ABSTRACT
Translation is fundamental for many biologic processes as it enables cells to rapidly respond to stimuli without requiring de novo mRNA synthesis. The mammalian/mechanistic target of rapamycin (mTOR) is a key regulator of translation. Although mTOR affects global protein synthesis, translation of a subset of mRNAs appears to be exceptionally sensitive to changes in mTOR activity. Recent efforts to catalog these mTOR-sensitive mRNAs resulted in conflicting results. Whereas ribosome-profiling almost exclusively identified 5′-terminal oligopyrimidine (TOP) mRNAs as mTOR-sensitive, polysome-profiling suggested that mTOR also regulates translation of non-TOP mRNAs. This inconsistency was explained by analytical and technical biases limiting the efficiency of ribosome-profiling in detecting mRNAs showing differential translation. Moreover, genome-wide characterization of 5′UTRs of non-TOP mTOR-sensitive mRNAs revealed 2 subsets of transcripts which differ in their requirement for translation initiation factors and biologic functions. We summarize these recent advances and their impact on the understanding of mTOR-sensitive translation.
KEYWORDS: eIFs, mRNA translation, mTOR, nanoCAGE, polysome profiling, ribosome profiling, UTR
Abbreviations
- eIFs
Eukaryotic translation initiation factors
- mTOR
mammalian/mechanistic target of rapamycin
- RPF
ribosome protected fragments
- RNAseq
RNA-sequencing
- TOP
terminal oligopyrimidine
- UTR
untranslated region
- TSS
transcript start site
- NanoCAGE
nano cap analysis of gene expression
Selective regulation of mRNA translation via the mTOR pathway
Gene expression is modulated at multiple levels including transcription, mRNA-splicing, -export, -stability, -translation and protein-stability.1 Each regulatory layer contributes to the repertoire and levels of expressed proteins. Modulation of mRNA-translation and/or protein-stability allow cells to rapidly adjust their proteomes in response to external and internal cues without altering mRNA levels.2,3 As a result, protein levels do not always reflect steady-state mRNA abundance.4-7 Moreover, it is thought that only a fraction of all cellular mRNA is translated at a given moment.8-10 Indeed, although still highly debated,11 mRNA translation has been suggested to modulate protein levels to a similar extent as transcription and has therefore emerged as a principal post-transcriptional mechanism affecting the proteome.6,12 Consistently, translational control plays central roles in pivotal biologic processes including control of the immune system, cell proliferation and development; and diseases including cancer.13,14 Common to these contexts is that mRNA translation is selectively modulated to alter synthesis of specific subsets of proteins which are required to mount an optimal response to a variety of stimuli; and when dysregulated can lead to a wide array of pathologies.13 Thus, deciphering mechanisms by which translation efficiencies of individual mRNAs are reprogrammed in response to stimuli and/or in normal vs. dysfunctional cells is crucial for a more complete understanding of many biologic phenomena.
mRNA translation can be divided into 4 phases – initiation, elongation, termination and ribosome recycling.15 To date, the best described examples of modulation of translational efficiencies occur at the rate-limiting initiation step, i.e. the efficiency of ribosome recruitment to mRNA.16 In mammals initiation is facilitated by multiple eukaryotic translation initiation factors (eIFs) including the eIF4F complex. eIF4F recruits mRNA to the ribosome and consists of the mRNA cap binding subunit eIF4E, the scaffolding protein eIF4G and the DEAD box RNA helicase eIF4A.16 The mechanistic/mammalian target of rapamycin (mTOR) complex 1 (mTORC1) stimulates assembly of the eIF4F complex by phosphorylating and inactivating the 4E-binding proteins (4E-BP1, 2 and 3) which otherwise prevent eIF4E:eIF4G interaction and thereby eIF4F complex assembly.17-21 Although eIF4E is required for cap-dependent translation of all nuclear-encoded mRNAs, some transcripts are dramatically more sensitive to changes in eIF4E levels and/or availability.22-29 Such mRNAs are commonly referred to as ”eIF4E-sensitive” and encode key proteins which stimulate proliferation (e.g. cyclins, ODC1, c-MYC) and survival (e.g., BCL-2 family members), many of which have been implicated in cancer.22-26 eIF4E-sensitive translation is in part mediated by long and complex 5′ untranslated regions (UTR).30 These mRNAs have a higher requirement for eIF4A helicase unwinding activity as compared with other cellular mRNAs.30,31 eIF4A activity is bolstered significantly within the eIF4F complex,32-34 whereby eIF4E acts as a rate limiting factor for eIF4F assembly.30 eIF4E-sensitivity of the mRNAs with long and structured 5′UTRs is therefore thought to stem from eIF4E-dependent recruitment of eIF4A and stimulation of eIF4A's activity.33 Accordingly, recent ribosome profiling studies in mammalian cells revealed that eIF4A inhibitors preferentially suppress translation of mRNAs that harbor long 5′UTRs enriched in complex structures, including G-quadraplexes,35-37 although this was recently disputed for the class of eIF4A inhibitors belonging to the rocaglate family.38 Intriguingly, in contrast to mammals, yeast ded1 (ortholog of mammalian DDX3 helicase) but not eIF4A appears to play a predominant role in stimulating translation of mRNAs with highly structured 5′UTRs.39 Given the key role of the mTOR pathway and eIF4E in many biologic contexts and human diseases including cancer, neurologic diseases, diabetes and metabolic syndrome, there has been a considerable interest in applying genome-wide approaches to obtain a complete catalog of mRNAs that are sensitive to changes in mTOR activity and thereby eIF4E availability.
