Edwardsiella tarda is an intracellular Gram-negative pathogen with typical characteristics of the Enterobacteriaceace family.1 It causes gastroenteritis, myonecrosis, cellulitis, gas gangrene and generalized infections in humans.2,3 In aquaculture, E. tarda-induced edwardsiellosis has caused disastrous losses to fish cultures in many countries in Europe, Asian, and America.3 E. tarda is equipped with sophisticated regulatory systems, including two-component systems, quorum-sensing system, secretory systems, and regulators such as Fur and Eha.4-9 The crosstalks between these systems/regulators allow E. tarda to sense changes in the environment and regulate the expression of virulence genes. However, the interaction mechanism between E. tarda and its host remains largely unknown.
MicroRNAs (miRNA) are short, conserved noncoding RNAs that transcriptionally and post-transcriptionally regulate gene expression. miRNAs have been identified in diverse organisms including plants, worms, flies, shrimp, fish, frogs, and mammals and play important roles in a wide range of cellular processes.10 Extensive studies on different types of miRNAs, such as those of the let-7 family, miR-143 family, miR-146 family, and miR-148 family, have been reported.11-14 Involvement of miRNAs in microbial infection was first observed with viral and parasitic pathogens.15,16 In bacteria, a pioneering study in the plant Arabidopsis thaliana revealed that a host miRNA (miR-393a) was induced by infection with the pathogen Pseudomonas syringae, which repressed the receptor for the hormone auxin.17 Since then, more and more studies have demonstrated that host miRNA expression can be altered in response to various bacterial pathogens.18-20 As a result, it has been accepted that host miRNAs act as a key player in the molecular interplay between bacterial pathogens and host cells. However, no such studies have been reported with E. tarda. In this study, we examined half-smooth tongue sole (Cynoglossus semilaevis) miRNAs associated with E. tarda infection through high-throughput sequencing and assessed the role of miRNAs in E. tarda infection.
Healthy tongue sole (average weight 13.7 g) were purchased from a commercial fish farm in Shandong Province, China, and maintained at 20 °C in aerated seawater. Before experimentation, the fish were acclimatized in the laboratory for 2 weeks and verified to be free of pathogens in the liver, kidney, and spleen as reported previously.21 E. tarda was cultured in LB medium at 28 °C to mid-logarithmic phase and resuspended in PBS as reported previously.21 Tongue sole were divided randomly into 2 groups and injected intraperitoneally (i.p.) with 100 μL bacterial suspension (1 × 105 CFU/mL) or PBS. At 0, 12, and 24 h post-infection (hpi), fish (5/time point) were killed, and spleen was collected under aseptic conditions. Equal amounts of tissues from 5 fish at each time-point were pooled together and immediately stored in liquid nitrogen. The experiment was performed 3 times, and the 3 sets of samples thus generated for each time point (0 h, 12 h, and 24 h) were each subjected to small RNA sequencing.
Small RNA isolation, library construction, and high-throughput sequencing were performed by Beijing Genomics Institute (BGI), Shenzhen, China. Small RNA library (18–30 nt) was prepared with TruSeq Small RNA Sample Prep Kits (Illumina, San Diego, USA), and single-end sequencing (50 bp) was performed on an Illumina Hiseq4000 by BGI. Sequencing reads and miRNAs identification were analyzed same to our previous study.22 Spleens of tongue sole at different infection times were used for total RNA extraction and cDNA synthesis.22 Quantitative real-time PCR (qRT-PCR) was performed as reported previously.22 The abundance of miRNAs was normalized relative to that of U6 RNA as reported previously.23 The expression level of mRNA was analyzed with qRT-PCR with 60S ribosomal protein L18a as the internal control.24
TargetScan 6.2 and miRanda were used for the prediction of miRNA target genes. TargetScan was used to search for miRNA seed matches (nucleotides 2–8 from the 5′ end of miRNA) in 3′-UTRs of tongue sole mRNAs, and the parameter of TargetScan was set as score >50. miRanda was used to match the entire miRNA sequences, and its parameter was set as free energy < −20 kcal/mol. By combining the results of the 2 algorithms, the overlaps were calculated. Enrichment analysis of the predicted target genes was conducted with GO and KEGG pathway.
miRNA mimics were synthesized by RiboBio (Guangzhou, China). The negative control and mimic control were designed and synthesized by RiboBio. miRNA agomirs and antagomirs are chemically engineered oligonucleotides especially suitable for in vivo analysis. Agomir and antagomir negative controls were also designed and synthesized. For each miRNA-target gene, a small interfering RNA (siRNA) was designed and synthesized by RiboBio.
