ABSTRACT
Ribosomes are responsible for translating the genome, in the form of mRNA, into the proteome in all organisms. Biogenesis of ribosomes in eukaryotes is a complex process involving numerous remodeling events driven in part by the concerted actions of hundreds of protein assembly factors. A major challenge in studying eukaryotic ribosome assembly has, until recently, been a lack of structural data to facilitate understanding of the conformational and compositional changes the pre-ribosome undergoes during its construction. Cryo-electron microscopy (cryo-EM) has begun filling these gaps; recent advances in cryo-EM have enabled the determination of several high resolution pre-ribosome structures. This review focuses mainly on lessons learned from the study of pre-60S particles purified from yeast using the assembly factor Nog2 as bait. These Nog2 particles provide insight into many aspects of nuclear stages of 60S subunit assembly, including construction of major 60S subunit functional centers and processing of the ITS2 spacer RNA.
Keywords: Assembly intermediates, high resolution cryo-EM, remodeling, ribosome biogenesis
Introduction
Ribosomes are the macromolecular machines responsible for producing proteins in all organisms. Biosynthesis of ribosomes is a highly complex process requiring the coordinated modification, processing, and folding of ribosomal RNA (rRNA) and binding of ribosomal proteins (r-proteins).1-7 Ribosome assembly is facilitated by protein assembly factors (AFs); while prokaryotes have relatively few AFs, eukaryotes rely upon hundreds of these proteins to accurately and efficiently carry out ribosome biosynthesis. Eukaryotic ribosome construction has been most extensively studied in the yeast Saccharomyces cerevisiae, hereafter referred to simply as yeast. Human homologs of most yeast AFs have been identified, suggesting a high level of conservation in ribosome assembly between yeast and humans.
Ribosome assembly in yeast begins in the nucleolus with transcription of the long 35S precursor rRNA (pre-rRNA), which is co-transcriptionally bound by many AFs and r-proteins. Through a series of endo- and exonucleolytic processing events, the 35S pre-rRNA is ultimately processed into 3 mature rRNAs (18S, 5.8S, and 25S) (Fig. 2A). The pre-5S rRNA is transcribed and processed separately into 5S rRNA. While these pre-rRNA processing events occur, r-proteins bind and become stably associated with the nascent ribosomal subunits, AFs enter and exit, carrying out various functions, and the immature subunits transit from the nucleolus to the nucleoplasm and are then exported to the cytoplasm for final maturation (Fig. 1A).
Figure 2.
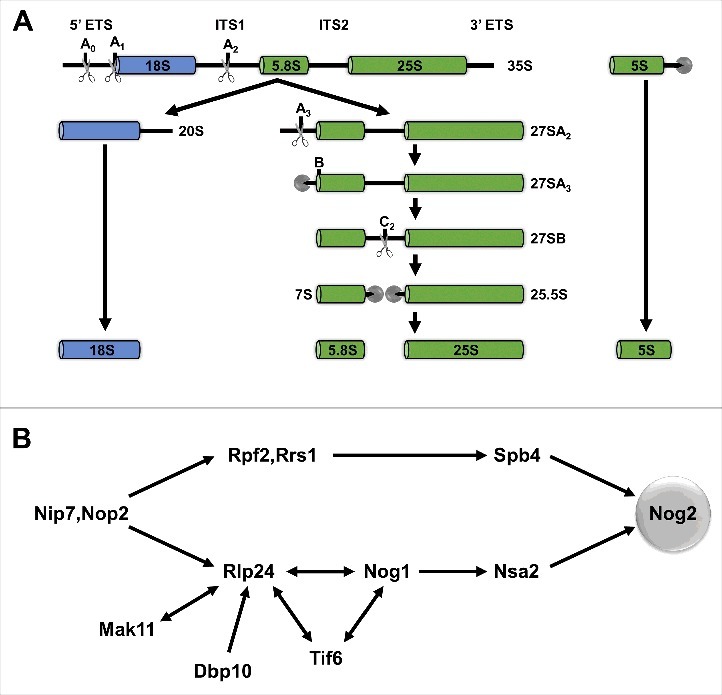
Yeast pre-rRNA processing and hierarchical recruitment of B-factors. (A) The pre-rRNA processing pathway in yeast. (B) Assembly of B-factors begins early in 60S subunit assembly with binding of Nip7 and Nop2 to the pre-60S subunit. The remaining B-factors assemble in 2 parallel pathways, ending with recruitment of Nog2.
Figure 1.