Controversy regarding the repertoire of mRNAs showing mTOR-sensitive translation
Three recent studies using pharmacological inhibitors of mTOR (including the allosteric mTOR inhibitor rapamycin, active-site mTOR inhibitors and the biguanide metformin, which inhibits mTOR indirectly via AMP-activated kinase-dependent and independent mechanisms) were performed to catalog the transcriptome-wide set of mRNAs showing mTOR-sensitive translation.40-42 Unexpectedly, these studies reached radically different conclusions. Two studies using ribosome-profiling suggested that mTOR almost exclusively regulates translation of mRNAs harboring a 5′ terminal oligopyrimidine (5′ TOP) motif.40,41 The TOP motif consists of a cytosine (C) directly after the mRNA cap followed by a stretch of 4–15 pyrimidines and is mainly found in mRNAs encoding for components of the translational machinery including ribosomal proteins, poly (A) binding protein (PABP) and eukaryotic elongation factor 2 (eEF2).43 The mTOR-sensitivity of TOP mRNAs was recognized over a decade ago,44 but this appeared to be largely eIF4E-independent.44-46 In contrast, the conclusion that TOP mRNAs would essentially be the only targets of mTOR was unexpected, especially as translation of mRNAs encoding growth, proliferation, survival and tumor-promoting proteins, such as cyclins,47 ornithine decarboxylase (ODC1),48 vascular endothelial growth factor (VEGF)49 or c-MYC50 had been shown to be sensitive to alterations in eIF4E levels, which is a major mediator of mTOR-dependent translational control. Moreover, the conclusion drawn by the 2 aforementioned studies,40,41 that the effects of mTOR on TOP mRNA translation is chiefly mediated via 4E-BPs was in conflict with previously reported findings which showed that TOP mRNA translation is not eIF4E dependent.46 Moreover, a recent report revealed that under conditions when mTOR signaling is modulated by physiologic stimuli, TOP mRNA translation is regulated via an 4E-BP-independent mechanism.45 Indeed, it appears that mTOR regulates TOP mRNA translation via La-related protein 1 (LARP1).51,52 In stark contrast to ribosome-profiling studies, polysome-profiling suggested that several non TOP mRNAs including those that were previously identified as eIF4E-senstive (e.g., cyclins, ODC1) exhibit mTOR-sensitive translation. The cohort of mTOR sensitive mRNAs also contained those encoding mitochondria related proteins.42 Indeed, modulation of mitochondria related mRNAs by mTOR was further functionally evaluated which revealed that the mTOR/4E-BP/eIF4E axis coordinates energy expenditure by the mRNA translation machinery with mitochondrial ATP production.53 In conclusion, strikingly disparate catalogs of mRNAs that are translated in an mTOR-dependent fashion were captured using ribosome- vs. polysome-profiling, which suggested biases of these approaches in the detection of mTOR-sensitive mRNAs.
Ribosome-profiling introduces biases in identification of mTOR-sensitive translation
Because translational efficiency is primarily regulated at the initiation step, efficiently translated mRNAs are associated with more ribosomes than inefficiently translated mRNAs leading to more proteins being synthesized.54 This tenet underpins current transcriptome-wide approaches to study changes in translational efficiency, including polysome- and ribosome-profiling. During polysome-profiling efficiently translated mRNAs (commonly those associated with more than 3 ribosomes) are isolated and quantified using DNA microarrays or, more recently, RNA-sequencing (RNAseq). Polysome-profiling therefore directly assesses changes in translation efficiency from an “mRNA perspective,” by physically separating efficiently and non-efficiently translated mRNA molecules by ultracentrifugation on sucrose gradients (Fig 1A).54 During ribosome-profiling, ribosome protected fragments (RPF), i.e., RNA fragments protected by the ribosome from RNase-mediated degradation, are isolated and quantified using RNAseq (Fig 1B).55 Thus, in contrast to polysome-profiling, ribosome-profiling has a “ribosome perspective” wherein translational efficiency is determined indirectly by counting the number of RPFs from both efficiently and inefficiently translated mRNAs. Although several factors affect the performance of ribosome-profiling,56,57 this technique holds a great promise by providing unprecedented single nucleotide resolution of ribosome positioning on the mRNA.58,59 In contrast, polysome-profiling does not reveal ribosomal location on the mRNA, but it allows isolation of intact mRNAs from the polysomes that can be further studied (see 5′UTR profiling below).
Figure 1.
