Abstract
Lung ischemia-reperfusion (IR) injury contributes to post-transplant complications, including primary graft dysfunction. Decades of reports show that reactive oxygen species generated during lung IR contribute to pulmonary vascular endothelial barrier disruption and edema formation, but the specific target molecule(s) that “sense” injury-inducing oxidant stress to activate signaling pathways culminating in pathophysiologic changes have not been established. This review discusses evidence that mitochondrial DNA (mtDNA) may serve as a molecular sentinel wherein oxidative mtDNA damage functions as an upstream trigger for lung IR injury. First, the mitochondrial genome is considerably more sensitive than nuclear DNA to oxidant stress. Multiple studies suggest that oxidative mtDNA damage could be transduced to physiologic dysfunction by pathways that are either a direct consequence of mtDNA damage per se or involve formation of proinflammatory mtDNA damage-associated molecular patterns. Second, transgenic animals or cells overexpressing components of the base excision DNA repair pathway in mitochondria are resistant to oxidant stress–mediated pathophysiologic effects. Finally, published and preliminary studies show that pharmacologic enhancement of mtDNA repair or mtDNA damage–associated molecular pattern degradation suppresses reactive oxygen species–induced or IR injury in multiple organs, including preclinical models of lung procurement for transplant. Collectively, these findings point to the interesting prospect that pharmacologic enhancement of DNA repair during procurement or ex vivo lung perfusion may increase the availability of lungs for transplant and reduce the IR injury contributing to primary graft dysfunction.
Keywords: lung ischemia-reperfusion injury, mitochondrial DNA damage, mitochondrial DNA, damage-associated molecular patterns
Lung transplant is the only cure for end-stage lung diseases. Worldwide, almost 30,000 lungs were authorized for procurement, but only 10 to 15% of donated lungs were deemed suitable for transplant (1). The low proportion of lungs ultimately transplanted is due to a selection process that relies on highly conservative criteria and physician experience to identify those lungs most likely to achieve a positive clinical outcome. Among the challenges confronting the lung procurement process, two are particularly prominent. First, the number of transplantable lungs remains low, resulting in long wait times and unnecessary morbidity and mortality, and second, despite the rigorous nature of the lung selection process, 10 to 20% of lung transplants are complicated by primary graft dysfunction (PGD).
Outcomes of lung transplant depend on a complex interplay between donor health, premortem treatments, causes of death, recipient condition, and many other factors. One pathway that seems to be critical to overall lung transplant success is ischemia-reperfusion (IR) injury. In this context, mitochondrial dysfunction, and specifically increased mitochondrial reactive oxygen species (ROS) production (2, 3), is characteristic of IR injury, but ROS production has been considered to be a nonspecific effect of a dysfunctional respiratory chain interacting with oxygen during reperfusion (4–6). Recently, however, increased mitochondrial ROS production driving IR injury has been ascribed to activation of metabolic response pathways conserved across multiple organs wherein an expanded succinate pool, rapidly reoxidized by succinate dehydrogenase, drives extensive oxygen radical generation by reverse electron transport at mitochondrial complex I (7). In the context of PGD, the IR injury–related increase in ROS generation almost certainly occurs in resident and itinerant lung cells and triggers a complex cascade of inflammatory events resulting in pulmonary edema, hypoxemia, and alveolar damage in the first 72 hours (8). The targets of increased ROS generated in the setting of lung IR injury have not been thoroughly elucidated.
The introduction of ex vivo lung perfusion (EVLP) has the potential to alleviate the shortage of lungs available for transplant and perhaps to reduce the incidence of PGD (9). Particularly for marginally suitable lungs, EVLP provides a means to engender a degree of recovery from premortem and procurement-related insults as well as an opportunity to employ quantitative indices of physiologic performance as part of the selection criteria (10). In addition, use of EVLP raises the possibility of applying pharmacotherapeutic strategies between lung procurement and transplant, also with the dual aims of increasing the number of physiologically acceptable lungs available for transplant and minimizing risk of PGD (11). Against this background, this focused review describes evidence that ROS-induced mitochondrial DNA (mtDNA) damage may be a key event initiating lung cell dysfunction in the setting of IR injury, and highlights the possible utility of agents enhancing mtDNA repair or promoting degradation of mtDNA fragments for increasing the availability of lungs for transplant and reducing the severity of PGD.
mtDNA as a Sentinel Molecule in Oxidant-mediated Injury
Among the potential molecular targets of ROS produced in IR injury, the mitochondrial genome is particularly interesting for several reasons. First, mtDNA, structurally similar to bacterial DNA, is about 50-fold more sensitive than the nuclear genome to oxidative damage (12–14). Second, studies in cultured cells reveal a conspicuous association between mtDNA damage and ROS-mediated cell death; cell types displaying a slower rate of mtDNA repair are more prone to oxidant-induced mtDNA damage and cytotoxicity than cell types that rapidly repair mtDNA, which are resistant to oxidant-induced mtDNA damage and cell death (13, 15, 16). Third, transgenic enhancement of mtDNA repair blocks IR and other forms of cellular injury.
It is fair to point out that transgenic strategies to explore the specific involvement of oxidative mtDNA damage are challenging because the key pathway repairing such damage—the base excision repair pathway—is present in both nuclear and mitochondrial compartments; thus, total cell knockdown or overexpression of pathway components exerts highly complex effects not easily attributable to modulation of oxidative damage in one genome or the other (17). However, authors of several recent reports have employed transgenic mice deficient in the first enzyme of base excision repair 8-oxoguanine DNA glycosylase (Ogg1), a mixed-function DNA glycosylase that excises the common oxidative base damage product 8-oxoguanine, with the enzyme selectively reconstituted in mitochondria (18–20). In these instances, Ogg1 reconstitution in mitochondria, despite the absence of Ogg1-dependent repair in the nucleus, inhibits pathophysiologic responses to developmental, environmental, or pathologic stressors.
