ABSTRACT
Since the hypermucoviscous variants of Klebsiella pneumoniae were first reported, many cases of primary liver abscesses and other invasive infections caused by this pathogen have been described worldwide. Hypermucoviscosity is a phenotypic feature characterized by the formation of a viscous filament ≥5 mm when a bacterial colony is stretched by a bacteriological loop; this is the so-called positive string test. Hypermucoviscosity appears to be associated with this unusual and aggressive type of infection, and therefore, the causal strains are considered hypervirulent. Since these first reports, the terms hypermucoviscosity and hypervirulence have often been used synonymously. However, new evidence has suggested that hypermucoviscosity and hypervirulence are 2 different phenotypes that should not be used synonymously. Moreover, it is important to establish that a negative string test is insufficient in determining whether a strain is or is not hypervirulent. On the other hand, hypervirulence- and hypermucoviscosity-associated genes must be identified, considering that these phenotypes correspond to 2 different phenomena, regardless of whether they can act in synergy under certain circumstances. Therefore, it is essential to quickly identify the genetic determinants behind the hypervirulent phenotype to develop effective methodologies that can diagnose in a prompt and effective way these hypervirulent variants of K. pneumoniae.
KEYWORDS: hypermucoviscosity, hypervirulence, invasive infections, Klebsiella pneumoniae, regulator of mucoid phenotype, serotypes
Introduction
In the mid-1980s, a new hypervirulent variant of Klebsiella pneumoniae was described for the first time in Taiwan.1 This new variant of hypervirulent K. pneumoniae (hvKpn) differs from the classical K. pneumoniae (cKpn) in its ability to produce invasive infections even in apparently healthy adults. The hvKpn variants tend to spontaneously form abscesses (especially liver ones) and develop metastases (with or without secondary bacteraemia), particularly in the brain and eyes, from the primary sites of infection.2-4 These clinical features, which are rarely caused by cKpn, together with a hypermucoviscous phenotype appearing when they are grown in various culture media (determined by a positive string test) are the hallmark of hvKpn variants.
Since the hypermucoviscous clones of K. pneumoniae were described for the first time, they have been associated with hypervirulence, although no locus or loci have been clearly identified as being responsible for this trait. At first, capsular serotypes K1 and K2 were described as highly pathogenic to humans; they were more frequently associated with this phenotype and characterized by producing mucous colonies. However, hvKpn clones with a hypermucoviscous phenotype and belonging to different capsular serotypes have been now described worldwide.5-7 Hypermucoviscous clones belonging to the same capsular serotype do not produce a homogeneous pattern of infection despite sharing the same virulence factor profile.6,8
After the first cases of invasive infections caused by K. pneumoniae were reported, the hypermucoviscous phenotype was considered a marker of hypervirulence. This partnership is still widely accepted. Furthermore, it is common for these terms to be considered synonyms in the medical literature. However, is the hypermucoviscous phenotype an indicator of hypervirulence? Based on the information currently available, the aim of this review was to explore this question and identify unresolved questions that need to be investigated for a better understanding of hypermucoviscosity and hypervirulence.
Hypermucoviscous and hypervirulent variants of Klebsiella pneumoniae (hmvKpn/hvKpn)
Since they were described for the first time, K. pneumoniae hypermucoviscous clones have been considered hypervirulent.9 Like all capsulated bacteria, K. pneumoniae produces mucoid colonies in a nutritive medium. This mucoid phenotype differs from a hypermucoviscous phenotype in that hypermucoviscosity is defined by the formation of a viscous filament ≥5 mm after stretching a Klebsiella spp colony with a loop on an agar plate in the so-called string test (Fig. 1). Not all mucoid colonies of K. pneumoniae show a positive string test. This phenomenon highligths a distinct difference between mucoid capsular strains and the hypermucoviscous variants.10 The first reports from the Far East indicated that these variants could produce invasive infections causing primary liver, prostate, bone, kidney, and lung abscesses; some cases of necrotizing fasciitis have also been described.2,4,11-13 Metastatic complications to the brain and eyes from these primary sites of infection have also been documented.2,14-16 Therefore, the invasive capability and the hypermucoviscous phenotype account for the main differences between the hvKpn and cKpn variants (Fig. 2).2 However, the hypermucoviscous K. pneumoniae phenotype without a hypervirulence genotype has been poorly studied. The infections caused by these hypervirulent/hypermucoviscous K. pneumoniae (hvKpn/hmvKpn) variants have been observed in immunocompromised Asian patients and diabetics with poor glycemic control. Moreover, most of these strains belonged to the K1 serotype, the most prevalent one in the Far Eastern region.2,16,17,18 In addition to the serotype K1/K2, other serotypes have also been associated with the hvKpn/hmvKpn phenotypes.6,19 Similarly, hvKpn/hmvKpn strains are sensitive to several classes of antibiotics, except for ampicillin, to which K. pneumoniae is intrinsically resistant. Although the rate of mortality associated with these variants is low, the disabling consequences resulting from the invasion of the central nervous system and the eye are usually devastating.2 Since most of hvKpn variants are sensitive to several classes of antimicrobials, the timely and prompt administration of appropriate antibiotics avoids the development of catastrophic disabilities and death.20 The main differences between cKpn and hvKpn are summarized in Table 1.
Figure 1.
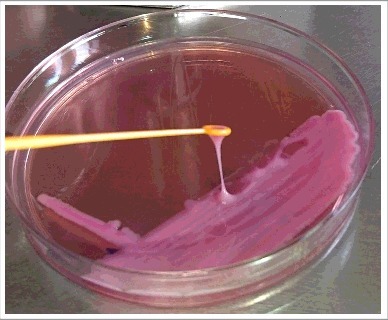
String test. A hypermucoviscous phenotype is seen when a viscous filament ≥5 mm is produced after stretching a K. pneumoniae colony with a loop on an agar plate (positive string test).
Figure 2.
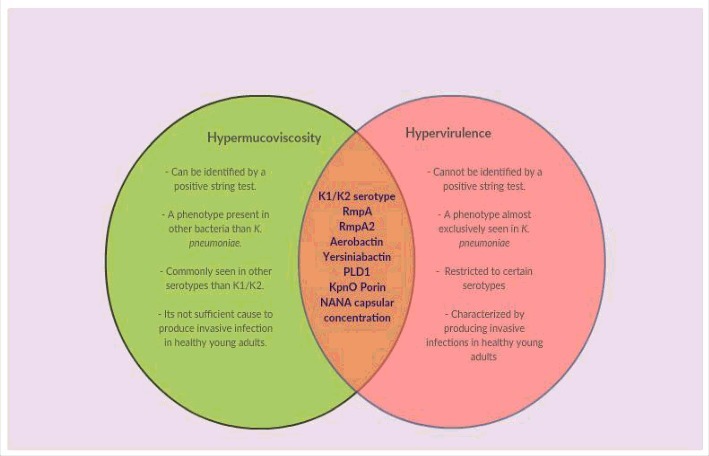
Venn diagram showing differences and similarities between hypermucoviscous and hypervirulent phenotypes. New evidence suggests that hypermucoviscosity and hypervirulence are 2 different phenomena. The green region shows the main characteristics of the hypermucoviscous phenotype that differ from those of the hypervirulent phenotype. The intersection highlights the elements that have been strongly associated with both phenotypes.
Table 1.
