ABSTRACT
We investigated the genetic background and microbiological features of T6SS-positive Acinetobacter baumannii isolates and clinical impact of the T6SS in patients with A. baumannii bacteremia. One hundred and 62 A. baumannii isolates from patients with bacteremia in 2 tertiary-care hospitals in Korea were included in this study. Approximately one-third (51/162, 31.5%) of the A. baumannii clinical isolates possessed the hcp gene, and the hcp-positive isolates were found in several genotypes in multilocus sequence typing. The expression and secretion of Hcp protein varied among the clinical isolates. A. baumannii isolates with detectable Hcp secretion (T6SS+) could better outcompete Escherichia coli compared with T6SS- isolates, including hcp-negative and inactivated hcp-positive isolates. In addition, T6SS+ isolates showed higher biofilm-forming activity and better survival in the presence of normal human serum than the T6SS– isolates. T6SS+ isolates were more frequently detected in patients with catheter-related bloodstream infection, haematopoietic stem cell transplant recipients, and patients receiving immunosuppressive agents. However, T6SS was not a prognostic factor for mortality. Our results suggest that the T6SS of A. baumannii is associated with virulence and contributes to infections in immunocompromised patients and those with implanted medical devices.
KEYWORDS: biofilm formation, catheter-related bloodstream infection, hcp, serum resistance, type VI secretion system (T6SS)
Introduction
Protein secretion systems are characteristic components on the cell surface of gram-negative bacteria. In many bacteria, these systems export effector proteins, which are often mediators of virulence factors. The type VI secretion system (T6SS) has been recently shown to be important for microbial communication within human hosts and the environment.1 These systems were first identified in Vibrio cholerae and Pseudomonas aeruginosa, and they were shown to play a role in the defense against eukaryotic hosts.2,3 Since then, many bacterial species, including P. aeruginosa, Pseudomonas syringae, Vibrio parahaemolyticus, V. cholerae, Burkholderia thailandensis, Serratia marcescens, and Citrobacter rodentium, have been shown to use a T6SS to mediate interbacterial antagonism and increase their fitness in competition with other bacteria.4
The T6SS is structurally and functionally similar to a bacteriophage tail,4,5 and it consists of 2 membrane-associated subunits with cytoplasmic elements, an anchoring assembly and an assembly with components that structurally resemble the bacteriophage sheath, tube, and tail spike proteins. These cooperate to translocate effector proteins across the envelope of the cell and inject them into the target cell membrane through a contractile phage tail-like apparatus.5,6 The phage-like assembly has 3 components, hemolysin coregulated protein (Hcp), which forms a tubular structure that is secreted out of the cell, valine-glycine repeat G (VgrG), which forms the tip of this structure and has effector activity or promotes effector secretion, and TssB and TssC, which form a sheath that contracts to provide the energy for effector transport.7 Since Hcp and VgrG are shed into the extracellular milieu by activation of the system, they serve as molecular markers of a functional T6SS. Several studies have shown that the T6SS transports bacteriolytic effector proteins into target cells in a contact-dependent manner.8-10 However, the role of T6SS beyond that of bacterial antagonism is unclear.
Recently, T6SS-encoding genes were identified in Acinetobacter baumannii,11 and T6SS-positive A. baumannii strains were shown to have cytotoxic activity against competing bacteria.12-15 However, the mechanisms of T6SS-dependent effectors, functions of the effectors within target cells, and regulatory systems in A. baumannii remain largely unknown. In some bacterial isolates, the T6SS was identified but was not activated. It was recently reported that the T6SS of A. baumannii was repressed by a repressor encoded by tetR, which is placed in a plasmid.15
In this study, we aimed to investigate the role of the T6SS in A. baumannii both in and around the human host. The function of T6SS was assessed by in vitro experiments, including competition, biofilm formation, and serum resistance assays. In addition, we conducted a retrospective study to determine its clinical impact in patients with A. baumannii bacteremia.
Materials and methods
Bacterial isolates
A total of 228 clinical isolates were collected from patients with Acinetobacter bacteremia during the period from January 2012 to December 2015 in 2 tertiary-care hospitals in South Korea, Samsung Medical Center (Seoul) and Samsung Changwon Hospital (Changwon). Using partial rpoB gene sequences (468 bp), A. baumannii was distinguished from other Acinetobacter species.16,17 Of the 228 isolates, 162 (71.1%) were identified as A. baumannii and were analyzed in this study.
Antimicrobial susceptibility testing
In vitro antimicrobial susceptibility testing was performed for all A. baumannii isolates using the broth microdilution method according to the guidelines of the Clinical and Laboratory Standard Institute (CLSI).18 Nine antimicrobial agents were tested, including imipenem, ceftazidime, ampicillin-sulbactam, gentamicin, piperacillin-tazobactam, ciprofloxacin, trimethoprim-sulfamethoxazole, colistin, and tigecycline. CLSI susceptibility interpretive criteria were used.18 Escherichia coli ATCC 25922, Staphylococcus aureus ATCC 29213, and P. aeruginosa ATCC 27853 were used as control strains.
Multilocus sequence typing (MLST)
All A. baumannii clinical isolates were genotyped by MLST using the Oxford scheme as described previously.19 The sequence of each allele was compared with sequences in PubMLST databases by BLAST, and sequence types (STs) were designated according to the allelic profiles. Newly identified allelic profiles and STs were submitted to the PubMLST database and approved (http://pubmlst.org/abaumannii). Based on the MLST data, the genetic relationships and clonal complexes (CCs) of the isolates were analyzed and represented in a minimum-spanning tree using the PHYLOViZ program.20
Detection of the hcp gene and quantitative reverse transcription PCR (qRT-PCR) to determine hcp expression levels
To identify strains containing T6SS, the hcp gene was amplified with the primers hcp-F (5′-TGCTGAGCGTGTTGAACATT-3′) and hcp-R (5′-ACGTTTATCGCCATTTGCAC-3′). hcp expression levels were determined by qRT-PCR using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) on the QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems) with primers, hcp-QF (5′-TGCTGAGCGTGTTGAACATT-3′) and hcp-QR (5′-ACGTTTATCGCCATTTGCAC-3′). These primers for the amplification and qRT-PCR were designed based on the sequences of A. baumannii strain ACICU (GenBank accession number CP000863), using Primer 3 online software. Expression of the rpoB gene was assessed in parallel to calculate the fold changes according to the threshold cycle (CT) method.21 The experiments were repeated with 3 independent cultures, each tested in duplicate.
We assayed the presence of tetR in the isolates in which hcp did not express despite the presence of hcp gene using the primers tetR18-F (5′-ATGACTAAAGTTATTTCAAAAAGAAAAACC-3′) and tetR18-R (5′-AGCCTCAAATGTCTGAATTAGACTC-3′) as described previously.15 The presence of T6SS was confirmed by detection of another gene in T6SS, tssM, with primers tssM-F (5′-GCAAAGACGTCTACAACAGTTAGAC-3′) and tssM-R (5′-TTGCTGATCATCACGTTTACGAAC-3′), in addition to hcp gene.