The 3′-UTRs of tongue sole mRNAs were amplified by PCR with the primer pairs listed in Table S1. The PCR products were inserted into the luciferase vector pMIR-REPORTER (Thermo Fisher Scientific, Waltham, USA) at the Spe I/Hind III or Pme I site, resulting in pMIR-mRNA-3′-UTR, which contain the firefly luciferase gene with the pMIR-REPORTER-3′-UTR replaced by the 3′-UTRs of the miRNA-targeted mRNAs. To construct overexpression plasmids of mRNAs, the coding sequences of tongue sole mRNAs were each amplified with primers listed in Table S1. The PCR product of each mRNA was inserted into vector pCN325 at the EcoRV site, resulting in plasmid pCN-mRNA. 293T human embryonic kidney epithelial cells were cultured as reported previously.26 Transfection was performed as reported previously.26 Luciferase activity assay was performed as reported previously.26
Tongue sole were injected i.p. with E. tarda (104 CFU/fish) plus the agomir (2 μg agomir/1 g fish) of each of the 7 downregulated miRNA or agomir negative control. To maintain the in vivo level of the agomir, the fish were re-injected with miRNA agomir at 24 hpi as reported previously.26 At 48 hpi, spleen was collected, and bacterial number in spleen was determined by plate count as reported previously.25 The effect of the antagomirs of 3 upregulated miRNAs on bacterial replication was determined similarly. To examine the dependence of miRNA activity on target genes, tongue sole were infected as above with E. tarda in the presence of miRNA agomir but plus the overexpression plasmid pCN-mRNA. At 48 hpi, the bacterial number in spleen was determined as above; tongue sole were infected as above with E. tarda in the presence of miRNA antagomir but plus the siRNA of each miRNA-target gene. At 48 hpi, the bacterial number in spleen was determined as above.
All experiments were performed at least 3 times, and statistical analyses were performed using SPSS 17.0 software (IBM-SPSS Inc., Chicago, USA). Data were analyzed with analysis of variance, and statistical significance was defined as P < 0.05.
Tongue sole were infected with E. tarda for 0 h, 12 h, or 24 h. Three small-fragment RNA libraries corresponding to the 3 time-points were constructed and subjected to high-throughput sequence analysis. After filtering the low-quality tags and removing adaptor sequences, 356,319, 344,950, and 302,667 small fragment reads were obtained in the 0 hpi, 12 hpi, and 24 hpi libraries, respectively; 54.6%, 51.5%, and 58.4% of the reads in the 0 hpi, 12 hpi, and 24 hpi libraries, respectively, were mapped to known miRNAs in miRBase 21.0. In the 3 libraries, 0.03% (0 hpi), 0.05% (12 hpi), and 0.03% (24 hpi) of the reads mapped to the hairpin structures of the tongue sole genome (Associated accession code: AGRG01000000) were identified as novel miRNAs (Fig. S1A). In total, 350 miRNAs were identified, including 5 novel miRNAs (Table S2). Among these miRNAs, 51.7%, 49.3%, and 50.73% from the 0 hpi, 12 hpi, and 24 hpi libraries, respectively, were 22-nt in sequence, which was the main length in length distribution (Fig. S1B).
The expression profile of the 350 miRNAs was normalized with TPM (Transcripts Per Kilobase Million) and then tested for significant difference among different time points with Bonferroni correction. The results showed that 266 miRNAs were expressed at all examined time point (0 hpi, 12 hpi, and 24 hpi) (Fig. 1A). Compared to 0 hpi, 311 miRNAs showed significant (> 2-fold and P < 0.01) differences in expression after bacterial infection (Fig. 1B). For convenience, these differentially expressed miRNAs were named DEmiRNA. Following E. tarda infection, 121 and 167 miRNAs were significantly upregulated and downregulated at 12 hpi, respectively; 97 and 187 miRNAs were significantly upregulated and downregulated at 24 hpi, respectively (Fig. 1C and D, Table S3). Eighty-nine miRNAs displayed continuously upregulated expression, while 154 miRNAs displayed steadily downregulated expression at 2 different time points after bacterial infection (Fig. 1C and D, Table S3).
Figure 1.
