Planarians are freshwater non-parasitic flatworms that have fascinated researchers for more than 200 years.1 Planarians have incredible capacities for whole body regeneration. While humans or mice have only limited potential to regenerate their tissues (mainly skin, liver and bone marrow), the planarian flatworm is able to regenerate a complete organism including its surprisingly complex brain from a tiny piece of its body within 7–10 days.2-5 This is due to the fact that different to humans or mice who have a very limited number of stem cells distributed in stem cell niches in very specific regions of the body, planarians possess many adult somatic stem cells called neoblasts distributed along the whole planarian body. Neoblasts account for approximately 25% of the total number of cells in the parenchyma6 and include a population of pluripotent stem cells.7 Often planarians present homologs for human genes that are not present either in Drosophila melanogaster or Caenorhabditis elegans. Technological advances to aid in the study of planarians include genomics, transcriptomics (including single-cell transcriptomics), proteomics, high-throughput RNAi-screens, behavioral platforms (learning and memory), and drug toxicity assays.8-20 Altogether the planarian model has become an excellent system for investigating in vivo stem cell biology and regeneration and it is an emerging biomedical model to study cancer and diverse human pathologies.21-25
Although the immune system has been described as an important modulator of wound repair and regeneration26-28 it is largely unexplored in planarians. It is likely that the mucus that covers the planarian body and whose production is greatly increased after wounding has anti-microbial properties.29,30 Proteomic analyses have shown that planarian mucus proteins display similarities with the ones in human mucosal secretions31 and that the introduction of bacterial endotoxins into planarian wounds induces several potential immunity related genes, some of which share sequence similarity with vertebrate innate immunity genes.32 Phagocytic activity has also been observed in planarians by means of electron microscopy where putative phagocytic cells called reticular cells have been observed phagocytosing bacteria and also cellular debris during regeneration.33,34 Finally, it has been discovered that the planarian genome contains many potential homologs of genes related to innate immunity and that some of them are activated during regeneration.35,36 Recently two publications have shown the potential of the planarian model to become an excellent system for investigating immunity. The first one comes also from the laboratory of Eric Ghigo37 in which they demonstrate that planarians are highly resistant to infection with bacteria pathogenic to Homo sapiens, C. elegans and/or D. melanogaster and thus are a good model to identify novel innate resistance mechanisms in humans. Indeed, after performing whole-transcriptome analysis coupled with an RNAi screen in planarians challenged with bacteria, they were able to identify the novel regulator of mammalian LC3-associated phagocytosis MORN2. The second study elucidated the composition of the planarian microbiome.38 They observed that culture conditions that increase susceptibility to tissue lesions or the process of wounding itself coincided with an increase in Proteobacteria. Interestingly, they found that the elucidated planarian microbiome has a distribution of bacterial phyla similar to the human lower intestinal microbiome. This phyla composition is not present in other invertebrate or vertebrate systems that are consolidated models for the study of immunity and host-microbe interactions.39-41
In this issue of Virulence, the laboratory of Eric Ghigo (Tsoumtsa et al.)42 set out to investigate how the circadian clock regulates the capacity of planarians to fight infection. The circadian clock is an internal time-keeping regulator present in most if not all of the cells of most organisms. It regulates many physiological processes in eukaryotes, from sleep/wake cycles, metabolism, body temperature, blood pressure or cardiovascular function in mammals to growth and photosynthesis in plants. The photoperiod is the most important Zeitgeber (time giver) for circadian oscillators in all investigated organisms so far. In mammals the central oscillator is the suprachiasmatic nucleus (SCN) located at the base of the hypothalamus in the brain, which is in charge of coordinating more peripheral oscillators present in the cells of many other organs. The