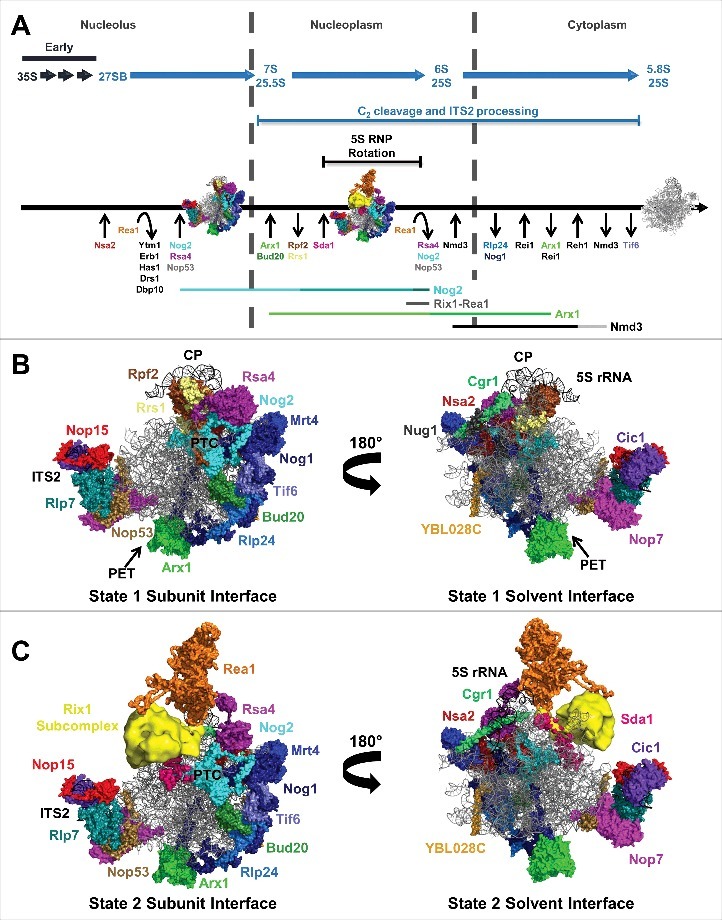
The yeast 60S subunit assembly pathway and AFs identified in state 1 and state 2 particles. (A) Timing of pre-rRNA processing events, entry and exit of AFs, and 5S RNP rotation. Also shown are the lifetimes of the Nog2-, Rix1-Rea1-, Arx1-, and Nmd3-bound pre-60S subunit assembly intermediates. The lifetimes of the state 1, 2, and 3 Nog2 particles are indicated by different shades of teal. The portions of the lifetimes of Arx1 and Nmd3 reported on by the Arx1 and Nmd3 particles are indicated by light green and gray, respectively. (B) The 19 AFs identified in the Nog2 state 1 particle (PDB ID 3JCT). The AF arc consists of 14 AFs stretching from the CP to the PET. Nop15, Rlp7, Nop53, Cic1, and Nop7 bind to ITS2. (C) The Nog2 state 2 particle.
The mature yeast large, or 60S, subunit, comprised of the 25S, 5.8S, and 5S rRNAs, plus 46 r-proteins, contains several functional centers necessary for translation of mRNA into protein.8 Two of these are the peptidyl transferase center (PTC), the major catalytic site of the 60S subunit where peptide bond formation occurs, and the polypeptide exit tunnel (PET) through which nascent polypeptides travel to emerge from the translating ribosome.
In 2011, Ben-Shem and colleagues determined the crystal structure of the mature yeast 60S subunit.9 This gave researchers a first detailed view of rRNA folding and of where r-proteins are bound to the rRNA core in mature 60S subunits. However, structural information regarding the binding sites of AFs and the conformational changes that pre-rRNA undergoes during the assembly process remained largely elusive until recent years. Before the advent of new direct electron detectors for cryo electron microscopy (cryo-EM), methods available to study pre-60S subunit structure included rRNA structure probing,10 yeast 2- and 3-hybrid assays,11 negative stain electron microscopy,12,13 crosslinking of AFs to rRNA,14 isolation of AF subcomplexes,15 and reconstitution of AFs with the mature 60S subunit.16 Each of these approaches has limitations, and none can provide the level of structural detail that has now been attained with cryo-EM.
The first cryo-EM reconstruction of a pre-60S subunit was published in 2012 by Bradatsch and colleagues, who solved the structure of a late nuclear Arx1 particle to 11.9 Å resolution.17 Since then, the structures of Rix1-Rea1, Nog2, and Nmd3 particles have also been solved.18-20 It is important to note that these particles are actually non-homogenous mixtures of various pre-60S subunit assembly intermediates to which a given AF is bound. The cryo-EM structures of the Arx1, Rix1-Rea1, Nog2, and Nmd3 particles represent the major state or states present in the mixture of assembly intermediates. The structures of these 4 particles report on overlapping late stages of 60S subunit assembly (Fig. 1A). These 60S subunit assembly intermediates were the first to be investigated due to their increased stability and decreased complexity compared with earlier particles.
The cryo-EM structures of Nog2 particles are windows into nuclear stages of 60S subunit assembly
This review focuses primarily on principles that have emerged from studying the structures of Nog2-bound 60S subunit assembly intermediates. Nog2 is a GTPase AF present in the pre-60S subunit during nuclear stages of assembly.22-24 This period of 60S subunit biosynthesis is marked by the entry and exit of numerous AFs as well as several other major structural rearrangements (Fig. 1A).22,23,25 A series of 3 consecutive 60S subunit assembly intermediates were identified in the pool of Nog2 particles; the cryo-EM structures of these intermediates, termed states 1–3, present a detailed view of many remodeling events that occur during the lifetime of Nog2-bound assembly intermediates.19 States 1 and 2 were solved to nominal resolutions of 3.08 Å and ∼6.6 Å respectively. Mapping of protein binding sites and interactions in the Nog2 particles was guided by chemical crosslinking coupled with mass spectrometry (XL-MS), which, together with the development of new direct electron detectors,21 contributed to the high resolution at which the structures were solved.