Western blotting
Cloning and overexpression of histidine-tagged Hcp was performed as described previously,12,13 with a few modifications. The hcp gene of A. baumannii ATCC 19606 was amplified using primers Hcp-(N)F (5′-CCAACCATGGGCATGAAAGATATATACGTTGAGTT-3′) and Hcp-(H)R (5′-CCAAAAGCTTAGCTGCGTAAGAAGCTGTAT-3′) and cloned into the pET28a vector to generate a C-terminal histidine-tagged fusion of the Hcp protein. The resultant plasmid, pET28-Hcp (C-His), was electroporated into Escherichia coli BL21 cells. The histidine-tagged Hcp protein was purified with the QIAexpress Ni-NTA Fast Start Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Purified Hcp protein was sent to AbFrontier antibody services (Young In Frontier Co. Ltd., Seoul, Korea) to generate rabbit-derived polyclonal antibodies.
To prepare cell-free supernatants for the Hcp secretion assay, overnight bacterial cultures were inoculated 1:100 into 20 mL of fresh Luria-Bertani (LB) broth and grown to stationary phase. After approximately 4 h, bacteria were harvested by centrifugation, and the culture supernatants were collected and filtered through 0.22-µm syringe filters (Millipore Corporation, Billerica, MA, USA) to obtain cell-free supernatants. The cleared supernatants were concentrated by filtration through Amicon Ultra-15 10K centrifugal filters (Merck Millipore, Germany) according to the manufacturer's protocol. The proteins in the recovered concentrated supernatants were analyzed by SDS-PAGE. OD600-normalized volumes of whole cells or supernatants were loaded onto 12% SDS-PAGE gels for separation, and the contents of the gels were transferred to nitrocellulose membranes. Membranes were blocked with 5% skim milk for 1 h and probed with polyclonal rabbit anti-Hcp (1:5000; Young In Frontier Co. Ltd.) and rabbit monoclonal anti-RNA polymerase β-subunit (1:2000; Abcam, Cambridge, UK) antibodies. A horseradish peroxidase-conjugated goat anti-rabbit IgG (1:10000; Abcam) was used to visualize Hcp along with SuperSignal west Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific., Rockford, IL, USA) according to the manufacturer's instructions.
In vitro competition assay
The relative fitness of A. baumannii isolates against E. coli strain MG1655 was determined by calculating an in vitro competitive index (CI) using a described previously method13 with some modifications. Overnight cultures of the A. baumannii isolates and the E. coli strain were inoculated to obtain a 0.5 McFarland standard and diluted 1:50 in 10 mL of LB broth. The diluted cultures were then mixed at a 2:1 predator:prey ratio and incubated at 37°C and 180 rpm for 20 h. The number of cells for each strain was determined by spreading serial 10-fold dilutions onto LB agar plates with or without 100 mg/L ampicillin. The CI was defined as the ratio of ampicillin-resistant CFUs (A. baumannii) to ampicillin-susceptible CFUs (E. coli). Eight independent competition experiments were performed.
Biofilm formation assay
To assess biofilm formation, 96-well microtiter plate assays were performed as described previously,22 with minor modifications. Briefly, overnight cultures were normalized by the OD600 and diluted 1:100 in LB supplemented with 0.5% glucose. A 200-μL aliquot of this suspension was inoculated into the wells of a 96-well flat-bottom polystyrene plate and incubated for 18 h at 37°C. Sterile LB was added to one well, which served as a control. After incubation, planktonic bacteria were removed, and each well was gently washed and air-dried. Biofilm bacteria remaining in the well were stained with 0.5% crystal violet for 15 min and suspended in 200 μL of 95% ethanol. The OD600 was then measured with a microplate reader, yielding a measure of biofilm formation relative to the control. Experiments were performed with 3 independent cultures, each tested in duplicate.
Serum resistance assay
Serum resistance assays were performed as described previously,23 with minor modifications. Briefly, overnight bacterial cultures were diluted 1:100 into 10 mL of fresh LB medium and incubated until the bacterial suspension reached an OD600 of 0.5. Then, a 1-mL aliquot of the culture was washed with phosphate-buffered saline (PBS) and resuspended in 1 mL of PBS. Next, 100 μL of the bacterial suspension was added and mixed with 300 μL of normal human serum (NHS). After mixing, the serum-bacteria suspensions were incubated at 37°C for 3 h. To calculate the serum bactericidal effect, a 100-µL aliquot was taken from each suspension before and after the 3-h incubation period, serially diluted, and plated. The serum bactericidal effect was expressed as the ratio of the CFUs in the serum-bacteria suspension to the CFUs in a bacterial suspension without NHS. All experiments were performed in triplicate, and results are expressed as percent survival.
Clinical data collection
We retrospectively reviewed the electronic medical records of patients with A. baumannii bacteremia. Patients under 18 y of age were excluded. Data were collected, including demographics, underlying diseases, comorbidities, acquisition of infection, antimicrobial regimens, and outcomes. The severity of illness was measured with the Pitt bacteremia score and Charlson weighted index of comorbidity (WIC).24,25 Drug resistance, multidrug-resistance (MDR), extensive drug resistance (XDR), and pandrug-resistance (PDR) were defined as described by Magiorakos et al.26 Neutropenia was defined as an absolute neutrophil count of fewer than 500 neutrophils/mm3. Acute kidney injury (AKI) was defined as an absolute increase in serum creatinine equal to or greater than 0.3 mg/dL or a percentage increase in serum creatinine equal to or greater than 50% from the baseline level within 48 h.27 Hospital-acquired infection was defined as an infection that occurred more than 48 h after admission to the hospital.28 The primary source of bacteremia was determined by the investigators based on medical records. Primary bacteremia was considered when no infection focus was diagnosed. All-cause mortality at 14 and 30 d was calculated from the day of blood sampling on which bacteremia was identified. The definition of appropriate antibiotics was an antimicrobial regimen that included at least one parenteral antibiotic active against A. baumannii in vitro. As an observational study, antimicrobial regimens were not standardized but selected by primary care physicians.
Statistical analysis
Student's t-test and one-way ANOVA were used to compare continuous variables. The χ2 and Fisher's exact test were used to compare categorical variables. A stepwise logistic regression analysis was used to control for potential confounding factors. Variables with a P value less than 0.1 in the univariate analysis were included in the multivariate logistic regression model to control for confounding factors and identify independent risk factors for mortality. All P values were 2-tailed, and for all analyses, a P value less than 0.05 was considered statistically significant. Data were analyzed with SPSS version 18.0 (SPSS Inc., Chicago, IL, USA).
Results
The prevalence of T6SS in A. baumannii clinical isolates
Among the 162 A. baumannii clinical isolates, 51 isolates (31.5%) contained an hcp gene. MDR and XDR were identified in 144 (88.9%) and 132 (81.5%) isolates, respectively (Table 1). One isolate was resistant to all antimicrobial agents tested in this study. The carbapenem resistance rate was 84.6% (137 isolates), of the resistant isolates, 99.6% (132/137) and 73.7% (101/137) were susceptible to colistin and tigecycline, respectively. The hcp gene was detected in 33.6% of carbapenem-resistant A. baumannii (CRAB) isolates (46/137) and 34.8% of XDR isolates (36/132). Antimicrobial resistance rates in hcp-positive (hcp+) and hcp-negative (hcp–) isolates were not significantly different, although resistance rates to all antibiotics, except tigecycline, were slightly higher in hcp+ isolates (Table 1). hcp– isolates were also negative to tssM gene.
Table 1.