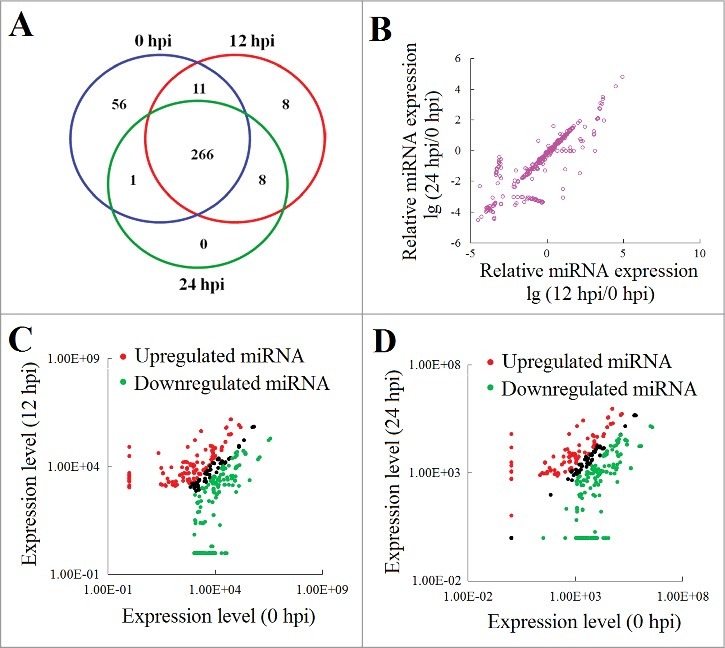
miRNA expression during Edwardsiella tarda infection. (A) Venn diagram showing miRNAs expression at different hours post infection (hpi). The numbers inside the diagram stand for the numbers of miRNAs. (B) Differential expression of miRNAs after bacterial infection. (C and D) Scatter plot of the miRNA expression levels at 12 hpi (C) and 24 hpi (D) in comparison with that at 0 hpi. Red and green spots represent miRNAs significantly upregulated and downregulated, respectively.
To investigate the effect of the miRNA on target mRNAs, global change in gene expression associated with E. tarda infection was examined. Based on the RNA-Seq data after RPKM (Reads Per Kilobase Million) normalization, it was found that the expressions of 1,110 mRNAs (reads >100) were significantly (> 2-fold and P < 0.01) altered during E. tarda infection (Fig. S2). Specifically, 442 and 275 mRNAs were significantly upregulated and downregulated, respectively, at 12 hpi, while 497and 313 mRNAs were significantly upregulated and downregulated, respectively, at 24 hpi (Fig. S2). For convenience, these differentially expressed mRNAs were named DEmRNA. Of these DEmRNA, 287 and 130 showed continuous increase and decrease in expression from 12 to 24 hpi, respectively.
It is well known that miRNAs negatively regulate the expression of their target mRNAs. Thus, the prediction of the target sites of DEmiRNA was performed by examining the upregulated miRNA and the downregulated mRNA at the same time-point; likewise, the downregulated miRNA and the upregulated mRNA were examined as well. The results showed that at 12 hpi, 114 upregulated miRNAs and 160 downregulated miRNAs were predicted to target 324 downregulated mRNAs and 274 upregulated mRNAs, respectively; at 24 hpi, 86 upregulated miRNAs were predicted to target 186 downregulated genes, and 170 downregulated miRNAs were predicted to target 319 upregulated genes (Table S4). In particular, the upregulated cse-miR-143, cse-miR-143–3p, cse-miR-16–5p, cse-miR-16b, cse-miR-17, cse-miR-30b-5p, cse-miR-30d-5p, cse-miR-30e-5p, and cse-miR-458 were predicted to target the same autophagy-related gene atg2b at 12 hpi, and the upregulated cse-miR-8192–3p, cse-miR-26c, cse-miR-26a-5p, cse-miR-26a, cse-miR-26–5p, cse-miR-148a-5p, and cse-miR-101b-3p were predicted to target the same autophagic vacuole membrane protein (WIPI1) gene at 24 hpi (Table S4). At 12 hpi and 24 hpi, 4 downregulated miRNAs, i.e. miR-146a, miR-146a-5p, miR-146b, and miR-5100, were predicted to target the upregulated IL-1β gene, and 11 downregulated miRNAs, i.e., miR-122, miR-122–5p, miR-143–5p, miR-199b, miR-221, miR-221–3p, miR-23a, miR-23a-3p, miR-23b, miR-4206–3p, and miR-5606–5p, were predicted to target the upregulated signal transducer and activator of transcription 1 (STAT1) gene (Table S4). The downregulated let-7 family members (cse-let-7, cse-let-7b, cse-let-7c, cse-let-7d, cse-let-7e, and cse-let-7 g) were predicted to target the upregulated IL-12R2 (Table S4). In total, 298 DEmiRNAs were predicted to target 802 DEmRNAs.
