ABSTRACT
Planarians, which are non-parasitic flatworms, are highly resistant to bacterial infections. To better understand the mechanisms underlying this resistance, we investigated the role of the circadian machinery in the anti-bacterial response of the freshwater planarian Schmidtea mediterranea. We identified Smed-Tim from S. mediterranea as a homolog of the mammalian clock gene Tim. We showed via RNA interference that Smed-Tim is required for the anti-microbial activities of Schmidtea mediterranea against Staphylococcus aureus infection during the light/dark cycle. Indeed, S. aureus infection leads to the expression of Smed-Tim, which in turn promotes Smed-Traf6 and Smed-morn2, but not Smed-p38 MAPK expression, 2 master regulators of planarian anti-microbial responses.
KEYWORDS: anti-bacterial response, MORN2, planarians;, S. aureus, Tim
Introduction
Non-vertebrates such as fruit flies and nematodes have provided insight into the conserved mechanisms in host-pathogen interactions. The immortal Plathyhelminthes planaria, which is known for its ability to regenerate any part of its body,1 can be a useful tool to identify and characterize evolutionarily conserved anti-bacterial mechanisms due to its ability to eliminate a large spectrum of human pathogens.2 However, the mechanisms that govern planarian immunity remain largely unknown and require investigation.
To better understand the anti-microbial capacity of freshwater planarians, we investigated the contribution of the genes from the internal time keeping system, the circadian clock, to this process. Planarians, similar to numerous other organisms, have a circadian clock.3,4 It has been suggested that genes from the circadian cycle might play a role in the modulation of antibacterial immunity of non-vertebrates and vertebrates.5-7 Indeed, mice with Aryl Hydrocarbon Receptor Nuclear Translocator-Like (Arntl)-1 knockdown are more susceptible to Listeria monocytogenes infection8 than wild type mice are. In non-vertebrates such as Drosophila melanogaster, deletion of the clock gene Period-2 (Per)-2 causes an increase in susceptibility to Pseudomonas aeruginosa, Streptococcus pneumonia, and L. monocytogenes infections, whereas deletion of the clock genes Circadian Locomotor Output Cycles Kaput (Clock), Cycle (Cyc, a homolog of Hs-Arntl-1), and, although discussed, Timeless (Tim) could causes an increase in resistance to Pseudomonas aeruginosa.6,9
Based on these findings, we sought to determine the role of the circadian clock genes Arntl-1, Tim, Per-2, and Clock in the ability of the freshwater planarian Schmidtea mediterranea to cope with bacterial infections. In this report, we identified Schmidtea mediterranea (Smed)-TIM, the planarian homolog of Homo sapiens (Hs)-TIM, as a component of the Clock machinery of planarians, required for anti-microbial activities involving Smed-traf6 and Smed-morn2 in the planarian against Staphylococcus aureus during the light/dark cycle but not during the D/D cycle.
Results
First, we have searched for homologs of the Homo sapiens (Hs)-Arntl-1, Hs-Tim, Hs-Per-2, and Hs-Clock genes in the S. mediterranea genome using the S. mediterranea transcriptome database PlanMine (http://planmine.mpi-cbg.de/planmine/begin.do)10 and the S. mediterranea genome database SmedGD (http://smedgd.neuro.utah.edu/).11 We were unable to find sequences with significant homologies to Hs-Clock and Hs-Per-2 in either database, which suggests that Clock and Per-2 are absent from S. mediterranea. In contrast, TBLASTN analysis provided 3 sequences (lcl|dd_Smed_v6_9171_0_1, e-value 4e−77; lcl|dd_Smed_v6_17731_0_1, e-value 4e−28; lcl|dd_Smed_v6_17537_0_1, e-value 4e−22) with significant alignment to Hs-ARNTL-1 (NP_001025443.1) and 2 sequences (lcl|dd_Smed_v6_19257_0_2, e-value 2e−82; lcl|dd_Smed_v6_17957_0_1, e-value 9e−71) with significant alignment to Hs-TIM (NP_003911.2). Using FGENESH+ (http://www.softberry.com/), we predicted the Schmidtea mediterranea gene Smed-Arntl-1 as being homologous to Hs-Arntl-1 and Smed-Tim as being homologous to Hs-Tim. Sequence conservation analysis with BLASTx and PRALINE12 showed a 52% similarity at the protein level (68% coverage, e-value 1e−142) for the predicted Smed-ARNTL-1 and Hs-ARNTL-1 (Fig. 1A) and a 31% similarity at the protein level (99% coverage, e-value 4e−72) for the predicted Smed-TIM and Hs-TIM (Fig. 1B). The use of cNLS Mapper software (http://nls-mapper.iab.keio.ac.jp/) identify the presence of a nuclear localization sequence (SRTKRRKSI) at the position 519, in the amino acid sequence of Smed-TIM (Fig. S1).
Figure 1.