In a comparatively larger number of studies, however, researchers have examined the effect of selectively increasing mitochondrial Ogg1 activity on pathophysiologic responses to oxidant stress. Using transgenic constructs overexpressing the enzyme linked to a mitochondrial targeting sequence, it has been shown that enhanced mtDNA repair prevents ROS-mediated mtDNA damage, cytotoxicity, and apoptosis evoked by exogenous ROS in rat-cultured lung endothelial cells and other cell types (15, 21–24). Similarly, overexpression of mitochondria-targeted Ogg1 protects against asbestos-induced cytotoxicity in human lung adenocarcinoma cells (25–27), thus implicating mtDNA integrity in fibrotic responses of the lung to this environmental toxin. Although the presence of mtDNA damage in this system has yet to be established, provocative evidence shows that overexpression of mutant Ogg1 deficient in mtDNA repair activity, acting in concert with aconitase, retains its ability to suppress asbestos-induced cytotoxicity. This observation suggests that the mitochondria-targeted DNA glycosylase may exert protective effects via a mechanism unrelated to mtDNA repair per se (26, 27).
Given the salutary effects of overexpressing mitochondria-targeted DNA repair glycosylases, we developed recombinant fusion proteins with the goal of pharmacologically enhancing mtDNA repair (28). The proteins consist of a TAT sequence to facilitate cellular uptake, the mitochondrial targeting sequence from manganese superoxide dismutase, and one of two DNA glycosylases, either the mammalian Ogg1 or the bacterial endonuclease III. In rodent models, these agents prevent and/or reverse oxidant-induced lung injury, IR-induced dysfunction in myocardial infarction and stroke, ROS-mediated pathology in ventilator-induced lung injury, hyperoxic lung injury in the newborn, and bacteria-induced acute lung injury and multiple organ system failure (28–33). Of particular relevance to this report, our preliminary studies also suggest that the mitochondria-targeted Ogg1 fusion protein attenuates IR-mediated lung injury (28, 34). In all of the instances noted above where mtDNA integrity has been measured, the fusion protein consistently decreased the extent of oxidative mtDNA damage without impacting nuclear DNA integrity. Collectively, these observations support the interesting idea that a pharmacologic strategy directed toward enhancement of mtDNA repair applied in concert with EVLP could be useful in terms of salvaging marginal lungs for transplant.
In part on the basis of the observations summarized above, it has been proposed that mtDNA integrity serves as a molecular sentinel linking oxidant stress to cellular responses, including both death of the damaged cell and transmission of alarm signals to nearby or distant neighbors (35). There is also a teleologic argument for such a function of the mitochondrial genome: Because mtDNA has the same chemical composition as the nuclear genome, and because the former is more sensitive to oxidative damage than the latter, cytotoxicity triggered by damage to the mitochondrial genome would seem to be an effective strategy for eliminating cells threatened with nuclear DNA mutations before they are able to replicate.
Potential Mechanisms Linking mtDNA Damage to Lung IR Injury
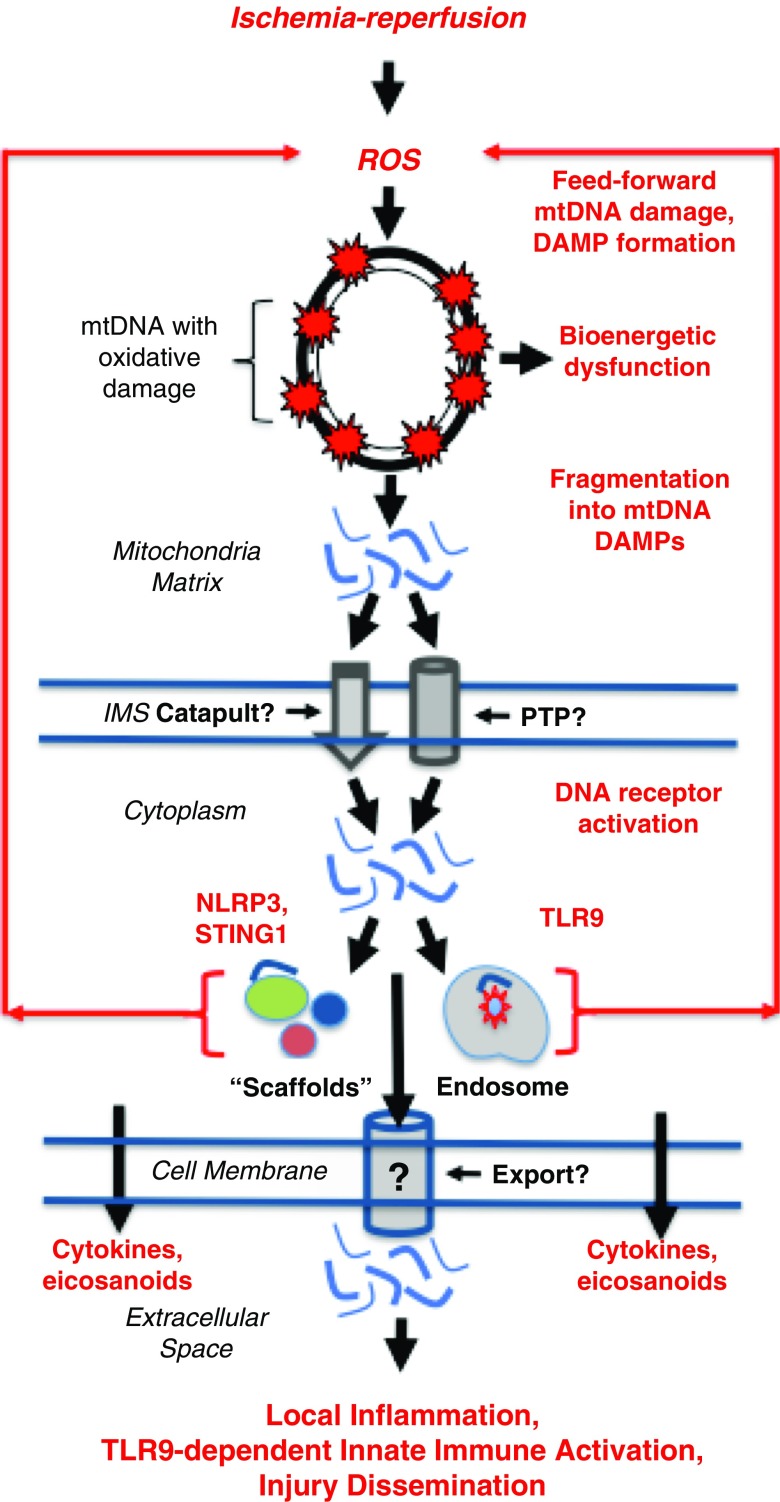