The DEmiRNA target genes identified above were subjected to GO analysis, and the resulting top 12 significantly enriched GO terms of biologic processes are shown in Fig. S3. Some genes of innate immunity, such as IL-1β, IL-8, IL-10, CXC chemokine, CXCR3, and CC chemokine, were enriched with 12 hpi-upregulated mRNAs, while other genes of innate immunity, such as dusp3, peli2, and TRAF2, were enriched with 12 hpi-downregulated mRNAs. Likewise, some apoptosis-related genes, such as caps1, caps8, card9, igfbp1, and ddit4, were enriched with 12 hpi-upregulated mRNAs, while other some apoptosis-related genes, such as ypel3, TP53INP1, and mdm2, were enriched with 12 hpi-downregulated mRNAs (Table S4).
To examine whether there existed negative expressional correlations between DEmiRNAs and their target genes, 30 DEmiRNAs with high differential expressions at 12 hpi and 24 hpi and 21 corresponding DEmRNAs were selected for expression-correlation analysis. Based on a one-to-one target relationship, the 30 DEmiRNAs and 21 DEmRNAs formed 46 miRNA–mRNA pairs, and for each pair, the miRNA targeted the mRNA at both 12 hpi and 24 hpi. The 46 pairs of miRNA–mRNA were determined for expression during E. tarda infection by qRT-PCR. The results showed that negative expression correlations were detected in 36 miRNA–mRNA pairs, which contained 24 miRNAs and 15 target mRNAs (Fig. 2A).
Figure 2.
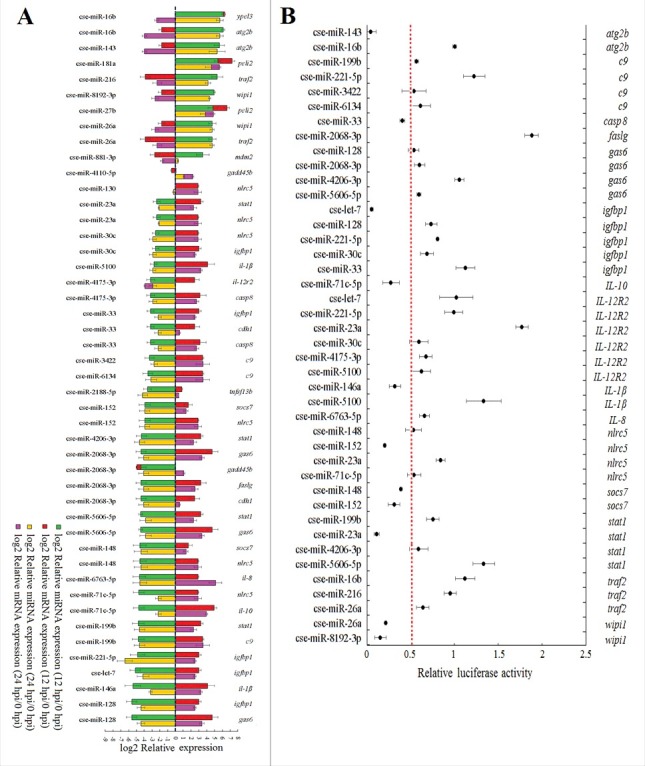
Quantitative real time PCR (qRT-PCR) analysis of the expression of selected miRNAs and their target mRNAs during Edwardsiella tarda infection (A) and validation of miRNA targets (B). (A)Tongue sole were infected with E. tarda for 0 h, 12 h, and 24 h, and miRNA and mRNA expressions in spleen were determined by qRT-PCR. Expression levels relative to 0 hpi were expressed as bars. Bars stacked together indicate no negative correlation between miRNAs and mRNAs. The names of the mRNAs and miRNAs are shown above and below the bars, respectively. (B) 293T cells were co-transfected with different miRNA mimics and their target mRNA-3′-UTR reporter plasmids. Relative luciferase activity was measured at 24 h after transfection. The experiments were performed 3 times, and values are shown as means ± SEM.