Main differences between cKpn and hvKpn strains.
|
Feature |
cKpn |
hvKpn |
| Infections | Urinary tract infections, pneumonia, bloodstream infections, wound surgical infections and meningitis, mainly in alcoholics, elderly adults, patients undergoing invasive procedures or have significant comorbidities and immunocompromised patients. These infections are mainly nosocomial-acquired | Liver, lungs, kidney, bones, prostate abscesses and necrotizing fasciitis have been already described. Invasive infections to eye and central nervous system from this primaries sites of infection even in healthy adults. These infections are usually acquired in community. |
| Antimicrobial susceptibility | Multidrug-resistant strains represent the main threat in this variants of K. pneumoniae. Strains harboring extended-spectrum β-lactamases (ESBLs) and carbapenemases genes are a clear example. | The first cases of these variants showed a susceptibility phenotype to several classes of antimicrobials. Worryingly, hypervirulent and multi-drug resistant strains have now been described. hvKpn strains harboring ESBLs, carbapenemases or mcr-1 genes have been described. |
| Virulence-associated genes | Mainly enterobactin, aerobactin, yersiniabactin and salmochelin. | rmpA, rmpA2, aerobactin, yersiniabactin, pld1, KpnO porin and higher content of capsular sialic acid |
Over the years, infections caused by hvKpn/hmvKpn K. pneumoniae have been detected all over the world. Unlike the first cases reported, the most affected people have been young adults without apparent immunodeficiency and people of Asian descent.6,21-26 Infections in children have also been described,27,28 and cases of invasive infections in animals have been reported.29,30 Lee et al. (2016) tried to determine whether the development of invasive infection was due to genetic predisposition or geospecific strain acquisition. They analyzed 70 patients living in Singapore and belonging to 4 ethnic groups (Chinese, Malay, Indian and Caucasian) from which demographic and clinical profile information was recorded. In these groups, type 2 diabetes mellitus was the comorbidity more frequently found at a lower prevalence in the Chinese group. Lee et al. (2016) found that the Chinese had a higher probability of being infected and developing invasive infection by K. pneumoniae K1 serotype than did non-Chinese groups. In accordance with this result, the Chinese were rarely infected by non-K1 serotypes compared with non-Chinese ethnic groups. This particular result may be because the Chinese have a higher rate of intestinal carriage of K. pneumoniae K1 serotype compared with Malay, Indian and Caucasian groups. On the other hand, non-Chinese groups were more frequently infected by K2 and other non-K1 serotypes, but the development of invasive infection was associated with some comorbidity, especially type 2 diabetes mellitus. In conclusion, environmental factors (diet, water and direct contact with K. pneumoniae K1 carriers) that lead to the acquisition of K. pneumoniae with certain serotypes, genetic predisposition and comorbidities represent the principal factors determining the differential susceptibility to the development of invasive infection, independent of living in the same geographical area.31
Thus, hypervirulence in K. pneumoniae could be defined as the ability of the bacteria to produce invasive infections (metastatic dissemination) after a primary focus of infection in apparently healthy adults. The emergence of multidrug-resistant strains with hvKpn/hmvKpn phenotypes is a cause for concern.32-35 Siu et al. (2014) have shown that the transferability of plasmidic blaKPC (KPC-2 and KPC-3) into a virulent K. pneumoniae K2 serotype isolated from a liver abscess is possible. Despite the plasmid harboring carbapenemase, the recipient strain conserved its virulence attributes and gained a multidrug-resistant phenotype.36 Zhang et al. (2015) have shown the emergence of a K1 hypervirulent strain of K. pneumoniae harboring blaKPC-2 carbapenemase in China.35 The hypervirulent K1 K. pneumoniae strain harboring plasmid-mediated colistin resistance mcr-1 gene has also been identified, which is even more alarming.37 So far, the number of strains showing multidrug-resistant and hypervirulent phenotypes has constantly increased.
Virulence factors behind the hypervirulent/hypermucoviscous phenotype: From polysaccharide capsule to nowhere?
Due to the characteristic hypermucoviscous aspect of the colonies produced by these hvKpn variants, it was proposed that the polysaccharide capsule was the main virulence factor responsible for the hypervirulent phenotype.38 As observed in all encapsulated bacteria, the polysaccharide capsule represents a major virulence factor in K. pneumoniae. The capsule acts as a protective shield that protects bacteria from phagocytosis by immune cells, avoiding the bactericidal action of complement and antimicrobial peptides by preventing its deposition on the bacterial surface. Furthermore, the capsule blocks the bacterial opsonization with antibodies and prevents the activation of the innate immune response.39-41 In accordance with this mechanism, acapsular variants of K. pneumoniae seem to be less virulent than capsular ones.40,41 Moreover, the polysaccharide capsule is responsible for the appearance of mucus (but not necessarily the hypermucoviscous phenotype) when the microorganism is grown in different culture media.2
So far, 78 serotypes have been described, of which K1, K2, K5, K20, K54 and K57 have been identified as the most frequent, pathogenic and prevalent in human infections caused by K. pneumoniae. Among them, serotype K1 is predominant in Asia, and serotype K2 has been more frequently isolated in America and Europe; these serotypes are the most pathogenic to humans.42 The genes involved in the synthesis and transport of the capsular antigen in K. pneumoniae are coded at the cps locus. Among others, this locus contains the genes required for the polymerization and translocation of the capsular antigen, and this operon is located within the bacterial chromosome.43-45
The magA gene (“mucoviscosity-associated gene A”) was identified as the gene responsible for the hypermucoviscous phenotype by means of site-directed mutagenesis studies of the genes present in the locus.46,47 Subsequent studies showed that the magA gene encodes a polysaccharide polymerase enzyme that is specific to capsular serotype K1 and is responsible for the formation of the characteristic structure of the capsule.48 It was later discovered that this polymerase is specific to each of the currently known 78 capsular serotypes. Therefore, mucoviscosity-associated gene magA was renamed as wzy_K1, and this nomenclature was applied to the remaining serotypes.49 For example, the polymerase gene specific to serotype K2 is called wzy_K2, the polymerase gene specific to serotype K3 is called wzy_K3, and so forth.48,50 This polymerase is essential for the synthesis of the capsule. Deletion of the wzy gene prevents the production of the polysaccharide capsule.51 These features, along with the classical limitations of serotyping, have turned molecular typing by wzy-PCR into the gold standard for the characterization K. pneumoniae clinical isolates.52,53 However, strains with the hypervirulent/hypermucoviscous phenotype that do not belong to the K1 serotype (wzy_K1 (magA)-negative) have also been described.54 Therefore, another virulence gene (or genes), different from those present in the cps cluster, could be associated with the hypervirulent/hypermucoviscous phenotype.
Capsular polysaccharide (CPS) in Klebsiella is similar to Escherichia coli group I CPS in primary structure and biosynthesis mechanisms.55 In E. coli, the 2-component system (TCS) RcsBCD plays an important role as a regulator system for the synthesis of colonic acid, which simultaneously functions as a model for group I CPS biosynthesis. In this system, protein RcsC acts a sensor kinase, which is autophosphorylated when it senses environmental stimuli. It subsequently transfers the stimuli to RcsD, a phosphotransfer histidine-containing protein, which is located in the inner membrane. Later, the signal is relayed to RcsB, a cytoplasmic response regulator protein. When RcsB is phosphorylated, it interacts with RcsA, which acts as an auxiliary transcriptional regulator. Finally, RcsA binds to promoters in the cps cluster, and then the biosynthesis of colonic acid capsule is activated.55,56,57
In K. pneumoniae, the mucoid phenotype A2 regulator (rmpA2) gene and the truncated form of the mucoid phenotype A regulator (rmpA) gene apparently act as positive transcriptional regulators of the capsular polysaccharide synthesis; thus, they code for the counterparts of the RcsA protein found in E. coli, leading to the association of capsular hyperproduction with hypermucoviscosity and hypervirulence.56 Both can be encoded either in a chromosome or in a plasmid, and more than one copy can be found in a bacterial cell. Hsu et al.58 studied a K1 K. pneumoniae strain harboring 2 rmpA genes (plasmidic and chromosomal) and one copy of rmpA2 (plasmidic).58 Through isogenic gene deletions and complementation experiments, they found that only plasmidic rmpA was able to increase the expression of cps genes and CPS production. On the other hand, cps expression was inhibited by plasmidic rmpA2 but not chromosomal rmpA. Surprisingly, the authors found that although the plasmidic rmpA promoted cps gene expression and increased CPS production, the plasmidic ΔrmpA mutants did not show any differences in serum resistance, proliferation in non-immune human serum or virulence in a murine model.59 A high prevalence of both genes, combined or alone, was associated with strains that had a hypervirulent/hypermucoviscous phenotype.56,58,60 For example, plasmid pLVPK (219-Kb) harbors the rmpA and rmpA2 genes, as well as others that code for virulence factors, such as several siderophore systems and genes associated with resistance to tellurite, copper, silver and lead. This virulence plasmid has been associated with virulent strains of K. pneumoniae.61 Likewise, numerous studies have reported that the presence of some of these transcriptional regulators, regardless of the capsular serotype, is strongly associated with hypervirulent/hypermucoviscous K. pneumoniae.58,62 However, similar to the wzy_K1 gene, the strains lacking the rmpA/rmpA2 genes with the hypervirulent/hypermucoviscous phenotype were also described.6,46 Moreover, strains positive for the rmpA and rmpA2 genes and without a hypermucoviscous phenotype or capsular serotypes K1 or K2 were also isolated.6 Some authors suggest that frameshift mutations (insertion or deletion) in the rmpA/rmpA2 genes in these strains can produce altered RmpA/RmpA2 proteins that lead to a non-hypermucoviscous phenotype.63 Therefore, hypervirulent/hypermucoviscous K. pneumoniae strains that do not belong to capsular serotypes K1 or K2 and that may or may not harbor the rmpA/rmpA2 genes represent a great enigma regarding the genetic factor or factors responsible for its particular phenotype.