There are at least two general pathways by which oxidative mtDNA damage adversely impacts cell and organ function (Figure 1). First, damage to the mitochondrial genome directly disrupts transcription of mtDNA-encoded RNA, thus reducing the availability of electron transport chain components. Ensuing limitations in ATP production can lead to a bioenergetic crisis and cell death (36). Impaired electron transport chain efficiency also alters the cellular redox state, with the potential to initiate ROS-driven proinflammatory signaling (24, 37–39). Second, oxidative mtDNA damage also adversely impacts cell viability and physiology by promoting fragmentation of the molecule into so-called mtDNA damage-associated molecular patterns (DAMPs) (40, 41). As discussed subsequently, mtDNA DAMPs may lead to enhanced ROS generation and function as “alarm signals” or second messengers in IR and other forms of injury (42–44). In support of the importance of mtDNA DAMPs in human disease, observational studies using quantitative reverse transcriptase–polymerase chain reactions to quantify selected mtDNA fragments in plasma from patients with severe illness (45, 46) or injury (47) as well as patients at risk for PGD after lung transplant (48) show that their abundance is predictive of clinical outcomes.
Figure 1.
Postulated feedforward cycle linking oxidative mitochondrial DNA (mtDNA) damage to regenerative formation of mtDNA damage-associated molecular patterns (DAMPs) culminating in self-propagating activation of proinflammatory stimulator of interferon genes 1 (STING1), Nod-like receptor protein 3 (NLRP3), and Toll-like receptor 9 (TLR9) receptors. Critical events linking mtDNA damage to ischemia-reperfusion injury are indicated in red. The authors postulate that this general mechanism may contribute to lung ischemia-reperfusion injury and that its interruption by pharmacologic strategies to suppress mtDNA damage or degrade mtDNA DAMPs could be applied in concert with ex vivo lung perfusion as a means to increase the number of lungs available for transplant and reduce the severity of primary graft dysfunction. Text in italic on left side of figure specifies intracellular cellular “domains” or structures. Text or symbols in red denote key events in injury and injury propagation. See text for details. IMS = inner membrane space; PTP = permeability transition pore; ROS = reactive oxygen species.
There are many fundamental questions about the structures of mtDNA DAMPs, their mechanisms of action, and pathways of intra- and extra-cellular trafficking. To illustrate the first of these deficiencies, conventional methods of measuring mtDNA DAMPs employing quantitative reverse transcriptase–polymerase chain reactions to detect sequences of about 200 bp in length shed no light on their sequence composition or mechanism of formation. Indeed, it was recently shown that the average size of circulating mtDNA sequences in normal human subjects was 30–75 bp (49). Adding to the complexity, the mechanism of mtDNA DAMP formation also is unknown. As noted above, although oxidative damage appears to trigger fragmentation of the molecule into mtDNA DAMPs (40, 41), it is unclear whether mtDNA fragmentation occurs randomly, thus leading to formation of an unpredictable spectrum of mtDNA DAMP species, or whether damage is targeted to oxidant-sensitive regions of the mitochondrial genome (50), thus engendering formation of mtDNA DAMP species of predictable sequence composition. Knowing whether the sequence characteristics of mtDNA DAMP species are random or predictable is essential for understanding how they interact with effector molecules. For example, the degree of deoxyribose backbone curvature (51), the presence of specific motifs (52–54), DNase-mediated digestion (55, 56), and the presence of oxidized bases in the mtDNA sequence (57, 58) all may impact the ability of mtDNA fragments to interact with their known receptors. These considerations point to the unsettling prospect that previous studies may have failed to detect the most prevalent and/or biologically active mtDNA DAMP species.