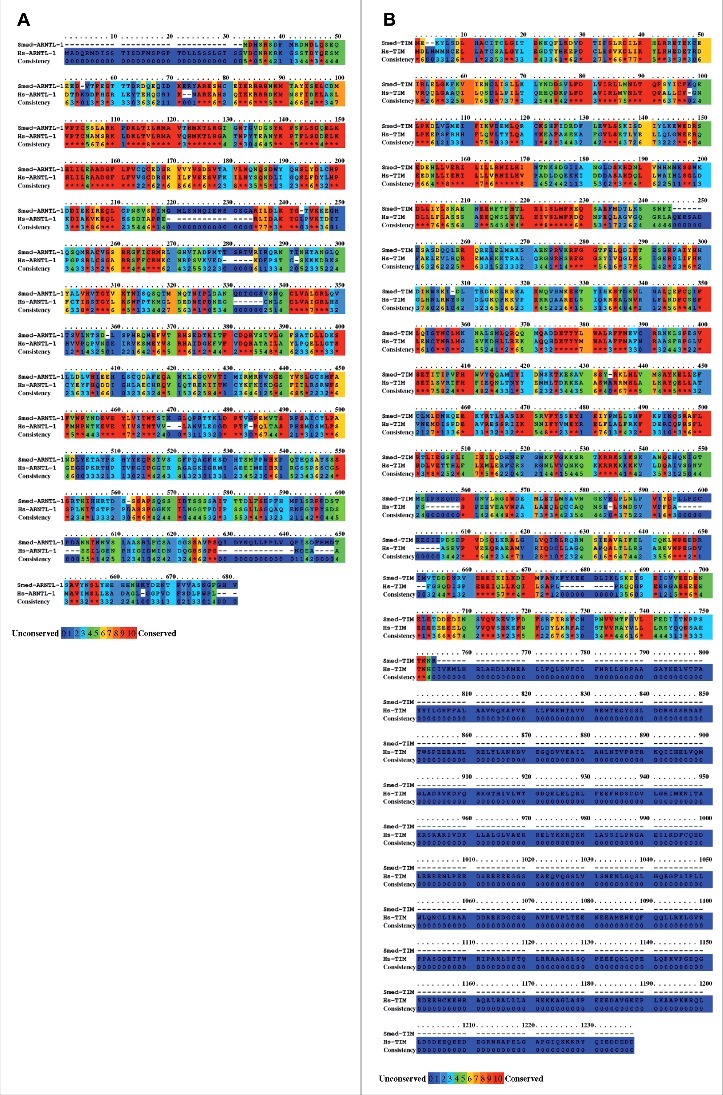
Predicted amino acid sequence of Smed-ARNTl-1 and Smed-TIM. Alignment of (A) Smed-ARNTL-1 with Hs-ARNTL-1 (NP_001025443.1), and (B) Smed-TIM with Hs-TIM (NP_003911.2) were performed using the PRALINE multiple sequence alignment (Center for Integrative Bioinfomatics, http://www.ibi.vu.nl/programs/pralinewww/) setup with default parameters. The results are color-coded for amino acid conservation and the scoring scheme works from 0, for the least conserved alignment position, to 10, for the most conserved alignment position.
Second, we have investigated if Smed-Tim and Smed-Arntl-1 played a role in the anti-microbial response of the planarian S. mediterranea. Animals were subjected to either a light/dark (L/D) cycle (12/12 hours) or to a dark/dark (D/D) cycle (12/12 hours) and were then genetically deprived of Smed-Arntl-1 and Smed-Tim function via RNA interference. Planarians were then exposed to S. aureus by feeding. By the direct measurement of colony-forming units (CFUs), we evaluated bacterial clearance at 3, 6, and 9 d post-feeding. We observed that animals silenced for Smed-Arntl-1, or Smed-Tim (Fig. S2A) did not show any particular phenotype (Fig. 2A), and their viability was not affected (Fig. S2B). To note, under the light/light (L/L) cycle (12/12 hours), animals did not eat enough to be sufficiently infected by S. aureus compared with the L/D and D/D conditions to proceed for experiments. After feeding there was 1.32 × 106 less bacteria in worms under the L/L condition compared with animals under the L/D and the D/D condition (Fig. S2C). Consistent with previous studies, we found that the control S. mediterranea eGFP (RNAi) worms had the ability to resolve S. aureus infection in 6 d,2 independently of the light cycle (Fig. 2B). Worms that were subjected to the D/D cycle and knocked down for Smed-Arntl-1 and Smed-Tim eliminated S. aureus in 6 days, similarly to the control eGFP (RNAi). Under the L/D condition, planarians silenced for Smed-Arntl-1, eliminated S. aureus in 6 d (Fig. 2C). In worms silenced for Smed-Tim expression under the L/D condition, we found that the elimination of S. aureus was significantly delayed because it took 9 d instead of 6 d to observe the complete elimination of S. aureus (Fig. 2C). Indeed, at 3 d post-infection 3.5 × 10e4 ± 2.2 × 10e4 S. aureus CFU/animal were detected in eGFP (RNAi) control worms and 1.9 × 10e6 ± 1.3 × 10e6 S. aureus CFU/animal in Smed-Tim (RNAi) planarians. After 6 d of infection, bacteria were not detected in the control worms (eGFP RNAi), in contrast to the Smed-Tim (RNAi) planarians (2.3 × 10e2 ± 1.2 × 10e2 CFU/animal). At day 9, bacteria were not detected in S. mediterranea that had been silenced for Smed-Tim expression (Fig. 2C). To note, silencing Smed-Tim does not significantly affect the capacity of worms to ingest bacteria (1.9 × 10e7 ± 2.2 × 10e7 CFU/animal) compared with the control (1.3 × 10e7 ± 1.2 × 10e7 CFU/animal) at day 0, respectively (Fig. 2D). Thus, the delay in the elimination of S. aureus that was observed in Smed-Tim (RNAi) knockdown worms cannot be explained by an alteration in the ability to feed. Taken together, these data suggest that planarians living in the L/D condition require the expression of Smed-Tim to have an efficient antibacterial immune response. For the rest of the study, we focus on Smed-Tim because in contrast to Smed-Arntl-1, it plays a role in the anti-microbial response of planarians against S. aureus.