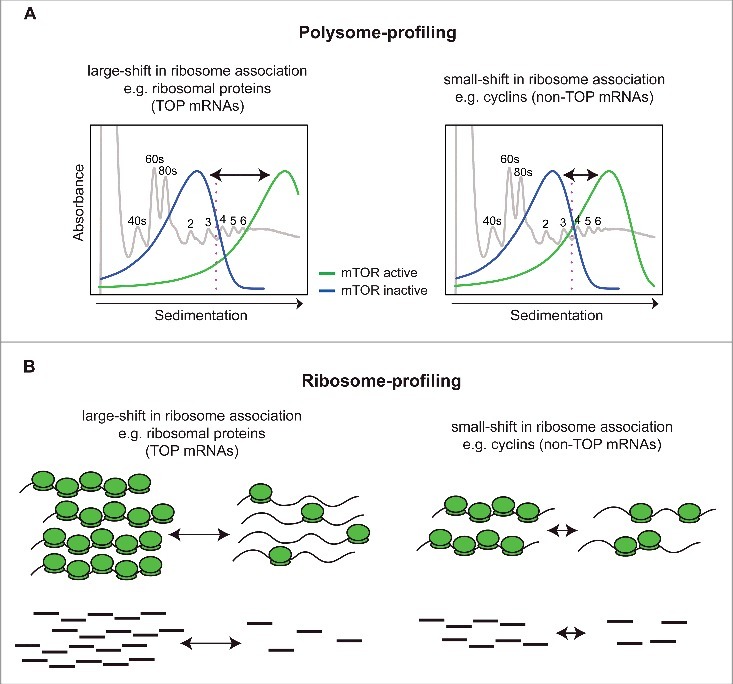
Schematics illustrating properties underlying a bias toward identification of mRNAs that show large shift in translational efficiency as differentially translated when applying ribosome-profiling. (A) Shown in gray is a model UV absorbance profile from a polysome-preparation where ribosome subunits (40S or 60S), monosomes (80S) or polysomes (i.e., mRNAs associated with >1 ribosome) are separated on sucrose gradients by ultracentrifugation. In polysome-profiling, translational efficiency is measured by quantifying the amount of mRNA that is efficiently translated (i.e., associated with >3 ribosomes, which is indicated by a pink dotted line). Transcripts differ in their basal translational efficiency (green). Left panel represents TOP mRNAs that are associated with the heaviest polysomes and are thus more efficiently translated than non-TOP mRNA (e.g., cyclin) which are associated with intermediate polysomes. Upon mTOR inhibition (blue), TOP mRNAs shift more dramatically than non-TOP mRNAs (i.e., shifts are indicated by arrows). (B). In ribosome-profiling, the amount of mRNA fragments protected by ribosomes (RPFs) is quantified using RNAseq and compared between conditions. This generates radically different fold-changes for TOP and non-TOP mRNAs, as the former are much more abundant and exhibit larger shifts. Such fold-changes are directly proportional to the magnitude of the mRNA shifts leading to a bias favoring identification of TOP mRNAs as differentially translated. This bias is more pronounced when fold-change based vs. statistical analysis is performed.60
We recently showed that a key difference between polysome- and ribosome-profiling occurs when mRNAs that exhibit different magnitudes in their changes in translational efficiency are studied in parallel. For example, a treatment with a pharmacological inhibitor may alter ribosome association of a class of mRNAs from a mean of 4 to 2 ribosomes, while others may shift from 7 to 1 ribosomes. The differences in shifts reflect intrinsic properties of these mRNAs and are thus not expected to directly mirror their relative importance for the biology or pathology studied. In ribosome profiling translation efficiency is inferred indirectly based on the number of RPFs, and therefore the size of the shifts in ribosome association will result in directly proportional effects on fold-change estimates (i.e., 2 vs 7-fold from the example above). This effect is much less pronounced in polysome-profiling studies, as herein translation efficiency is estimated directly from mRNAs associated with heavy polysomes (i.e., >3 ribosomes) (Fig. 1A-B).60 Moreover, mRNAs that show large shifts in translational efficiency and are also very abundant will cause a seemingly global effect on translation. As a result, changes in translation of mRNAs that show less dramatic shifts in translational efficiency are masked. This is because RNAseq applied during polysome- or ribosome-profiling generates relative quantification, whereby changes in translational efficiency are compared with the ‘mean change’ in global translation.61 Smaller fold-changes of low abundant mRNAs will therefore be strongly down weighted. Altogether, this indicates that ribosome-profiling is biased toward detecting changes in translation of highly abundant mRNAs that exhibit large shifts in polysome association but is considerably less sensitive in identifying those mRNAs with smaller shifts and lower abundance (Fig 1B).60
A prototypical example of a very abundant mRNA-class that shows a large shift in translational efficiency upon changes in mTOR activity are TOP mRNAs.43 Many other mRNAs, such as those encoding for cyclins or mitochondria-related proteins, show relatively smaller shifts and are expressed at a much lower level as compared with TOP mRNAs.42,60 Thus, application of ribosome profiling to conditions when these 2 mRNA populations change their translational efficiency at the same time (high expression/big shift vs. moderate expression/moderate shift) leads to preferential identification of TOP mRNAs (Fig. 1B).60 These technical differences, together with the issues related to sequencing depth discussed below, appear to explain why ribosome profiling studies essentially only identified TOP mRNAs as mTOR sensitive,40,41 while polysome profiling revealed a more diverse set of genes including TOP and non-TOP mRNAs (e.g., those encoding cyclins and mitochondria-related proteins).42
An additional source of bias in ribosome-profiling studies of mTOR-sensitive translation40,41 likely stems from the sequencing depth. Optimal RNA sequencing depth allows reliable quantification across all expression levels and conditions.62 When performing ribosome-profiling this is an important consideration, as highly expressed genes that are also very efficiently translated (e.g., TOP mRNAs) will dominate the sequencing library and hence constitute a large proportion of the RPF RNAseq reads. Sub-optimal RNAseq depth greatly influences the signal to variance relationship (low number of RNAseq reads is associated with increased noise in the data set), preventing identification of differential translation for less expressed, and less efficiently translated mRNAs.60 Indeed, ribosome-profiling studies of mTOR-dependent translation had relatively low sequencing depth and accordingly almost exclusively detected effects on translation of the most abundant cellular mRNAs including TOP mRNAs.40,41,60
Altogether, the disparity between the repertoires of mTOR-regulated mRNAs observed between ribosome- and polysome-profiling studies appears to stem from technical biases and insufficient sequencing depth which limited detection of mTOR-dependent changes in the translatome in studies which used ribosome-profiling.