circadian rhythm is maintained through 24 hours oscillating transcriptional-translational feedback loops that regulate the rhythmic expression of thousands of genes, which is translated into rhythms in metabolism and behavior.43,44 Currently it is known that RNA-level post-transcription mechanisms, such as mRNA polyadenylation45 and RNA methylation,46 are at least as important regulators of the circadian clock as the activation/repression of transcription. The vertebrate primary feedback loop includes among others the transcription factors BMAL1 (also called in vertebrates ARNTL1 or Cycle in Drosophila), CLOCK, PERIOD (PER1, PER2 and PER3) and two CRYPTOCHROME proteins (CRY1 and CRY2).47,48 These components are not always all present in other peripheral oscillators of other tissues or other organisms or even if expressed, other transcription factors may play their role. For instance, in Drosophila the function of CRY is performed by Timeless (Tim),49 although in mammals there is a TIMELESS-LIKE protein, which has other functions in addition to regulating the circadian clock. In the same way, in the forebrain of mice, NPAS acts as a functional substitute for CLOCK.43
Although the circadian rhythm may be an important regulator of diverse physiological functions in planarians, it remains largely unstudied. The evidences for a planarian circadian rhythm are few. One evidence is the presence of a 24-hour cycle of melatonin production. In vertebrates, one of the main functions of melatonin is as a hormonal output of the circadian clock in the SCN in the brain. Circadian clock genes in the SCN control melatonin production through signaling to the pineal gland, which secretes melatonin that signals in photoperiod to the other peripheral clocks in the body.47 It has been suggested that planarians possess a circadian clock controlling melatonin synthesis, since melatonin and both its precursor serotonin and its synthesizing enzymes N-acetyltransferase (NAT) and hydroxyindole-O-methyltransferase (HIOMT) fluctuate following a diurnal variation, peaking during the dark period.50-52 In addition, it has been shown that planarian asexual reproduction through fission follows photoperiodicity in a day-night rhythm with reproduction occurring preferentially at night. Furthermore, fissioning is inhibited when maintaining planarians under continuous light.53,54 It has also been hypothesized that melatonin release from the brain may mediate the influence of environmental photoperiods on fissioning since amputation of the planarian head breaks this rhythm and increases the fissioning rate50
Tsoumtsa et al. seek for circadian genes in the planarian genome that regulate immunity by searching for those whose downregulation was previously linked in other model systems to an increase in the susceptibility to bacteria infection. They found homologs for Arntl1 (Smed-arntl-1 in planarians) and for Timeless (Smed-tim in planarians) but they could not find either Clock or Per-2. Thus, at least Clock may have been lost in the planarian lineage, since a Clock gene has been reported in the cnidarian Nematostella vectensis.55 A requirement for those genes being part of the circadian clock is that their expression shows a 24 hours expression cycle. Indeed, they observed that Smed-tim expression changes during light/dark (L/D) conditions with a maximum peak of expression in the dark phase in a similar way to what happens in Drosophila.56 On the other hand, when planarians were entrained to a L/D condition and then exposed to a dark/dark (D/D) condition they showed a desynchronized Smed-tim expression. Although clear daily oscillations in melatonin production are present in many protostomes including planarians, a clear association between circadian clock genes and melatonin production has been done only in vertebrates and recently also in the cnidarian Nematostella vectensis.57 A possible experiment in planarians that would clarify this relationship would consist in trying to reset the circadian pattern of expression of Smed-tim in D/D conditions by exposing animals to melatonin, as reported in Nematostella.57 Altogether these are very promising results that further support the idea of a circadian clock in planarians. It would be interesting to obtain the transcriptional profile of planarians at different time points during the L/D cycle. This would help to identify which physiological processes in addition to asexual reproduction, the circadian clock is regulating in planarians and to better understand the workings of the planarian circadian clock.