mtDNA DAMPs interact with three proinflammatory receptor types (Figure 1). Those accumulating in the cytosol interact autogenously with the cytoplasmic receptors stimulator of interferon genes 1 (STING1) and Nod-like receptor protein 3 (NLRP3). mtDNA DAMPs exported into the extracellular environment can access the proinflammatory endosomal Toll-like receptor 9 (TLR9) in autocrine or paracrine manners. It is unclear whether the two cytoplasmic receptor populations can be accessed by extracellular mtDNA (43). All three mtDNA receptors are capable of evoking cytokine and eicosanoid generation, whereas the NLRP3 initiates pyroptosis as well (43, 59). The NLRP3 and TLR9 receptors have both been directly implicated in IR injury (60). Interestingly, mtDNA DAMP formation and proinflammatory ROS production may interact in a feedforward or regenerative manner. In bacteria-induced lung injury, for example, mtDNA DAMPs released into the extracellular environment in response to mitochondrial genome damage trigger additional TLR9-dependent oxidant stress, which leads to more mtDNA damage and continued mtDNA DAMP generation. The NLRP3 inflammasome also may be involved in this regenerative pathway; as noted above, NLRP3 is activated by mtDNA, and its recruitment to the mitochondrial outer membrane surface is associated with enhanced mitochondrial ROS production (61). It is thus tempting to speculate that an mtDNA damage–dependent feedforward cycle could propagate injury even after removal of the initiating stimulus.
How mtDNA fragments are released from mitochondria and trafficked to specific targets is not completely understood. Studies on isolated, ROS-stressed mitochondria show that mtDNA DAMP release is size dependent and occurs via the mitochondrial permeability transition pore (41), which is intriguing because the transition pore is centrally involved in the induction of ROS-mediated IR injury (62, 63). mtDNA nucleoids, which are nucleoprotein complexes containing machinery necessary for mtDNA transcription, translation, and replication, are linked to the cytoskeleton via actomyosin filaments that span both inner and outer membranes (64). This relationship between nucleoids and motor proteins points to a pathway by which mtDNA could be actively “catapulted” from the organelle in a manner similar to the extracellular release mechanism for mtDNA DAMPs in eosinophils (65), as discussed subsequently.
Intracellular trafficking of mtDNA fragments from damaged mitochondria to mtDNA receptors also is largely unexplored. Trafficking could be purely “functional;” that is, certain mtDNA sequences have a particular affinity for a specific type of mtDNA receptor. Some evidence suggests that this may be the case for the NLRP3 inflammasome, which was found in association exclusively with D-loop sequences in cardiomyocytes, despite the prospect that other mtDNA fragments were present in the cytosol (66). Another possibility, also not well studied, is that mitochondria are actively translocated to subcellular domains such that mtDNA fragments are released in close proximity to their molecular targets. In general support of this concept, transport of mitochondria to perinuclear, plasma membrane, or endoplasmic reticular regions has been implicated in ROS-mediated mitochondrial retrograde signaling (67), calcium buffering in the vicinity of immunologic synapses (68), and triggering endoplasmic reticulum calcium release (69), respectively. The prospect that mitochondrial redeployment contributes to the actions of mtDNA fragments in IR injury derives indirect support from the finding that active translocation of mitochondria to the nucleus–endoplasmic reticulum interface creates an optimal environment for NLRP3 activation (70), perhaps by targeting delivery of stimulatory mtDNA DAMPs. Mechanisms of mtDNA DAMP export into the extracellular domain also are unexplored, although the process governing their release from stimulated eosinophils is ROS dependent and occurs extremely quickly by a “catapult-like” mechanism (65).
Prospects for mtDNA as a Pharmacologic Target in Patients with Lung Transplants
Accumulating evidence derived from cultured cells, isolated organs, intact animals, and human patients supports the concept that mtDNA may serve a sentinel function in ROS-mediated disorders, using pathways whereby oxidative mtDNA damage initiates proinflammatory and cytotoxic signaling. In an extension of this idea, it is also tempting to suggest that pharmacologic enhancement of mtDNA repair or degradation could be therapeutically advantageous in the specific context of lung transplants. For example, as discussed above, enhanced mtDNA repair initiated during lung procurement or EVLP might have the potential to increase the availability of lungs for transplant and perhaps reduce the incidence or severity of PGD. Mechanisms of action of enhanced mtDNA repair include not only suppression of the direct consequences of impaired mitochondrial genomic function but also prevention of mtDNA DAMP release (40). The prospect that such experimental “mtDNA repair agents” could find application in lung transplantation or any clinical setting is unknown. However, there is another pharmacologic strategy already available that takes advantage of a repurposed drug to disrupt at least some of the consequences of mtDNA damage. Specifically, DNase I, used for many years to improve mucus clearance in patients with cystic fibrosis (71) or, more recently, to treat pleural infections (72, 73), has been shown in experimental situations to degrade mtDNA DAMPs and inhibit their biologic effects (40, 74). In addition, whereas DNase I used for cystic fibrosis or pleural infection is administered locally, early studies on its potential benefit in lupus nephritis demonstrated that systemic administration of the enzyme was both safe and effective in degrading circulating DNA (75, 76). Against this background, we believe that agents enhancing mtDNA repair or mtDNA DAMP degradation, used either alone or in combination (as has been done in experimental myocardial infarction [29]), should be explored as adjunctive pharmacologic therapy coupled with EVLP to increase the number of lungs available for transplant and reduce the severity of PGD.
Supplementary Material
Acknowledgments
Acknowledgment
The authors thank Dr. Glenn L. Wilson, Exscien Corp., Louisville, Kentucky, and Drs. Mykhaylo Ruchko and Viktor Pastukh, University of South Alabama, Mobile, Alabama, for their expert advice.
Footnotes
Supported by National Institutes of Health grants R41 HL114225 and R01 HL113614 (M.N.G.), and K08 GM109113 (J.D.S.).