An interesting observation in the state 1 and 2 structures is that the solvent interface, or “back,” of the pre-60S subunit is largely devoid of AFs (Fig. 1B,C).19 In contrast, the subunit interface is decorated with numerous AFs. This suggests that the subunit interface, containing the PTC, is assembled later than the solvent interface, consistent with previous observations of hierarchical assembly of 60S subunits.26 In state 1, an arc of 14 AFs that bind to the subunit interface of the pre-60S subunit between the central protuberance (CP) and the PET was identified (Fig. 1B). A second cluster of 5 AFs binds proximal to the internal transcribed spacer 2 (ITS2), the pre-rRNA sequence that separates the mature 5.8S and 25S rRNAs (Fig. 1B and 2A). Furthermore, in state 2, which is highly similar to the previously determined Rix1-Rea1 structure,18 the binding sites of Sda1, Rea1, and the Rix1 subcomplex were identified (Fig. 1C).
These AF binding sites provide insight into construction of 60S subunit functional centers, namely the PET and PTC, as well as ITS2 cleavage and removal, all of which are major remodeling events that occur during the lifetime of Nog2 particles. Knowing at high resolution where these AFs are located on the pre-60S subunit not only facilitates the formation of new hypotheses and models, some of which are discussed here, but it also validates and clarifies previous results.
Insights into the importance of Nog2 in 27SB pre-rRNA processing and PTC construction
The GTPase AF Nog2 has previously been established as a key protein in 27SB pre-rRNA processing,22,27 which is initiated by cleavage of ITS2 at the C2 site (Fig. 2A). Nog2 is recruited to the pre-60S subunit only once 2 parallel assembly pathways converge and all of the other AFs required for 27SB pre-rRNA processing (“B-factors”) have bound (Fig. 2B).27 These B-factors are AFs that, when depleted or mutated, prevent C2 cleavage and further ITS2 processing. Entry of Nog2 into the pre-60S subunit is one of the last remodeling events preceding C2 cleavage. The cryo-EM structures of Nog2 particles showed that Nog2 acts as a hub protein centered at the PTC; it interacts directly with the nearby B-factors Rpf2, Nog1, and Nsa2 as well as the later-acting AFs Rsa4 and Bud20 and numerous rRNA helices in 25S rRNA domains II, IV, and V. Nog2 may also interact with the B-factor Dbp10, as crosslinking data suggest that Dbp10 also binds near the PTC.28
These structural data explain the previous observation of hierarchical recruitment of B-factors.27,29 Tif6, Rlp24, and Nog1 are interdependent for binding to the pre-60S subunit, consistent with the finding that they are tightly clustered in the AF arc (Fig. 1B,C and 2B). Furthermore, Nog1 was found to bind to Nsa2, providing an explanation for why Nsa2 cannot stably associate with the pre-60S subunit in the absence of Tif6, Rlp24, or Nog1. While Spb4 was not identified in any of the Nog2 particle cryo-EM structures, it may also interact with Nog2, or it might stabilize interactions between Rpf2/Rrs1 and Nog2, which could explain why Nog2 cannot assemble in the absence of Rpf2, Rrs1, or Spb4. Another possibility is that Spb4, which is an RNA helicase, may remodel the pre-60S subunit to allow association of Nog2. A remaining question, however, is how entry of Nog2 into the pre-60S subunit is coordinated with cleavage at the C2 site.
Like Nog2, Nog1 is a GTPase AF23,30 that interacts with many AFs, r-proteins, and rRNA helices; the N-terminal 4-helical-bundle domain and GTPase domain of Nog1 are located near Nog2 and the PTC, while the C-terminus of Nog1 stretches along the entire AF arc (Figs. 1B,C and 3A,C).19 The GTPase AF Nug1 is also bound to the PTC near Nog2 and Nog1 (Fig. 3A).19,28 Nsa2, which has no predicted enzymatic activity, also forms numerous contacts with the PTC.19,31 It is possible that the extensive interactions of these 3 GTPases and Nsa2 with the PTC are necessary during late nuclear stages of assembly to construct the PTC, to protect it from premature binding with translation factors, or to signal that it has achieved a particular level of maturation. For example, Nog1 and Nsa2 may sense whether the PTC is properly constructed and prevent entry of Nog2 if it is not.
Figure 3.