The main focus of Tsoumtsa et al. is to find out the relationship between the circadian clock genes and infection. Emerging evidence indicates that the immune system is under tight regulation of the circadian clock. It has been observed that perturbations of circadian rhythms impair immune function and thus health. For instance, epidemiological studies on shift-workers have shown that working during the night increases the risk for several cancers, obesity, diabetes, and cardiovascular problems. It is known that neural and endocrine signals transmit circadian information to immune tissues and that immune cells express molecular clock components to mediate immune responses including NK cytotoxicity, phagocytosis, and inflamation.58 In vertebrates the use of clock genes mutant mice exposed to bacterial infection has demonstrated their role in immune function. It is known that circadian clock genes regulate the expression of several transcription factors involved in the immune system, including Stat3 and Stat5, Egr1 and NF-kB, and also control NK cell and macrophage activities.44,58 In invertebrates there are also many evidences showing that circadian clock genes control the immune system. These are based on Drosophila mutants for clock genes, which have been or not infected with bacteria. For instance, in Drosophila the resistance to Staphylococcus pneumoniae oscillates daily. However, these oscillations are absent in Tim mutants. Indeed, it was shown that Tim regulates phagocytosis, with a maximum activity at night. While Tim mutants have normal phagocytic activity during the day, they lack the night peak.56 Tsoumtsa et al. followed a similar approach to the one described here. The authors kept planarians under a L/D cycle or under a D/D cycle and then downregulated Smed-tim by RNA interference (RNAi). Planarians were then fed with Staphylococcus aureus. The authors could observe that Smed-tim RNAi treated planarians took longer than controls to eliminate S. aureus when kept under L/D conditions. Under D/D conditions the time to clear S. aureus was similar to controls. They concluded that Smed-tim is necessary for efficiently eliminate S. aureus under L/D conditions (Fig. 1). The authors also sought into the mechanism that Smed-tim signals to clear S. aureus. They found that the expression of antimicrobial genes p38 map-kinase, traf6 and morn2 in planarians are significantly induced after S. aureus infection. However, Smed-tim RNAi planarians had a significant decrease only in the expression of morn2 and traf6 and only under L/D conditions. Open questions remains as to if Smed-tim regulates resistance to other bacteria and whether Smed-arntl-1 has any antibacterial activity even though it does not regulate resistance to S. aureus. It would also be interesting to know whether phagocytosis is the main process regulated by Smed-tim and whether a peak of phagocytosis exists during the night.
Figure 1.
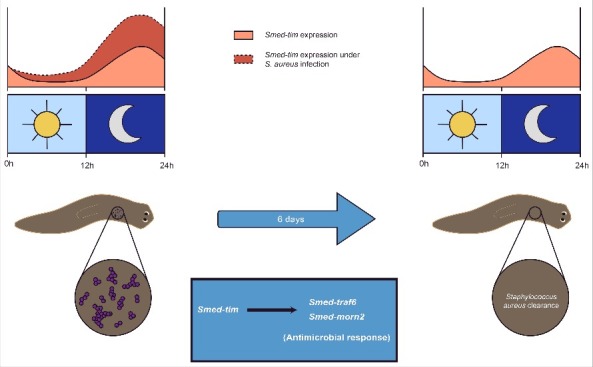
The antimicrobial response of planarians against Staphylococcus aureus is under the control of timeless. During light/dark cycles, planarians are able to eliminate S. aureus in 6 days through the induction of Smed-tim. Smed-tim promotes the antibacterial response by regulating Smed-traf6 and Smed-morn2.
The study of Tsoumtsa et al. uncovers a function of the gene timeless as a member of the planarian circadian clock and regulator of immune response to S. aureus in planarians. The paper opens up for further work not only into the relationship between the circadian clock and immunity but also into the functions and mechanisms of the circadian clock in planarians. It also contributes in consolidating the planarian system as a model for the study of immunity.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- [1].Morgan TH. Experimental studies of the regeneration of Planaria maculata. Archiv für Entwickelungsmechanik der organismen 1898; 7:364-97; http://dx.doi.org/ 10.1007/BF02161491 [DOI] [Google Scholar]