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Valapour M, Skeans MA, Smith JM, Edwards LB, Cherikh WS, Uccellini K, Israni AK, Snyder JJ, Kasiske BL. OPTN/SRTR 2015 Annual Data Report: Lung. American J Transplant. 2017;17(Suppl1):357–424. doi: 10.1111/ajt.14129. [DOI] [PubMed] [Google Scholar]
- 2.Sommer SP, Sommer S, Sinha B, Wiedemann J, Otto C, Aleksic I, Schimmer C, Leyh RG. Ischemia-reperfusion injury–induced pulmonary mitochondrial damage. J Heart Lung Transplant. 2011;30:811–818. doi: 10.1016/j.healun.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 3.Ferrari RS, Andrade CF. Oxidative stress and lung ischemia-reperfusion injury. Oxid Medicine Cell Longev. 2015;2015:590987. doi: 10.1155/2015/590987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burwell LS, Nadtochiy SM, Brookes PS. Cardioprotection by metabolic shut-down and gradual wake-up. J Mol Cell Cardiol. 2009;46:804–810. doi: 10.1016/j.yjmcc.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eltzschig HK, Eckle T. Ischemia and reperfusion—from mechanism to translation. Nat Med. 2011;17:1391–1401. doi: 10.1038/nm.2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yellon DM, Hausenloy DJ. Myocardial reperfusion injury. N Engl J Med. 2007;357:1121–1135. doi: 10.1056/NEJMra071667. [DOI] [PubMed] [Google Scholar]
- 7.Chouchani ET, Pell VR, Gaude E, Aksentijević D, Sundier SY, Robb EL, Logan A, Nadtochiy SM, Ord ENJ, Smith AC, et al. Ischaemic accumulation of succinate controls reperfusion injury through mitochondrial ROS. Nature. 2014;515:431–435. doi: 10.1038/nature13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Perrot M, Liu M, Waddell TK, Keshavjee S. Ischemia-reperfusion-induced lung injury. Am J Respir Crit Care Med. 2003;167:490–511. doi: 10.1164/rccm.200207-670SO. [DOI] [PubMed] [Google Scholar]
- 9.Cypel M, Yeung JC, Liu M, Anraku M, Chen F, Karolak W, Sato M, Laratta J, Azad S, Madonik M, et al. Normothermic ex vivo lung perfusion in clinical lung transplantation. N Engl J Med. 2011;364:1431–1440. doi: 10.1056/NEJMoa1014597. [DOI] [PubMed] [Google Scholar]
- 10.Zamora MR. Updates in lung transplantation. Clin Transpl. 2012:185–192. [PubMed] [Google Scholar]
- 11.Cypel M, Liu M, Rubacha M, Yeung JC, Hirayama S, Anraku M, Sato M, Medin J, Davidson BL, de Perrot M, et al. Functional repair of human donor lungs by IL-10 gene therapy. Sci Transl Med. 2009;1:4ra9. doi: 10.1126/scitranslmed.3000266. [DOI] [PubMed] [Google Scholar]
- 12.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci USA. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grishko V, Solomon M, Wilson GL, LeDoux SP, Gillespie MN. Oxygen radical-induced mitochondrial DNA damage and repair in pulmonary vascular endothelial cell phenotypes. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1300–L1308. doi: 10.1152/ajplung.2001.280.6.L1300. [DOI] [PubMed] [Google Scholar]
- 14.Ballinger SW, Patterson C, Yan CN, Doan R, Burow DL, Young CG, Yakes FM, Van Houten B, Ballinger CA, Freeman BA, et al. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res. 2000;86:960–966. doi: 10.1161/01.res.86.9.960. [DOI] [PubMed] [Google Scholar]
- 15.LeDoux SP, Druzhyna NM, Hollensworth SB, Harrison JF, Wilson GL. Mitochondrial DNA repair: a critical player in the response of cells of the CNS to genotoxic insults. Neuroscience. 2007;145:1249–1259. doi: 10.1016/j.neuroscience.2006.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollensworth SB, Shen C, Sim JE, Spitz DR, Wilson GL, LeDoux SP. Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic Biol Med. 2000;28:1161–1174. doi: 10.1016/s0891-5849(00)00214-8. [DOI] [PubMed] [Google Scholar]
- 17.Sampath H. Oxidative DNA damage in disease--insights gained from base excision repair glycosylase-deficient mouse models. Environ Mol Mutagen. 2014;55:689–703. doi: 10.1002/em.21886. [DOI] [PubMed] [Google Scholar]
- 18.Wang W, Esbensen Y, Kunke D, Suganthan R, Rachek L, Bjørås M, Eide L. Mitochondrial DNA damage level determines neural stem cell differentiation fate. J Neurosci. 2011;31:9746–9751. doi: 10.1523/JNEUROSCI.0852-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuzefovych LV, Kahn AG, Schuler MA, Eide L, Arora R, Wilson GL, Tan M, Rachek LI. Mitochondrial DNA repair through OGG1 activity attenuates breast cancer progression and metastasis. Cancer Res. 2016;76:30–34. doi: 10.1158/0008-5472.CAN-15-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuzefovych LV, Schuler AM, Chen J, Alvarez DF, Eide L, Ledoux SP, Wilson GL, Rachek LI. Alteration of mitochondrial function and insulin sensitivity in primary mouse skeletal muscle cells isolated from transgenic and knockout mice: role of OGG1. Endocrinology. 2013;154:2640–2649. doi: 10.1210/en.2013-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dobson AW, Grishko V, LeDoux SP, Kelley MR, Wilson GL, Gillespie MN. Enhanced mtDNA repair capacity protects pulmonary artery endothelial cells from oxidant-mediated death. Am J Physiol Lung Cell Mol Physiol. 2002;283:L205–L210. doi: 10.1152/ajplung.00443.2001. [DOI] [PubMed] [Google Scholar]