Figure 2.
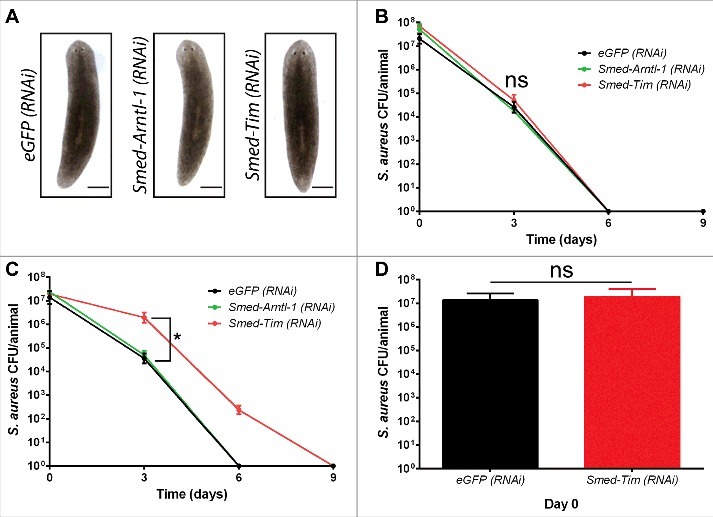
Smed-TIM silencing leads to a delay in S. aureus elimination by the planarians. (A) S. mediterranea were silenced for eGFP (control), Smed-Arntl-1, and Smed-Tim. Silenced animals do not present a particular phenotype (representative micrographs of 2 experiments; 10 animals per experimental conditions; scale bars, 150 mm). The efficiency of the gene silencing by RNAi in the S. mediterranea was determined using RT-qPCR (Figure S2A). (B, C) The S. mediterranea eGFP (RNAi), the S. mediterranea Smed-Arntl-1 (RNAi), and S. mediterranea Smed-Tim (RNAi) under D/D cycle (B) or under L/D cycle (C) were fed with S. aureus (1 × 10e9 bacteria), and the CFUs per animal were counted over time. In the L/D condition, the silencing of Smed-Tim via RNAi delayed the clearance of S. aureus by S. mediterranea. The results are expressed as the mean ± SD (10 animals per time point, number of experiments = 3, *p < 0.05). (D) The amount of bacteria ingested by the S. mediterranea eGFP (RNAi) and the S. mediterranea Smed-Tim (RNAi) under L/D cycle, after feeding (Time 0 day) was determined by counting the CFUs. The invalidation of Smed-Tim does not affect significantly the capacity of worms to ingest bacteria compared with S. mediterranea eGFP (RNAi). The results are expressed as the mean ± SD (10 animals per time point, number of experiments = 3).
Third, we wondered if Smed-Tim is a component of the planarians' circadian clock. To this purpose, we assessed the cycle of Smed-Tim expression under both conditions L/D and D/D condition (Fig. 3A and Fig. 3B). Under the L/D condition, we observed a Smed-Tim expression cycle (Fig. 3A). The increase in Smed-Tim expression is observed in the dark phase of the L/D condition between Circadian Time (CT) 12 and CT 24, and CT 36 and CT 48, with a maximum fold change of 1.20 ± 0.33 at CT 20 and 0.96 ± 0.16 at CT 44 (Fig. 3B). In contrast, under D/D condition, we observed a loss of synchronisation which lead to a phase shift called free run (Fig. 3B).13 Interestingly, worms infected and submitted to L/D cycle had a level of Smed-Tim mRNA which showed a 2.3-fold increase at CT 20 (Fig. 3C). Then, we visualized the expression of Smed-Tim mRNA by in situ hybridization (ISH) in planarians infected or not by S. aureus. In the planarian tissues, we observed that Smed-Tim is weakly expressed (Fig. 3D and Fig. S3) as in several other organisms.14-17 Next, we investigated the level of production of the melatonin in planarians, a hallmark of circadian rhythm functionality.18 As described previously3 in planarians under L/D condition, we observed an increase in the production of melatonin at CT 18 (Fig. 3E), in the same concentration range than previously observed.3 In animals silenced for Smed-Tim we find that the melatonin level showed a significant 3-fold increase between CT 14 and 22 compared with the control animals eGFP (RNAi) (Fig. 3E). Taken together, these data suggest that Smed-Tim might play a role in the circadian clock machinery regulation, and thus we can hypothesize that Smed-Tim is a component of the circadian clock of planarians.
Figure 3.