To further examine the relationship between the miRNAs and their predicted target mRNAs, the mimics of 24 miRNAs were synthesized, and 15 target reporter plasmids (pMIR-mRNA-3′-UTRs) were constructed that contain a firefly luciferase reporter linked to the 3′-UTRs of the target mRNAs. 293T cells were co-transfected with miRNA mimics and their potential target reporter plasmids at a one-to-one correspondence. A total of 42 miRNA-mRNA pairs were thus examined in 293T cells. The results showed that luciferase activity was significantly reduced in cells transfected with cse-miR-143 mimic plus pMIR-atg2b-3′-UTR, cse-miR-33 mimic plus pMIR-casp8–3′-UTR, cse-let-7 mimic plus pMIR-igfbp1–3′-UTR, cse-miR-71c-5p mimic plus pMIR-IL-10–3′-UTR, cse-miR-146a mimic plus pMIR-IL-1β−3′-UTR, cse-miR-152 mimic plus pMIR-nlrc5–3′-UTR, cse-miR-152 mimic plus pMIR-socs7–3′-UTR, cse-miR-148 mimic plus pMIR-socs7–3′-UTR, cse-miR-23a mimic plus pMIR-stat1–3′-UTR, cse-miR-26a mimic plus pMIR-wipi1–3′-UTR, and cse-miR-8192–3p mimic plus pMIR-wipi1–3′-UTR (Fig. 2B). In contrast, no apparent reduction in luciferase activity was detected in the cells transfected with other miRNAs plus corresponding mRNA 3′-UTRs (Fig. 2B).
Ten of the above miRNAs with confirmed targets were selected, and their effects on E. tarda infection were examined. Of these miRNAs, 3 (cse-miR-143, cse-miR-26a, and cse-miR-8192–3p) and 7 (cse-miR-146a, cse-miR-148, cse-miR-152, cse-miR-23a, cse-miR-33, cse-miR-71c-5p, cse-let-7) were significantly upregulated and downregulated, respectively, during E. tarda infection. qRT-PCR showed that when tongue sole were treated with the antagomir of each of the 3 upregulated miRNAs, the expressions of the target genes were upregulated; similarly, when tongue sole were treated with the agomir of each of the 7 downregulated miRNAs, the expressions of the target genes were downregulated (Fig. S4), suggesting that the antagomirs and agomirs could target at the corresponding miRNA target genes. To examine the effects of the miRNAs on E. tarda infection, tongue sole were inoculated with E. tarda in the presence/absence of each of the antagomirs and agomirs, and bacterial loads in spleen were subsequently determined at 48 hpi. The results showed that the numbers of bacteria recovered from the fish treated with cse-miR-146a agomir, cse-miR-71c-5p agomir, cse-let-7 agomir, and cse-miR-26a antagomir were significantly increased (Fig. 3). In contrast, the bacterial recoveries from the fish treated with cse-miR-148 agomir, cse-miR-152 agomir, cse-miR-23a agomir, and cse-miR-143 antagomir were significantly reduced (Fig. 3). The bacterial recoveries from the fish treated with cse-miR-33 agomir and cse-miR-8192 antagomir were not significantly changed (data not shown).
Figure 3.
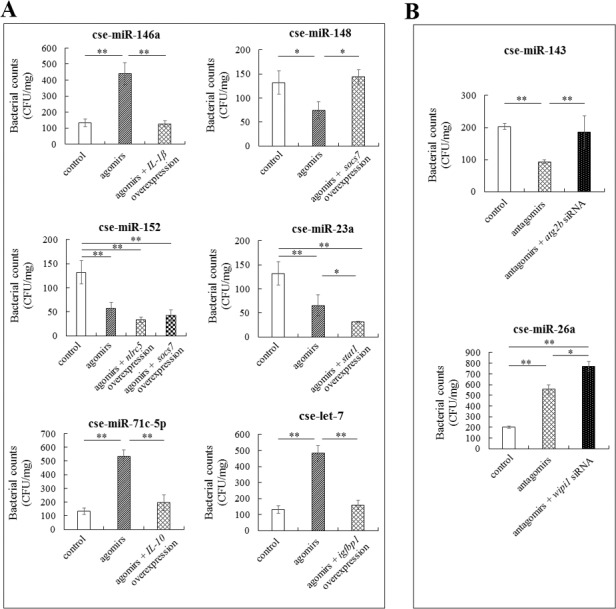
Effects of miRNAs and miRNA target genes on Edwardsiella tarda infection. (A) Tongue sole were infected with E. tarda in the presence of the agomir of each of the downregulated miRNAs, or agomir plus the plasmid that overexpresses the target gene of the miRNA. Bacterial numbers in spleen were determined at 48 h post-infection (hpi). (B) Tongue sole were infected with E. tarda in the presence of the antagomir of each of the upregulated miRNAs, or antagomir plus the siRNA against the target genes of the miRNAs. Bacterial numbers in spleen were determined as above. The experiments were performed 3 times, and values are shown as means ± SEM. *P < 0.05, **P < 0.01.