Sialic acid, also known as N-acetylneuraminic acid (NANA), is a monosaccharide that is present in the K. pneumoniae polysaccharide capsule. Recent studies have shown that the hypervirulent/hypermucoviscous variants of K. pneumoniae have a significantly higher concentration of sialic acid in their capsular extracts than do those strains that do not display the hypermucoviscous phenotype.64 Moreover, these strains have a phagocytosis-resistance phenotype, in contrast to those strains with smaller concentrations of sialic acid in their capsules and without a hypermucoviscous phenotype. Sialic acid in the K. pneumoniae polysaccharide capsule inhibits the infected host's innate immune response by modulating the inflammatory responses. In addition, the depletion of sialic acid in the capsules produced by K. pneumoniae may result in a loss of hypermucoviscosity and, therefore, a decrease in the resistance to phagocytosis by neutrophils.64 Given that only a small number of studies have analyzed the concentration of sialic acid present in the capsules of these hypervirulent/hypermucoviscous K. pneumoniae variants, it would be interesting to study a larger number of hypermucoviscous strains to determine whether the concentration of sialic acid plays a fundamental role in the expression of the hypervirulent phenotype.
As was noted above, the genetic factors associated with the synthesis of the polysaccharide capsule were the first analyzed. However, the role of the virulence factors described in cKpn was also studied. Among these, the siderophore system was the most analyzed. Siderophores play an important role in the ability of K. pneumoniae to cause invasive disease in human hosts with a functional immune system.65 Russo et al. (2014) found that hypervirulent/hypermucoviscous variants of K. pneumoniae produced more siderophores than did the classical (not hypermucoviscous) ones.65 In addition, they found that aerobactin was the most-produced siderophore under certain experimental conditions and that the increase in siderophore production was not related to gene copy number.65 Moreover, they found that aerobactin was the primary factor in a conditioned medium that increased the growth/survival of bacteria in human ascites or serum. An increase in the virulence of an in vivo infection of a mouse pulmonary challenge model was also identified.66 It is noteworthy that these effects were not observed in the case of salmochelin, enterobactin and yersiniabactin siderophores.65,66 These data suggest that aerobactin plays an important role as a fundamental virulence factor in the development of the hypervirulence of K. pneumoniae. In silico studies performed by Holt et al. (2015) showed a higher prevalence of siderophores in the invasive variants of K. pneumoniae than in the classical K. pneumoniae. In this study, yersiniabactin was the most prevalent siderophore found in the hypervirulent/hypermucoviscous variants of K. pneumoniae. Unlike the findings made by Russo et al. (2014), in relation to aerobactin, Holt et al. (2015) found that yersiniabactin showed no significant association between any of the other siderophores and the expression of the invasive phenotype.67
Searching for the genetic determinants behind the hypervirulent phenotype has identified new factors associated with virulence. A phospholipase D family protein (PLD1) encoded by the pld1 gene that is located within a type 6 secretion system (T6SS) locus has been described by Lery et al (2014). These authors found that, in an intranasal murine model of infection, Δpld1 mutants were avirulent in comparison with wild-type strains, while virulence was restored in complemented strains.68 In concordance, phospholipase D family proteins have been identified as key factors for virulence in Legionella pneumophila because they promote host cell invasion and consequent bacterial dissemination.69 Interestingly, an in silico analysis of pld1 in 171 Klebsiella genomes found that this gene was widely distributed in K. pneumoniae subsp rhinoscleromatis, the only intracellular subspecies of the Klebsiella genus.68
In bacteria, porins constitute one of the major proteins of the outer membrane (OM). These also called OM proteins (OMPs), which interact with the bacterial environment and act as pores through which certain molecules can diffuse. Moreover, these proteins have been implicated in bacterial pathogenesis. A novel OMP, the KpnO porin, was identified by Srinivasan et al (2012). These authors found that ΔkpnO strains produced less capsular polysaccharide, which was reflected in non-mucoid and smaller colonies in comparison with the hypervirulent wild-type strains. In addition, using a Caenorhabditis elegans infection model, the authors found that kpnO mutants were less virulent than the hypervirulent wild-type strains. Complemented strains recovered virulent and hypermucoviscous phenotypes.70 As we observed, a large number of virulence factors have been associated with the hypervirulent phenotype; thus, hypervirulence seems to be a consequence of a complex interplay of several genetic determinants rather than a phenomenon originated by a single gene.
Hypervirulence-associated clonal groups
Bialek-Davenet et al. (2014) analyzed 694 highly conserved genes using a core-genome multilocus sequence typing scheme (cgMLST). These authors reported that serotype K1 hypervirulent variants belonged to clonal group 23 (CG23), where ST23 was predominant. Serotype K2 hypervirulent variants, on the other hand, belonged to the following clonal groups: CG86 (the most widely distributed), CG65, CG375 and CG380. These results suggest that invasive infection-causing serotype K2 isolates are more genetically diverse than serotype K1 ones.71 Interestingly, these researchers found that tellurite resistance (due to the presence of the terW gene) is strongly associated with CG23, CG65 and CG86.72 Globally, CG23 (associated with the K1 serotype) is the most prevalent CG associated with hypervirulence, followed by CG86. To date, all these clonal groups have been described as causal agents of invasive infections around the world.17,71
Hypermucoviscosity and hypervirulence: Two different but (at times) complementary phenotypes?
In 2014 in China, Luo et al. (2014) analyzed 51 non-repetitive K. pneumoniae clinical isolates causing primary liver abscesses (PLAs).73 The samples were obtained from the drainage of these PLAs. Patients from both sexes were included in the study, and their mean age was 53 y. Most isolates belonged to the K1 serotype, accounting for 39% (20/51) of the isolates; 31% (16/51) belonged to the K2 serotype, and 29% (15/51) belonged to non-K1/K2 isolates. Interestingly, approximately 29% (15/51) of the clinical isolates causing PLAs did not show the characteristic hypermucoviscous phenotype that was associated with the hypervirulent K. pneumoniae variant. Almost all clinical isolates belonging to the K2 serotype (93.8% (15/16)) displayed the hypermucoviscous phenotype. Nevertheless, 70% (14/20) of the clinical isolates belonging to the K1 serotype showed the hypermucoviscous phenotype, whereas only 46.7% (7/15) of the non-K1/K2 clinical isolates showed a positive string test. In accordance with other studies, K. pneumoniae clinical isolates belonging to the K1 serotype were grouped into clonal complex 23 (CC23); those belonging to the K2 serotype were grouped into CC65, and non-K1/K2 isolates were grouped into minor clonal complexes.67,71,74 In addition, virulence genes, such as rmpA and aerobactin, kfu, and allS siderophore genes showed a diverse distribution among all clonal complexes. Overall, these results suggest that the hypervirulent phenotype is not dependent on hypermucoviscosity. Therefore, hypervirulence should be defined not only by the phenotype but also by the genotype and the clinical characteristics of the infection. In other words, hypervirulence goes beyond a capsular serotype and a positive string test.73
Similar results were reported in Spain, where Cubero et al. (2015) analyzed 878 cases of bacteremia caused by K. pneumoniae.6 Only 5.4% (53/878) of the isolates collected from 50 patients displayed the hypermucoviscous phenotype identified through string test. Among these, 16/53 (30.2%) were K1 serotype and rmpA-gene positive, 12/53 (22.6%) were non-K1 serotype and rmpA gene-positive, and 25/53 (47.2%) were non-K1 serotype and rmpA gene-negative. Cases of bacteraemia due to hypermucoviscous were more frequent in men with a mean age of 67 y and were community acquired. The vast majority of these patients had an underlying disease. Moreover, these researchers identified that some rmpA gene-positive strains belonging to the K1 serotype did not display the hypermucoviscous phenotype. Nevertheless, these strains were able to produce an invasive infection. Surprisingly, they found a small number of non-hypermucoviscous isolates that did not harbor rmpA genes and did not belong to the K1 serotype causing liver abscesses, a type of infection that is commonly restricted to K1/K2, rmpA-positive and hmvKpn isolates. These results were in agreement with the observations reported by Luo et al. (2014) in the sense that hypermucoviscosity detected through the string test is not enough to render an isolate hypervirulent.73 According to these findings, hypermucoviscosity and hypervirulence are 2 separate entities, and a hypervirulent phenotype is only determined by genetic traits, in particular (but not solely) by the transcriptional regulator of the cps cluster, rmpA. Cubero et al. (2015) also identified rmpA-negative invasive strains belonging to the diverse capsular serotypes.6
In Japan, Togawa et al. (2014) studied 83 bloodstream infection cases caused by K. pneumoniae.75 Thirty-eight percent (32/83) of the isolates were classified as community-acquired infections and 61% (51/83) as hospital-acquired infections. These authors reported that the mucoid phenotypes (colonies with a higher degree of mucoviscosity than other cKpn but with a negative string test) and hypermucoviscous phenotypes were more prevalent in the community-acquired K. pneumoniae isolates. As expected, septic shock was more frequently observed in isolates with mucoid and hypermucoviscous phenotypes than in non-hypermucoviscous isolates. However, no statistically significant differences were observed when mucoid isolates were compared with non-mucoid ones. Interestingly, a statistically significant association was seen between liver abscess formation and the mucoid phenotype. Surprisingly, liver abscess formation was not significantly associated with hypermucoviscosity. This finding is contrary to the idea that the hypermucoviscous phenotype is strongly associated with the development of liver abscesses.75 This phenomenon, which is rarely described, needs to be studied more deeply for a better understanding of the bacterial genetic determinants and the host factors that could be responsible for the hypervirulent phenotype in K. pneumoniae.