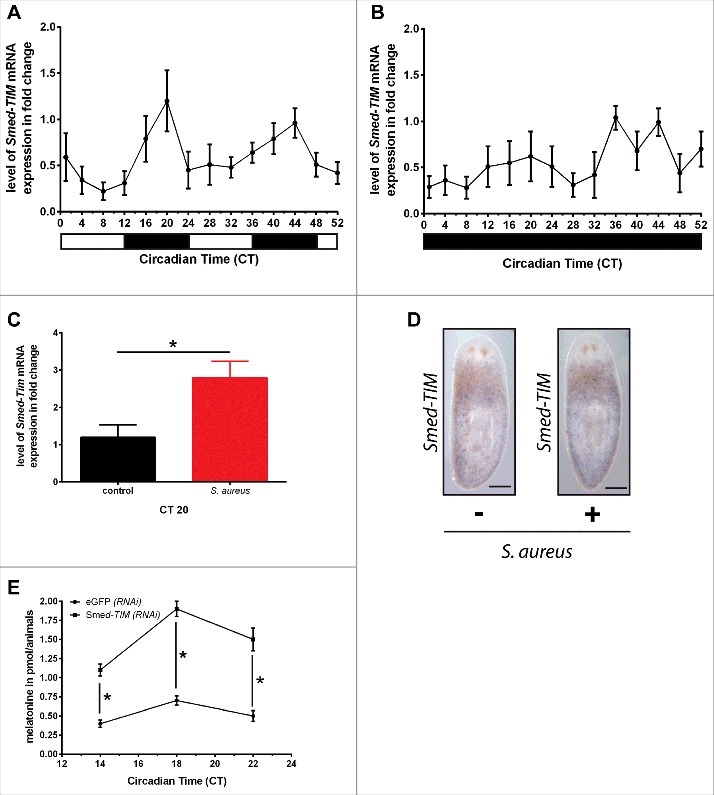
Smed-Tim is a gene of the planarians circadian clock. (A) Planarians were submitted to the condition L/D and then Smed-Tim expression was evaluated using RT-qPCR. Planarians have a maximum of Tim expression in dark phase. The results are presented as the mean ± SD (3 worms per time point processed individually in triplicate, number of experiments = 3). (B) Planarians were submitted to D/D cycle and Smed-Tim expression was evaluated using RT-qPCR. Worms submitted to D/D condition have a desynchronized Smed-Tim expression cycle. The results are presented as the mean ± SD (3 worms per time point processed individually in triplicate, number of experiments = 3). (C) The expression of Smed-Tim was evaluated at CT20 in worms submitted to L/D condition and infected by S. aureus. (D) Whole-mount in situ hybridization analysis of the expression of Smed-Tim in control and S. aureus infected animals were fixed at CT 20 (representative micrographs of 2 experiments; 5 animals per experimental conditions; scale bars, 250 µm). See also Figure S3. (E) Melatonin level was determined between CT14 and CT22 in animals eGFP (RNAi) and Smed-Tim (RNAi) under the L/D cycle level was assessed in planarian controls or silenced for Smed-Tim via RNAi. The results are presented as the mean ± SD (30 worms per time point, number of experiments = 2, *p < 0.05).
Finally, we analyze whether Smed-Tim silencing affects the expression of the planarians' antimicrobial gene responses such as p38 MAP-Kinase and Traf6, that transduces signals of pathogen perception,19,20 and morn2, that controls the LC3-associated phagocytosis of bacteria for their destruction.2 The expression of Smed-Morn2 (Fig. 4A), Smed-Traf6 (Fig. 4B) and Smed-p38 MAPK (Fig. 4C) mRNA is significantly induced in worms from 6 hours to 36 hours following S. aureus infection, with a maximum fold change of around 4 to 12 hours post infection. The silencing of Smed-Tim diminished the mRNA expression of Smed-morn2 by 69% (Fig. 4D) and Smed-Traf6 by 75% (Fig. 4E) compared with the control animals infected by S. aureus and maintained in L/D cycle. In contrast, p38 MAP-kinase mRNA expression is not chnaged (Fig. 4F). To note, in planarians under D/D condition and infected by S. aureus, the silencing of Smed-Tim does not affect the expression of Smed-morn2 (Figure. S4A), Smed-Traf6 (Figure. S4B) and Smed-p38 MAP-kinase mRNA (Figure. S4C). Taken together, our data suggest that Smed-Tim regulates the capacity of planarians to kill S. aureus by modulating the antimicrobial genes such as Smed-Traf6 and Smed-morn2.
Figure 4.