Antibiotic resistance rates of A. baumannii isolates according to the presence of the hcp gene.
| Number of resistant isolates (%) |
|||
|---|---|---|---|
| Antimicrobial agent | Total (n = 162) | hcp+ (n = 51) | hcp– (n = 111) |
| Imipenem | 137 (84.6) | 46 (90.2) | 91 (82.0) |
| Ceftazidime | 141 (87.0) | 47 (92.2) | 94 (84.7) |
| Ampicillin/sulbactam | 138 (85.2) | 46 (90.2) | 92 (82.9) |
| Piperacillin/tazobactam | 140 (86.4) | 46 (90.2) | 94 (84.7) |
| Gentamicin | 129 (79.6) | 45 (88.2) | 84 (75.7) |
| Ciprofloxacin | 138 (85.2) | 46 (90.2) | 92 (82.9) |
| Trimethoprim/sulfamethoxazole | 143 (88.3) | 48 (94.1) | 95 (85.6) |
| Colistin | 6 (3.7) | 3 (5.9) | 3 (2.7) |
| Tigecycline | 15 (9.3) | 3 (5.9) | 12 (10.8) |
In the MLST, 35 distinct sequence types (STs) were identified among the 162 A. baumannii clinical isolates (Table 2 and Fig. 1). One hundred 32 isolates (81.5%) of 11 STs belonged to CC191, corresponding to global clone 2 (GC2), and all CC191 isolates were resistant to imipenem. Only 5 non-GC2 isolates (ST229 and ST491) were resistant to carbapenem; thus, 96.4% of carbapenem-resistant isolates belonged to GC2 (Table 2). There might be no correlation between the expression levels of hcp gene and ST Table S1.
Table 2.
Genotypes and distribution of A. baumannii isolates.
| ST |
Allelic profilea |
Total (n = 162) (%) |
hcp+ (n = 51) (%) |
hcp–(n = 111) (%) |
Imipenem resistance (%) |
|---|---|---|---|---|---|
| GC2 | 132 (81.5) | 41 (31.1) | 91 (68.9) | 132 (100) | |
| ST191 | 1–3–3–2–2–94–3 | 66 (40.7) | 66 (100) | 66 (100) | |
| ST208 | 1–3–3–2–2–97–3 | 14 (8.6) | 13 (92.9) | 1 (7.1) | 14 (100) |
| ST357 | 1–12–3–2–2–145–3 | 11 (6.8) | 11 (100) | 11 (100) | |
| ST368 | 1–3–3–2–2–140–3 | 6 (3.7) | 6 (100) | 6 (100) | |
| ST369 | 1–3–3–2–2–106–3 | 11 (6.8) | 3 (27.3) | 8 (72.7) | 11 (100) |
| ST451 | 1–3–3–2–2–142–3 | 13 (8.0) | 13 (100) | 13 (100) | |
| ST784 | 1–3–3–2–2–107–3 | 6 (3.7) | 6 (100) | 6 (100) | |
| ST1114 | 1–12–3–2–2–79–3 | 1 (0.6) | 1 (100) | 1 (100) | |
| ST1141 | 1–3–3–104–2–140–3 | 1 (0.6) | 1 (100) | 1 (100) | |
| ST1144 | 1–3–3–102–2–94–3 | 2 (1.2) | 2 (100) | 2 (100) | |
| ST1316 | 1–35–3–2–2–107–3 | 1 (0.6) | 1 (100) | 1 (100) | |
| Others | |||||
| ST681 | 1–102–59–28–1–83–45 | 1 (0.6) | 1 (100) | ||
| ST734 | 1–1–5–59–28–4–157–45 | 1 (0.6) | 1 (100) | ||
| ST1319 | 1–15–59–28–4–269–45 | 1 (0.6) | 1 (100) | ||
| ST503 | 1–62–80–28–35–164–45 | 1 (0.6) | 1 (100) | ||
| ST1077 | 1–62–80–28–35–178–45 | 1 (0.6) | 1 (100) | ||
| ST435 | 1–54–80–28–48–127–45 | 1 (0.6) | 1 (100) | ||
| ST552 | 21–35–2–28–1–145–4 | 1 (0.6) | 1 (100) | ||
| ST1318 | 21–35–2–28–1–268–4 | 1 (0.6) | 1 (100) | ||
| ST711 | 21–12–2–28–1–109–4 | 1 (0.6) | 1 (100) | ||
| ST1262 | 21–35–2–28–22–23–4 | 1 (0.6) | 1 (100) | ||
| ST447 | 1–15–13–12–4–106–2 | 1 (0.6) | 1 (100) | ||
| ST1313 | 1–15–13–12–4–94–2 | 1 (0.6) | 1 (100) | ||
| ST1317 | 1–15–13–12–4–163–112 | 1 (0.6) | 1 (100) | ||
| ST1315 | 1–1–13–12–92–16–2 | 1 (0.6) | 1 (100) | ||
| ST751 | 33–31–2–28–1–83–5 | 1 (0.6) | 1 (100) | ||
| ST1181 | 33–31–2–28–1–144–5 | 1 (0.6) | 1 (100) | ||
| ST229 | 1–15–2–28–1–107–32 | 4 (2.5) | 4 (100) | 4 (100) | |
| ST491 | 10–53–4–11–4–98–5 | 1 (0.6) | 1 (100) | 1 (100) | |
| ST373 | 1–12–12–11–4–103–3 | 5 (3.1) | 5 (100) | ||
| ST613 | 15–48–58–42–36–54–41 | 2 (1.2) | 2 (100) | ||
| ST1131 | 60–17–140–1–4–103–107 | 1 (0.6) | 1 (100) | ||
| ST1312 | 1–53–135–12–1–110–110 | 1 (0.6) | 1 | ||
| ST1314 | 21–15–2–28–35–193–111 | 1 (0.6) | 1 (100) | ||
| ST1320 | 1–102–80–85–28–214–113 | 1 (0.6) | 1 |
Note.
gltA-gyrB-gdhB-recA-cpn60-gpi-rpoD.
Figure 1.
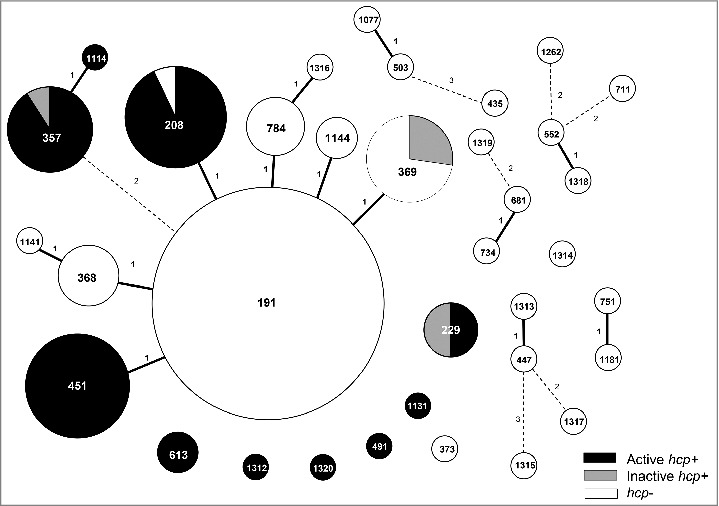
Diagram of the distribution of the hcp gene according to sequence type (ST). The numbers in the circles are the STs. The size of each circle represents the proportion of each ST, which are the marks within the circles. Solid line linkages indicate a single-locus variant, and dotted-line linkages indicate differences in 2 or 3 alleles.