To examine whether the anti-/pro-bacterial invasion effects observed above with the miRNAs were dependent on their target genes, tongue sole were infected with E. tarda in the presence of each of the agomirs of 6 downregulated miRNAs (cse-miR-146a, cse-miR-148, cse-miR-152, cse-miR-23a, cse-miR-71c-5p, and cse-let-7) plus the plasmid that overexpresses the target gene of the miRNA. The results showed that for cse-miR-146a, cse-miR-71c-5p, and cse-let-7, overexpression of their corresponding target genes significantly reduced the pro-bacterial invasion effects of these miRNAs; for cse-miR-148 and cse-miR-23a, overexpression of their corresponding target genes significantly reduced and enhanced their anti-bacterial invasion effects, respectively; for cse-miR-152, overexpression of one target gene (nlrc5) significantly enhanced the anti-bacterial invasion effect of the miRNA, whereas overexpression of another target gene (socs7) had no significant impact on the effect of the miRNA (Fig. 3).
In the same fashion, we also examined the target gene-dependence of 2 upregulated miRNAs (cse-miR-143 and cse-miR-26a). For this purpose, siRNAs against the target genes of cse-miR-143 and cse-miR-26a were synthesized, and these siRNAs were proved to significantly inhibit the expression of the target genes of cse-miR-143 and cse-miR-26a (atg2b and wipi1, respectively) (data not shown). Tongue sole were infected with E. tarda in the presence of the antagomir of cse-miR-143 or the antagomir plus the siRNA against atg2b. Subsequent bacterial recovery showed that, compared with the bacterial recovery from fish treated with the antagomir, the bacterial recovery from fish treated with the antagomir plus siRNA was significantly increased (Fig. 3). Similar results were obtained with the siRNA against the target gene of cse-miR-26a (Fig. 3).
In this study, we made the first investigation on the global profile and potential role of E. tarda-induced host miRNAs in a teleost model. We identified 311 host miRNAs (DEmiRNAs) that were significantly altered in expression during the course of E. tarda infection. Interestingly, the number of downregulated DEmiRNAs was far greater than that of upregulated DEmiRNAs. These results suggest that with respect to host interaction, E. tarda may behave more like a virus than an extracellular pathogen, which is in line with the intracellular replication capacity of E. tarda.5,27 The results of correlation analysis indicate that most of the predicted target mRNAs are indeed negatively regulated by their respective miRNAs, and that in tongue sole, miRNA–mRNA interaction networks are dynamically and temporally regulated by E. tarda infection. It is of note that, as revealed by GO and KEGG enrichment analysis, the target gene repertoire of DEmiRNAs were abundant with immune-related genes, suggesting that miRNAs are important regulators of E. tarda-induced immune response.28 The results of DEmiRNAs's potential roles in E. tarda infection indicate that temporal expressions of DEmiRNAs are vital to the immune defense against E. tarda, and that DEmiRNAs exert their effects on E. tarda infection through regulating the expression of target genes. Of these DEmiRNAs, E. tarda infection downregulated cse-let-7, which caused upregulated expression of the target gene igfbp1, and downregulated cse-miR-146a expression, which enhanced the expression of the target gene IL-1β, reduced E. tarda infection. miR-148 was found to be downregulated during E. tarda infection, which led to augmented expression of the immune suppressor gene sosc7 and, consequently, reduced E. tarda infection, most likely due to the inhibition of cytokine-induced signaling by sosc7.29 It is noteworthy that the upregulated cse-miR-143 and cse-miR-152 targeted the autophagy-related gene atg2b and the NOD-like receptor (NLR) gene nlrc5, respectively, resulting in elevated E. tarda invasion into fish tissues. Since NLR and autophagy are involved in recognition/elimination of intracellular bacterial pathogens,30,31 it is possible that upregulation of cse-miR-143 and cse-miR-152 may be a virulence strategy of E. tarda to evade host defense by suppressing the relevant pathways through hijacked miRNAs.
In conclusion, we provided the first global profile of E. tarda-induced host miRNAs in a teleost system. We detected 311 tongue sole miRNAs regulated by E. tarda infection and confirmed the target genes of 10 miRNAs through integrative analysis of differentially expressed miRNAs and mRNAs. Our results indicate that miRNAs affect E. tarda infection through regulating the expression of their target genes, which include immune genes of diverse functions, notably immune suppression and autophagy. These observations suggest that miRNAs are not only important players in host defense but may also serve as targets for E. tarda manipulation of the host defense system.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the National Natural Science Foundation of China (41476138 and 31330081), the Scientific and Technological Innovation Project and the AoShan Talents Program Supported by Qingdao National Laboratory for Marine Science and Technology (No.2015ASKJ02 and No. 2015ASTP), and the Taishan Scholar Program of Shandong Province.
