE, Holland SM, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med. 2010;182:970–976. doi: 10.1164/rccm.201002-0310OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirsaeidi M, Machado RF, Garcia JG, Schraufnagel DE. Nontuberculous mycobacterial disease mortality in the United States, 1999–2010: a population-based comparative study. PLoS One. 2014;9:e91879. doi: 10.1371/journal.pone.0091879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Adjemian J, Olivier KN, Prevots DR. Nontuberculous mycobacteria among patients with cystic fibrosis in the United States: screening practices and environmental risk. Am J Respir Crit Care Med. 2014;190:581–586. doi: 10.1164/rccm.201405-0884OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adjemian J, Prevots DR, Gallagher J, Heap K, Gupta R, Griffith D. Lack of adherence to evidence-based treatment guidelines for nontuberculous mycobacterial lung disease. Ann Am Thorac Soc. 2014;11:9–16. doi: 10.1513/AnnalsATS.201304-085OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spaulding AB, Lai YL, Olivier KN, Zelazny A, Kadri SS, Prevots DR, Adjemian J. Prevalence of nontuberculous mycobacterial infections among patients at 151 US hospitals, 2009–2013 [abstract] Am J Respir Crit Care Med. 2016;193:A3014. [Google Scholar]
- 12.American Hospital Association. 2017 ed. Chicago: American Hospital Association; 2017. AHA hospital statistics. [accessed 2017 Mar 2]. Available from: http://www.aha.org/products-services/aha-hospital-statistics.shtml. [Google Scholar]
- 13.Premier. Charlotte, NC: Premier, Inc.; 2017. Premier healthcare database: data that informs and performs. [accessed 2017 Mar 2]. Available from: https://www.premierinc.com/transforming-healthcare/healthcare-performance-improvement/premier-research-services/ [Google Scholar]
- 14.United States Census Bureau. Washington, DC: Author; 2010. Maps & data, 2010. [accessed 2016 Jul 1]. Available from: https://www.census.gov/geo/maps-data/ [Google Scholar]
- 15.Prevots DR, Marras TK. Epidemiology of human pulmonary infection with nontuberculous mycobacteria: a review. Clin Chest Med. 2015;36:13–34. doi: 10.1016/j.ccm.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adjemian J, Frankland TB, Daida YG, Honda JR, Olivier KN, Zelazny A, Honda S, Prevots DR. Epidemiology of nontuberculous mycobacterial lung disease and tuberculosis, Hawaii, USA. Emerg Infect Dis. 2017;23:439–447. doi: 10.3201/eid2303.161827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Good RC, Snider DE., Jr Isolation of nontuberculous mycobacteria in the United States, 1980. J Infect Dis. 1982;146:829–833. doi: 10.1093/infdis/146.6.829. [DOI] [PubMed] [Google Scholar]
- 18.O’Brien RJ, Geiter LJ, Snider DE., Jr The epidemiology of nontuberculous mycobacterial diseases in the United States: results from a national survey. Am Rev Respir Dis. 1987;135:1007–1014. doi: 10.1164/arrd.1987.135.5.1007. [DOI] [PubMed] [Google Scholar]
- 19.Bodle EE, Cunningham JA, Della-Latta P, Schluger NW, Saiman L. Epidemiology of nontuberculous mycobacteria in patients without HIV infection, New York City. Emerg Infect Dis. 2008;14:390–396. doi: 10.3201/eid1403.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cassidy PM, Hedberg K, Saulson A, McNelly E, Winthrop KL. Nontuberculous mycobacterial disease prevalence and risk factors: a changing epidemiology. Clin Infect Dis. 2009;49:e124–e129. doi: 10.1086/648443. [DOI] [PubMed] [Google Scholar]
- 21.Smith GS, Ghio AJ, Stout JE, Messier KP, Hudgens EE, Murphy MS, Pfaller SL, Maillard J-M, Hilborn ED. Epidemiology of nontuberculous mycobacteria isolations among central North Carolina residents, 2006–2010. J Infect. 2016;72:678–686. doi: 10.1016/j.jinf.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 22.Satyanarayana G, Heysell SK, Scully KW, Houpt ER. Mycobacterial infections in a large Virginia hospital, 2001–2009. BMC Infect Dis. 2011;11:113. doi: 10.1186/1471-2334-11-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henkle E, Hedberg K, Schafer S, Novosad S, Winthrop KL. Population-based incidence of pulmonary nontuberculous mycobacterial disease in Oregon 2007 to 2012. Ann Am Thorac Soc. 2015;12:642–647. doi: 10.1513/AnnalsATS.201412-559OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Winthrop KL, Baxter R, Liu L, McFarland B, Austin D, Varley C, Radcliffe L, Suhler E, Choi D, Herrinton LJ. The reliability of diagnostic coding and laboratory data to identify tuberculosis and nontuberculous mycobacterial disease among rheumatoid arthritis patients using anti-tumor necrosis factor therapy. Pharmacoepidemiol Drug Saf. 2011;20:229–235. doi: 10.1002/pds.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olivier KN, Weber DJ, Wallace RJ, Jr, Faiz AR, Lee JH, Zhang Y, Brown-Elliot BA, Handler A, Wilson RW, Schechter MS, et al. Nontuberculous Mycobacteria in Cystic Fibrosis Study Group. Nontuberculous mycobacteria. I: Multicenter prevalence study in cystic fibrosis. Am J Respir Crit Care Med. 2003;167:828–834. doi: 10.1164/rccm.200207-678OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.