- [2].Aboobaker AA. Planarian stem cells: A simple paradigm for regeneration. Trends Cell Biol 2011; 21:304-11; PMID:21353778; http://dx.doi.org/ 10.1016/j.tcb.2011.01.005 [DOI] [PubMed] [Google Scholar]
- [3].Elliott SA, Sanchez Alvarado A. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip Rev Dev Biol 2013; 2:301-26; PMID:23799578; http://dx.doi.org/ 10.1002/wdev.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gentile L, Cebria F, Bartscherer K. The planarian flatworm: An in vivo model for stem cell biology and nervous system regeneration. Dis Model Mech 2011; 4:12-9; PMID:21135057; http://dx.doi.org/ 10.1242/dmm.006692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gonzalez-Estevez C. Autophagy meets planarians. Autophagy 2009; 5:290-7; PMID:19164934; http://dx.doi.org/ 10.4161/auto.5.3.7665 [DOI] [PubMed] [Google Scholar]
- [6].Baguñà J. Mitosis in the intact and regenerating planarian Dugesia mediterranea n. sp. II. Mitotic studies during regeneration and a possible mechanism of blastema formation. J Exp Zool 1976; 195:65-80; http://dx.doi.org/ 10.1002/jez.1401950107 [DOI] [Google Scholar]
- [7].Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science 2011; 332:811-6; PMID:21566185; http://dx.doi.org/ 10.1126/science.1203983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Robb SM, Gotting K, Ross E, Sanchez Alvarado A. SmedGD 2.0: The schmidtea mediterranea genome database. Genesis 2015; 53:535-46; PMID:26138588; http://dx.doi.org/ 10.1002/dvg.22872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Abril JF, Cebria F, Rodriguez-Esteban G, Horn T, Fraguas S, Calvo B, Bartscherer K, Saló E. Smed454 dataset: Unravelling the transcriptome of schmidtea mediterranea. BMC Genomics 2010; 11:731; PMID:21194483; http://dx.doi.org/ 10.1186/1471-2164-11-731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Blythe MJ, Kao D, Malla S, Rowsell J, Wilson R, Evans D, Jowett J, Hall A, Lemay V, Lam S, et al.. A dual platform approach to transcript discovery for the planarian schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology. PLoS One 2010; 5:e15617; PMID:21179477; http://dx.doi.org/ 10.1371/journal.pone.0015617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].van Wolfswinkel JC, Wagner DE, Reddien PW. Single-cell analysis reveals functionally distinct classes within the planarian stem cell compartment. Cell Stem Cell 2014; 15:326-39; PMID:25017721; http://dx.doi.org/ 10.1016/j.stem.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Fernandez-Taboada E, Rodriguez-Esteban G, Salo E, Abril JF. A proteomics approach to decipher the molecular nature of planarian stem cells. BMC Genomics 2011; 12:133; PMID:21356107; http://dx.doi.org/ 10.1186/1471-2164-12-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Boser A, Drexler HC, Reuter H, Schmitz H, Wu G, Scholer HR, Gentile L, Bartscherer K. SILAC proteomics of planarians identifies Ncoa5 as a conserved component of pluripotent stem cells. Cell Rep 2013; 5:1142-55; PMID:24268775; http://dx.doi.org/ 10.1016/j.celrep.2013.10.035 [DOI] [PubMed] [Google Scholar]
- [14].Reddien PW, Bermange AL, Murfitt KJ, Jennings JR, Sanchez Alvarado A. Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev Cell 2005; 8:635-49; PMID:15866156; http://dx.doi.org/ 10.1016/j.devcel.2005.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Inoue T, Kumamoto H, Okamoto K, Umesono Y, Sakai M, Sanchez Alvarado A, Agata K. Morphological and functional recovery of the planarian photosensing system during head regeneration. Zoolog Sci 2004; 21:275-83; PMID:15056922; http://dx.doi.org/ 10.2108/zsj.21.275 [DOI] [PubMed] [Google Scholar]