Recently in China, Wu et al (2017) conducted a retrospective study to investigate the phenotypic and genotypic characteristics of 76 K. pneumoniae strains causing bloodstream and at least one another site infection during the same period of infection. Of these 76 strains, 28 were considered as hypervirulent and the remaining 48 as classic variants. Because there is no consensus about the definition of hypervirulence, the authors used 3 microbiological characteristics that have been associated to it: A positive string test, a positive PCR amplification of rmpA gen and a positive PCR amplification of aerobactin gen. Those strains with at least 2 of these indicators were considered as hypervirulent. They found that 2 of 28 strains classified as hypervirulent did not show the hypermucosvicous phenotype. However, up to 90% of them were positive to rmpA and aerobactin genes. Interestengly, 2 strains classified as classic variants of K. pneumoniae harbored rmpA gen which usually has been associated only to hypervirulent variants. Although none of them expressed the hypermucoviscous phenotype and only less than 10% harbored the aerobactin gen. Regarding to the development of metastasis (invasive infection), which was defined by the authors as infections occurred at 2 or more different infection sites in one patient with bacteremia and without injury or former invasive medical procedure around the infection sites. Two metastatic infection cases were identified in homo-cKpn (cKpn strains isolated from different specimens sources from the same patient with the same PFGE pattern).Surprisingly, with respect to metastasis development, no differences were observed between the homo-hvKpn and the homo-cKpn strains.76
Hypermucoviscosity has not only been described in K. pneumoniae clinical isolates but also in Klebsiella quasipneumoniae subsp. similipneumoniae (initially classified as K. pneumoniae through automated methods) with a hypermucoviscous phenotype by Garza-Ramos et al. (2016). This identification was performed using whole genome sequencing and a subsequent analysis of the rpoB gene, and it was confirmed by average nucleotide identity (ANI). The string test was positive, and the isolate was identified as an extended-spectrum β-lactamase producer encoding the extended-spectrum β-lactamase (ESBL) SHV-12. Both in silico analysis and PCR assays showed negative results for the rmpA and rmpA2 genes. Regarding the “classical” virulence factors, this strain was positive for enterobactin, urease, UDP galacturonate 4-epimerase, glycosyltransferases, pili, and several adhesins.77 Simultaneously, a previous study (personal communication, August 2015) analyzed a hypermucoviscous K2 serotype K. pneumoniae obtained from liver abscess drainage that was positive for the rmpA and rmpA2 genes. The PCR assay to detect the terW gen was also positive. Furthermore, Southern hybridization showed that this strain harbored 2 copies of the rmpA gene: one was located in the bacterial chromosome, and the other one was plasmidic. Southern hybridization against the rmpA2 gene showed its plasmidic location. Subsequent whole genome sequencing indicated that this strain harbors the virulence plasmid pLVPK. Siderophore-associated genes iucA, iroB and entB were also detected in silico and by PCR. Finally, yersiniabactin gene irp2 was not present in this strain. Once identified, these 2 clinical isolates with a hypermucoviscous phenotype were subject to an analysis of virulence through resistance to non-immune human serum, biofilm formation, and survival rate assays in a murine model by intraperitoneal inoculation of 1 × 104 CFU/mL of both strains. Despite its hypermucoviscous phenotype, the K. quasipneumoniae subsp similipneumoniae isolate was highly sensitive to the non-immune human serum; it was sensitive to neutrophil phagocytosis and was unable to kill the infected mice after a 14-day follow-up (personal communication, August 2015). On the other hand, the K2 serotype K. pneumoniae clinical isolate with a hypermucoviscous phenotype was resistant to the components of the non-immune human serum; it evaded neutrophil phagocytosis and killed mice 2 d after inoculation (personal communication, August 2015). No differences were observed with respect to biofilm production: both hypermucoviscous isolates were classified as non-producers, which is consistent with previous studies.
Moreover, other studies have described the hypermucoviscous phenotype in the Klebsiella genus. Arena et al. (2015) reported the whole genome sequence of a Klebsiella quasipneumoniae subsp quasipneumoniae clinical isolate displaying a hypermucoviscous phenotype. This strain was isolated from a bloodstream infection, and it had a new capsular wzi allele. In silico screening of virulence genes showed the absence of the rmpA and rmpA2 genes. However, this strain harbored other virulence genes, such as the allABCDRS operon (associated with allantoin metabolism), the kfuABC system, and the mrkABCDFHIJ fimbriae cluster.78 Cases of PLAs and subsequent endophthalmitis produced by K. quasipneumoniae subsp quasipneumoniae have also been described. Brisse et al. (2016) reported the whole genome sequence of a K. quasipneumoniae subsp quasipneumoniae isolate that showed a cps gene cluster associated with a capsular serotype K1. Interestingly, this virulent strain harbored a 76-Kb DNA genomic island, which is closely related to the integrative and conjugative element (ICE) ICEKp1, a mobile genetic element described previously in the hypervirulent/hypermucoviscous strain of K. pneumoniae NTUH-K2044. The mobile genetic element contained the rmpA gene and genes encoding for the siderophores yersiniabactin and salmochelin. These data suggest that the natural horizontal acquisition of gene clusters associated with interspecies virulence in the Klebsiella genus is a concerning possibility. In addition, the clusters mrkABCDFHIJ and fimABCDEFGHI (encoding type III and type I fimbriae, respectively) and the iutA gene (encoding the aerobactin receptor) were also identified in the bacterial genome.79 In these 2 reports, both K. quasipneumoniae subsp quasipneumoniae strains were susceptible to several classes of antimicrobials, and only β-lactamase blaOKP (a β-lactamase characteristic of K. quasipneumoniae subsp quasipneumoniae) was detected.78-80
The hypermucoviscous phenotype has also been observed in Klebsiella variicola. K. variicola is a Gram-negative rod bacterium closely related to K. pneumoniae and K. quasipneumoniae species. It was initially described as endophytic and pathogenic to humans and subsequently identified as a symbiont in leaf-cutting ants. In the last year, K. variicola has been broadly described as a human pathogen in different parts of the world.81-85 Garza-Ramos et al. (2015) sequenced the whole genome of a K. variicola hypermucoviscous clinical isolate obtained from the sputum of an elderly man in Mexico. The in silico search of the rmpA, rmpA2 and wzy_K1 (magA) genes was negative, but the search for virulence genes, such as those involved in iron capture systems (iroN, iutA, entB and kfuABC), bacterial metabolism (uge, ureA and wabG) and adhesion (mrkABCDFHIJ) was positive. A 100% nucleotide identity between this strain and the serotype harboring the wzc-932 variant was also found.86 Currently, hypermucoviscosity has been described even in other bacterial genera, particularly in the uropathogenic Escherichia coli (UPEC).87 Therefore, these results could indicate that the hypermucoviscous phenotype might be a feature that can be acquired horizontally through a mobile genetic element. However, the results found in the Klebsiella species suggest that hypermucoviscosity per se is not enough to confer hypervirulence to a bacterial strain and that other bacterial factors are required for the development of hypervirulence.