References
- [1].Janda JM, Abbott SL. Infections associated with the genus Edwardsiella: the role of Edwardsiella tarda in human disease. Clin Infect Dis. 1993; 17:742-48. PMID:8268359 http://www.jstor.org/stable/4457376 [DOI] [PubMed] [Google Scholar]
- [2].Schlenker C, Surawicz CM. Emerging infections of the gastrointestinal tract. Best Pract Res Clin Gastroenterol. 2009; 23:89-99. doi: 10.1016/j.bpg.2008.11.014. PMID:19258189 [DOI] [PubMed] [Google Scholar]
- [3].Mohanty BR, Sahoo PK. Edwardsiellosis in fish: a brief review. J Biosci. 2007; 32(7):1331-44 [DOI] [PubMed] [Google Scholar]
- [4].Park SB, Aoki T, Jung TS. Pathogenesis of and strategies for preventing Edwardsiella tarda infection in fish. Vet Res. 2012; 43:67. doi: 10.1186/1297-9716-43-67. PMID:19258189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Leung KY, Siame BA, Tenkink BJ, Noort RJ, Mok YK. Edwardsiella tarda—virulence mechanisms of an emerging gastroenteritis pathogen. Microbes Infect. 2012; 14:26-34. doi: 10.1016/j.micinf.2011.08.005. PMID:21924375 [DOI] [PubMed] [Google Scholar]
- [6].Yi J, Xiao SB, Zeng ZX, Lu JF, Liu LY, Laghari ZA, Nie P, Yu HB, Xie HX. EseE of Edwardsiella tarda augments secretion of translocon protein EseC and expression of the escC-eseE operon. Infect Immun. 2016; 84:2336-44. doi: 10.1128/IAI.00106-16. PMID:27271743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gao D, Li Y, Xu Z, Sheng A, Zheng E, Shao Z, Liu N, Lu C. The role of regulator Eha in Edwardsiella tarda pathogenesis and virulence gene transcription. Microb Pathog. 2016; 95:216-23. doi: 10.1016/j.micpath.2016.03.010. PMID:27038844 [DOI] [PubMed] [Google Scholar]
- [8].Hu YH, Sun L. The global regulatory effect of Edwardsiella tarda Fur on iron acquisition, stress resistance, and host infection: A proteomics-based interpretation. J Proteomics. 2016; 140:100-10. doi: 10.1016/j.jprot.2016.04.005. PMID:27102497 [DOI] [PubMed] [Google Scholar]
- [9].Chen H, Yang D, Han F, Tan J, Zhang L, Xiao J, Zhang Y, Liu Q. The bacterial T6SS effector EvpP prevents NLRP3 inflammasome activation by inhibiting the Ca2+-dependent MAPK-Jnk pathway. Cell Host Microbe. 2017; 21:47-58. doi: 10.1016/j.chom.2016.12.004. PMID:28081443 [DOI] [PubMed] [Google Scholar]
- [10].Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005; 132:4653-62. doi: 10.1242/dev.02073. PMID:16224045 [DOI] [PubMed] [Google Scholar]
- [11].Wolter JM, Le HH, Linse A, Godlove VA, Nguyen TD, Kotagama K, Lynch A, Rawls A, Mangone M. Evolutionary patterns of metazoan microRNAs reveal targeting principles in the let-7 and miR-10 families. Genome Res. 2017; 27:53-63. doi: 10.1101/gr.209361.116. PMID:27927717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Almeida MI, Calin GA. The miR-143/miR-145 cluster and the tumor microenvironment: unexpected roles. Genome Med. 2016; 8:29. doi: 10.1186/s13073-016-0284-1. PMID:26988859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu R, Liu C, Chen D, Yang WH, Liu X, Liu CG, Dugas CM, Tang F, Zheng P, Liu Y, et al.. FOXP3 controls an miR-146/NF-κB negative feedback loop that inhibits apoptosis in breast cancer cells. Cancer Res. 2015; 75:1703-13. doi: 10.1158/0008-5472.CAN-14-2108. PMID:25712342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu XY, He YJ, Yang QH, Huang W, Liu ZH, Ye GR, Tang SH, Shu JC. Induction of autophagy and apoptosis by miR-148a through the sonic hedgehog signaling pathway in hepatic stellate cells. Am J Cancer Res. 2015; 5:2569-89. PMID:26609469 [PMC free article] [PubMed] [Google Scholar]
- [15].Cullen B R. Viruses and microRNAs: RISCy interactions with serious consequences. Genes Dev. 2011; 25:1881-94. doi: 10.1101/gad.17352611. PMID:21896651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Hakimi M, Cannella D. Apicomplexan parasites and subversion of the host cell microRNA pathway. Trends Parasitol. 2011; 27:481-6. doi: 10.1016/j.pt.2011.07.001. PMID:21840260 [DOI] [PubMed] [Google Scholar]