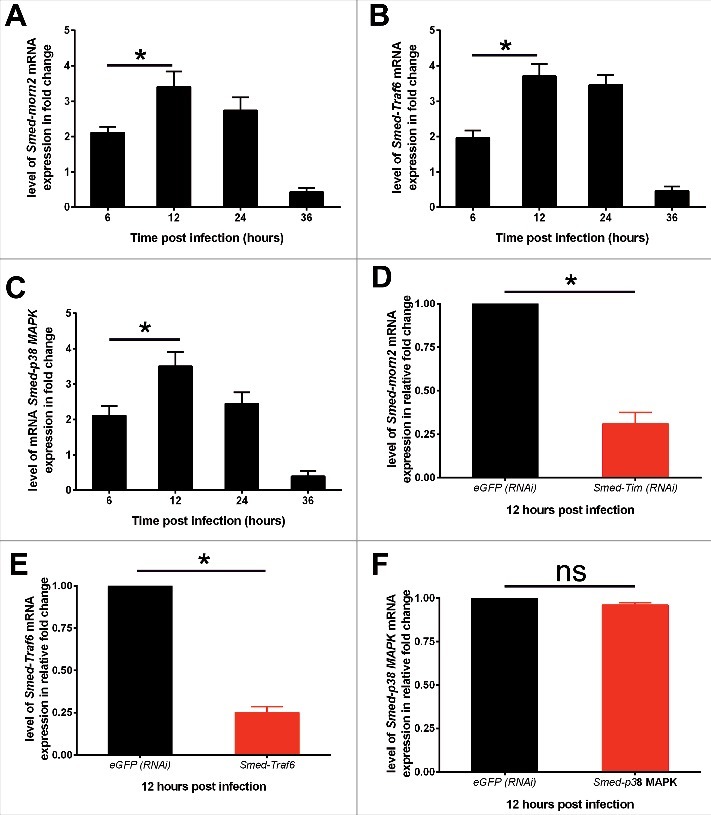
Smed-Tim controls the expression of Smed-morn2 and Smed-Traf6. (A, B, C) The level of expression of (A) Smed-morn2, (B) Smed-Traf6, and (C) Smed-p38 MAPK mRNA in planarians under L/D condition and challenged with S. aureus was assessed by RT-qPCR. The results are presented as the mean ± SD (3 worms per time point processed individually in triplicate, number of experiments = 3, *p < 0.05). (D, E, F) Planarians under L/D condition were silenced for Smed-Tim via RNAi, then infected with S. aureus, and the level of mRNA expression of Smed-morn2 (D), Smed-Traf6 (E), and Smed-p38 MAPK (F) was evaluated using RT-qPCR. The expression of Smed-Morn2 and Smed-Traf6 was significantly diminished in animals which have been silenced for Smed-Tim under L/D cycle. The results are presented as the mean ± SD (3 animals per time point processed individually in triplicate, number of experiments = 3, *p < 0.05).
Discussion
In this work, we have analyzed the contribution of the circadian clock component in planarian antimicrobial activity. We have identified in planarians the homologs of human Tim and Arntl-1, however we haven't identified homologs for Clock and Per gene. The molecular components of the planarian clock machinery may differ from those in evolved organisms such as mammals. Indeed, as previously shown in sponges, Clock, which is the main component of the circadian machinery in bilaterians, is absent from the Schmidtea mediterranea genome and transcriptome databases. Per-2 is also absent from these databases and is not found in the starlet sea anemone Nematostella vectensis, though it has a functional circadian clock that remains driven by light cues.21 It is possible to hypothesize that Clock and Per-2 genes were lost from the planarian lineage. Nevertheless, the presence of 2 others genes, Arntl-1 and Tim, suggests the existence of a circadian clock machinery in planarians. This is reinforced by the capacity of planarians to produce melatonin (3 and in this study). We have investigated the implication of these 2 genes in the anti-microbial response of planarians against S. aureus. In contrast to Smed-Arntl-1, we observed that Smed-Tim played a role in the anti-bacterial activities of planarians, since it silencing via RNAi delayed S. aureus elimination. The silencing of Smed-Arntl-1 did not induce the death of infected worms. In other model organisms, the knock down of Arntl-1 changed significantly the survival rate of these models during bacterial infections. Indeed, in mice, silencing Arntl-1 prevented a decrease in the survival rate after infection with L. monocytogenes.8 Arntl-1 (−/−) fruit flies were more resistant to Pseudomonas aeruginosa.6 Therefore, the involvement of Arntl-1 during the response to bacteria could be a function of either the model organism or the bacterial species.
TIM is a key component of the circadian clock in Drosophila, but whether the mouse homolog is involved in the circadian rhythm remains controversial.22 Moreover, numerous non-clock functions for timeless have been described.23,24 TIM has been reported to play a central role in the control of D. melanogaster susceptibility to bacterial infection.24 Indeed, TIM is required for D. melanogaster to resist Streptococcus pneumoniae and L. monocytogenes infections.9 A contradictory study has shown that TIM expression is deleterious when D. melanogaster is infected with P. aeruginosa.6 Here we unravel the idea that TIM is involved in the antibacterial potential of the flatworm under the L/D condition. The silencing of Smed-Tim delayed bacterial elimination, but the planarians remained able to eliminate bacteria. Thus, silencing Smed-Tim is not deleterious for planarians that are infected with S. aureus. We cannot exclude the possibility that Smed-Tim silencing could be deleterious or does not exert any action on planarians infected with pathogens other than S. aureus because of the pathogen-dependent actions of Tim.24 Interestingly, several pieces of our data suggest that Smed-Tim is might be a component of the circadian cycle of planarians. Indeed, we have observed that in L/D condition there is a cycle of Smed-Tim expression, and that Smed-Tim has a maximum expression in the dark phase compared with what is observed in anopheles species and drosophila.24,25
The production of melatonin has been defined as a hallmark of circadian rhythm functionality, and a perturbation of the circadian clock machinery lead to a dysregulation of the melatonin production.18 We do not observe an inhibition of melatonin production in animals silenced for Smed-TIM as expected. However, we find a deregulation of the melatonin production in animals silenced for Smed-TIM, characterized by an increase in the level of melatonin production in planarians, allowing us to hypothesize a potential link between Smed-Tim and circadian clock in planarians. Finally, we analyze whether Smed-Tim silencing regulates the expression of the antimicrobial genes of planarians, such as p38 MAP-kinase and Traf6, that transduces signals of pathogen perception,19,20 and morn2, that controls the LC3-associated phagocytosis of bacteria for their destruction.2 Importantly, we characterized the critical involvement of Smed-Tim in the induction of Smed-Traf6 and Smed-morn2 mRNA expression in response to S. aureus in the L/D condition. Interestingly, in the D/D condition; condition in which Smed-Tim cycle is in free run, the planarians' antimicrobial genes tested here are not regulated by Smed-Tim. This underscores the importance of a 12/12 Light and dark cycle in the anti-microbial capacity of planarians and that Smed-TIM is in upstream of Smed-TRAF6 and Smed-MORN2.