Over time, new evidence has arisen suggesting that the string test is not an effective means of determining whether an isolate is hypervirulent. However, we cannot discard the string test as a probe that give us a warning signal that indicates the possibility that a clinical isolate could be hypervirulent. Until now, string test together with the clinical data and molecular probes, for example the PCR identification of rmpA gen, represent the best preliminary way to detect and diagnose infections produced by hypervirulent isolates. Therefore, considering the most important advances in this field, we propose that the hypermucoviscous phenotype is not a reliable indicator of hypervirulence. We also postulate that the search for the genetic traits responsible for the hypervirulent and hypermucoviscous phenotypes should be made considering these phenotypes as 2 different phenomena that may act as 2 synergistic phenotypes (only in some cases) to produce strains that display a hypervirulent phenotype.
Concluding remarks/Future perspective
It has long been widely accepted that K. pneumoniae isolates displaying a hypermucoviscous phenotype are responsible for causing invasive infections in apparently healthy people that are rarely produced by enteric Gram-negative microorganisms. In addition, due to the apparent association between this phenotype and the clinical manifestations of the infection, these variants have been called hypervirulent. In medical literature, the terms hypervirulent and hypermucoviscous are commonly used as synonyms. Moreover, a positive string test has been considered as indicative of hypervirulence. However, recent findings show that in at least 3 Klebsiella spp bacterial species, a hypermucoviscous phenotype per se is not sufficient to induce a hypervirulent state, and the identification of hypervirulent K. pneumoniae variants that do not express the hypermucoviscous phenotype but still can cause invasive infections in healthy people is concerning. Thus, the absence of hypermucoviscosity is not an appropriate way to exclude hypervirulence. Molecular tests to detect virulence factors, such as aerobactin, yersiniabactin and the rmpA/rmpA2 genes associated with this phenotype, and the clinical manifestations of the disease are necessary. However, a simple molecular test is not enough to detect hypervirulence.
Several studies have indicated the K1 and K2 capsular serotypes (despite the fact that hypervirulence is associated with clones rather than with serotypes74) or transcriptional regulators, such as the rmpA/rmpA2 genes, and even to some siderophores, such as aerobactin and yersiniabactin, as the responsible factors for the hypervirulent phenotype. However, several studies have reported the existence of non-K1/K2, rmpA/rmpA2-negative strains with a different distribution of siderophores that cause invasive infections. Therefore, until the genetic determinants associated with hypervirulence are clearly identified, we cannot assert or rule out that a clinical isolate is hypervirulent. Next-generation DNA sequencing techniques, bioinformatics and transcriptomics represent powerful tools that could help us identify the elements underlying the hypervirulence and hypermucoviscous phenotypes. All of these methodologies could also help us understand whether (under certain circumstances) these phenotypes act synergistically or whether they definitely correspond to 2 different entities. Since hypermucoviscosity is not a perfect marker of hypervirulence, the available literature concerning this theme would be imperfect too.
On the other hand, most available studies have focused on the analysis of bacteria. Few studies have analyzed host factors that could favor the development of the invasive infection, for example, age and health status. The in-depth study of the host-associated factors, such as the major histocompatibility complex (MHC) variants that are more frequently associated with such infections, eating habits, nutritional status, and gut microbiota composition, could provide important information to enhance our understanding of the hypervirulence phenomenon. The rapid spread of these hypervirulent variants around the world and the emergence of multidrug-resistant hypervirulent strains require us to act quickly in such a way that we can appropriately face this great threat to global public health. Finally, the evidence exposed in this review suggests that hypermucoviscosity and hypervirulence are 2 different phenotypes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
J.C.C.N. is a Ph.D. fellow at Consejo Nacional de Ciencia y Tecnología (CONACYT).
References
- [1].Fung CP, Chang FY, Lee SC, Hu BS, Kuo BI, Liu CY, Ho M, Siu LK. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut 2002; 50(3):420-4; PMID:11839725; http://dx.doi. org/ 10.1136/gut.50.3.420 (accessed March27, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pomakova DK, Hsiao CB, Beanan JM, Olson R, MacDonald U, Keynan Y, Russo TA. Clinical and phenotypic differences between classic and hypervirulent Klebsiella pneumonia: an emerging and under-recognized pathogenic variant. Eur J Clin Microbiol Infect Dis 2012; 31(6):981-9; PMID:21918907; http://dx.doi.org/ 10.1007/s10096-011-1396-6 [DOI] [PubMed] [Google Scholar]
- [3].Kawai T. Hypermucoviscosity: an extremely sticky phenotype of Klebsiella pneumoniae associated with emerging destructive tissue abscess syndrome. Clin Infect Dis 2006; 42(10):1359-61; PMID:16619145; http://dx.doi.org/ 10.1086/503429 [DOI] [PubMed] [Google Scholar]
- [4].Lee HC, Chuang YC, Yu WL, Lee NY, Chang CM, Ko NY, Wang LR, Ko WC. Clinical implications of hypermucoviscosity phenotype in Klebsiella pneumoniae isolates: Association with invasive syndrome in patients with community-acquired bacteraemia. J Intern Med 2006; 259(6):606-14; PMID:16704562; http://dx.doi.org/ 10.1111/j.1365-2796.2006.01641.x [DOI] [PubMed] [Google Scholar]
- [5].Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis 2007; 45(3):284-93; PMID:17599305; http://dx.doi.org/ 10.1086/519262 [DOI] [PubMed] [Google Scholar]
- [6].Cubero M, Grau I, Tubau F, Pallarés R, Dominguez MA, Liñares J, Ardanuy C. Hypervirulent Klebsiella pneumoniae clones causing bacteraemia in adults in a teaching hospital in Barcelona, Spain (2007-2013). Clin Microbiol Infect 2015; 22(2):154-60; PMID:26454059 [DOI] [PubMed] [Google Scholar]
- [7].Zhang Y, Sun J, Mi C, Li W, Zhao S, Wang Q, Shi D, Liu L, Ding B, Chang YF, et al.. First report of two rapid-onset fatal infections caused by a newly emerging hypervirulent K. Pneumonia ST86 strain of serotype K2 in China. Front Microbiol 2015; 6:721; PMID:26257712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Qu T, Zhou J, Jiang Y, Shi KR, Li B, Shen P, Wei ZQ, Yu YS. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis 2015; 15:161; PMID:25886859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fang FC, Sandler N, Libby SJ. Liver Abscess Caused by magA + Klebsiella pneumoniae in North America. J Clin Microbiol 2005; 43(2):991-3; PMID:15695726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hagiya H, Watanabe N, Maki M, Murase T, Otsuka F. Clinical utility of string test as a screening method for hypermucoviscosity-phenotype K lebsiella pneumoniae. Acute Med Surg 2014; 1(4):245-6; http://dx.doi.org/ 10.1002/ams2.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu KH, Lee HC, Chuang YC, Tu CA, Chang K, Lee NY, Kos WC. Prostatic abscess in southern Taiwan: another invasive infection caused predominantly by Klebsiella pneumoniae. J Microbiol Immunol Infect 2003; 36(1):31-6; PMID:12741730 (accessed March28, 2016). [PubMed] [Google Scholar]