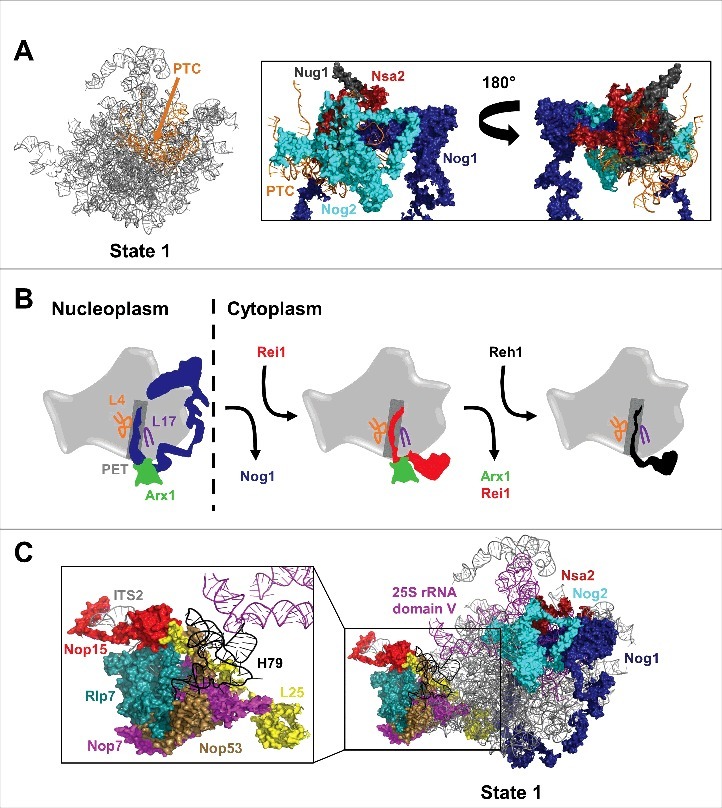
New insights into construction and inspection of the PTC and PET and ITS2 cleavage. (A) Nog2, Nog1, Nug1, and Nsa2 form extensive interactions with the PTC during late nuclear stages of 60S subunit assembly. Only 69 amino acids of the N-terminus of Nug1 were resolved in state 1. (B) Nog1, Rei1, and Reh1 consecutively probe the PET during 60S subunit assembly. Nog1 and Rei1 interact with Arx1 at the PET exit. (C) L25 extends from the exit of the PET to ITS2, interacting with Nop53, Nop7, Rlp7, and Nop15. Domain V of 25S rRNA stretches from the PTC to the ITS2 region. H79 of domain V interacts with Nop53 and Rlp7.
Furthermore, the tight association of Nog2 with the PTC suggests that Nog2 could act as a placeholder, with its departure being necessary for further maturation of the PTC. Indeed, the export factor Nmd3 assembles onto the pre-60S subunit following release of Nog2 and exhibits partially overlapping binding sites with Nog2.20,24,32 This indicates that the PTC is constructed in a hierarchical fashion, with inspection by AFs at successive structural checkpoints. Furthermore, it suggests that assembly and inspection of the PTC are coupled to nuclear export of pre-60S subunits. It is also of note that 3 GTPase AFs, YphC, YsxC, and RbgA, participate in PTC construction in Bacillus subtilis, perhaps indicating a conserved requirement for GTPase activity when building or inspecting the PTC.33-35
Nog1, Rei1, and Reh1: Sequential PET inspector AFs
Intriguingly, the C-terminal ∼75 amino acids of Nog1 are inserted into the PET in Nog2 particles (Figs. 1B,C and 3B,C).19 Similarly, the AFs Rei1 and Reh1 have been shown to probe the PET during later, cytoplasmic stages of 60S subunit assembly.20,36 Release of Nog1 from the pre-60S subunit is coupled to entry of Rei1,37,38 and Reh1 enters the pre-60S subunit following release of Rei1,20 suggesting that the PET is probed by an AF for most of the assembly pathway (Fig. 3B). While it has been proposed that these PET inspector AFs could aid in construction of the PET or check that it has been properly constructed as part of a quality control checkpoint, it remains a mystery why the C-termini of Nog1, Rei1, and Reh1 are present in the PET. Additionally, it is as of yet unclear why there are 3 AFs that sequentially inspect the PET throughout 60S subunit assembly.
The portions of the C-terminal domains of Nog1, Rei1, and Reh1 that are inserted into the PET have strikingly similar secondary structures.19,20,36 Furthermore, each of these AFs probes the tunnel deeply enough that their inserted C-terminal domains nearly reach through the subunit to the PTC. Of the 3 tunnel inspector AFs, only Nog1 has an N-terminal domain bound at the PTC, although interactions between Rei1 and Sdo1 may facilitate communication between Rei1 and the PTC.39 The function of the GTPase domain of Nog1 remains unknown, but it is tempting to speculate that Nog1 GTP binding or hydrolysis may serve as a signal that could be propagated through Nog1 along the AF arc or even between the nearby N- and C-termini of Nog1. In this way, Nog1 may be able to coordinate maturation of the PTC and PET, or signal that these major functional centers have been constructed.
Further complicating the potential roles of Nog1 in 60S subunit assembly is the finding that Nog1, like Rei1, interacts with Arx1 at the exit of the PET (Fig. 3B). While Rei1 is thought to play a role in release of Arx1 in the cytoplasm, it is unclear if the interaction between Arx1 and Nog1 is significant. It also remains to be seen whether interactions between Nog1, Rei1, and Reh1 and the r-proteins lining the PET, including L4 (uL4) and L17 (uL22), are important for 60S subunit assembly (Fig. 3B).
Insights into coordination of C2 cleavage and construction of the PTC and PET
A portion of ITS2 itself was resolved for the first time in the Nog2 cryo-EM structures.19 Counterintuitively, the B-factors required for cleavage of 27SB pre-rRNA at the C2 site and further ITS2 processing are concentrated in the AF arc, rather than binding to ITS2 (Figs. 1B,C and 2A). There may be some exceptions to this rule, including the DEAD-box helicases Has1 and Drs1; mutations in these AFs have been shown to block C2 cleavage, and they are thought to bind on or near ITS2.40-42 Nevertheless, there is no simple explanation as to why AFs located at the CP and along the AF arc are necessary for cleavage of ITS2. One emerging theme is that 3 major remodeling events seem to be required for C2 cleavage to occur: removal of AFs bound proximal to ITS2, stable association of the r-proteins surrounding the PET exit, and binding of several AFs to the nascent PTC.43 It is unclear in what order these events occur, but it seems likely that there is a complicated checkpoint that ensures proper construction of the PTC and PET before the irreversible steps of C2 cleavage and ITS2 removal.