- [16].Inoue T, Hoshino H, Yamashita T, Shimoyama S, Agata K. Planarian shows decision-making behavior in response to multiple stimuli by integrative brain function. Zoological Lett 2015; 1:7; PMID:26605052; http://dx.doi.org/ 10.1186/s40851-014-0010-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hagstrom D, Cochet-Escartin O, Collins EM. Planarian brain regeneration as a model system for developmental neurotoxicology. Regeneration (Oxf) 2016; 3:65-77; PMID:27499880; http://dx.doi.org/ 10.1002/reg2.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Shomrat T, Levin M. An automated training paradigm reveals long-term memory in planarians and its persistence through head regeneration. J Exp Biol 2013; 216:3799-810; PMID:23821717; http://dx.doi.org/ 10.1242/jeb.087809 [DOI] [PubMed] [Google Scholar]
- [19].Pagan OR, Baker D, Deats S, Montgomery E, Tenaglia M, Randolph C, Kotturu D, Tallarida C, Bach D, Wilk G, et al.. Planarians in pharmacology: Parthenolide is a specific behavioral antagonist of cocaine in the planarian girardia tigrina. Int J Dev Biol 2012; 56:193-6; PMID:22451007; http://dx.doi.org/ 10.1387/ijdb.113486op [DOI] [PubMed] [Google Scholar]
- [20].Plusquin M, Stevens AS, Van Belleghem F, Degheselle O, Van Roten A, Vroonen J, Blust R, Cuypers A, Artois T, Smeets K. Physiological and molecular characterisation of cadmium stress in Schmidtea mediterranea. Int J Dev Biol 2012; 56:183-91; PMID:22451006; http://dx.doi.org/ 10.1387/ijdb.113485mp [DOI] [PubMed] [Google Scholar]
- [21].Gonzalez-Estevez C, Felix DA, Smith MD, Paps J, Morley SJ, James V, Sharp TV, Aboobaker AA. SMG-1 and mTORC1 act antagonistically to regulate response to injury and growth in planarians. PLoS Genet 2012; 8:e1002619; PMID:22479207; http://dx.doi.org/ 10.1371/journal.pgen.1002619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Oviedo NJ, Pearson BJ, Levin M, Sanchez Alvarado A. Planarian PTEN homologs regulate stem cells and regeneration through TOR signaling. Dis Model Mech 2008; 1:131-43; discussion 41; PMID:19048075; http://dx.doi.org/ 10.1242/dmm.000117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pearson BJ, Sanchez Alvarado A. Regeneration, stem cells, and the evolution of tumor suppression. Cold Spring Harb Symp Quant Biol 2008; 73:565-72; PMID:19150962; http://dx.doi.org/ 10.1101/sqb.2008.73.045 [DOI] [PubMed] [Google Scholar]
- [24].Thi-Kim Vu H, Rink JC, McKinney SA, McClain M, Lakshmanaperumal N, Alexander R, Sánchez Alvarado A. Stem cells and fluid flow drive cyst formation in an invertebrate excretory organ. Elife 2015; 4: e07405; PMID:26057828; http://dx.doi.org/ 10.7554/eLife.07405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stubenhaus BM, Dustin JP, Neverett ER, Beaudry MS, Nadeau LE, Burk-McCoy E, He X, Pearson BJ, Pellettieri J. Light-induced depigmentation in planarians models the pathophysiology of acute porphyrias. Elife 2016; 5:e14175; PMID:27240733; http://dx.doi.org/ 10.7554/eLife.14175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Godwin JW, Brockes JP. Regeneration, tissue injury and the immune response. J Anat 2006; 209:423-32; PMID:17005015; http://dx.doi.org/ 10.1111/j.1469-7580.2006.00626.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Murawala P, Tanaka EM, Currie JD. Regeneration: The ultimate example of wound healing. Semin Cell Dev Biol 2012; 23:954-62; PMID:23059793; http://dx.doi.org/ 10.1016/j.semcdb.2012.09.013 [DOI] [PubMed] [Google Scholar]
- [28].Godwin JW, Rosenthal N. Scar-free wound healing and regeneration in amphibians: Immunological influences on regenerative success. Differentiation 2014; 87:66-75; PMID:24565918; http://dx.doi.org/ 10.1016/j.diff.2014.02.002 [DOI] [PubMed] [Google Scholar]