Profiling 5′UTRs unraveled 2 distinct subsets of mTOR-sensitive mRNAs encoding proteins with different cellular functions
mRNAs exhibit different translational properties, which is in part conferred by their 5′UTR features.2,22,24,60,63,64 A number of regulatory elements in 5′UTRs are implicated in translational control of gene expression including upstream open reading frames (uORFs), stem loops (e.g., the iron-responsive element; IRE) and internal ribosome-entry sites (IRESes) which allow cap-independent translation (reviewed in Hinnebusch et al.30). In addition, it is thought that the position of these regulatory elements and/or structural features relative to the mRNA cap or the initiation codon, as well as the length of the 5′UTR play a major role in determining translation efficiency (reviewed in Hinnebusch et al.30). Analysis of 5′UTRs, however, relies on data repositories such as RefSeq and UCSC, which provide a wealth of data and tools to understand the genomic contexts of many species. These data repositories, thereby, offer a framework to interrogate and interpret observed changes in translational efficiency in the context of e.g., 5′UTRs. It is widely thought, however, that these databases contain a repertoire of 5′UTRs that may not necessarily reflect those that are expressed in the cell of interest, thereby potentially leading to faulty conclusions about the role of 5′UTRs in translational control. Several initiatives to accurately pinpoint transcription start sites (TSSs) including the FANTOM Consortium (The FANTOM consortium and the RIKEN PMI and CLST [DGT] 2014) or TSS-seq65,66 are currently ongoing but information is still missing for most cell lines and tissues, especially if one considers that 5′UTRs in the same cell line may be context dependent due to e.g., stress induced alternative transcription site selection or alternative splicing.67 Thus, for precise understanding of the relationship between 5′UTR features and translational control, both TSSs and translational efficiency should be determined in the same cell line. Importantly, ribosome-profiling does not allow for such assessment, as the nuclease digestion degrades both 5′ and 3′ UTRs of mRNA. In contrast, polysome-profiling can be readily coupled with techniques such as Nano Cap Analysis of Gene Expression (nanoCAGE),68 an RNAseq library construction method that allows nucleotide resolution mapping of TSSs and, as a result, 5′UTRs. Application of this approach to mTOR-sensitive translation unveiled that nearly 30% of 5′ UTRs were substantially shorter than those indicated in the RefSeq database.60 Moreover, nanoCAGE revealed 2 distinct non-TOP, mTOR-sensitive mRNA subsets: one whose members harbor long 5′UTRs encoding cell-cycle and survival-promoting proteins and a second whose members have extremely short 5′UTR (< 30 nucleotides) encoding proteins with mitochondrial function. These 2 subsets could not have been separated using current databases, as these suggested radically different 5′UTRs than the ones identified by nanoCAGE for the subset of mitochondrial-related mRNAs with very short 5′UTRs.60
The distinct subsets of mTOR-sensitive mRNAs require different translation initiation factors and encode for proteins participating in discrete cellular processes
The first subset of mRNAs with long 5′UTRs identified by nanoCAGE (including cyclins, BCL-2, MCL1, BIRC5) supports the long-held idea that downstream of mTOR, changes in eIF4E availability primarily affect mRNAs with long and complex 5′UTRs that critically depend on eIF4A activity for efficient translation.30,31 In contrast, mitochondrial-related mRNAs with short 5′UTRs (such as ATP5O, ATP5G1, NDUF6, UQCC2) do not seem to fit into this model.60 A subset of mRNAs with short 5′ UTRs harboring a Translation Initiatior of Short 5′UTR (TISU) element (SAASATGGCGGC, in which S is C or G) which is enriched for mRNAs encoding proteins with mitochondrial functions, however, was previously show to have eIF4E sensitive but eIF4A insensitive translation.69-71 Consistently, we revealed differential requirements of the 2 mTOR-sensitive mRNA subsets: those with long 5′UTR are both eIF4E- and eIF4A-sensitive while those with short 5′UTR are eIF4E-, but not eIF4A-sensitive. Surprisingly, this sensitivity of short mRNAs appeared to be more dependent on the length of the 5′UTR than the presence of the TISU element. Moreover, the dichotomy in eIF4A sensitivity between short and long 5′UTRs of mTOR-sensitive mRNAs appear to in part account for the different responses to mTOR or eIF4A inhibitors. mTOR inhibitors affect translation of mRNAs with both very short (encoding for mitochondria related proteins) and long (encoding for pro-survival proteins) 5′UTRs60 which reduces mitochondrial53 activity but concomitantly downregulates energy consumption by the translation machinery resulting in metabolic dormancy and a cytostatic effect.42 eIF4A inhibitors, on the other hand, display strong cytotoxic effect as they reduce the translation of pro-survival mRNAs with long 5′UTRs, without affecting the translation of short 5′UTR mRNAs encoding for proteins with mitochondrial function, which leads to mitochondria dysfunction and apoptosis.60 In addition, eIF4A inhibitors, unlike mTOR inhibitors, reduce autophagy which removes depolarized mitochondria, thereby further bolstering their pro-apoptotic effects.60