- 22.Ruchko M, Gorodnya O, LeDoux SP, Alexeyev MF, Al-Mehdi AB, Gillespie MN. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L530–L535. doi: 10.1152/ajplung.00255.2004. [DOI] [PubMed] [Google Scholar]
- 23.Rachek LI, Grishko VI, Musiyenko SI, Kelley MR, LeDoux SP, Wilson GL. Conditional targeting of the DNA repair enzyme hOGG1 into mitochondria. J Biol Chem. 2002;277:44932–44937. doi: 10.1074/jbc.M208770200. [DOI] [PubMed] [Google Scholar]
- 24.Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol. 2008;294:C413–C422. doi: 10.1152/ajpcell.00362.2007. [DOI] [PubMed] [Google Scholar]
- 25.Kim SJ, Cheresh P, Jablonski RP, Williams DB, Kamp DW. The role of mitochondrial DNA in mediating alveolar epithelial cell apoptosis and pulmonary fibrosis. Int J Mol Sci. 2015;16:21486–21519. doi: 10.3390/ijms160921486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SJ, Cheresh P, Williams D, Cheng Y, Ridge K, Schumacker PT, Weitzman S, Bohr VA, Kamp DW. Mitochondria-targeted Ogg1 and aconitase-2 prevent oxidant-induced mitochondrial DNA damage in alveolar epithelial cells. J Biol Chem. 2014;289:6165–6176. doi: 10.1074/jbc.M113.515130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panduri V, Liu G, Surapureddi S, Kondapalli J, Soberanes S, de Souza-Pinto NC, Bohr VA, Budinger GR, Schumacker PT, Weitzman SA, et al. Role of mitochondrial hOGG1 and aconitase in oxidant-induced lung epithelial cell apoptosis. Free Radic Biol Med. 2009;47:750–759. doi: 10.1016/j.freeradbiomed.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chouteau JM, Obiako B, Gorodnya OM, Pastukh VM, Ruchko MV, Wright AJ, Wilson GL, Gillespie MN. Mitochondrial DNA integrity may be a determinant of endothelial barrier properties in oxidant-challenged rat lungs. Am J Physiol Lung Cell Mol Physiol. 2011;301:L892–L898. doi: 10.1152/ajplung.00210.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang XM, Cui L, White J, Kuck J, Ruchko MV, Wilson GL, Alexeyev M, Gillespie MN, Downey JM, Cohen MV. Mitochondrially targeted endonuclease III has a powerful anti-infarct effect in an in vivo rat model of myocardial ischemia/reperfusion. Basic Res Cardiol. 2015;110:3. doi: 10.1007/s00395-014-0459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon R. Post-conditioning and reperfusion injury in the treatment of stroke. Dose Response. 2014;12:590–599. doi: 10.2203/dose-response.14-026.Simon. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hashizume M, Mouner M, Chouteau JM, Gorodnya OM, Ruchko MV, Potter BJ, Wilson GL, Gillespie MN, Parker JC. Mitochondrial-targeted DNA repair enzyme 8-oxoguanine DNA glycosylase 1 protects against ventilator-induced lung injury in intact mice. Am J Physiol Lung Cell Mol Physiol. 2013;304:L287–L297. doi: 10.1152/ajplung.00071.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee YL, Obiako B, Gorodnya OM, Ruchko MV, Kuck JL, Pastukh VM, Wilson GL, Simmons JD, Gillespie MN. Mitochondrial DNA damage initiates acute lung injury and multi-organ system failure evoked in rats by intra-tracheal Pseudomonas aeruginosa. Shock. 2017;48:54–60. doi: 10.1097/SHK.0000000000000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebb SA, Decoux A, Waggoner A, Wilson GL, Gillespie MN. Mitochondrial DNA damage mediates hyperoxic dysmorphogenesis in rat fetal lung explants. Neonatology. 2013;103:91–97. doi: 10.1159/000342632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mulekar S, Obiako BO, Gorodnya O, Ruchko M, Hill JK, Wilson GL, Gillespie MN. Pharmacologic protection of mitochondrial (mt) DNA integrity may afford a new strategy for suppressing hypothermic pneumoplegia-induced lung ischemia-reperfusion injury [abstract] Am J Respir Crit Care Med. 2013;187:A2208. [Google Scholar]
- 35.Schumacker PT, Gillespie MN, Nakahira K, Choi AM, Crouser ED, Piantadosi CA, Bhattacharya J. Mitochondria in lung biology and pathology: more than just a powerhouse. Am J Physiol Lung Cell Mol Physiol. 2014;306:L962–L974. doi: 10.1152/ajplung.00073.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piantadosi CA, Suliman HB. Mitochondrial dysfunction in lung pathogenesis. Annu Rev Physiol. 2017;79:495–515. doi: 10.1146/annurev-physiol-022516-034322. [DOI] [PubMed] [Google Scholar]
- 37.Yu EP, Bennett MR. The role of mitochondrial DNA damage in the development of atherosclerosis. Free Radic Biol Med. 2016;100:223–230. doi: 10.1016/j.freeradbiomed.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 38.Pinto M, Pickrell AM, Wang X, Bacman SR, Yu A, Hida A, Dillon LM, Morton PD, Malek TR, Williams SL, et al. Transient mitochondrial DNA double strand breaks in mice cause accelerated aging phenotypes in a ROS-dependent but p53/p21-independent manner. Cell Death Differ. 2017;24:288–299. doi: 10.1038/cdd.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Akhmedov AT, Marín-García J. Mitochondrial DNA maintenance: an appraisal. Mol Cell Biochem. 2015;409:283–305. doi: 10.1007/s11010-015-2532-x. [DOI] [PubMed] [Google Scholar]