- [12].Lim KH, Tan YM, Chow PK. Liver abscess metastasizing to prostate and lung. J R Soc Med 2002; 95(11):554-5; PMID:12411623; http://dx.doi.org/ 10.1258/jrsm.95.11.554. (accessed March28, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Keller JJ, Tsai MC, Lin CC, Lin YC, Lin HC. Risk of infections subsequent to pyogenic liver abscess: A nationwide population-based study. Clin Microbiol Infect 2013; 19(8):717-22; PMID:23034092; http://dx.doi.org/ 10.1111/1469-0691.12027 [DOI] [PubMed] [Google Scholar]
- [14].Chiu CT, Lin DY, Liaw YF. Metastatic septic endophthalmitis in pyogenic liver abscess. J Clin Gastroenterol 1988; 10(5):524-7; PMID:3053874; http://dx.doi.org/ 10.1097/00004836-198810000-00009 (accessed March27, 2016). [DOI] [PubMed] [Google Scholar]
- [15].Keynan Y, Rubinstein E. Endogenous endophthalmitis caused by hypermucoviscous Klebsiella pneumoniae: an emerging disease in Southeast Asia and beyond. Curr Infect Dis Rep 2008; 10(5):343-5; PMID:18687194; http://dx.doi.org/ 10.1007/s11908-008-0055-2 (accessed March28, 2016). [DOI] [PubMed] [Google Scholar]
- [16].Patel G, Shah N, Sharma R. Pyogenic liver abscess, bacteremia, and meningitis with hypermucoviscous klebsiella pneumoniae: An unusual case report in a human T-cell lymphotropic virus positive patient of caribbean origin in the United States. Case Rep Infect Dis 2013; 2013:676340; PMID:24490092; http://dx.doi.org/ 10.1155/2013/676340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Turton JF, Englender H, Gabriel SN, Turton SE, Kaufmann ME, Pitt TL. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. J Med Microbiol 2007; 56(Pt 5):593-7; PMID:17446279; http://dx.doi.org/ 10.1099/jmm.0.46964-0 [DOI] [PubMed] [Google Scholar]
- [18].Chung DR, Lee HR, Lee SS, Kim SW, Chang HH, Jung SI, Oh MD, Ko KS, Kang CI, Peck KR, et al.. Evidence for clonal dissemination of the serotype K1 Klebsiella pneumoniae strain causing invasive liver abscesses in Korea. J Clin Microbiol 2008; 46(12):4061-3; PMID:18971367; http://dx.doi.org/ 10.1128/JCM.01577-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chuang YC, Lee MF, Yu WL. Mycotic aneurysm caused by hypermucoviscous Klebsiella pneumoniae serotype K54 with sequence type 29: an emerging threat. Infection 2013; 41(5):1041-4; PMID:23508461; http://dx.doi.org/ 10.1007/s15010-013-0447-6 [DOI] [PubMed] [Google Scholar]
- [20].Namikawa H, Yamada K, Fujimoto H, Oinuma KI, Tochino Y, Takemoto Y, Kaneko Y, Shuto T, Kakeya H. Two unusual cases of successful treatment of hypermucoviscous Klebsiella pneumoniae invasive syndrome. BMC Infect Dis 2016; 16(1):680; PMID:27852233; http://dx.doi.org/ 10.1186/s12879-016-2011-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Frazee BW, Hansen S, Lambert L. Invasive infection with hypermucoviscous Klebsiella pneumoniae: multiple cases presenting to a single emergency department in the United States. Ann Emerg Med 2009; 53(5):639-42; PMID:19135282; http://dx.doi.org/ 10.1016/j.annemergmed.2008.11.007 [DOI] [PubMed] [Google Scholar]
- [22].Fierer J, Walls L, Chu P. Recurring Klebsiella pneumoniae pyogenic liver abscesses in a resident of San Diego, California, due to a K1 strain carrying the virulence plasmid. J Clin Microbiol 2011; 49(12):4371-3; PMID:21998428; http://dx.doi.org/ 10.1128/JCM.05658-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pastagia M, Arumugam V. Klebsiella pneumoniae liver abscesses in a public hospital in Queens, New York. Travel Med Infect Dis 2008; 6(4):228-33; PMID:18571114; http://dx.doi.org/ 10.1016/j.tmaid.2008.02.005 [DOI] [PubMed] [Google Scholar]
- [24].Sachdev DD, Yin MT, Horowitz JD, Mukkamala SK, Lee SE, Ratner AJ. Klebsiella pneumoniae K1 liver abscess and septic endophthalmitis in a U.S. resident. J Clin Microbiol 2013; 51(3):1049-51; PMID:23303492; http://dx.doi.org/ 10.1128/JCM.02853-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Coutinho RL, Visconde MF, Descio FJ, Nicoletti AG, Pinto FC, Silva AC, Rodrigues-Costa F, Gales AC, Furtado GH. Community-acquired invasive liver abscess syndrome caused by a K1 serotype Klebsiella pneumoniae isolate in Brazil: a case report of hypervirulent ST23. Memórias do Inst Oswaldo Cruz 2014; 109(7):970-1; PMID:25411006; http://dx.doi.org/ 10.1590/0074-0276140196 (accessed March28, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vila A, Cassata A, Pagella H, Amadio C, Yeh KM, Chang FY, Siu LK. Appearance of Klebsiella pneumoniae liver abscess syndrome in Argentina: case report and review of molecular mechanisms of pathogenesis. Open Microbiol J 2011; 5:107-13; PMID:22145012; http://dx.doi.org/ 10.2174/1874285801105010107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yoon DH, Jeon YJ, Bae EY, Jeong DC, Kang JH. Liver abscess due to Klebsiella pneumoniae in a healthy 12-year-old boy. Korean J Pediatr 2013; 56(11):496-9; PMID:24348663; http://dx.doi.org/ 10.3345/kjp.2013.56.11.496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kwon JM, Jung HL, Shim JW, Kim DS, Shim JY, Park MS. Klebsiella pneumoniae liver abscess in an immunocompetent child. Korean J Pediatr 2013; 56(9):407-10; PMID:24223603; http://dx.doi.org/ 10.3345/kjp.2013.56.9.407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Jang S, Wheeler L, Carey RB, Jensen B, Crandall CM, Schrader KN, Jessup D, Colegrove K, Gulland FM. Pleuritis and suppurative pneumonia associated with a hypermucoviscosity phenotype of Klebsiella pneumoniae in California sea lions (Zalophus californianus). Vet Microbiol 2010; 141(1-2):174-7; PMID:19709820; http://dx.doi.org/ 10.1016/j.vetmic.2009.07.032 [DOI] [PubMed] [Google Scholar]
- [30].Roe WD, Rogers L, Pinpimai K, Dittmer K, Marshall J, Chilvers BL. Septicaemia and meningitis caused by infection of New Zealand sea lion pups with a hypermucoviscous strain of Klebsiella pneumoniae. Vet Microbiol 2015; 176(3-4):301-8; PMID:25682024; http://dx.doi.org/ 10.1016/j.vetmic.2015.01.019 [DOI] [PubMed] [Google Scholar]
- [31].Lee R, Molton JS, Wyres KL, Gorrie C, Wong J, Hoh CH, Teo J, Kalimuddin S, Lye DC, Archuleta S, et al.. Differential host susceptibility and bacterial virulence factors driving Klebsiella liver abscess in an ethnically diverse population. Sci Rep 2016; 6:29316; In press.(February) [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lin JC, Siu LK, Fung CP, Yeh KM, Chang FY. Nosocomial liver abscess caused by extended-spectrum beta-lactamase-producing Klebsiella pneumoniae. J Clin Microbiol 2007; 45(1):266-9; PMID:17093025; http://dx.doi.org/ 10.1128/JCM.01413-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Su SC, Siu LK, Ma L, Yeh KM, Fung CP, Lin JC, Chang FY. Community-acquired liver abscess caused by serotype K1 Klebsiella pneumoniae with CTX-M-15-type extended-spectrum beta-lactamase. Antimicrob Agents Chemother 2008; 52(2):804-5; PMID:18056273; http://dx.doi.org/ 10.1128/AAC.01269-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yao B, Xiao X, Wang F, Zhou L, Zhang X, Zhang J. Clinical and molecular characteristics of multi-clone carbapenem-resistant hypervirulent (hypermucoviscous) Klebsiella pneumoniae isolates in a tertiary hospital in Beijing, China. Int J Infect Dis 2015; 37:107-12; PMID:26141415; http://dx.doi.org/ 10.1016/j.ijid.2015.06.023 [DOI] [PubMed] [Google Scholar]
- [35].Zhang Y, Zeng J, Liu W, Zhao F, Hu Z, Zhao C, Wang Q, Wang X, Chen H, Li H, et al.. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect 2015; 71(5):553-60; PMID:26304687; http://dx.doi.org/ 10.1016/j.jinf.2015.07.010 [DOI] [PubMed] [Google Scholar]