The hcp gene was detected in A. baumannii isolates of 11 STs (Table 2), and in 41 of 132 A. baumannii GC2 isolates (31.1%), which is similar to the prevalence among all A. baumannii isolates (33.6%). However, the hcp gene was not distributed evenly across clones, instead was present in specific clones; all isolates of ST357 (11 isolates) and ST451 (13 isolates) were hcp+, and all isolates of ST208 except one (13 isolates) were hcp+ (Table 2; Fig. 1). All 4 isolates of ST229 in non-GC2 harbored the hcp gene. In contrast, all isolates of ST191 (66 isolates), ST368 (6 isolates), and ST784 (6 isolates) were hcp–. Only ST208 and ST369 included both hcp+ and hcp– isolates.
hcp expression and secretion of Hcp protein
To assess the expression of the hcp gene, qRT-PCR was performed on 51 hcp+ A. baumannii clinical isolates, and expression levels were compared with that of the reference strain ATCC 19606, which is known to express and secrete Hcp protein.11,13 hcp transcript levels in the clinical isolates ranged from 0.0008- to 1.5-fold (mean, 0.18-fold) of the level in ATCC 19606 (Fig. 2). We identified 6 isolates with mRNA levels 100-fold lower than that of ATCC 19606. These isolates with low hcp expression that likely have non-functional T6SS belonged to ST369, ST229, and ST357 (Fig. 1).
Figure 2.
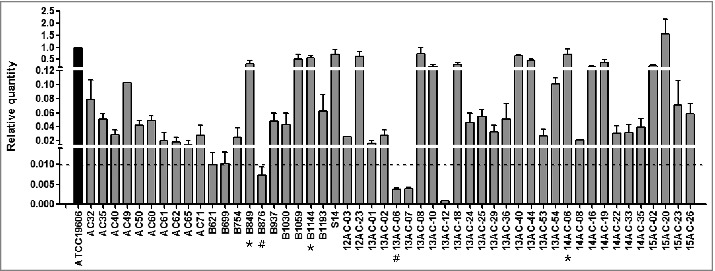
Relative hcp transcript levels in A. baumannii isolates as measured by qRT PCR. hcp transcription levels are shown relative to that in A. baumannii ATCC 19606. The dashed line indicates 0.01-fold the expression levels of hcp in the reference strain, ATCC19606. The asterisk (*) and pound (#) symbols indicate the isolates with high and low hcp expression, respectively, that were used in the western blotting, competition, biofilm formation, and serum resistance assays.
We assessed the secretion of Hcp protein in hcp+ isolates with a rabbit-derived polyclonal antibody generated against purified Hcp protein from reference strain ATCC 19606. For western blotting, we selected 5 isolates, 3 showing high hcp expression (B849, B1144, and 14AC-06; * in Fig. 2) and 2 showing hcp mRNA levels 100-fold lower than that of ATCC 19606 (B876 and 13AC-06; # in Fig. 2). Since RNA polymerase (RNAP) was used as a control, its detection in only whole cells suggested that the presence of Hcp in supernatants was due to exportation by the bacterium rather than cell lysis. Hcp was detected in whole cells and in concentrated culture supernatants of the 3 isolates showing high hcp expression, but not in the 2 isolates with low hcp expression (Fig. 3). These results showed that A. baumannii clinical isolates with high hcp expression have an active T6SS (T6SS+), whereas isolates with low hcp expression have an inactive T6SS (T6SS–) under these conditions. Thus, we performed further analyses by classifying the isolates as T6SS+ and T6SS–.
Figure 3.
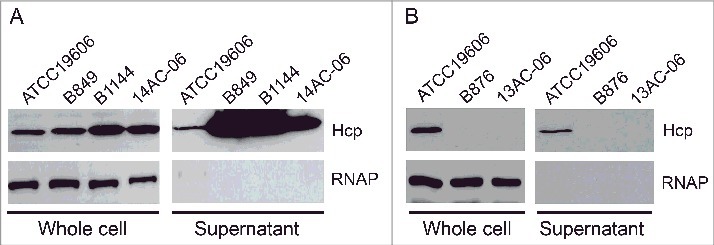
Detection of Hcp, with RNA polymerase (RNAP) as a lysis control. (A) Three isolates showing high hcp transcript levels; (B) 2 isolates showing low hcp transcript levels, (i.e., less than 0.01-fold that of ATCC 19606). Immunoblots show Hcp in whole cells and concentrated culture supernatants prepared from cultures of clinical isolates.
Microbiological characteristics of T6SS+ isolates
To understand the microbiological characteristics of T6SS+ A. baumannii isolates, we selected 8 A. baumannii clinical isolates, 3 T6SS+ isolates (B849, B1144, and 14AC-06) and 5 T6SS- isolates including 2 inactive hcp+ (B876 and 13AC-06) and 3 hcp– isolates (AC12, B764, and 13AC-45; Table 3), and we performed competition, biofilm formation, and serum resistance assays using these isolates.
Table 3.
Profiles of A. baumannii isolates used in western blotting and analyses of microbiologic characteristics.
| MIC (μg/mL) of antimicrobial agents b |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strain | Genotype | Date of isolation | Site of Infectiona | IMI | CAZ | A/S | P/T | GEN | CIP | SXT | COL | TGC |
| T6SS+ (active hcp+) | ||||||||||||
| B849 | ST357 | 2013 SEP 3 | SSTI | >64 | >64 | 64/32 | <256/4 | >64 | >64 | >32/608 | 2 | 1 |
| B1144 | ST357 | 2014 JUL 2 | CRBSI | >64 | >64 | 64/32 | <256/4 | >64 | >64 | >32/608 | 1 | 1 |
| 14AC-06 | ST208 | 2014 JAN 9 | VAP | >64 | >64 | >64/32 | <256/4 | >64 | 64 | >32/608 | 1 | 1 |
| T6SS– (inactive hcp+) | ||||||||||||
| B876 | ST357 | 2013 SEP 14 | CRBSI | 64 | >64 | >64/32 | <256/4 | 2 | >64 | >32/608 | 2 | 4 |
| 13AC-06 | ST369 | 2013 MAR 3 | CRBSI | >64 | >64 | 64/32 | <256/4 | >64 | 64 | >32/608 | 1 | 1 |
| T6SS– (hcp–) | ||||||||||||
| AC12 | ST191 | 2013 MAY 14 | IAI | >64 | >64 | >64/32 | <256/4 | 8 | >64 | >32/608 | 1 | 2 |
| B764 | ST191 | 2013 JUL 25 | IAI | >64 | >64 | >64/32 | <256/4 | >64 | >64 | >32/608 | 1 | 4 |
| 13AC-45 | ST369 | 2013 SEP 22 | Primary bacteremia | >64 | >64 | >64/32 | <256/4 | >64 | >64 | >32/608 | 1 | 1 |
Notes.