- 40.Kuck JL, Obiako BO, Gorodnya OM, Pastukh VM, Kua J, Simmons JD, Gillespie MN. Mitochondrial DNA damage-associated molecular patterns mediate a feed-forward cycle of bacteria-induced vascular injury in perfused rat lungs. Am J Physiol Lung Cell Mol Physiol. 2015;308:L1078–L1085. doi: 10.1152/ajplung.00015.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.García N, Chávez E. Mitochondrial DNA fragments released through the permeability transition pore correspond to specific gene size. Life Sci. 2007;81:1160–1166. doi: 10.1016/j.lfs.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 42.Mills EL, Kelly B, O’Neill LAJ. Mitochondria are the powerhouses of immunity. Nat Immunol. 2017;18:488–498. doi: 10.1038/ni.3704. [DOI] [PubMed] [Google Scholar]
- 43.Krysko DV, Agostinis P, Krysko O, Garg AD, Bachert C, Lambrecht BN, Vandenabeele P. Emerging role of damage-associated molecular patterns derived from mitochondria in inflammation. Trends Immunol. 2011;32:157–164. doi: 10.1016/j.it.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Nakahira K, Hisata S, Choi AM. The roles of mitochondrial damage-associated molecular patterns in diseases. Antioxid Redox Signal. 2015;23:1329–1350. doi: 10.1089/ars.2015.6407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakahira K, Kyung SY, Rogers AJ, Gazourian L, Youn S, Massaro AF, Quintana C, Osorio JC, Wang Z, Zhao Y, et al. Circulating mitochondrial DNA in patients in the ICU as a marker of mortality: derivation and validation. PLoS Med. 2013;10:e1001577. doi: 10.1371/journal.pmed.1001577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L, Xie L, Zhang Q, Cai X, Tang Y, Wang L, Hang T, Liu J, Gong J. Plasma nuclear and mitochondrial DNA levels in acute myocardial infarction patients. Coron Artery Dis. 2015;26:296–300. doi: 10.1097/MCA.0000000000000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmons JD, Lee YL, Mulekar S, Kuck JL, Brevard SB, Gonzalez RP, Gillespie MN, Richards WO. Elevated levels of plasma mitochondrial DNA DAMPs are linked to clinical outcome in severely injured human subjects. Ann Surg. 2013;258:591–596, discussion 596–598. doi: 10.1097/SLA.0b013e3182a4ea46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mohsen I, Strah H, Scozzi D, Huang HJ, Kreisel D, Krupnick A, Puyo C, Hachem RR, Alouch A, Trulock E, et al. Circulating mitochondrial DNA is elevated in lung transplant recipient patients with immediate early primary graft dysfunction [abstract] Am J Respir Crit Care Med. 2014;189:A1605. [Google Scholar]
- 49.Zhang R, Nakahira K, Guo X, Choi AM, Gu Z. Very short mitochondrial DNA fragments and heteroplasmy in human plasma. Sci Rep. 2016;6:36097. doi: 10.1038/srep36097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clark DW, Phang T, Edwards MG, Geraci MW, Gillespie MN. Promoter G-quadruplex sequences are targets for base oxidation and strand cleavage during hypoxia-induced transcription. Free Radic Biol Med. 2012;53:51–59. doi: 10.1016/j.freeradbiomed.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Y, Berke IC, Modis Y. DNA binding to proteolytically activated TLR9 is sequence-independent and enhanced by DNA curvature. EMBO J. 2012;31:919–931. doi: 10.1038/emboj.2011.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen J, Nag S, Vidi PA, Irudayaraj J. Single molecule in vivo analysis of Toll-like receptor 9 and CpG DNA interaction. PLoS One. 2011;6:e17991. doi: 10.1371/journal.pone.0017991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuznetsov NV. The design and structure-functional properties of DNA-based immunomodulatory sequences. Methods Mol Biol. 2013;986:41–56. doi: 10.1007/978-1-62703-311-4_3. [DOI] [PubMed] [Google Scholar]
- 54.Herzner AM, Wolter S, Zillinger T, Schmitz S, Barchet W, Hartmann G, Bartok E, Schlee M. G-rich DNA-induced stress response blocks type-I-IFN but not CXCL10 secretion in monocytes. Sci Rep. 2016;6:38405. doi: 10.1038/srep38405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chan MP, Onji M, Fukui R, Kawane K, Shibata T, Saitoh S, Ohto U, Shimizu T, Barber GN, Miyake K. DNase II-dependent DNA digestion is required for DNA sensing by TLR9. Nat Commun. 2015;6:5853. doi: 10.1038/ncomms6853. [DOI] [PubMed] [Google Scholar]
- 56.Miyake K, Shibata T, Ohto U, Shimizu T. Emerging roles of the processing of nucleic acids and Toll-like receptors in innate immune responses to nucleic acids. J Leukoc Biol. 2017;101:135–142. doi: 10.1189/jlb.4MR0316-108R. [DOI] [PubMed] [Google Scholar]