Concluding remarks
In conclusion, analytical and technical biases should be taken into consideration when performing transcriptome-wide analysis of translational regulation. Moreover, the limitations of databases for e.g., 5′ UTRs can obscure significant findings due to a lack in accuracy or lack of information for a specific experimental model. Therefore, choosing the appropriate experimental methodology coupled with a tailored analysis can address important gaps in knowledge pertinent to the regulation of translation. Factors that limit these advances include suboptimal study design, lack of appropriate quality control and incorrect statistical methods - which are all inherent difficulties of today's science, and were in fact predicted to undermine systems biology approaches.72 Inappropriate strategies such as limited replication which does not allow application of statistical methods, adopting analytical approaches to identify a set of favorite genes that fit the hypothesis (i.e., cherry picking), or simply a lack of understanding of data analysis methodology can compromise validity of conclusions and study reproducibility.73 We have previously demonstrated that using the ratio between polysome-associated mRNA or RPFs to cytosolic mRNA (commonly denoted as translation efficiency or TE score) for analysis of differential translation results in spurious correlation thereby favoring false positive and negative findings.74 Furthermore, we showed that using a fold-changes based analysis is similarly inappropriate, especially when assessing differential translation using ribosome-profiling.60 Nonetheless, both approaches are commonly applied, thereby indicating the lack of a consensus regarding optimal data analysis. Similarly, a recent advance in quality evaluation of ribosomal profiling data revealed factors pertinent to the validity of ribosome profiling data that are also likely to skew the interpretation of the results.56 Collectively, a consensus on guidelines for study design, quality control and data analysis appears to be required to improve translatome analysis, and thus to advance the understanding of translation regulation in health and disease.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
LM is supported by a post-doc fellowship from the Swedish Child Cancer Society. LF is supported by VCA Mid-Career Research Fellowship (MCRF16007) and grant from Cancer Australia (1084546). IT is supported by CIHR Young Investigator Award and research in his laboratory is funded by grants from Prostate Cancer Canada (D2015–13), Cancer Research Society (01713), Canadian Cancer Society Research Institute (CCSRI-703816), Canadian Institutes for Health Research (MOP-363027), Terry Fox Research Institute (TFF-116128) and National Institutes of Health (R01CA202021). OL and research in his laboratory is supported by the Wallenberg Academy Fellow's program, the Swedish Research Council, the Swedish Cancer Society, The Cancer Society in Stockholm, and the Strategic Research Program in Cancer. OL, LF and IT are supported by STINT.
References
- 1.Alberts, B, et al. Molecular Biology of the Cell, Vol 1, Sixth edition. edn (New York, NY: Garland Science, Taylor and Francis Group, 2014), ISBN 9780815344322. [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; https://doi.org/ 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanov P, Anderson P. Post-transcriptional regulatory networks in immunity. Immunol Rev 2013; 253:253-72; PMID:23550651; https://doi.org/ 10.1111/imr.12051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between Protein and mRNA Abundance in Yeast. Mol Cell Biol 1999; 19:1720-1730; PMID:10022859; https://doi.org/ 10.1128/MCB.19.3.1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abreu R, de S, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst 2009; 5:1512-26; PMID:20023718; https://doi.org/ 10.1039/b908315d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristensen AR, Gsponer J, Foster LJ. Protein synthesis rate is the predominant regulator of protein expression during differentiation. Mol Syst Biol 2013; 9:689; PMID:24045637; https://doi.org/ 10.1038/msb.2013.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012; 13:227-32; PMID:22411467; https://doi.org/ 10.1038/nrg3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geyer PK, Meyuhas O, Perry RP, Johnson LF. Regulation of ribosomal protein mRNA content and translation in growth-stimulated mouse fibroblasts. Mol Cell Biol 1982; 2:685-93; PMID:14582163; https://doi.org/ 10.1128/MCB.2.6.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shields D. Association of messenger ribonucleic acid with mammalian microsomal membranes: characterization by analysis of cell-free translation products. Biochemistry (Mosc.) 1979; 18:2622-7; PMID:444481; https://doi.org/560911 10.1021/bi00579a029 [DOI] [PubMed] [Google Scholar]