In conclusion, here we identified the planarian homolog to Hs-Tim and revealed that Smed-TIM is a gene of the circadian cycle of planarians, and that Smed-Tim regulates the expression of downstream antimicrobial genes such as Smed-morn2 and Smed-Traf6 expression during S. aureus infection.
Methods
Materials and methods
Planarians
Planarians belonging to the species Schmidtea mediterranea (ClW4) were used. Planarians were maintained in static culture as described previously26 in autoclaved water at 20°C and fed once per week with calf liver. Animals were starved for at least 1 week before the experiments. Water was changed every 2 days, and planarians were maintained without antibiotics. Before experiments, worms were subjected to the L/D cycles (12/12 h) for 4 weeks.
Bacteria
Staphylococcus aureus (ATCC25923) was grown on blood agar plates (BioMerieux SA) at 37°C.
Worm feeding with bacteria
S. mediterranea were fed with S. aureus using a protocol adapted from a dsRNA feeding method27 as described previously.2 Briefly, S. aureus (1 × 10e9) were suspended in homogenized liver, mixed with ultra-low-gelling-temperature agar and red food coloring, and allowed to solidify on ice. Room-temperature, solidified food was fed to planarians. After 2 hr (defined as day 0) of feeding, the planarians were extensively washed.
CFU counting
As described previously,2 S. mediterranea were collected and homogenized in PBS. The lysate was passed 5 times through a sterile syringe with a 29G needle to disrupt planarian tissue clumps, and CFUs were counted after plating 10 µl of serial dilutions onto blood agar plates (BioMerieux SA).
Gene prediction
To identify a planarian homolog of Hs-ARNTL-1 (NP_001025443.1, NM_001030272.2), Hs-Per-2 (NP_073728, NM_022817.2), Hs-TIM (NP_003911.2, NM_003920.3), and Hs-CLOCK (NP_001254772.1, NM_001267843.1) we examined, via TBLASTN (e-value 1e−5), the S. mediterranea transcriptome database PlanMine (http://planmine.mpi-cbg.de/planmine/begin.do)10 and the S. mediterranea genome database SmedGD (http://smedgdkc03.stowers.org/).11 We identified sequences producing a significant alignment with the genes of interest. The top BLAST hit was used to predict Smed-Tim and Smed-Arntl-1 via FGENESH+ (http://www.softberry.com/). Homology at the protein level between predicted Smed-Tim and Hs-Tim, or Smed-Arntl-1 and Hs-Arntl-1 was analyzed using BLAST (see supplemental data online for predicted sequence). The conservation scoring was performed by PRALINE (http://www.ibi.vu.nl/)12 using default parameters. The results are color-coded for amino acid conservation and the scoring scheme works from 0, for the least conserved alignment position, to 10, for the most conserved alignment position. Nuclear localization sequence in Smed-TIM was identified using cNLS Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) with the following setup: cut of score 6.
Cloning
To generate the Smed-Tim and Smed-Arntl-1 RNAi molecules, cDNA from S. mediterranea was amplified via PCR using primers Tim (left primer TGGAGAGTGGCTGGAAGGAT, right primer CGAAGAAATGCGCGACTCTG), Arntl-1 (left primer CAGAAGGTACCACGACCGAT, right primer TGATTAAGGACGGCAGTGACT) designed with Primer3 (http://primer3.sourceforge.net/) and containing attB recombination sequences (CATTACCATCCCG). PCR products were cloned27 and sequences verified on a 3130XL Applied machine, and Blast against the predicted gene sequence. The dsRNA in silico accuracy prediction was defined as follows. Targeted transcript sequences were extracted between the 3′ end of the 5′ primer and the 5′ end of the 3′ primer used for cloning. The extracted sequences were then cut into 21 mers using a sliding window of 1 nucleotide. All possible RNAi sequences for Smed-Tim and Smed-Arntl-1 were then generated, and each putative RNAi sequence was aligned to the planarian transcriptome using BLAST28 with a word size of 21; only perfect matches were considered. For each transcript for which an RNAi was designed, theoretical target accuracy was calculated based on the number of RNAi sequences matching the target divided by the total number of generated RNAi sequences. The number of theoretical off-target events was equal to 0, thus giving a target accuracy of 100%, which strongly suggests a gene-specific effect but does not exclude the possibility of an off-target effect (Table. S1 and S2).2
Delivery of dsRNAs
dsRNAs were delivered to S. mediterranea as described previously.2,27,29 Briefly, animals were submitted to 3 rounds of RNAi feeding (1 every 3 days), then 3 d after the last RNAi feeding, worms were challenged by S. aureus.2 The quality of Smed-Arntl-1 and Smed-Tim knock down was controlled via Real-time RTqPCR as described elsewhere.2,30 Primers used for RTqPCR were for Smed-Arntl-1 (left primer ACGTGGAACTGGTAATACTGGT, right primer AAAACCATCAGCAGCCTCCA) and for Smed-Tim (left primer GGATTCTGAGCCGGTGGATT, right primer AATCCGATCTTCTTCCGGCC). The expression of the Smed-p38 MAPK, Smed-Traf6, and Smed-morn2 was evaluated via RTqPCR using the following primers: Smed-Morn2 (CGTCAAGGGAAAGGTATTAGCG, GTCGCCTTCATATTTTGCACCA), Smed-p38 MAPK (GCGAGGCAGACAGATGAAGA, GCGTGTAAACAATTCGGCCA), and Smed-Traf6 (ACCCCACAATCATAAGGGCA, AACTCCATTTTGTGCCCGGT).