SSTI, skin and soft tissue infection; CRBSI, catheter-related bloodstream infection; VAP, ventilator-associated pneumonia; IAI, intra-abdominal infection
MIC, minimum inhibitory concentration; IMI, imipenem; CAZ, ceftazidime; A/S, ampicillin-sulbactam; P/T, piperacillin-tazobactam; GEN, gentamicin; CIP, ciprofloxacin; SXT, trimethoprim-sulfamethoxazole; COL, colistin; TGC, tigecycline
To determine whether T6SS+ isolates could better outcompete other bacteria, each A. baumannii isolate was mixed with an ampicillin-susceptible E. coli MG1655 strain at a 2:1 ratio and incubated. Then, surviving A. baumannii and E. coli MG1655 CFUs were counted by spotting serial dilutions on LB agar plates with and without ampicillin. T6SS+ isolates (mean CI, 4.15) caused a considerable reduction in E. coli MG1655 counts in competition assays compared with T6SS– isolates (mean CI, 0.9; P = 0.006; Fig. 4A).
Figure 4.
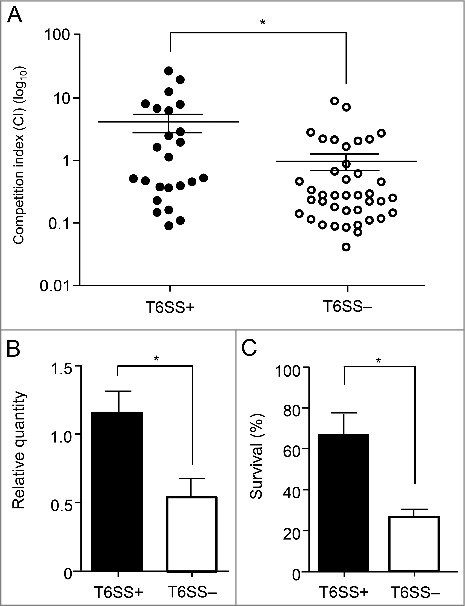
Results of competition, biofilm formation, and serum resistance assays. These results came from 3 T6SS+ and 2 T6SS- isolates. (A) Relative in vitro competition indexes (CIs) for A. baumannii isolates by the presence of T6SS (T6SS+ vs. T6SS–). The CI values shown are the number of A. baumannii divided by the number of E. coli MG1655. Each circle represents the CI of each in vitro replicate. (B) Relative quantity of biofilm in A. baumannii isolates compared with that of ATCC 19606. (C) Serum resistance in A. baumannii isolates of each T6SS+ and T6SS– replicate. Serum resistance as a viability ratio (colony forming units [CFUs] of serum-bacterial suspension/ CFUs of bacterial suspension without normal human serum). The error bars indicate the standard deviations. *, Statically significant difference (P < 0.05).
Relative biofilm-forming activity was estimated for T6SS+ and T6SS– strains and compared with that of ATCC 19606, and each isolate was tested in triplicate (Fig. 4B). The T6SS+ isolates formed significantly more biofilm mass than the T6SS– isolates (mean ± standard deviation [SD], 1.15 ± 0.51 vs. 0.54 ± 0.51; P = 0.010). The survival of A. baumannii in the presence of human serum also differed significantly among the 2 groups (Fig. 4C). The T6SS+ isolates showed a mean survival rate of 67% after a 3-h incubation with normal human serum (NHS), and the survival rate of the T6SS– isolates was 26% (P < 0.001).
Among the T6SS– isolates, inactive hcp+ and hcp– isolates showed no significant difference in terms of CI, biofilm formation, and serum resistance (Fig. S1).
Clinical impact of T6SS in patients with A. baumannii bacteremia
The clinical data analysis included 162 patients with A. baumannii bacteremia. Most patients (92.0%) had hospital-acquired infections. More than half had bacteremic episodes in the ICU (61.1%) and one or more immunocompromising conditions (67.3%). Owing to the high rate of carbapenem resistance (88.7%), one-third of the patients (35.2%) were treated with appropriate antibiotics within 48 h.
Table 4 shows the clinical manifestations of patients infected with T6SS+ and T6SS– A. baumannii isolates. Among the patients, 45 (28.4%) had T6SS+ infections. Compared to patients with T6SS– infections, more patients with T6SS+ infections had haematopoietic stem cell transplantation (HSCT; 13.3% vs. 3.4%, P = 0.029), treatment with immunosuppressive agents (62.2% vs. 40.2%, P = 0.014), and catheter-related bloodstream infection (44.4% vs. 17.9%, P = 0.001). The rates of intra-abdominal infections and concurrent polymicrobial bacteremia did not differ between the 2 groups. There was also no difference in mortality rates between the T6SS+ and T6SS– groups.
Table 4.
Clinical characteristics of patients with T6SS+ A. baumannii bacteremia.a
| Total(n = 162) | T6SS+(n = 45, 28.4%) | T6SS–(n = 117, 71.3%) | P-value | |
|---|---|---|---|---|
| Age, years | 62.2 ± 14.7 | 61.6 ± 12.5 | 62.5 ± 15.5 | 0.710 |
| Male gender | 94 (58.0) | 27 (60.0) | 67 (57.3) | 0.859 |
| Underlying disease | ||||
| Immunocompromised status | 109 (67.3) | 35 (77.8) | 74 (63.2) | 0.093 |
| Solid tumor | 47 (29.0) | 11 (24.1) | 36 (30.8) | 0.449 |
| Hematologic malignancy | 32 (19.8) | 10 (22.2) | 22 (18.8) | 0.662 |
| HSCT | 10 (6.2) | 6 (13.3) | 4 (3.4) | 0.029c |
| Immunosuppressive agents | 75 (46.3) | 28 (62.2) | 47 (40.2) | 0.014 |
| Neutropenia | 32 (19.8) | 8 (17.8) | 24 (20.5) | 0.827 |
| Chronic renal disease | 52 (32.1) | 14 (31.1) | 38 (32.5) | 1.000 |
| Hemodialysis | 17 (10.5) | 8 (17.8) | 9 (7.7) | 0.084c |
| Congestive heart failure | 23 (14.2) | 4 (8.9) | 19 (16.2) | 0.317 |
| Liver cirrhosis | 18 (11.1) | 5 (11.1) | 13 (11.1) | 1.000 |
| Diabetes mellitus | 52 (32.1) | 14 (31.1) | 38 (32.5) | 1.000 |
| Comorbid conditions | ||||
| Septic shock | 80 (49.4) | 21 (46.7) | 59 (50.4) | 0.727 |
| Acute kidney injury | 68 (42.0) | 16 (35.6) | 52 (44.4) | 0.375 |
| Mechanical ventilation care b | 81 (50.0) | 15 (33.3) | 53 (45.3) | 0.214 |
| Surgery within 30 d | 45 (27.8) | 12 (26.7) | 33 (28.2) | 1.000 |
| Pitt bacteremia score | 3.3 ± 2.9 | 3.1 ± 2.6 | 3.4 ± 3.0 | 0.515 |
| Charlson WIC | 3.2 ± 2.6 | 3.4 ± 2.9 | 3.1 ± 2.4 | 0.525 |
| Site of infection | ||||
| Pneumonia | 59 (36.4) | 12 (26.7) | 47 (40.2) | 0.145 |
| CRBSI | 41 (25.3) | 20 (44.4) | 21 (17.9) | 0.001 |
| Intra-abdominal infection | 34 (21.0) | 4 (8.9) | 30 (25.6) | 0.303 |
| Primary bacteremia | 17 (10.5) | 4 (8.9) | 13 (11.1) | 0.782c |
| Skin and soft tissue infection | 8 (4.9) | 4 (8.9) | 4 (3.4) | 0.219c |
| Urinary tract infection | 3 (1.9) | 1 (2.2) | 2 (1.7) | 1.000 |
| Polymicrobial bacteremia | 32 (19.8) | 9 (20.0) | 23 (19.7) | 1.000 |
| Bacteremic episodes in ICU | 99 (61.1) | 29 (64.4) | 70 (59.8) | 0.719 |
| Hospital-acquired infection | 149 (92.0) | 44 (97.8) | 105 (89.7) | 0.114c |
| Appropriate antibiotics ≤ 48 h | 57 (35.2) | 16 (35.6) | 41 (35.0) | 1.000 |
| Antibiotic resistance | ||||
| MDR | 144 (88.9) | 41 (91.1) | 103 (88.0) | 0.781 |
| CR | 137 (84.6) | 40 (88.9) | 97 (82.9) | 0.468 |
| XDR | 132 (81.5) | 40 (88.9) | 92 (78.6) | 0.176 |
| 30-day mortality | 94 (58.0) | 25 (55.6) | 69 (59.0) | 0.725 |
Notes.