- 10.Laskey RA, Mills AD, Gurdon JB, Partington GA. Protein synthesis in oocytes of Xenopus laevis is not regulated by the supply of messenger RNA. Cell 1977; 11:345-51; PMID:560911; https://doi.org/ 10.1016/0092-8674(77)90051-4 [DOI] [PubMed] [Google Scholar]
- 11.Li JJ, Biggin MD. Gene expression. Statistics requantitates the central dogma. Science 2015; 347:1066-7; PMID:25745146; https://doi.org/ 10.1126/science.aaa8332 [DOI] [PubMed] [Google Scholar]
- 12.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 2011; 473:337-42; PMID:21593866; https://doi.org/ 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 13.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov 2015; 14:261-78; PMID:25743081; https://doi.org/ 10.1038/nrd4505 [DOI] [PubMed] [Google Scholar]
- 14.Piccirillo CA, Bjur E, Topisirovic I, Sonenberg N, Larsson O. Translational control of immune responses: from transcripts to translatomes. Nat Immunol 2014; 15:503-11; PMID:24840981; https://doi.org/ 10.1038/ni.2891 [DOI] [PubMed] [Google Scholar]
- 15.Hershey JW, Sonenberg N, Mathews MB. Principles of translational control: an overview. Cold Spring Harb Perspect Biol 2012; 4:a011528; PMID:23209153; https://doi.org/ 10.1101/cshperspect.a011528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinnebusch AG. The Scanning Mechanism of Eukaryotic Translation Initiation. Annu Rev Biochem 2014; 83:779-812; PMID:24499181; https://doi.org/ 10.1146/annurev-biochem-060713-035802 [DOI] [PubMed] [Google Scholar]
- 17.von Manteuffel SR, Gingras AC, Ming XF, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen-activated protein kinase. Proc Natl Acad Sci U S A 1996; 93:4076-80; PMID:8633019; https://doi.org/ 10.1073/pnas.93.9.4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara K, Yonezawa K, Kozlowski MT, Sugimoto T, Andrabi K, Weng QP, Kasuga M, Nishimoto I, Avruch J. Regulation of eIF-4E BP1 phosphorylation by mTOR. J Biol Chem 1997; 272:26457-63; PMID:9334222; https://doi.org/ 10.1074/jbc.272.42.26457 [DOI] [PubMed] [Google Scholar]
- 19.Burnett PE, Barrow RK, Cohen NA, Snyder SH, Sabatini DM. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci U S A 1998; 95:1432-7; PMID:9465032; https://doi.org/ 10.1073/pnas.95.4.1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 1999; 13:1422-37; PMID:10364159; https://doi.org/ 10.1101/gad.13.11.1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 2001; 15:2852-64; PMID:11691836; https://doi.org/ 10.1101/gad.887201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koromilas AE, Lazaris-Karatzas A, Sonenberg N. mRNAs containing extensive secondary structure in their 5′ non-coding region translate efficiently in cells overexpressing initiation factor eIF-4E. EMBO J 1992; 11:4153-8; PMID:1396596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zimmer SG, DeBenedetti A, Graff JR. Translational control of malignancy: the mRNA cap-binding protein, eIF-4E, as a central regulator of tumor formation, growth, invasion and metastasis. Anticancer Res 2000; 20:1343-51; PMID:10928042 [PubMed] [Google Scholar]
- 24.Graff JR, Konicek BW, Carter JH, Marcusson EG. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res 2008; 68:631-4; PMID:18245460; https://doi.org/ 10.1158/0008-5472.CAN-07-5635 [DOI] [PubMed] [Google Scholar]
- 25.Roux PP, Topisirovic I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb Perspect Biol 2012; 4:a012252; PMID:22888049; https://doi.org/ 10.1101/cshperspect.a012252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pelletier J, Graff J, Ruggero D, Sonenberg N. Targeting the eIF4F translation initiation complex: a critical nexus for cancer development. Cancer Res 2015; 75:250-63; PMID:25593033; https://doi.org/ 10.1158/0008-5472.CAN-14-2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larsson O, Li S, Issaenko OA, Avdulov S, Peterson M, Smith K, Bitterman PB, Polunovsky VA. Eukaryotic translation initiation factor 4E induced progression of primary human mammary epithelial cells along the cancer pathway is associated with targeted translational deregulation of oncogenic drivers and inhibitors. Cancer Res 2007; 67:6814-24; PMID:17638893; https://doi.org/ 10.1158/0008-5472.CAN-07-0752 [DOI] [PubMed] [Google Scholar]
- 28.Larsson O, Perlman DM, Fan D, Reilly CS, Peterson M, Dahlgren C, Liang Z, Li S, Polunovsky VA, Wahlestedt C, et al.. Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res 2006; 34:4375-86; PMID:16936314; https://doi.org/ 10.1093/nar/gkl558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mamane Y, Petroulakis E, Martineau Y, Sato TA, Larsson O, Rajasekhar VK, Sonenberg N. Epigenetic activation of a subset of mRNAs by eIF4E explains its effects on cell proliferation. PloS One 2007; 2:e242; PMID:17311107; https://doi.org/ 10.1371/journal.pone.0000242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 2016; 352:1413-6; PMID:27313038; https://doi.org/ 10.1126/science.aad9868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA N Y N 2001; 7:382-94; PMID:11333019;https://doi.org/26113725 10.1017/S135583820100108X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.García-García C, Frieda KL, Feoktistova K, Fraser CS, Block SM. RNA BIOCHEMISTRY. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science 2015; 348:1486-8; PMID:26113725; https://doi.org/ 10.1126/science.aaa5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feoktistova K, Tuvshintogs E, Do A, Fraser CS. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci U S A 2013; 110:13339-44; PMID:23901100; https://doi.org/ 10.1073/pnas.1303781110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pause A, Méthot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 1994; 13:1205-15; PMID:8131750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Modelska A, Turro E, Russell R, Beaton J, Sbarrato T, Spriggs K, Miller J, Gräf S, Provenzano E, Blows F, et al.. The malignant phenotype in breast cancer is driven by eIF4A1-mediated changes in the translational landscape. Cell Death Dis 2015; 6:e1603; PMID:25611378; https://doi.org/ 10.1038/cddis.2014.542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL, Fanidi A. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol 2014; 15:476; PMID:25273840; https://doi.org/ 10.1186/s13059-014-0476-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolfe AL, Singh K, Zhong Y, Drewe P, Rajasekhar VK, Sanghvi VR, Mavrakis KJ, Jiang M, Roderick JE, Van der Meulen J, et al.. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature 2014; 513:65-70; PMID:25079319; https://doi.org/ 10.1038/nature13485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwasaki S, Floor SN, Ingolia NT. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature 2016; 534:558-61; PMID:27309803; https://doi.org/ 10.1038/nature17978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res 2015; 25:1196-205; PMID:26122911; https://doi.org/ 10.1101/gr.191601.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al.. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 2012; 485:55-61; PMID:22367541; https://doi.org/ 10.1038/nature10912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109-13; PMID:22552098; https://doi.org/ 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larsson O, Morita M, Topisirovic I, Alain T, Blouin MJ, Pollak M, Sonenberg N. Distinct perturbation of the translatome by the antidiabetic drug metformin. Proc Natl Acad Sci U S A 2012; 109:8977-82; PMID:22611195; https://doi.org/ 10.1073/pnas.1201689109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyuhas O, Kahan T. The race to decipher the top secrets of TOP mRNAs. Biochim Biophys Acta 2015; 1849:801-11; PMID:25234618; https://doi.org/ 10.1016/j.bbagrm.2014.08.015 [DOI] [PubMed] [Google Scholar]
- 44.Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol Cell Biol 2001; 21:8671-83; PMID:11713299; https://doi.org/ 10.1128/MCB.21.24.8671-8683.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miloslavski R, Cohen E, Avraham A, Iluz Y, Hayouka Z, Kasir J, Mudhasani R, Jones SN, Cybulski N, Rüegg MA, et al.. Oxygen sufficiency controls TOP mRNA translation via the TSC-Rheb-mTOR pathway in a 4E-BP-independent manner. J Mol Cell Biol 2014; 6:255-66; PMID:24627160; https://doi.org/ 10.1093/jmcb/mju008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shama S, Avni D, Frederickson RM, Sonenberg N, Meyuhas O. Overexpression of initiation factor eIF-4E does not relieve the translational repression of ribosomal protein mRNAs in quiescent cells. Gene Expr 1995; 4:241-52; PMID:7787416 [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenwald IB, Kaspar R, Rousseau D, Gehrke L, Leboulch P, Chen JJ, Schmidt EV, Sonenberg N, London IM. Eukaryotic translation initiation factor 4E regulates expression of cyclin D1 at transcriptional and post-transcriptional levels. J Biol Chem 1995; 270:21176-80; PMID:7673150; https://doi.org/ 10.1074/jbc.270.36.21176 [DOI] [PubMed] [Google Scholar]
- 48.Fagan RJ, Lazaris-Karatzas A, Sonenberg N, Rozen R. Translational control of ornithine aminotransferase. Modulation by initiation factor eIF-4E. J Biol Chem 1991; 266:16518-23; PMID:1909329 [PubMed] [Google Scholar]
- 49.Kevil CG, De Benedetti A, Payne DK, Coe LL, Laroux FS, Alexander JS. Translational regulation of vascular permeability factor by eukaryotic initiation factor 4E: Implications for tumor angiogenesis. Int J Cancer 1996; 65:785-90; PMID:8631593; https://doi.org/ 10.1002/(SICI)1097-0215(19960315)65:6%3c785::AID-IJC14%3e3.0.CO;2-3 [DOI] [PubMed] [Google Scholar]
- 50.DeBenedetti A, Joshi B, Graff JR, Zimmer SG. CHO cells transformed by the translation factor eIF-4E display increased c-Myc expression, but require overexpression of Max for tumorigenicity. Mol Cell Differ 1994; 2:347-71. CODEN: MCDIEL; ISSN: 1065-3074. [Google Scholar]