The results were normalized by the expression of the control housekeeping gene Smed-ef2.31
In situ hybridization and probe synthesis
Whole-mount in situ hybridization was performed as described previously.32,33 All animals were 1 to 2 mm in length and size-matched between the experimental and control groups for S. mediterranea infection. The animals were imaged using a Leica M165FC stereomicroscope (Leica, Heidelberg). The images were processed using Adobe Photoshop CS5 software, and figures were assembled using Adobe Illustrator Artwork 15.0.
Melatonin assay
The melatonin level was determined using Melatonin ELISA KIT (Emelca Bioscience). Briefly, worms (30 per time point, size 10 mm to 7 mm) were lysed in phosphate buffer salt solution complemented with proteases inhibitors (Roche), then samples were done according to the manufacturer's recommendation.
Statistical analysis
The results are expressed as the mean ± SD and were analyzed using the nonparametric Mann-Whitney U test. Differences were considered significant at p < 0.05.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank R. Taware (CNRS UMR 7278) for his technical help.
Funding
This work has been supported by the CNRS (PEPS to E.G.), the Scientific Cooperation Foundation “Infectiopole Sud” and the Regional Council P.A.C.A (2009, Technical Facility to E.G.). LL.T. is a fellow of “Infectiopole Sud” and of the “Agence Universitaire de la Francophonie.” C.T. is a fellow of the French Ministry of Research and Technology, and of “Fondation pour la Recherche Médicale” (FRM FDT20160435255). The funding sources had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation. The authors declare no competing financial interests.
References
- [1].Elliott SA, Sanchez Alvarado A. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip Rev Dev Biol 2013; 2:301-26; PMID:23799578; http://dx.doi.org/ 10.1002/wdev.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Abnave P, Mottola G, Gimenez G, Boucherit N, Trouplin V, Torre C, Conti F, Ben Amara A, Lepolard C, Djian B, et al.. Screening in planarians identifies MORN2 as a key component in LC3-associated phagocytosis and resistance to bacterial infection. Cell Host Microbe 2014; 16:338-50; PMID:25211076; http://dx.doi.org/ 10.1016/j.chom.2014.08.002 [DOI] [PubMed] [Google Scholar]
- [3].Itoh MT, Shinozawa T, Sumi Y. Circadian rhythms of melatonin-synthesizing enzyme activities and melatonin levels in planarians. Brain Res 1999; 830:165-73; PMID:10350570 [DOI] [PubMed] [Google Scholar]
- [4].Itoh MT, Igarashi J. Circadian rhythm of serotonin levels in planarians. Neuroreport 2000; 11:473-6; PMID:10718297 [DOI] [PubMed] [Google Scholar]
- [5].Curtis AM, Bellet MM, Sassone-Corsi P, O'Neill LA. Circadian clock proteins and immunity. Immunity 2014; 40:178-86; PMID:24560196; http://dx.doi.org/ 10.1016/j.immuni.2014.02.002 [DOI] [PubMed] [Google Scholar]
- [6].Lee JE, Edery I. Circadian regulation in the ability of drosophila to combat pathogenic infections. Curr Biol 2008; 18:195-9; PMID:18261909; http://dx.doi.org/ 10.1016/j.cub.2007.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Nakao A. Temporal regulation of cytokines by the circadian clock. J Immunol Res 2014; 2014:614529; PMID:24809063; http://dx.doi.org/ 10.1155/2014/614529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Nguyen KD, Fentress SJ, Qiu Y, Yun K, Cox JS, Chawla A. Circadian gene Bmal1 regulates diurnal oscillations of Ly6C(hi) inflammatory monocytes. Science 2013; 341:1483-8; PMID:23970558; http://dx.doi.org/ 10.1126/science.1240636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shirasu-Hiza MM, Dionne MS, Pham LN, Ayres JS, Schneider DS. Interactions between circadian rhythm and immunity in drosophila melanogaster. Curr Biol 2007; 17:R353-5; PMID:17502084 [DOI] [PubMed] [Google Scholar]
- [10].Brandl H, Moon H, Vila-Farre M, Liu SY, Henry I, Rink JC. PlanMine–a mineable resource of planarian biology and biodiversity. Nucleic Acids Res 2016; 44:D764-73; PMID:26578570; http://dx.doi.org/ 10.1093/nar/gkv1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Robb SM, Gotting K, Ross E, Sanchez Alvarado A. SmedGD 2.0: The schmidtea mediterranea genome database. Genesis 2015; 53:535-46; PMID:26138588; http://dx.doi.org/ 10.1002/dvg.22872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Simossis VA, Heringa J. PRALINE: A multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res 2005; 33:W289-94; PMID:15980472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hughes AT, Piggins HD. Disruption of daily rhythms in gene expression: The importance of being synchronised. Bioessays 2014; 36:644-8; PMID:24832865; http://dx.doi.org/ 10.1002/bies.201400043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Tischkau SA, Barnes JA, Lin FJ, Myers EM, Barnes JW, Meyer-Bernstein EL, Hurst WJ, Burgoon PW, Chen D, Sehgal A, et al.. Oscillation and light induction of timeless mRNA in the mammalian circadian clock. J Neurosci 1999; 19:RC15; PMID:10366653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: Transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J Comp Neurol 2000; 422:66-94; PMID:10842219 [DOI] [PubMed] [Google Scholar]