Data are expressed as no. (%) of patients, unless otherwise indicated. Continuous variables expressed as mean and standard deviation. Bold indicates P < 0.05. HSCT, haematopoietic stem cell transplantation; WIC, weighted index of comorbidity; CRBSI, catheter-related bloodstream infection; ICU, intensive care unit; MDR, multidrug-resistant; CR, carbapenem-resistant; XDR, extensively drug-resistant;
Mechanical ventilation care before bacteremia onset;
Fisher's exact test
Patients with T6SS+ CRAB infections also had higher frequencies of HSCT (15.0% vs. 3.1%, P = 0.018), treatment with immunosuppressive agents (60.0% vs. 40.2%, P = 0.040), and catheter-related bloodstream infection (50.0% vs. 21.6%, P = 0.002), as well as a lower frequency of intra-abdominal infection than patients with T6SS– CRAB infections (10.0% vs. 30.9%, P = 0.036; Table S2). There was no significant difference in the mortality rates between patients with T6SS+ and T6SS– CRAB infections. The variables associated with mortality of patients with A. baumannii bacteremia were also analyzed. In multivariate analysis, hematologic malignancy (OR, 2.945; 95% CI, 1.523–5.692; P = 0.001), acute kidney injury (OR, 2.977; 95% CI, 1.214–7.304; P = 0.017), higher Pitt bacteremia score (OR, 1.359; 95% CI, 1.130–1.635; P = 0.001), and pneumonia (OR, 2.441; 95% CI, 1.049–5.678; P = 0.038) were prognostic factors associated with 30-day mortality (Table S3). However, T6SS+ was not related to 30-day mortality.
Discussion
A previous study reported that T6SS genes are conserved among several A. baumannii strains regardless of T6SS activity.12 However, another comparative study, based on genome sequences, showed that the T6SS locus is only present in isolates of a particular clade.29 In the present study, the hcp gene, an essential component of the T6SS, was detected in about one-third of A. baumannii isolates from bloodstream infections, which supports the latter study. In addition, our results indicate that the hcp gene (i.e., the T6SS locus) is present mainly in specific clones. Except ST208 and ST369, all isolates of a specific clone contained the hcp gene, and none of isolates of other clone contained the gene. Specifically, no isolates of ST191, a presumed ancestral clone of GC2, contained a T6SS. Thus, we speculated that the T6SS locus was incorporated independently into A. baumannii isolates during the divergence of ST191 into other clones. Sporadic T6SSs were found in minor clones of singletons, such as ST491, ST613, ST1131, ST1312, and ST1320, which also suggests independent introduction. Further investigation based on whole genomes would verify it.
It has been reported that the T6SS is usually silenced and is only activated under stress responses, such as nutrient limitation, cell damage, and ecological competition [9]. In addition, a recent study reported that the T6SS of A. baumannii was activated by loss of a MDR plasmid containing its repressor, tetR.15 When we assayed 51 hcp+ isolates for the presence of tetR, only 4 isolates (AC40, B621, B699, and B754) possessed tetR. All of the tetR-positive isolates belonged to ST229, and the T6SS in these isolates was either inactive or showed low activity since the hcp expression levels in these isolates were more than 30-fold lower than that in ATCC 19606. However, tetR was not detected in other hcp+ isolates with inactive T6SS. That is, inactivation of T6SS by a MDR plasmid containing tetR may be specific to a certain strain, and additional regulatory systems may be associated with the secretion of Hcp, as suggested by Repizo et al.14 In addition, a trade-off between antibiotic resistance and T6SS was not identified in our study. Although it was recently shown that the T6SS leads to DNA release and induces horizontal gene transfer, which may contribute to the spread of antibiotic resistance in V. cholera,30 we did not find any correlation between antibiotic resistance and T6SS in this study. Thus, the mechanisms underlying the repression of T6SS in A. baumannii should be investigated further.
In this study, we observed that T6SS+ isolates better outcompeted E. coli compared with T6SS–isolates, including both hcp– and inactivated hcp+ isolates. The competition index of 2 isolates containing inactivated hcp gene did not differ from that of the hcp– isolates. T6SS-mediated bacterial killing has been demonstrated in several bacterial species, including A. baumannii.9,12,31-34 However, only a few studies included multiple clinical isolates. Our results, based on several T6SS+, inactivated hcp+, and hcp– clinical isolates, statistically support T6SS-mediated bacterial killing.
Our results also showed the biofilm-forming activity and resistance to human serum of T6SS+ A. baumannii isolates. The correlation between high biofilm-forming activity and high resistance to human serum in T6SS+ isolates has been well documented,35 since growth in a biofilm allows bacteria to evade the host immune response.36 Consistent with the increased biofilm formation in T6SS+ isolates, patients with T6SS+ isolates had significantly more frequent catheter-related bloodstream infections. A. baumannii controls biofilm formation in response to cell density through quorum sensing,37 and T6SS is often induced under high cell density.38,39 In addition, resistance to human serum in vitro might be responsible for the bacteremia in patients treated with immunosuppressive agents who had compromised immune systems.40 These findings point to the potential roles of T6SS beyond antagonism, which could be used for defense when the cells acquire spatial segregation after clonal expansion.4 Our results suggest that T6SS are beneficial for host colonization and survival in A. baumannii.
However, it is not clear whether the high biofilm-forming activity and serum resistance of T6SS+ isolates is due to the function of the T6SS itself. Several studies have demonstrated that the T6SS functions in biofilm-forming activity in several bacterial species;41-44 however, some bacteria do not require the T6SS for biofilm formation.45 The A. baumannii reference strain ATCC 17978 has a functional T6SS, but does not form biofilms.13,14 Repizo et al.14 showed that the T6SS of an environmental A. baumannii isolate, DSM30011, was not required for biofilm formation through experiments with a tssM-deletion mutant. Thus, it is unclear whether the high biofilm-forming activity of T6SS+ isolates in this study was due to the presence of the T6SS or other features. In addition, the possibility that the high proportion of catheter-related bloodstream infection in the T6SS+ patient group may be due to the enhanced characteristics of certain clones rather than the T6SS itself cannot be excluded. Higher biofilm formation in a certain A. baumannii clone was previously reported.46 However, we also found that the differences in clinical characteristics, such a higher rate of immunosuppression and a lower rate of pneumonia and intra-abdominal infection in T6SS+ cases. High rate of immunosuppression in T6SS+ cases may be partially due to high resistance to human serum. Clinically, pneumonia and intra-abdominal infections often include diverse bacterial species.47 However, our study included only bacteremic patients rather than patients with chronic infections who were likely to be colonized by mature communities of A. baumannii. Moreover, T6SS was less frequent in cases of polymicrobial bacteremia or ventilation. In addition, the 30-day mortality rates in patients with A. baumannii bloodstream infections was higher in the T6SS+ group; however, the difference was not significant (35.3% vs. 28.7%, P = 0.396). T6SS was not a prognostic factor for mortality in patients with A. baumannii bacteremia. Thus, additional studies should be performed to determine if the T6SS itself is involved in the virulence of bacteria, including A. baumannii.