It is possible that Nog1 is important to facilitate communication across the pre-60S subunit to coordinate these remodeling events. However, Nog1 does not interact directly with Nop7, Nop15, Nop53, Rlp7, or Cic1, the AFs bound to ITS2 in state 1. While it is simple to imagine how remodeling events at the PET and PTC could be linked, an explanation is less forthcoming as to how release of the ITS2 AFs is coordinated with PET and PTC construction. One possibility is that the r-proteins surrounding the PET could provide a link between the tunnel and ITS2. The N-terminal domain of the r-protein L25 (uL23) extends from the ring of r-proteins surrounding the PET to the ITS2 foot region. Interestingly, the tip of this N-terminal tail of L25 is in close proximity to Nop15, Nop53, and Rlp7 and the extension also interacts with Nop7 (Fig. 3C). Perhaps, then, the N-terminal region of L25 could provide a link between the PET and the ITS2 region.
Another possible explanation that has been put forth is that 25S rRNA domain V could serve as a link between ITS2 and the PTC.44 The AFs that bind to the nascent PTC are predominantly domain V binding AFs; at the ITS2 foot, Nop53 and, to a lesser extent, Rlp7, interact with H79 in domain V of 25S rRNA (Fig. 3C). Perhaps removal of AFs from ITS2 could initiate a conformational change via Nop53 or Rlp7 that is transmitted to the PTC along domain V, allowing Nsa2, Nog2, Rsa4, and other PTC binding AFs to associate. The opposite could also be true, with binding of PTC AFs being transmitted along domain V to signal release of ITS2 AFs. A structural observation in support of this notion is that the L1 stalk, a flexible component of domain V, oscillates between inward and outward positions.19 In the state 1 structure, the L1 stalk is in an outward position, but in the state 2 structure it is in an inward conformation.
Stepwise rotation of the 5S RNP requires entry and exit of several AFs
In 2014, Leidig and colleagues discovered that the 5S ribonucleoprotein (RNP), consisting of 5S rRNA and the r-proteins L5 (uL18) and L11 (uL5), is initially assembled onto the pre-60S subunit such that it is rotated almost 180° from its mature conformation (Fig. 4).25 The Nog2 particle cryo-EM structures revealed that rotation of the 5S RNP occurs in a stepwise manner.19 In state 1 the 5S RNP has not begun rotating, while in state 3 it is in an almost mature conformation. Between state 1 and state 2, the AFs Rpf2 and Rrs1 exit the pre-60S subunit (Fig. 1). These AFs may anchor the 5S RNP in its immature position, and their removal appears necessary for rotation to occur. However, it is unclear how removal of Rpf2 and Rrs1 is triggered. The state 1 structure revealed that the C-terminus of Rpf2 interacts with Nog2. It remains to be seen if these interactions with a GTPase are significant in the release of Rpf2 and Rrs1 and in 5S RNP rotation.
Figure 4.
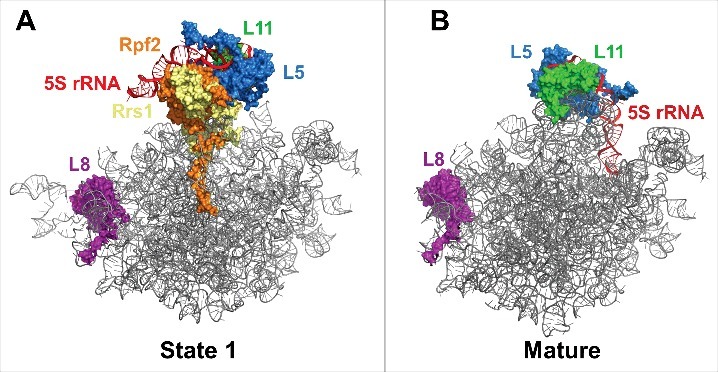
Rotation of the 5S RNP. (A) The pre-rotated position of the 5S RNP in state 1 (PDB ID 3JCT). (B) The rotated position of the 5S RNP in the mature 60S subunit (PDB ID 4V88). R-protein L8 is shown to demonstrate that the state 1 and mature structures are in the same orientation.
While the 5S RNP acquires a near-mature conformation in state 2, additional assembly events are required to complete its movement into the mature position. Further rotation of the 5S RNP may require binding of Sda1, Rea1, and the Rea1 adaptor complex composed of Ipi1, Rix1 (Ipi2), and Ipi3. These AFs were identified in state 2 but not in state 1 (Fig. 1B,C).19 Therefore, removal of Rpf2 and Rrs1 and initial swiveling of the 5S RNP may be necessary to allow association of Sda1, Rea1, and the Rix1 subcomplex with the pre-60S subunit. Binding of these AFs may stabilize the rotated position into which the 5S RNP moves following removal of Rpf2 and Rrs1. Furthermore, Rea1 is a AAA-ATPase; ATP hydrolysis by Rea1 powers the removal of Rsa4 near the end of nuclear stages of 60S subunit assembly (Fig. 1A).13,18 This removal of Rsa4 is likely necessary to allow the 5S RNP to complete its rotation into the mature position.