- 57.Ermakov AV, Konkova MS, Kostyuk SV, Izevskaya VL, Baranova A, Veiko NN. Oxidized extracellular DNA as a stress signal in human cells. Oxid Medicine Cell Longev. 2013;2013:649747. doi: 10.1155/2013/649747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kostyuk SV, Tabakov VJ, Chestkov VV, Konkova MS, Glebova KV, Baydakova GV, Ershova ES, Izhevskaya VL, Baranova A, Veiko NN. Oxidized DNA induces an adaptive response in human fibroblasts. Mutat Res. 2013;747-748:6–18. doi: 10.1016/j.mrfmmm.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 59.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guo Z, Yu S, Chen X, Ye R, Zhu W, Liu X. NLRP3 is involved in ischemia/reperfusion injury. CNS Neurol Disord Drug Targets. 2016;15:699–712. doi: 10.2174/1871527315666160321111829. [DOI] [PubMed] [Google Scholar]
- 61.Yu J, Nagasu H, Murakami T, Hoang H, Broderick L, Hoffman HM, Horng T. Inflammasome activation leads to caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci USA. 2014;111:15514–15519. doi: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morciano G, Bonora M, Campo G, Aquila G, Rizzo P, Giorgi C, Wieckowski MR, Pinton P. Mechanistic role of mPTP in ischemia-reperfusion injury. Adv Exp Med Biol. 2017;982:169–189. doi: 10.1007/978-3-319-55330-6_9. [DOI] [PubMed] [Google Scholar]
- 64.Reyes A, He J, Mao CC, Bailey LJ, Di Re M, Sembongi H, Kazak L, Dzionek K, Holmes JB, Cluett TJ, et al. Actin and myosin contribute to mammalian mitochondrial DNA maintenance. Nucleic Acids Res. 2011;39:5098–5108. doi: 10.1093/nar/gkr052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yousefi S, Gold JA, Andina N, Lee JJ, Kelly AM, Kozlowski E, Schmid I, Straumann A, Reichenbach J, Gleich GJ, et al. Catapult-like release of mitochondrial DNA by eosinophils contributes to antibacterial defense. Nat Med. 2008;14:949–953. doi: 10.1038/nm.1855. [DOI] [PubMed] [Google Scholar]
- 66.Oka T, Hikoso S, Yamaguchi O, Taneike M, Takeda T, Tamai T, Oyabu J, Murakawa T, Nakayama H, Nishida K, et al. Mitochondrial DNA that escapes from autophagy causes inflammation and heart failure. Nature. 2012;485:251–255. doi: 10.1038/nature10992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Al-Mehdi AB, Pastukh VM, Swiger BM, Reed DJ, Patel MR, Bardwell GC, Pastukh VV, Alexeyev MF, Gillespie MN. Perinuclear mitochondrial clustering creates an oxidant-rich nuclear domain required for hypoxia-induced transcription. Sci Signal. 2012;5:ra47. doi: 10.1126/scisignal.2002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quintana A, Schwarz EC, Schwindling C, Lipp P, Kaestner L, Hoth M. Sustained activity of calcium release-activated calcium channels requires translocation of mitochondria to the plasma membrane. J Biol Chem. 2006;281:40302–40309. doi: 10.1074/jbc.M607896200. [DOI] [PubMed] [Google Scholar]
- 69.Mironov SL, Ivannikov MV, Johansson M. [Ca2+]i signaling between mitochondria and endoplasmic reticulum in neurons is regulated by microtubules: from mitochondrial permeability transition pore to Ca2+-induced Ca2+ release. J Biol Chem. 2005;280:715–721. doi: 10.1074/jbc.M409819200. [DOI] [PubMed] [Google Scholar]
- 70.Misawa T, Takahama M, Kozaki T, Lee H, Zou J, Saitoh T, Akira S. Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol. 2013;14:454–460. doi: 10.1038/ni.2550. [DOI] [PubMed] [Google Scholar]
- 71.Shak S, Capon DJ, Hellmiss R, Marsters SA, Baker CL. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci USA. 1990;87:9188–9192. doi: 10.1073/pnas.87.23.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piccolo F, Pitman N, Bhatnagar R, Popowicz N, Smith NA, Brockway B, Nickels R, Burke AJ, Wong CA, McCartney R, et al. Intrapleural tissue plasminogen activator and deoxyribonuclease for pleural infection: an effective and safe alternative to surgery. Ann Am Thorac Soc. 2014;11:1419–1425. doi: 10.1513/AnnalsATS.201407-329OC. [DOI] [PubMed] [Google Scholar]
- 73.Rahman NM, Maskell NA, West A, Teoh R, Arnold A, Mackinlay C, Peckham D, Davies CW, Ali N, Kinnear W, et al. Intrapleural use of tissue plasminogen activator and DNase in pleural infection. N Engl J Med. 2011;365:518–526. doi: 10.1056/NEJMoa1012740. [DOI] [PubMed] [Google Scholar]
- 74.Simmons JD, Freno DR, Muscat CA, Obiako B, Lee YL, Pastukh VM, Brevard SB, Gillespie MN. Mitochondrial DNA damage associated molecular patterns in ventilator-associated pneumonia: prevention and reversal by intratracheal DNase I. J Trauma Acute Care Surg. 2017;82:120–125. doi: 10.1097/TA.0000000000001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Davis JC, Jr, Manzi S, Yarboro C, Rairie J, McInnes I, Averthelyi D, Sinicropi D, Hale VG, Balow J, Austin H, et al. Recombinant human DNase I (rhDNase) in patients with lupus nephritis. Lupus. 1999;8:68–76. doi: 10.1191/096120399678847380. [DOI] [PubMed] [Google Scholar]
- 76.Prince WS, Baker DL, Dodge AH, Ahmed AE, Chestnut RW, Sinicropi DV. Pharmacodynamics of recombinant human DNase I in serum. Clin Exp Immunol. 1998;113:289–296. doi: 10.1046/j.1365-2249.1998.00647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.