- [16].Barnes JW, Tischkau SA, Barnes JA, Mitchell JW, Burgoon PW, Hickok JR, Gillette MU. Requirement of mammalian timeless for circadian rhythmicity. Science 2003; 302:439-42; PMID:14564007 [DOI] [PubMed] [Google Scholar]
- [17].Uryu O, Tomioka K. Circadian oscillations outside the optic lobe in the cricket gryllus bimaculatus. J Insect Physiol 2010; 56:1284-90; PMID:20416318; http://dx.doi.org/ 10.1016/j.jinsphys.2010.04.010 [DOI] [PubMed] [Google Scholar]
- [18].Pevet P. The internal time-giver role of melatonin. A key for our health. Rev Neurol 2014; 170:646-52; http://dx.doi.org/ 10.1016/j.neurol.2014.05.008 [DOI] [PubMed] [Google Scholar]
- [19].Pang Q, Gao L, Hu W, An Y, Deng H, Zhang Y, Sun X, Zhu G, Liu B, Zhao B. De novo transcriptome analysis provides insights into immune related genes and the RIG-I-like receptor signaling pathway in the freshwater planarian (dugesia japonica). PloS One 2016; 11:e0151597; PMID:26986572; http://dx.doi.org/ 10.1371/journal.pone.0151597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Arnold CP, Merryman MS, Harris-Arnold A, McKinney SA, Seidel CW, Loethen S, Proctor KN, Guo L, Sánchez Alvarado A. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. Elife 2016; 5:1-36; PMID:27441386; http://dx.doi.org/ 10.7554/eLife.16793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Reitzel AM, Behrendt L, Tarrant AM. Light entrained rhythmic gene expression in the sea anemone nematostella vectensis: The evolution of the animal circadian clock. PloS One 2010; 5:e12805; PMID:20877728; http://dx.doi.org/ 10.1371/journal.pone.0012805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gotter AL, Manganaro T, Weaver DR, Kolakowski LF, Possidente B, Sriram S, MacLaughlin DT, Reppert SM. A time-less function for mouse timeless. Nat Neurosci 2000; 3:755-6; PMID:10903565 [DOI] [PubMed] [Google Scholar]
- [23].Beaver LM, Rush BL, Gvakharia BO, Giebultowicz JM. Noncircadian regulation and function of clock genes period and timeless in oogenesis of drosophila melanogaster. J Biol Rhythms 2003; 18:463-72; PMID:14667147 [DOI] [PubMed] [Google Scholar]
- [24].Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, Shirasu-Hiza MM. The circadian clock protein timeless regulates phagocytosis of bacteria in drosophila. PLoS Pathog 2012; 8:e1002445; PMID:22253593; http://dx.doi.org/ 10.1371/journal.ppat.1002445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rund SS, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector anopheles gambiae. Proc Natl Acad Sci U S A 2011; 108:E421-30; PMID:21715657; http://dx.doi.org/ 10.1073/pnas.1100584108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cebria F, Newmark PA. Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 2005; 132:3691-703; PMID:16033796 [DOI] [PubMed] [Google Scholar]
- [27].Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Developmental Cell 2005; 8:635-49; PMID:15866156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10; PMID:2231712 [DOI] [PubMed] [Google Scholar]
- [29].Sanchez Alvarado A, Newmark PA. Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc Natl Acad Sci U S A 1999; 96:5049-54; PMID:10220416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Forsthoefel DJ, James NP, Escobar DJ, Stary JM, Vieira AP, Waters FA, Newmark PA. An RNAi screen reveals intestinal regulators of branching morphogenesis, differentiation, and stem cell proliferation in planarians. Developmental Cell 2012; 23:691-704; PMID:23079596; http://dx.doi.org/ 10.1016/j.devcel.2012.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Solana J, Kao D, Mihaylova Y, Jaber-Hijazi F, Malla S, Wilson R, Aboobaker A. Defining the molecular profile of planarian pluripotent stem cells using a combinatorial RNAseq, RNA interference and irradiation approach. Genome Biol 2012; 13:R19; PMID:22439894; http://dx.doi.org/ 10.1186/gb-2012-13-3-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Pearson BJ, Eisenhoffer GT, Gurley KA, Rink JC, Miller DE, Sanchez Alvarado A. Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev Dyn 2009; 238:443-50; PMID:19161223; http://dx.doi.org/ 10.1002/dvdy.21849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].King RS, Newmark PA. In situ hybridization protocol for enhanced detection of gene expression in the planarian schmidtea mediterranea. BMC Dev Biol 2013; 13:8; PMID:23497040; http://dx.doi.org/ 10.1186/1471-213X-13-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