- [17].Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006; 312:436-9. doi: 10.1126/science.1126088. PMID:16627744 [DOI] [PubMed] [Google Scholar]
- [18].Staedel C, Darfeuille F. MicroRNAs and bacterial infection. Cell Microbiol. 2013; 15:1496-507. doi: 10.1111/cmi.12159. PMID:23795564 [DOI] [PubMed] [Google Scholar]
- [19].Sharbati J, Lewin A, Kutz-Lohroff B, Kamal E, Einspanier R, Sharbati S. Integrated microRNA-mRNA-analysis of human monocyte derived macrophages upon Mycobacterium avium subsp. hominissuis infection. PLoS One. 2011; 6:e20258. doi: 10.1371/journal.pone.0020258. PMID:21629653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xu X, Shen Y, Fu J, Lu L, Li J. Next-generation sequencing identified microRNAs that associate with motile aeromonad septicemia in grass carp. Fish Shellfish Immunol. 2015; 45:94-103. doi: 10.1016/j.fsi.2015.02.008. PMID:25698074 [DOI] [PubMed] [Google Scholar]
- [21].Li MF, Zhang J. CsTNF1, a teleost tumor necrosis factor that promotes antibacterial and antiviral immune defense in a manner that depends on the conserved receptor binding site. Dev Comp Immunol. 2016; 55:65-75. doi: 10.1016/j.dci.2015.10.010. PMID:26478190 [DOI] [PubMed] [Google Scholar]
- [22].Zhang BC, Zhang J, Sun L. In-depth profiling and analysis of host and viral microRNAs in Japanese flounder (Paralichthys olivaceus) infected with megalocytivirus reveal involvement of microRNAs in host-virus interaction in teleost fish. BMC Genomics. 2014; 15:878. doi: 10.1186/1471-2164-15-878. PMID:25297525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sha Z, Gong G, Wang S, Lu Y, Wang L, Wang Q, Chen S. Identification and characterization of Cynoglossus semilaevis microRNA response to Vibrio anguillarum infection through high-throughput sequencing. Dev Comp Immunol. 2014; 44:59-69. doi: 10.1016/j.dci.2013.11.014. PMID:24296438 [DOI] [PubMed] [Google Scholar]
- [24].Long H, Chen C, Zhang J, Sun L. Antibacterial and antiviral properties of tongue sole (Cynoglossus semilaevis) high mobility group B2 protein are largely independent on the acidic C-terminal domain. Fish Shellfish Immunol. 2014; 37:66-74. doi: 10.1016/j.fsi.2014.01.013. PMID:24468324 [DOI] [PubMed] [Google Scholar]
- [25].Li MF, Li J, Sun L. CsMAP34, a teleost MAP with dual role: A promoter of MASP-assisted complement activation and a regulator of immune cell activity. Sci Rep. 2016; 6:39287. doi: 10.1038/srep39287. PMID:28008939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang BC, Zhou ZJ, Sun L. pol-miR-731, a teleost miRNA upregulated by megalocytivirus, negatively regulates virus-induced type I interferon response, apoptosis, and cell cycle arrest. Sci Rep. 2016; 6:28354. doi: 10.1038/srep28354. PMID:27311682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhou ZJ, Sun L. Edwardsiella tarda-induced inhibition of apoptosis: a strategy for intracellular survival. Front Cell Infect Microbiol. 2016; 6:76. doi: 10.3389/fcimb.2016.00076. PMID:27471679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Chen JQ, Papp G, Szodoray P, Zeher M. The role of microRNAs in the pathogenesis of autoimmune diseases. Autoimmun Rev. 2016; 15:1171-80. doi: 10.1016/j.autrev.2016.09.003. PMID:27639156 [DOI] [PubMed] [Google Scholar]
- [29].Krebs DL, Hilton DJ. SOCS proteins: negative regulators of cytokine signaling. Stem Cells. 2001; 19:378-87. doi: 10.1634/stemcells.19-5-378. PMID:11553846 [DOI] [PubMed] [Google Scholar]
- [30].Jia K, Thomas C, Akbar M, Sun Q, Adams-Huet B, Gilpin C, Levine B. Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc Natl Acad Sci U S A. 2009; 106:14564-9. doi: 10.1073/pnas.0813319106. PMID:19667176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jo EK, Yuk JM, Shin DM, Sasakawa C. Roles of autophagy in elimination of intracellular bacterial pathogens. Front Immunol. 2013; 4:97. doi: 10.3389/fimmu.2013.00097. PMID:23653625 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