One-third of the A. baumannii clinical isolates in this study contained T6SS. A. baumannii isolates with a functional T6SS exhibited the capability to outcompete E. coli, biofilm-forming activity, and increased resistance to human serum in vitro. These phenotypes were also in agreement with the clinical features of T6SS+ in patients with A. baumannii bacteremia, including catheter-related bloodstream infections, receiving immunosuppressive agents, and HSCT recipients. However, it is unclear if these characteristics are due to T6SS itself.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT, and Future Planning (grant no. NRF-2016R1A2A2A05005075). JY Lee was partly supported by the Basic Science Research Program through the NRF funded by the Ministry of Education (grant no. NRF-2016R1A6A3A11931488).
References
- [1].Jani AJ, Cotter PA. Type VI secretion: not just for pathogenesis anymore. Cell Host Microbe 2010; 8:2-6; PMID:20638635; https://doi.org/ 10.1016/j.chom.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA 2006; 103:1528-33; PMID:16432199; https://doi.org/ 10.1073/pnas.0510322103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mougous JD, Cuff ME, Raunser S, Shen A, Zhou M, Gifford CA, Goodman AL, Joachimiak G, Ordoñez CL, Lory S, Walz T, Joachimiak A, Mekalanos JJ. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 2006; 312:1526-530; PMID:16763151; https://doi.org/ 10.1126/science.1128393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Russell AB, Peterson SB, Mougous JD. Type VI secretion system effectors: poisons with a purpose. Nat Rev Microbiol 2014; 12:137-48; PMID:24384601; https://doi.org/ 10.1038/nrmicro3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 2012; 483:182-6; PMID:22367545; https://doi.org/ 10.1038/nature10846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Clemens DL, Ge P, Lee BY, Horwitz MA, Zhou ZH. Atomic structure of T6SS reveals interlaced array essential to function. Cell 2015; 160:940-51; PMID:25723168; https://doi.org/ 10.1016/j.cell.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Silverman JM, Brunet YR, Cascales E, Mougous JD. Structure and regulation of the type VI secretion system. Annu Rev Microbiol 2012; 66:453-72; PMID:22746332; https://doi.org/ 10.1146/annurev-micro-121809-151619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 2011; 475:343-7; PMID:21776080; https://doi.org/ 10.1038/nature10244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Basler M, Ho BT, Mekalanos JJ. Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 2013; 152:884-94; PMID:23415234; https://doi.org/ 10.1016/j.cell.2013.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ho BT, Dong TG, Mekalanos JJ. A view to a kill: the bacterial type VI secretion system. Cell Host Microbe 2014; 15:9-21; PMID:24332978; https://doi.org/ 10.1016/j.chom.2013.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Henry R, Vithanage N, Harrison P, Seemann T, Coutts S, Moffatt JH, Nation RL, Li J, Harper M, Adler B, Boyce JD. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-beta-1,6-N-acetylglucosamine. Antimicrob Agents Chemother 2012; 56:59-69; PMID:22024825; https://doi.org/ 10.1128/AAC.05191-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Carruthers MD, Nicholson PA, Tracy EN, Munson RS Jr. Acinetobacter baumannii utilizes a type VI secretion system for bacterial competition. PLoS One 2013; 8:e59388; PMID:23527179; https://doi.org/ 10.1371/journal.pone.0059388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Weber BS, Miyata ST, Iwashkiw JA, Mortensen BL, Skaar EP, Pukatzki S, Feldman MF. Genomic and functional analysis of the type VI secretion system in Acinetobacter. PLoS One 2013; 8:e55142; PMID:23365692; https://doi.org/ 10.1371/journal.pone.0055142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Repizo GD, Gagne S, Foucault-Grunenwald ML, Borges V, Charpentier X, Limansky AS, Gomes JP, Viale AM, Salcedo SP. Differential role of the T6SS in Acinetobacter baumannii virulence. PLoS One 2015; 10:e0138265; PMID:26401654; https://doi.org/ 10.1371/journal.pone.0138265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Weber BS, Ly PM, Irwin JN, Pukatzki S, Feldman MF. A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci USA 2015; 112:9442-7; PMID:26170289; https://doi.org/ 10.1073/pnas.1502966112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].La Scola B, Gundi VA, Khamis A, Raoult D. Sequencing of the rpoB gene and flanking spacers for molecular identification of Acinetobacter species. J Clin Microbiol 2006; 44:827-32; PMID:16517861; https://doi.org/ 10.1128/JCM.44.3.827-832.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ko KS, Suh JY, Kwon KT, Jung SI, Park KH, Kang CI, Chung DR, Peck KR, Song JH. High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J Antimicrob Chemother 2007; 60:1163-7; PMID:17761499; https://doi.org/ 10.1093/jac/dkm305 [DOI] [PubMed] [Google Scholar]
- [18].Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. M100-S25. CLSI, Wayne, PA: 2015. [Google Scholar]
- [19].Bartual SG, Seifert H, Hippler C, Luzon MA, Wisplinghoff H, Rodriguez-Valera F. Development of a multilocus sequence typing scheme for characterization of clinical isolates of Acinetobacter baumannii. J Clin Microbiol 2005; 43:4382-90; PMID:16145081; https://doi.org/ 10.1128/JCM.43.9.4382-4390.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Francisco AP, Vaz C, Monteiro PT, Melo-Cristono J, Ramirez M, Carrico JA. PHYLOViZ: phhylogenetic inference and data visualization for sequence based tying methods. BMC Bioinformatics 2012; 13:87; PMID:22568821; https://doi.org/ 10.1186/1471-2105-13-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Coyne S, Guigon G, Courvalin P, Perichon B. Screening and quantification of the expression of antibiotic resistance genes in Acinetobacter baumannii with a microarray. Antimicrob Agents Chemother 2010; 54:333-40; PMID:19884373; https://doi.org/ 10.1128/AAC.01037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Merritt JH, Kadouri DE, O'Toole GA. Growing and analyzing static biofilms. Curr Protoc Microbiol 2005; Chapter 1:1B.1.1 -1B.1.17.; PMID:18770545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shin J, Ko KS. Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int J Antimicrob Agents 2015; 46:214-8; PMID:26116415; https://doi.org/ 10.1016/j.ijantimicag.2015.04.012 [DOI] [PubMed] [Google Scholar]