- [29].Pedersen KJ. Some features of the fine structure and histochemistry of planarian subepidermal gland cells. Zeitschrift für Zellforschung 1959; 50:121-42; http://dx.doi.org/ 10.1007/BF00350411 [DOI] [Google Scholar]
- [30].Pedersen KJ. Slime-secreting cells of planarians. Ann N Y Acad Sci 1963; 106:424-43; PMID:13942330; http://dx.doi.org/ 10.1111/j.1749-6632.1963.tb16655.x [DOI] [PubMed] [Google Scholar]
- [31].Bocchinfuso DG, Taylor P, Ross E, Ignatchenko A, Ignatchenko V, Kislinger T, Pearson BJ, Moran MF. Proteomic profiling of the planarian schmidtea mediterranea and its mucous reveals similarities with human secretions and those predicted for parasitic flatworms. Mol Cell Proteomics 2012; 11:681-91; PMID:22653920; http://dx.doi.org/ 10.1074/mcp.M112.019026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Altincicek B, Vilcinskas A. Comparative analysis of septic injury-inducible genes in phylogenetically distant model organisms of regeneration and stem cell research, the planarian schmidtea mediterranea and the cnidarian hydra vulgaris. Front Zool 2008; 5:6; PMID:18439314; http://dx.doi.org/ 10.1186/1742-9994-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Morita M, Best JB. Electron microscopic studies of planarian regeneration. II. Changes in epidermis during regeneration. J Exp Zool 1974; 187:345-73; PMID:4820343; http://dx.doi.org/ 10.1002/jez.1401870305 [DOI] [PubMed] [Google Scholar]
- [34].Morita M. Phagocytic response of planarian reticular cells to heat-killed bacteria. Hydrobiologia 1991; 227:193-9; http://dx.doi.org/ 10.1007/BF00027602 [DOI] [Google Scholar]
- [35].Zhou L, Wu S, Liu D, Xu B, Zhang X, Zhao B. Characterization and expression analysis of a trypsin-like serine protease from planarian dugesia japonica. Mol Biol Rep 2012; 39:7041-7; PMID:22314913; http://dx.doi.org/ 10.1007/s11033-012-1535-x [DOI] [PubMed] [Google Scholar]
- [36].Peiris TH, Hoyer KK, Oviedo NJ. Innate immune system and tissue regeneration in planarians: An area ripe for exploration. Semin Immunol 2014; 26:295-302; PMID:25082737; http://dx.doi.org/ 10.1016/j.smim.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Abnave P, Mottola G, Gimenez G, Boucherit N, Trouplin V, Torre C, Conti F, Ben Amara A, Lepolard C, Djian B, et al.. Screening in planarians identifies MORN2 as a key component in LC3-associated phagocytosis and resistance to bacterial infection. Cell Host Microbe 2014; 16:338-50; PMID:25211076; http://dx.doi.org/ 10.1016/j.chom.2014.08.002 [DOI] [PubMed] [Google Scholar]
- [38].Arnold CP, Merryman MS, Harris-Arnold A, McKinney SA, Seidel CW, Loethen S, Proctor KN, Guo L, Sánchez Alvarado A. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. Elife 2016; 5:e16793; PMID:27441386; http://dx.doi.org/ 10.7554/eLife.16793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Apidianakis Y, Rahme LG. Drosophila melanogaster as a model for human intestinal infection and pathology. Dis Model Mech 2011; 4:21-30; PMID:21183483; http://dx.doi.org/ 10.1242/dmm.003970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ermolaeva MA, Schumacher B. Insights from the worm: The C. elegans model for innate immunity. Semin Immunol 2014; 26:303-9; PMID:24856329; http://dx.doi.org/ 10.1016/j.smim.2014.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. ISME J 2011; 5:1595-608; PMID:21472014; http://dx.doi.org/ 10.1038/ismej.2011.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tsoumtsa LL, Torre C, Trouplin V, Coiffard B, Gimenez G, Mege JL, Ghigo E. Antimicrobial capacity of the freshwater planarians against S. aureus is under the control of timeless. Virulence 2017:1-10; PMID:28051908; http://dx.doi.org/ 10.1080/21505594.2016.1276689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: Organization and coordination of central and peripheral clocks. Annu Rev Physiol 2010; 72:517-49; PMID:20148687; http://dx.doi.org/ 10.1146/annurev-physiol-021909-135821 [DOI] [PubMed] [Google Scholar]