- 51.Tcherkezian J, Cargnello M, Romeo Y, Huttlin EL, Lavoie G, Gygi SP, Roux PP. Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5′TOP mRNA translation. Genes Dev 2014; 28:357-71; PMID:24532714; https://doi.org/ 10.1101/gad.231407.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fonseca BD, Zakaria C, Jia JJ, Graber TE, Svitkin Y, Tahmasebi S, Healy D, Hoang HD, Jensen JM, Diao IT, et al.. La-related Protein 1 (LARP1) Represses Terminal Oligopyrimidine (TOP) mRNA Translation Downstream of mTOR Complex 1 (mTORC1). J Biol Chem 2015; 290:15996-6020; PMID:25940091; https://doi.org/ 10.1074/jbc.M114.621730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morita M, Gravel SP, Chénard V, Sikström K, Zheng L, Alain T, Gandin V, Avizonis D, Arguello M, Zakaria C, et al.. mTORC1 Controls Mitochondrial Activity and Biogenesis through 4E-BP-Dependent Translational Regulation. Cell Metab 2013; 18:698-711; PMID:24206664; https://doi.org/ 10.1016/j.cmet.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 54.Gandin V, Sikström K, Alain T, Morita M, McLaughlan S, Larsson O, Topisirovic I. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp JoVE 2014; 87; PMID:24893926; https://doi.org/ 10.3791/51455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-Wide Analysis in Vivo of Translation with Nucleotide Resolution Using Ribosome Profiling. Science 2009; 324:218-23; PMID:19213877; https://doi.org/ 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Connor PB, Andreev DE, Baranov PV. Comparative survey of the relative impact of mRNA features on local ribosome profiling read density. Nat Commun 2016; 7:12915; PMID:27698342; https://doi.org/ 10.1038/ncomms12915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gerashchenko MV, Gladyshev VN. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res 2014; 42:e134; PMID:25056308; https://doi.org/ 10.1093/nar/gku671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andreev DE, O'Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res 2016; 45(2):513-26; PMID:27923997; https://doi.org/27015305 10.1093/nar/gkw1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ingolia NT. Ribosome Footprint Profiling of Translation throughout the Genome. Cell 2016; 165:22-33; PMID:27015305; https://doi.org/ 10.1016/j.cell.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al.. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res 2016; 26:636-48; PMID:26984228; https://doi.org/ 10.1101/gr.197566.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker MW, Rossi D, Peterson M, Smith K, Sikström K, White ES, Connett JE, Henke CA, Larsson O, Bitterman PB. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. J Clin Invest 2014; 124:1622-35; PMID:24590289; https://doi.org/ 10.1172/JCI71386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Larsson O, Nadon R. Re-analysis of genome wide data on mammalian microRNA-mediated suppression of gene expression. Translation 2013; 1:e24557; PMID:26824020; https://doi.org/ 10.4161/trla.24557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lodish HF. Model for the regulation of mRNA translation applied to haemoglobin synthesis. Nature 1974; 251:385-8; PMID:4421673; https://doi.org/ 10.1038/251385a0 [DOI] [PubMed] [Google Scholar]
- 64.Pelletier J, Sonenberg N. Insertion mutagenesis to increase secondary structure within the 5′ noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 1985; 40:515-26; PMID:2982496; https://doi.org/ 10.1016/0092-8674(85)90200-4 [DOI] [PubMed] [Google Scholar]
- 65.Suzuki Y, Sugano S. Construction of a full-length enriched and a 5′-end enriched cDNA library using the oligo-capping method. Methods Mol Biol Clifton NJ 2003; 221:73-91; PMID:12703735; https://doi.org/25378318 10.1385/1-59259-359-3:73 [DOI] [PubMed] [Google Scholar]
- 66.Suzuki A, Wakaguri H, Yamashita R, Kawano S, Tsuchihara K, Sugano S, Suzuki Y, Nakai K. DBTSS as an integrative platform for transcriptome, epigenome and genome sequence variation data. Nucleic Acids Res 2015; 43:D87-91; PMID:25378318; https://doi.org/ 10.1093/nar/gku1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Livingstone M, Sikström K, Robert PA, Uzé G, Larsson O, Pellegrini S. Assessment of mTOR-dependent translational regulation of interferon stimulated genes. PloS One 2015; 10:e0133482; PMID:26207988; https://doi.org/ 10.1371/journal.pone.0133482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salimullah M, Mizuho S, Plessy C, Carninci P. NanoCAGE: A high-resolution technique to discover and interrogate cell transcriptomes. Cold Spring Harb Protoc 2011; 2011:pdb.prot5559; PMID:21205859; https://doi.org/ 10.1101/pdb.erratum2011_01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Elfakess R, Dikstein R. A translation initiation element specific to mRNAs with very short 5′UTR that also regulates transcription. PloS One 2008; 3:e3094; PMID:18769482; https://doi.org/ 10.1371/journal.pone.0003094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 2011; 39:7598-609; PMID:21705780; https://doi.org/ 10.1093/nar/gkr484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 2015; 21:479-92; PMID:25738462; https://doi.org/ 10.1016/j.cmet.2015.02.010 [DOI] [PubMed] [Google Scholar]
- 72.Brenner S. Sillycon valley fever. Curr Biol 1999; 9:R671; PMID:10508625; https://doi.org/ 10.1016/S0960-9822(99)80432-1 [DOI] [PubMed] [Google Scholar]
- 73.Horrigan SK, Courville P, Sampey D, Zhou F, Cai S; Reproducibility Project: Cancer Biology . Replication Study: Melanoma genome sequencing reveals frequent PREX2 mutations. Elife 2017; 6:e21634; PMID:22622578; https://doi.org/21115840 10.1038/nature11071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Larsson O, Sonenberg N, Nadon R. Identification of differential translation in genome wide studies. Proc Natl Acad Sci U S A 2010; 107:21487-92; PMID:21115840; https://doi.org/ 10.1073/pnas.1006821107 [DOI] [PMC free article] [PubMed] [Google Scholar]