- [36].Siu LK, Huang DB, Chiang T. Plasmid transferability of KPC into a virulent K2 serotype Klebsiella pneumoniae. BMC Infect Dis 2014; 14(1):176; PMID:24678611; http://dx.doi.org/ 10.1186/1471-2334-14-176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Gu DX, Huang YL, Ma JH, Zhou HW, Fang Y, Cai JC, Hu YY, Zhang R. Detection of colistin resistance gene mcr-1 in hypervirulent Klebsiella pneumoniae and Escherichia coli isolates from an infant with diarrhea in China. Antimicrob Agents Chemother 2016; 60(8):5099-100; PMID:27270278; http://dx.doi.org/ 10.1128/AAC.00476-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee CH, Liu JW, Su LH, Chien CC, Li CC, Yang KD. Hypermucoviscosity associated with Klebsiella pneumoniae-mediated invasive syndrome: A prospective cross-sectional study in Taiwan. Int J Infect Dis 2010; 14(8):e688-92; PMID:20547084; http://dx.doi.org/ 10.1016/j.ijid.2010.01.007 [DOI] [PubMed] [Google Scholar]
- [39].Paczosa MK, Mecsas J. Klebsiella pneumoniae: Going on the Offense with a Strong Defense. Microbiol Mol Biol Rev 2016; 80(3):629-61; PMID:27307579; http://dx.doi.org/ 10.1128/MMBR.00078-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yoshida K, Matsumoto T, Tateda K, Uchida K, Tsujimoto S, Yamaguchi K. Role of bacterial capsule in local and systemic inflammatory responses of mice during pulmonary infection with Klebsiella pneumoniae. J Med Microbiol 2000; 49(11):1003-10; PMID:11073154; http://dx.doi.org/ 10.1099/0022-1317-49-11-1003 (accessed April24, 2017). [DOI] [PubMed] [Google Scholar]
- [41].Lawlor MS, Hsu J, Rick PD, Miller VL. Identification of Klebsiella pneumoniae virulence determinants using an intranasal infection model. Mol Microbiol 2005; 58(4):1054-73; PMID:16262790; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04918.x [DOI] [PubMed] [Google Scholar]
- [42].Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998; 11(4):589-603; PMID:9767057 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=88898&tool=pmcentrez&rendertype=abstract (accessed September 7, 2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rahn A, Drummelsmith J, Whitfield C. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J Bacteriol 1999; 181(7):2307-13; PMID:10094716 http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=93651&tool=pmcentrez&rendertype=abstract (accessed March28, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Arakawa Y, Wacharotayankun R, Nagatsuka T, Ito H, Kato N, Ohta M. Genomic organization of the Klebsiella pneumoniae cps region responsible for serotype K2 capsular polysaccharide synthesis in the virulent strain Chedid. J Bacteriol 1995; 177(7):1788-96; PMID:7896702; http://dx.doi.org/ 10.1128/jb.177.7.1788-1796.1995 (accessed March28, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pan YJ, Fang HC, Yang HC, Lin TL, Hsieh PF, Tsai FC, Keynan Y, Wang JT. Capsular polysaccharide synthesis regions in Klebsiella pneumoniae serotype K57 and a new capsular serotype. J Clin Microbiol 2008; 46(7):2231-40; PMID:18508935; http://dx.doi.org/ 10.1128/JCM.01716-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fang CT. A novel virulence gene in klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199(5):697-705; PMID:14993253; http://dx.doi.org/ 10.1084/jem.20030857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hunt JJ, Wang JT, Callegan MC. Contribution of mucoviscosity-associated gene A (magA) to virulence in experimental Klebsiella pneumoniae endophthalmitis. Invest Ophthalmol Vis Sci 2011; 52(9):6860-6; PMID:21791595; http://dx.doi.org/ 10.1167/iovs.11-7798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL. The function of wzy_K1 (magA), the serotype K1 polymerase gene in Klebsiella pneumoniae cps gene cluster. J Infect Dis 2010; 201(8):1268-9; PMID:20225957; http://dx.doi.org/ 10.1086/652183 [DOI] [PubMed] [Google Scholar]
- [49].Fang CT, Chuang YP, Shun CT, Chang SC, Wang JT. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med 2004; 199(5):697-705; PMID:14993253; http://dx.doi.org/ 10.1084/jem.20030857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yeh KM, Lin JC, Yin FY, Fung CP, Hung HC, Siu LK, Chang FY. Revisiting the importance of virulence determinant magA and its surrounding genes in Klebsiella pneumoniae causing pyogenic liver abscesses: exact role in serotype K1 capsule formation. J Infect Dis 2010; 201(8):1259-67; PMID:19785524; http://dx.doi.org/ 10.1086/606010 [DOI] [PubMed] [Google Scholar]
- [51].Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, Wang JT. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One 2013; 8(12):e80670; PMID:24349011; http://dx.doi.org/ 10.1371/journal.pone.0080670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yu WL, Fung CP, Ko WC, Cheng KC, Lee CC, Chuang YC. Polymerase chain reaction analysis for detecting capsule serotypes K1 and K2 of Klebsiella pneumoniae causing abscesses of the liver and other sites. J Infect Dis 2007; 195(8):1235-6; author reply 1236; PMID:17357063; http://dx.doi.org/ 10.1086/512686 [DOI] [PubMed] [Google Scholar]
- [53].Turton JF, Baklan H, Siu LK, Kaufmann ME, Pitt TL. Evaluation of a multiplex PCR for detection of serotypes K1, K2 and K5 in Klebsiella sp. and comparison of isolates within these serotypes. FEMS Microbiol Lett 2008; 284(2):247-52; PMID:18507682; http://dx.doi.org/ 10.1111/j.1574-6968.2008.01208.x [DOI] [PubMed] [Google Scholar]
- [54].Liu YM, Li B Bin, Zhang YY, Zhang W, Shen H, Li H, Cao B. Clinical and molecular characteristics of emerging hypervirulent Klebsiella pneumoniae bloodstream infections in mainland China. Antimicrob Agents Chemother 2014; 58(9):5379-85; PMID:24982067; http://dx.doi.org/ 10.1128/AAC.02523-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in E. coli. MolMicrobiol 1999; 31:1307-19; PMID:10200953 [DOI] [PubMed] [Google Scholar]
- [56].Cheng HY, Chen YS, Wu CY, Chang HY, Lai YC, Peng HL. RmpA regulation of capsular polysaccharide biosynthesis in Klebsiella pneumoniae CG43. J Bacteriol 2010; 192(12):3144-58; PMID:20382770; http://dx.doi.org/ 10.1128/JB.00031-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nassif X, Honoré N, Vasselon T, Cole ST, Sansonetti PJ. Positive control of colanic acid synthesis in Escherichia coli by rmpA and rmpB, two virulence-plasmid genes of Klebsiella pneumoniae. Mol Microbiol 1989; 3(10):1349-59. papers2://publication/uuid/35508EF8-6675-410C-982B-91213BA6567B; PMID:2693894; http://dx.doi.org/ 10.1111/j.1365-2958.1989.tb00116.x [DOI] [PubMed] [Google Scholar]
- [58].Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 2011; 157(Pt 12):3446-57; PMID:21964731; http://dx.doi.org/ 10.1099/mic.0.050336-0 [DOI] [PubMed] [Google Scholar]
- [59].Hsu CR, Lin TL, Chen YC, Chou HC, Wang JT. The role of Klebsiella pneumoniae rmpA in capsular polysaccharide synthesis and virulence revisited. Microbiology 2011; 157(12):3446-57; PMID:21964731; http://dx.doi.org/ 10.1099/mic.0.050336-0 [DOI] [PubMed] [Google Scholar]
- [60].Lai YC, Peng HL, Chang HY. RmpA2, an activator of capsule biosynthesis in Klebsiella pneumoniae CG43, regulates K2 cps gene expression at the transcriptional level. J Bacteriol 2003; 185(3):788-800; PMID:12533454; http://dx.doi.org/ 10.1128/JB.185.3.788-800.2003 (accessed March28, 2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene 2004; 337:189-98; PMID:15276215; http://dx.doi.org/ 10.1016/j.gene.2004.05.008 [DOI] [PubMed] [Google Scholar]
- [62].Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis 2006; 42(10):1351-8; PMID:16619144; http://dx.doi.org/ 10.1086/503420 [DOI] [PubMed] [Google Scholar]