- [24].Lesens O, Methlin C, Hansmann Y, Remy V, Martinot M, Bergin C, Meyer P, Christmann D. Role of comorbidity in mortality related to Staphylococcus aureus bacteremia: a prospective study using the Charlson weighted index of comorbidity. Infect Control Hosp Epidemiol 2003; 24:890-6; PMID:14700403; https://doi.org/ 10.1086/502156 [DOI] [PubMed] [Google Scholar]
- [25].Paterson DL, Ko WC, Von Gottberg A, Mohapatra S, Casellas JM, Goossens H, Mulazimoglu L, Trenholme G, Klugman KP, Bonomo RA, Rice LB, Wagener MM, McCormack JG, Yu VL. International prospective study of Klebsiella pneumoniae bacteremia: implications of extended-spectrum beta-lactamase production in nosocomial infections. Ann Intern Med 2004; 140:26-32; PMID:14706969; https://doi.org/ 10.7326/0003-4819-140-1-200401060-00008 [DOI] [PubMed] [Google Scholar]
- [26].Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18:268-81; PMID:21793988; https://doi.org/ 10.1111/j.1469-0691.2011.03570.x [DOI] [PubMed] [Google Scholar]
- [27].Mehta RL, Kellum JA, Shah SV, Molitoris BA, Ronco C, Warnock DG, Levin A; Acute Kidney Injury Network . Acute Kidney Injury Network: report of an initiative to improve outcomes in acute kidney injury. Crit Care 2007; 11:R31; PMID:17331245; https://doi.org/ 10.1186/cc5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Friedman ND, Kaye KS, Stout JE, McGarry SA, Trivette SL, Briggs JP, Lamm W, Clark C, MacFarquhar J, Walton AL, Reller LB, Sexton DJ. Health care-associated bloodstream infections in adults: a reason to change the accepted definition of community-acquired infections. Ann Intern Med 2002; 137:791-7; PMID:12435215; https://doi.org/ 10.7326/0003-4819-137-10-200211190-00007 [DOI] [PubMed] [Google Scholar]
- [29].Wright MS, Haft DH, Harkins DM, Perez F, Hujer KM, Bajaksouzian S, Benard MF, Jacobs MR, Bonomo RA, Adams MD. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. MBio 2014; 5:e00963-13; PMID:24449752; https://doi.org/ 10.1128/mBio.00963-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Borgeaud S, Metzger LC, Scrignari T, Blokesch M. The type VI secretion system of Vibrio cholera fosters horizontal gene transfer. Science 2015; 347:63-7; PMID:25554784; https://doi.org/ 10.1126/science.1260064 [DOI] [PubMed] [Google Scholar]
- [31].Brunet YR, Espinosa L, Harchouni S, Mignot T, Cascales E. Imaging type VI secretion-mediated bacterial killing. Cell Reports 2013; 3:36-41; PMID:23291094; https://doi.org/ 10.1016/j.celrep.2012.11.027 [DOI] [PubMed] [Google Scholar]
- [32].Bernardy EE, Turnsek MA, Wilson SK, Tarr CL, Hammer BK. Diversity of clinical and environmental isolates of Vibrio cholerae in natural transformation and contact-dependent bacterial killing indicative of type VI secretion system activity. Appl Environ Microbiol 2016; 82:2833-2842; PMID:26944842; https://doi.org/ 10.1128/AEM.00351-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Nat'l Acad Sci USA 2016; 113:3627-32; https://doi.org/ 10.1073/pnas.1522510113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Sana TG, Flaugnatti N, Lugo KA, Lam LH, Jacobson A, Baylot V, Durand E, Journet L, Cascales E, Monack DM. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc Natl Acad Sci USA 2016; 113:E5044-51; PMID:27503894; https://doi.org/ 10.1073/pnas.1608858113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].King LB, Swiatlo E, Swiatlo A, McDaniel LS. Serum resistance and biofilm formation in clinical isolates of Acinetobacter baumannii. FEMS Immunol Med Microbiol 2009; 55:414-21; PMID:19220466; https://doi.org/ 10.1111/j.1574-695X.2009.00538.x [DOI] [PubMed] [Google Scholar]
- [36].Gaddy JA, Actis LA. Regulation of Acinetobacter baumannii biofilm formation. Future Microbiol 2009; 4:273-8; PMID:19327114; https://doi.org/ 10.2217/fmb.09.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bhargava N, Sharma P, Capalash N. Quorum sensing in Acinetobacter: an emerging pathogen. Crit Rev Microbiol 2010; 36:349-60; PMID:20846031; https://doi.org/ 10.3109/1040841X.2010.512269 [DOI] [PubMed] [Google Scholar]
- [38].Lesic B, Starkey M, He J, Hazan R, Rahme LG. Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 2009; 155:2845-955; PMID:19497948; https://doi.org/ 10.1099/mic.0.029082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bernard CS, Brunet YR, Gueguen E, Cascales E. Nooks and crannies in type VI secretion regulation. J Bacteriol 2010; 192:3850-60; PMID:20511495; https://doi.org/ 10.1128/JB.00370-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Liao CH, Sheng WH, Chen YC, Hung CC, Wang JT, Chang SC. Predictive value of the serum bactericidal test for mortality in patients infected with multidrug-resistant Acinetobacter baumannii. J Infect 2007; 55:149-57; PMID:17376533; https://doi.org/ 10.1016/j.jinf.2007.01.015 [DOI] [PubMed] [Google Scholar]
- [41].Enos-Berlage JL, Guvener ZT, Keenan CE, McCarter LL. Genetic determinants of biofilm development of opaque and translucent Vibrio parahaemolyticus. Mol Microbiol 2005; 55:1160-82; PMID:15686562; https://doi.org/ 10.1111/j.1365-2958.2004.04453.x [DOI] [PubMed] [Google Scholar]
- [42].Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J Bacteriol 2008; 190:7523-31; PMID:18805985; https://doi.org/ 10.1128/JB.00945-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Zhang L, Hinz AJ, Nadeau JP, Mah TF. Pseudomonas aeruginosa tssC1 links type VI secretion and biofilm-specific antibiotic resistance. J Bacteriol 2011; 193:5510-3; PMID:21784934; https://doi.org/ 10.1128/JB.00268-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang L, Xu J, Xu J, Zhang H, He L, Feng J. TssB is essential for virulence and required for type VI secretion system in Ralstonia solanacearum. Microb Pathog 2014; 74:1-7; PMID:24972114; https://doi.org/ 10.1016/j.micpath.2014.06.006 [DOI] [PubMed] [Google Scholar]
- [45].Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 2010; 6:e1001068; PMID:20865170; https://doi.org/ 10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Na IY, Chung ES, Jung CY, Kim DH, Shin J, Kang KJ, Kim ST, Ko KS. Comparison of the virulence-associated phenotypes of five species of Acinetobacter baumannii complex. J Microbiol Biotechnol 2016; 26:182-90; https://doi.org/ 10.4014/jmb.1507.07076 [DOI] [PubMed] [Google Scholar]
- [47].Shah PM, Edwards BL, Dietch ZC, Guidry CA, Davies SW, Hennessy SA, Duane TM, O'Neill PJ, Coimbra R, Cook CH, Askari R, Popovsky K, Sawyer RG. Do polymicrobial intra-abdominal infections have worse outcomes than monomicrobial intra-abdominal infections. Surg Infect (Larchmt) 2016; 17:27-31; PMID:26397376; https://doi.org/ 10.1089/sur.2015.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