Although the Nog2 particle cryo-EM structures provide additional insight into the mechanism of 5S RNP rotation, the question remains why the “head” of the pre-60S subunit would be assembled backward. It has been proposed that rotation of the 5S RNP might function as a wrench, causing the surrounding 25S rRNA helices to move toward their mature conformation.25 Intriguingly, recent cryo-EM structures of Escherichia coli large ribosomal subunit assembly intermediates revealed that 5S rRNA also occupies a nonnative, rotated conformation in bacterial pre-50S subunits compared with mature 50S subunits.45 In E. coli, this immature orientation seems to be either highly transient or nonobligatory. Nevertheless, this conservation of 5S rRNA rotation from bacteria to eukaryotes suggests that there may be a common quality control checkpoint in large subunit assembly wherein rotation cannot occur until other assembly events have taken place. Perhaps in eukaryotes rotation of the 5S RNP serves as an assembly checkpoint before nuclear export; this could explain why 5S RNP maturation seems to be more highly regulated in eukaryotes than in prokaryotes.
Concluding remarks
In the few years since the first cryo-EM structure of a pre-60S subunit was determined, the availability of such rich structural data has revolutionized the study of ribosome assembly. Not only have these structures provided support and credibility to many previously proposed models, they are driving the formation of almost endless new hypotheses. As more cryo-EM structures become available, 2 themes seem to hold true. Seeing truly is believing, and yet these increasingly detailed structures also show how much about ribosome assembly remains a mystery. In the future, it seems likely that cryo-EM efforts will be aimed toward determining the structures of progressively earlier pre-60S subunits, as the currently available structures are all of late assembly intermediates. While amazing progress has been made, only the tip of the iceberg has been revealed.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the Woolford and Gao laboratories for helpful discussions and insights, and Joaquin Ortega and members of his laboratory for their comments on this manuscript.
Funding
This work was supported by National Institutes of Health grant R01GM028301 to J.L.W., the National Natural Science Foundation of China (31422016, 31470722, and 31630087 to N.G.), and the Roche/ARCS Foundation Scholar Award to S.B..
References
- 1.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA 2009;15(12):2083-104; PMID:19850913; https://doi.org/ 10.1261/rna.1792109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kressler D, Hurt E, Baβler J. Driving ribosome assembly. Biochimica et Biophysica Acta 2010; 1803(6):673-83; PMID:19879902; https://doi.org/ 10.1016/j.bbamcr.2009.10.009 [DOI] [PubMed] [Google Scholar]
- 3.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci 2010; 35(5):260-6; PMID:20137954; https://doi.org/ 10.1016/j.tibs.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woolford JL, Baserga SJ. Ribosome Biogenesis in the Yeast Saccharomyces cerevisiae. Genetics 2013; 195(3):643-81; PMID:24190922; https://doi.org/ 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernández-Pevida A, Kressler D, de la Cruz J. Processing of preribosomal RNA in Saccharomyces cerevisiae. Wiley Interdisciplinary Reviews: RNA 2015; 6(2):191-209; PMID:25327757; https://doi.org/ 10.1002/wrna.1267 [DOI] [PubMed] [Google Scholar]
- 6.Nerurkar P, Altvater M, Gerhardy S, Schutz S, Fischer U, Weirich C, Panse VG. Eukaryotic Ribosome Assembly and Nuclear Export. Int Rev Cell Mol Biol 2015; 319:107-40. PMID:26404467; https://doi.org/ 10.1016/bs.ircmb.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 7.de la Cruz J, Karbstein K, Woolford JL. Functions of Ribosomal Proteins in Assembly of Eukaryotic Ribosomes In Vivo. Ann Rev Biochem 2015; 84(1):93-129; PMID:25706898; https://doi.org/ 10.1146/annurev-biochem-060614-033917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilson DN, Cate JHD. The Structure and Function of the Eukaryotic Ribosome. Cold Spring Harbor Perspectives in Biology 2012; 4(5):a011536; PMID:22550233; https://doi.org/ 10.1101/cshperspect.a011536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Shem A, de Loubresse NG, Melnikov S, Jenner L, Yusupova G, Yusupov M. The Structure of the Eukaryotic Ribosome at 3.0 Å Resolution. Science 2011; 334(6062):1524-9; PMID:22096102; https://doi.org/ 10.1126/science.1212642 [DOI] [PubMed] [Google Scholar]