- [63].Yu WL, Lee MF, Tang HJ, Chang MC, Chuang YC. Low prevalence of rmpA and high tendency of rmpA mutation correspond to low virulence of extended spectrum β-lactamase-producing klebsiella pneumoniae isolates. Virulence 2015; 6(2):162-72; PMID:25830726; http://dx.doi.org/ 10.1080/21505594.2015.1016703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lee CH, Chang CC, Liu JW, Chen RF, Yang KD. Sialic acid involved in hypermucoviscosity phenotype of Klebsiella pneumoniae and associated with resistance to neutrophil phagocytosis. Virulence 2014; 5(6):673-9; PMID:25098744; http://dx.doi.org/ 10.4161/viru.32076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Russo TA, Olson R, MacDonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. Aerobactin mediates virulence and accounts for increased siderophore production under iron-limiting conditions by hypervirulent (hypermucoviscous) Klebsiella pneumoniae. Infect Immun 2014; 82(6):2356-67; PMID:24664504; http://dx.doi.org/ 10.1128/IAI.01667-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but Not Yersiniabactin, Salmochelin, or Enterobactin, Enables the Growth/Survival of Hypervirulent (Hypermucoviscous) Klebsiella pneumoniae Ex Vivo and In Vivo. Infect Immun 2015; 83(8):3325-33; PMID:26056379; http://dx.doi.org/ 10.1128/IAI.00430-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, et al.. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci 2015; 112(27):E3574-81; PMID:26100894; http://dx.doi.org/ 10.1073/pnas.1501049112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Lery LM, Frangeul L, Tomas A, Passet V, Almeida AS, Bialek-Davenet S, Barbe V, Bengoechea JA, Sansonetti P, Brisse S, et al.. Comparative analysis of Klebsiella pneumoniae genomes identifies a phospholipase D family protein as a novel virulence factor. BMC Biol 2014; 12(1):41; PMID:24885329; http://dx.doi.org/ 10.1186/1741-7007-12-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Newton HJ, Sansom FM, Dao J, McAlister AD, Sloan J, Cianciotto NP, Hartland EL. Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect Immun 2007; 75(12):5575-85; PMID:17893138; http://dx.doi.org/ 10.1128/IAI.00443-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Srinivasan VB, Venkataramaiah M, Mondal A, Vaidyanathan V, Govil T, Rajamohan G. Functional characterization of a novel outer membrane porin KpnO, regulated by PhoBR two-component system in Klebsiella pneumoniae NTUH-K2044. PLoS One 2012; 7(7):e41505; PMID:22848515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, Delannoy-Vieillard AS, Garin B, Le Hello S, Arlet G, Nicolas-Chanoine MH, et al.. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis 2014; 20(11):1812-20; PMID:25341126; http://dx.doi.org/ 10.3201/eid2011.140206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Passet V, Brisse S. Association of Tellurite Resistance with Hypervirulent Clonal Groups of Klebsiella pneumoniae: TABLE 1. J Clin Microbiol 2015; 53(4):1380-2; PMID:25631812; http://dx.doi.org/ 10.1128/JCM.03053-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect 2014; 20(11):O818-24; PMID:24804560 [DOI] [PubMed] [Google Scholar]
- [74].Brisse S, Fevre C, Passet V, Issenhuth-Jeanjean S, Tournebize R, Diancourt L, Grimont P. Virulent clones of Klebsiella pneumoniae: identification and evolutionary scenario based on genomic and phenotypic characterization. PLoS One 2009; 4(3):e4982; PMID:19319196; http://dx.doi.org/ 10.1371/journal.pone.0004982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Togawa A, Toh H, Onozawa K, Yoshimura M, Tokushige C, Shimono N, Takata T, Tamura K. Influence of the bacterial phenotypes on the clinical manifestations in Klebsiella pneumoniae bacteremia patients: A retrospective cohort study. J Infect Chemother 2015; 21(7):531-537; PMID:26002138; http://dx.doi.org/ 10.1016/j.jiac.2015.04.004 [DOI] [PubMed] [Google Scholar]
- [76].Wu H, Li D, Zhou H, Sun Y, Guo L, Shen D. Bacteremia and other body site infection caused by hypervirulent and classic Klebsiella pneumoniae. Microb Pathog 2017; 104:254-62; PMID:28132768; http://dx.doi.org/ 10.1016/j.micpath.2017.01.049 [DOI] [PubMed] [Google Scholar]
- [77].Garza-Ramos U, Silva-Sánchez J, Catalán-Nájera J, Barrios H, Rodríguez-Medina N, Garza-González E, Cevallos MA, Lozano L. Draft Genome Sequence of a Hypermucoviscous Extended-Spectrum- beta-Lactamase-Producing Klebsiella quasipneumoniae subsp. similipneumoniae Clinical Isolate. Genome Announc 2016; 4(4):e00475-16; PMID:27389261; http://dx.doi.org/ 10.1128/genomeA.00475-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Arena F, Henrici De Angelis L, Pieralli F, Di Pilato V, Giani T, Torricelli F, D'Andrea MM, Rossolini GM. Draft Genome Sequence of the First Hypermucoviscous Klebsiella quasipneumoniae subsp. quasipneumoniae Isolate from a Bloodstream. Genome Announc. 2015; 3(5):e00952-15. http://dx.doi.org/ 10.1128/genomeA.00952-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Breurec S, Melot B, Hoen B, Passet V, Schepers K, Bastian S, Brisse S.. Liver Abscess Caused by Infection with Community-Acquired Klebsiella quasipneumoniae subsp. quasipneumoniae. Emerg Infect Dis. 2016; 22(3):529-31. http://dx.doi.org/ 10.3201/eid2203.151466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hæggman S, Löfdahl S, Paauw a, Verhoef J, Brisse S, Lo S. Diversity and evolution of the class A chromosomal beta-lactamase gene in klebsiella pneumoniae diversity and evolution of the class A chromosomal beta-lactamase gene in klebsiella pneumoniae. Antimicrob Agents Chemother 2004; 48(7):2400-8; PMID:15215087; http://dx.doi.org/ 10.1128/AAC.48.7.2400-2408.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Brisse S, Passet V, Grimont PA. Description of Klebsiella quasipneumoniae sp. nov., a novel species isolated from human infections, with two subspecies Klebsiella quasipneumoniae subsp. quasipneumoniae subsp. nov. and Klebsiella quasipneumoniae subsp. similipneumoniae subsp. nov., and d. Int J Syst Evol Microbiol 2014; 64(Pt9):3146-52. http://dx.doi.org/ 10.1099/ijs.0.062737-0 [DOI] [PubMed] [Google Scholar]
- [82].Rosenblueth M, Martínez L, Silva J, Martínez-Romero E. Klebsiella variicola, a novel species with clinical and plant-associated isolates. Syst Appl Microbiol 2004; 27(1):27-35; PMID:15053318; http://dx.doi.org/ 10.1078/0723-2020-00261 [DOI] [PubMed] [Google Scholar]
- [83].Pinto-Tomas AA, Anderson MA, Suen G, Stevenson DM, Chu FS, Cleland WW, Weimer PJ, Currie CR. Symbiotic nitrogen fixation in the fungus gardens of leaf-cutting ants. Science 2009; 326(2009):1120-3; PMID:19965433; http://dx.doi.org/ 10.1126/science.1173036 [DOI] [PubMed] [Google Scholar]
- [84].Garza-Ramos U, Moreno-Dominguez S, Hernández-Castro R, Silva-Sanchez J, Barrios H, Reyna-Flores F, Sanchez-Perez A, Carrillo-Casas EM, Sanchez-León MC, Moncada-Barron D. Identification and characterization of imipenem-resistant Klebsiella pneumoniae and susceptible Klebsiella variicola isolates obtained from the same patient. Microb Drug Resist 2015; 22(3):179-84. 0(0):mdr.2015.0181; PMID:26571390 [DOI] [PubMed] [Google Scholar]
- [85].Maatallah M, Vading M, Kabir MH, Bakhrouf A, Kalin M, Nauclér P, Brisse S, Giske CG. Klebsiella variicola is a frequent cause of bloodstream infection in the Stockholm area, and associated with higher mortality compared to K. pneumoniae. PLoS One 2014; 9(11):e113539; PMID:25426853; http://dx.doi.org/ 10.1371/journal.pone.0113539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Garza-Ramos U, Silva-Sanchez J, Barrios H, Rodriguez-Medina N, Martínez-Barnetche J, Andrade V. Draft Genome Sequence of the First Hypermucoviscous Klebsiella variicola Clinical Isolate. Genome Announc 2015; 3(2):e01352-14; PMID:25858850; http://dx.doi.org/ 10.1128/genomeA.01352-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Bottone EJ. Hypermucoviscous phenotype expressed by an isolate of uropathogenic escherichia coli: An overlooked and underappreciated virulence factor. Clin Microbiol Newsl 2010; 32(11):81-5; http://dx.doi.org/ 10.1016/j.clinmicnews.2010.05.001 [DOI] [Google Scholar]