- [44].Tsoumtsa LL, Torre C, Ghigo E. Circadian control of antibacterial immunity: Findings from animal models. Front Cell Infect Microbiol 2016; 6:54; PMID:27242972; http://dx.doi.org/ 10.3389/fcimb.2016.00054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kojima S, Sher-Chen EL, Green CB. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev 2012; 26:2724-36; PMID:23249735; http://dx.doi.org/ 10.1101/gad.208306.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al.. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793-806; PMID:24209618; http://dx.doi.org/ 10.1016/j.cell.2013.10.026 [DOI] [PubMed] [Google Scholar]
- [47].Kalsbeek A, Perreau-Lenz S, Buijs RM. A network of (autonomic) clock outputs. Chronobiol Int 2006; 23:201-15; PMID:16687294; http://dx.doi.org/ 10.1080/07420520500464528 [DOI] [PubMed] [Google Scholar]
- [48].Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet 2006; 15(Spec No 2):R271-7; PMID:16987893; http://dx.doi.org/ 10.1093/hmg/ddl207 [DOI] [PubMed] [Google Scholar]
- [49].Gekakis N, Saez L, Delahaye-Brown AM, Myers MP, Sehgal A, Young MW, Weitz CJ. Isolation of timeless by PER protein interaction: Defective interaction between timeless protein and long-period mutant PERL. Science 1995; 270:811-5; PMID:7481773; http://dx.doi.org/ 10.1126/science.270.5237.811 [DOI] [PubMed] [Google Scholar]
- [50].Morita M, Hall F, Best JB, Gern W. Photoperiodic modulation of cephalic melatonin in planarians. J Exp Zool 1987; 241:383-8; PMID:3585272; http://dx.doi.org/ 10.1002/jez.1402410314 [DOI] [PubMed] [Google Scholar]
- [51].Itoh MT, Shinozawa T, Sumi Y. Circadian rhythms of melatonin-synthesizing enzyme activities and melatonin levels in planarians. Brain Res 1999; 830:165-73; PMID:10350570; http://dx.doi.org/ 10.1016/S0006-8993(99)01418-3 [DOI] [PubMed] [Google Scholar]
- [52].Ito H, Igarashi J. Circadian rhythm of serotonin levels in planarians. Neuroreport 2000; 11:473-6; PMID:10718297; http://dx.doi.org/ 10.1097/00001756-200002280-00009 [DOI] [PubMed] [Google Scholar]
- [53].Morita M, Best JB. Effects of photoperiods and melatonin on planarian asexual reproduction. J Exp Zool 1984; 231:273-82; http://dx.doi.org/ 10.1002/jez.1402310212 [DOI] [Google Scholar]
- [54].Sheiman IM, Sakharova N, Tiras Kh P, Shkutin MF, Isaeva VV. [Regulation of asexual reproduction in the planarian Dugesia tigrina]. Ontogenez 2003; 34:43-9; PMID:12625073 [PubMed] [Google Scholar]
- [55].Reitzel AM, Behrendt L, Tarrant AM. Light entrained rhythmic gene expression in the sea anemone Nematostella vectensis: The evolution of the animal circadian clock. PLoS One 2010; 5:e12805; PMID:20877728; http://dx.doi.org/ 10.1371/journal.pone.0012805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Stone EF, Fulton BO, Ayres JS, Pham LN, Ziauddin J, Shirasu-Hiza MM. The circadian clock protein timeless regulates phagocytosis of bacteria in drosophila. PLoS Pathog 2012; 8:e1002445; PMID:22253593; http://dx.doi.org/ 10.1371/journal.ppat.1002445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Peres R, Reitzel AM, Passamaneck Y, Afeche SC, Cipolla-Neto J, Marques AC, Martindale MQ. Developmental and light-entrained expression of melatonin and its relationship to the circadian clock in the sea anemone nematostella vectensis. Evodevo 2014; 5:26; PMID:25243057; http://dx.doi.org/ 10.1186/2041-9139-5-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Logan RW, Sarkar DK. Circadian nature of immune function. Mol Cell Endocrinol 2012; 349:82-90; PMID:21784128; http://dx.doi.org/ 10.1016/j.mce.2011.06.039 [DOI] [PubMed] [Google Scholar]


