Figure 3.
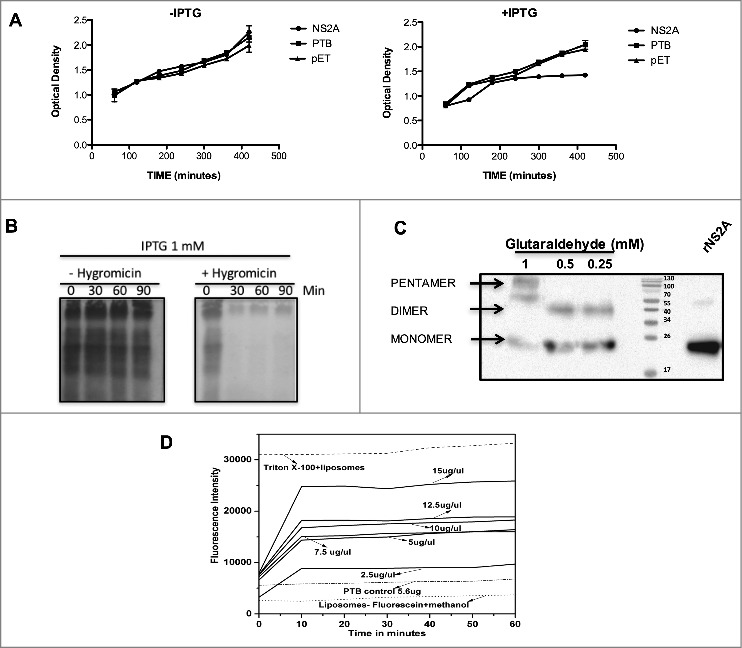
NS2A protein expression arrests the growth of Escherichia coli and affects the permeability of bacterial membranes and liposomes. (A) Bacterial membrane pET-NS2A permeability model. Growth curves of engineered E. coli BL21 pLys transformed with NS2A and PTB constructs in the absence (left) or presence (right) of IPTG. The cellular density of each bacterial culture was determined every 60 min after induction by measuring the optical density at 660 nm. (B) Cultures were induced with IPTG in the absence (−) or presence (+) of HygB, then subjected to metabolic labeling with [35S] Met-Cys and analyzed by SDS-PAGE at the indicated times. (C) Western blot analysis of NS2A oligomerisation in vitro. NS2A protein was incubated with different concentrations of glutaraldehyde (0.25, 0.5, and 1 mM) and then resolved by SDS-PAGE under reducing conditions. Oligomeric structures (dimers and pentamers) were detected using an anti-V5 antibody at a dilution of 1:5000. (D) Membrane disruption was examined by incubating liposome-encapsulated FITC (as a model for eukaryotic cell membranes) in solutions containing Triton X-100 (positive control), or NS2A (2.5–15 µg/µl). NS2A protein-mediated liposome disruption was monitored every 10 min by measuring the fluorescence emission of each mixture. Protein disruption studies were performed using 5 µL of liposome-encapsulated FITC to determine the baseline fluorescence intensity. Fluorescence was monitored using a Synergy H4 Fluorescence reader equipped with a FITC 485 excitation filter, a FITC 535 emission filter, and a 50/50 beam splitter. Fluorescence values were read every 30 s for 1 h at 37°C.