- 10.Hector RD, Burlacu E, Aitken S, Bihan TL, Tuijtel M, Zaplatina A, Cook AG, Granneman S. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res 2014; 42(19):12138-54; PMID:25200078; https://doi.org/ 10.1093/nar/gku815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCann KL, Charette JM, Vincent NG, Baserga SJ. A protein interaction map of the LSU processome. Gen Dev 2015; 29(8):862-75; PMID:25877921; https://doi.org/ 10.1101/gad.256370.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nissan TA, Galani K, Maco B, Tollervey D, Aebi U, Hurt E. A pre-ribosome with a tadpole-like structure functions in ATP-dependent maturation of 60S subunits. Mol Cell 2004; 15(2):295-301; PMID:15260980; https://doi.org/ 10.1016/j.molcel.2004.06.033 [DOI] [PubMed] [Google Scholar]
- 13.Ulbrich C, Diepholz M, Baßler J, Kressler D, Pertschy B, Galani K, Böttcher B, Hurt E. Mechanochemical Removal of Ribosome Biogenesis Factors from Nascent 60S Ribosomal Subunits. Cell 2009; 138(5):911-22; PMID:19737519; https://doi.org/ 10.1016/j.cell.2009.06.045 [DOI] [PubMed] [Google Scholar]
- 14.Granneman S, Petfalski E, Swiatkowska A, Tollervey D. Cracking pre-40S ribosomal subunit structure by systematic analyses of RNA-protein cross-linking. EMBO J 2010; 29(12):2026-36; PMID:20453830; https://doi.org/ 10.1038/emboj.2010.86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang J, Harnpicharnchai P, Jakovljevic J, Tang L, Guo Y, Oeffinger M, Rout MP, Hiley SL, Hughes T, Woolford JL. Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Gen Dev 2007; 21(20):2580-92; PMID:17938242; https://doi.org/ 10.1101/gad.1569307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greber BJ, Boehringer D, Montellese C, Ban N. Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit. Nat Struct Mol Biol 2012; 19(12):1228-33; PMID:23142985; https://doi.org/27251291 10.1038/nsmb.2425 [DOI] [PubMed] [Google Scholar]
- 17.Bradatsch B, Leidig C, Granneman S, Gnädig M, Tollervey D, Böttcher B, Beckmann R, Hurt E. Structure of the pre-60S ribosomal subunit with nuclear export factor Arx1 bound at the exit tunnel. Nat Struct Mol Biol 2012; 19(12):1234-41; PMID:23142978; https://doi.org/27251291 10.1038/nsmb.2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrio-Garcia C, Thoms M, Flemming D, Kater L, Berninghausen O, Baßler J, Beckmann R, Hurt E. Architecture of the Rix1–Rea1 checkpoint machinery during pre-60S-ribosome remodeling. Nat Struct Mol Biol 2015; 23:37-44; PMID:26619264; https://doi.org/27251291 10.1038/nsmb.3132 [DOI] [PubMed] [Google Scholar]
- 19.Wu S, Tutuncuoglu B, Yan K, Brown H, Zhang Y, Tan D, Gamalinda M, Yuan Y, Li Z, Jakovljevic J, et al.. Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes. Nature 2016; 534(7605):133-7; PMID:27251291; https://doi.org/ 10.1038/nature17942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma C, Wu S, Li N, Chen Y, Yan K, Li Z, Zheng L, Lei J, Woolford JL, Gao N. Structural snapshot of cytoplasmic pre-60S ribosomal particles bound with Nmd3, Lsg1, Tif6 and Reh1. Nat Struct Mol Biol 2017; 24(3):214-20; PMID:28112732; https://doi.org/25955078 10.1038/nsmb.3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Binshtein E, Ohi MD. Cryo-Electron Microscopy and the Amazing Race to Atomic Resolution. Biochemistry 2015; 54(20):3133-41; PMID:25955078; https://doi.org/ 10.1021/acs.biochem.5b00114 [DOI] [PubMed] [Google Scholar]
- 22.Saveanu C, Bienvenu D, Namane A, Gleizes PE, Gas N, Jacquier A, Fromont-Racine M. Nog2p, a putative GTPase associated with pre-60S subunits and required for late 60S maturation steps. EMBO J 2001; 20(22):6475-84; PMID:11707418; https://doi.org/ 10.1093/emboj/20.22.6475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saveanu C, Namane A, Gleizes P-E, Lebreton A, Rousselle J-C, Noaillac-Depeyre J, Gas N, Jacquier A, Fromont-Racine M. Sequential protein association with nascent 60S ribosomal particles. Mol Cell Biol 2003; 23(13):4449-60; PMID:12808088; https://doi.org/ 10.1128/MCB.23.13.4449-4460.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuo Y, Granneman S, Thoms M, Manikas R-G, Tollervey D, Hurt E. Coupled GTPase and remodelling ATPase activities form a checkpoint for ribosome export. Nature 2014; 505(7481):112-6; PMID:24240281; https://doi.org/ 10.1038/nature12731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leidig C, Thoms M, Holdermann I, Bradatsch B, Berninghausen O, Bange G, Sinning I, Hurt E, Beckmann R. 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle. Nat Commun 2014; 5:3491; PMID:24662372; https://doi.org/ 10.1038/ncomms4491 [DOI] [PubMed] [Google Scholar]
- 26.Gamalinda M, Ohmayer U, Jakovljevic J, Kumcuoglu B, Woolford J, Mbom B, Lin L, Woolford JL. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Gen Dev 2014; 28(2):198-210; PMID:24449272; https://doi.org/ 10.1101/gad.228825.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talkish J, Zhang J, Jakovljevic J, Horsey EW, Woolford JL. Hierarchical recruitment into nascent ribosomes of assembly factors required for 27SB pre-rRNA processing in Saccharomyces cerevisiae. Nucleic Acids Res 2012; 40(17):8646-61; PMID:22735702; https://doi.org/ 10.1093/nar/gks609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manikas R-G, Thomson E, Thoms M, Hurt E. The K+-dependent GTPase Nug1 is implicated in the association of the helicase Dbp10 to the immature peptidyl transferase centre during ribosome maturation. Nucleic Acids Res 2016; 44(4):1800-12; PMID:26823502; https://doi.org/ 10.1093/nar/gkw045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebreton A, Rousselle JC, Lenormand P, Namane A, Jacquier A, Fromont-Racine M, Saveanu C. 60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes. Nucleic Acids Res 2008; 36(15):4988-99; PMID:18658244; https://doi.org/ 10.1093/nar/gkn469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fuentes JL, Datta K, Sullivan SM, Walker A, Maddock JR. In vivo functional characterization of the Saccharomyces cerevisiae 60S biogenesis GTPase Nog1. Mol Genet Genomics 2007; 278(1):105-23; PMID:17443350; https://doi.org/ 10.1007/s00438-007-0233-1 [DOI] [PubMed] [Google Scholar]
- 31.Bassler J, Paternoga H, Holdermann I, Thoms M, Granneman S, Barrio-Garcia C, Nyarko A, Stier G, Clark SA, Schraivogel D, et al.. A network of assembly factors is involved in remodeling rRNA elements during preribosome maturation. J Cell Biol 2014; 207(4):481-98; PMID:25404745; https://doi.org/ 10.1083/jcb.201408111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol 2010; 189(7):1079-86; PMID:20584915; https://doi.org/ 10.1083/jcb.201001124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jomaa A, Jain N, Davis JH, Williamson JR, Britton RA, Ortega J. Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res 2014; 42(5):3419-35; PMID:24335279; https://doi.org/ 10.1093/nar/gkt1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ni X, Davis JH, Jain N, Razi A, Benlekbir S, McArthur AG, Rubinstein JL, Britton RA, Williamson JR, Ortega J. YphC and YsxC GTPases assist the maturation of the central protuberance, GTPase associated region and functional core of the 50S ribosomal subunit. Nucleic Acids Res 2016; 44(17):8442-55; PMID:27484475; https://doi.org/ 10.1093/nar/gkw678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li N, Chen Y, Guo Q, Zhang Y, Yuan Y, Ma C, Deng H, Lei J, Gao N. Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res 2013; 41(14):7073-83; PMID:23700310; https://doi.org/ 10.1093/nar/gkt423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Greber BJ, Gerhardy S, Leitner A, Aebersold R, Panse VG, Ban N, Boehringer D, Leulliot N, Aebersold R, Panse VG, et al.. Insertion of the Biogenesis Factor Rei1 Probes the Ribosomal Tunnel during 60S Maturation. Cell 2016; 164(1–2):91-102; PMID:26709046; https://doi.org/ 10.1016/j.cell.2015.11.027 [DOI] [PubMed] [Google Scholar]
- 37.Lebreton A, Saveanu C, Decourty L, Rain JC, Jacquier A, Fromont-Racine M. A functional network involved in the recycling of nucleocytoplasmic pre-60S factors. J Cell Biol 2006; 173(3):349-60; PMID:16651379; https://doi.org/ 10.1083/jcb.200510080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell 2010; 39(2):196-208; PMID:20670889; https://doi.org/ 10.1016/j.molcel.2010.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greber BJ. Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy. RNA 2016; 22(11):1643-62; PMID:27875256;https://doi.org/ 10.1261/rna.057927.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams CC, Jakovljevic J, Roman J, Harnpicharnchai P, Woolford JL. Saccharomyces cerevisiae nucleolar protein Nop7p is necessary for biogenesis of 60S ribosomal subunits. RNA 2002; 8(2):150-65; PMID:11911362; https://doi.org/ 10.1017/S1355838202010026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang L, Sahasranaman A, Jakovljevic J, Schleifman E, Woolford JL. Interactions among Ytm1, Erb1, and Nop7 Required for Assembly of the Nop7-Subcomplex in Yeast Preribosomes. Mol Biol Cell 2008; 19(7):2844-56; PMID:18448671; https://doi.org/ 10.1091/mbc.E07-12-1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dembowski Ja, Kuo B, Woolford JL. Has1 regulates consecutive maturation and processing steps for assembly of 60S ribosomal subunits. Nucleic Acids Res 2013; 41(16):7889-904; PMID:23788678; https://doi.org/ 10.1093/nar/gkt545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konikkat S, Biedka S, Woolford JL. The assembly factor Erb1 functions in multiple remodeling events during 60S ribosomal subunit assembly in S. cerevisiae. Nucleic Acids Res 2017; PMID:28115637; https://doi.org/ 10.1093/nar/gkw1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tutuncuoglu B, Jakovljevic J, Wu S, Gao N, Woolford JL. The N-terminal extension of yeast ribosomal protein L8 is involved in two major remodeling events during late nuclear stages of 60S ribosomal subunit assembly. RNA 2016; 22(9):1386-99; PMID:27390266; https://doi.org/ 10.1261/rna.055798.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis JH, Tan YZ, Carragher B, Potter CS, Lyumkis D, Williamson JR. Modular Assembly of the Bacterial Large Ribosomal Subunit. Cell 2016; 167(6):1610-22; PMID:27912064; https://doi.org/ 10.1016/j.cell.2016.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]


